Abstract
Viruses that infect bacteria are the most abundant biological agents on the planet and bacteria have evolved diverse defense mechanisms to combat these genetic parasites. One of these bacterial defense systems relies on a repetitive locus, referred to as a CRISPR (clusters of regularly interspaced short palindromic repeats). Bacteria and archaea acquire resistance to invading viruses and plasmids by integrating short fragments of foreign nucleic acids at one end of the CRISPR locus. CRISPR loci are transcribed and the long primary CRISPR transcript is processed into a library of small RNAs that guide the immune system to invading nucleic acids, which are subsequently degraded by dedicated nucleases. However, the development of CRISPR-mediated immune systems has not eradicated phages, suggesting that viruses have evolved mechanisms to subvert CRISPR-mediated protection. Recently, Bondy-Denomy and colleagues discovered several phage-encoded anti-CRISPR proteins that offer new insight into the ongoing molecular arms race between viral parasites and the immune systems of their hosts.
Keywords: phage, bacterial immunity, RNA-guided immunity, anti-CRISPR, viral suppressors of RNAi (VSR), viral suppressors of CRISPR (VSC)
Introduction
In many environments, viruses that infect bacteria (referred to as phages) are estimated to outnumber their hosts 10 to one, and these pervasive predators impose strong selective pressures on microbial populations. In response to these selective pressures, bacteria have evolved a diverse repertoire of phage defense mechanisms that target each stage of the viral life cycle.1 Historically, our understanding of immune systems in bacteria has been limited to innate defense systems such as receptor switching, restriction-modification and abortive-phage phenotypes. However, more recent studies have revealed that organisms from both the bacterial and archaeal domains of life have evolved RNA-guided adaptive immune systems that share conceptual similarity to RNA-interference (RNAi) systems in eukaryotes (Fig. 1).2-6 In both systems, long RNAs are processed into small non-coding RNAs that serve as sequence-specific guides for the detection and degradation of foreign nucleic acids (Fig. 1). These anti-viral defense systems impose strong selective pressures on phage populations, resulting in the emergence of mutant viruses capable of immune system evasion. This perpetual cycle of host adaptation and reciprocal counter adaptation by virus results in a rapid antagonistic co-evolution that has been likened to an escalating “arms race.”7,8
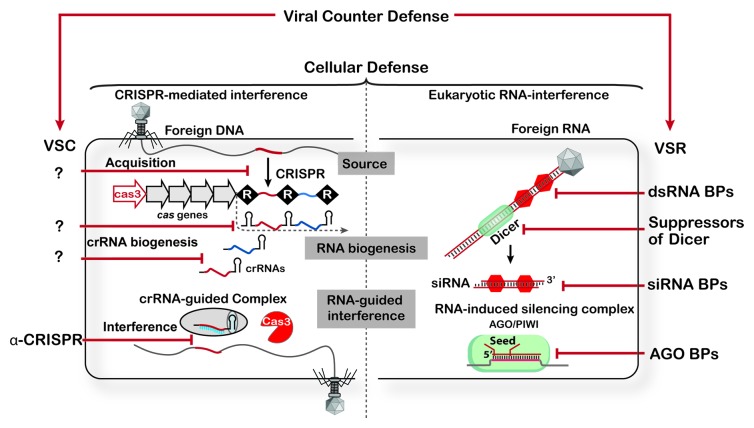
Figure 1. Each stage of RNA-guided immunity is a potential target for viral suppressors. Organisms from all domains of life (bacteria, archaea and eukaryotes) use small RNAs to target invading parasites, but the source of these RNAs, and the processing pathways that generate them are diverse. Viruses that infect eukaryotes have evolved a battery of counter defense mechanisms that target every stage of the RNAi pathway. Many viruses have evolved dsRNA-binding proteins (dsRNA BPs) that protect the dsRNA (e.g., B2 protein from Flock house virus) from degradation by Dicer. Although dsRNA BPs are probably the most common suppressor discovered so far, other viruses encode for proteins that selectively bind siRNAs (e.g., P19) and prevent them from being assembled into holo-RISC.40 The P38 protein from turnip crinkle virus contains two GW repeats and interacts directly with the AGO1 protein.41 In contrast to viral suppressors in eukaryotic systems, relatively little is known about suppressors of CRISPR-mediated defense. While the anti-CRISPR (α-CRISPR) proteins discovered by Bondy-Denomy et al. appear to block the immune system at a stage downstream of CRISPR RNA processing step, we anticipate that viruses infecting both bacteria and archaea have evolved suppression strategies that intervene at each stage of this process. For simplicity, this figure only depicts type I CRISPR-systems, though there are likely VSCs that target type II and III CRISPR systems as well.
Eukaryotic RNA-Interference
In eukaryotes, the term “RNA silencing” refers to a group of mechanistically related pathways that produce short non-coding RNAs for sequence-specific gene regulation. This is an evolutionarily conserved process that may have originally evolved as a defense mechanism against viruses and other foreign nucleic acids.9-12 Long double-stranded RNAs (dsRNAs) produced during a viral infection are recognized as antigens, and the dsRNA is processed into 21-nt small interfering RNAs (siRNAs) by a dedicated RNase III enzyme called Dicer.13,14 One strand of the siRNA is loaded into an Argonaute protein (AGO)-containing regulatory complex, where the siRNA serves as a sequence-specific guide that delivers the AGO nuclease to invading viral nucleic acids.15 To subvert this immune system, many viruses have evolved counter defensive strategies that suppress RNAi.16-18 Viral suppressors of RNAi (VSRs) are diverse in sequence and in their mechanism of immune system attenuation.16 Many VSRs circumvent antiviral RNAi by coating dsRNA and preventing it from coming into contact with the host’s RNAi machinery (dsRNA binding proteins), while other VSRs target steps downstream of dsRNA detection (Fig. 1).
CRISPR RNA-Guided Adaptive Immune Systems
CRISPRs are essential components of diverse nucleic-acid-based adaptive immune systems that are widespread in bacteria and archaea (Fig. 1). Each CRISPR locus consists of a series of short direct repeats that are separated by non-repetitive “spacer” sequences derived from foreign genetic elements. In response to viral or plasmid challenge, new spacer sequences are added to one end of the CRISPR locus and in this way CRISPR loci serve as molecular “vaccination cards” that maintain a chronological genetic record of prior encounters with foreign nucleic acid invaders. New sequence acquisition is the first step toward adaptive immunity, but acquisition alone does not provide long-term protection. CRISPR loci are transcribed, and the long primary transcript is processed into a library of short CRISPR-derived RNAs (crRNAs) that each contains a short sequence that was derived from and is complementary to a previously encountered invader. Each crRNA is bound by one or more CRISPR-associated proteins (Cas) and the resulting ribonucleoprotein complexes serve as surveillance systems that patrol the intracellular environment for foreign nucleic acid targets that are complementary to the crRNA-guide sequence.19-24 Viruses with mutations in specific regions of the target sequence escape detection by the crRNA-guided surveillance systems,25-28 and as a consequence of this selective pressure, CRISPR may play an influential role in maintaining sequence diversity in phage genomes.29-32 However, mutations are often associated with a fitness cost and alternative escape mechanisms are anticipated.
Viral Suppressers of CRISPR-Mediated Defense
To identify alternative phage escape strategies, Bondy-Denomy et al. recently screened a collection of 44 lysogens of P. aeruginosa PA14 for their susceptibility to infection by three different phages (JBD18, JBD25 and JBD67).33 These three phages are effectively eliminated by the CRISPR-mediated immune system in P. aeruginosa PA14 cells and are only able to replicate in PA14 strains where the CRISPR loci (∆CRISPR) or cas genes (∆cas) have been deleted. By challenging P. aeruginosa PA14 lysogens (lysogens are bacterial strains that contain an integrated copy of a phage genome) with these phages, the authors identified three different lysogenic strains (PA14JBD24, PA14MP29 and PA14JBD30) that are sensitive to infection by phage that were previously blocked by the CRISPR system. In fact, the PA14JDB30 strain (a PA14 strain lysogenized by phage JDB30) is as sensitive to infection by JBD18, JBD25 and JBD67 phages as the PA14 strain with a deleted CRISPR and cas locus.33 These results indicated that the integrated phage genome (i.e., prophage) might be interfering with the CRISPR-mediated defense system. To identify the putative immune system suppressor, the authors aligned the three different prophage genomes with related viral genomes. These genomes share high-sequence similarity and gene synteny; however, the prophage genomes associated with immunocompromised lysogens all contained several short open reading frames in one particular genomic region that were not found in prophage genomes associated with immunocompetent strains. These genes were suspected to be viral suppressors of the CRISPR-mediated defense system (VSC). To test this hypothesis, they cloned 17 of these genes and overexpressed the proteins in immunocompetent PA14 cells. Remarkably, seven of these genes convert phage-resistant PA14 cells to a phage-sensitive phenotype, but not all the suppressors were equally potent. Gene 35 from phage JBD30 encodes for a small (9 kDa) basic protein (pI = 8.1) that results in phage sensitivities similar to the CRISPR/cas deletion stain. Rigorous mutational studies confirmed that the suppressor is a protein rather than a non-coding RNA, and when this gene was cloned into the genome of a phage that was previously shown to be eliminated by the CRISPR system in PA14,25 the phage was no longer sensitive to CRISPR-mediated suppression. This may indicate that the suppressor protein is packaged and delivered to the cell along with the viral genome, or that the suppressor is transcribed and translated with efficiencies capable of rapidly neutralizing the immune system.
Mechanism of CRISPR-System Suppression
CRISPR-mediated adaptive immunity proceeds in three distinct stages: acquisition of foreign DNA into the CRISPR locus, CRISPR RNA biogenesis and CRISPR RNA-guided target interference (Fig. 1).34 Conceivably, a viral suppressor that interrupts any one of these stages could neutralize the immune system. To determine at what stage the viral suppressor is disarming the system, Bondy-Denomy et al. determined expression levels of the cas genes and the CRISPR loci in the lysogenic strains of PA14 that are sensitive to infections. Their results indicated that neither cas gene expression nor CRISPR RNA processing are affected by the suppressor.33 While the precise mechanism of these suppressors remains undetermined, these data suggest that the immune system is derailed at a stage downstream of CRISPR RNA processing. Possible mechanisms of the suppression include, but are not limited to: binding the mature crRNA and preventing it from being incorporated into the crRNA-guide surveillance complex, binding to the assembled crRNA-guided surveillance complex and preventing this complex from interacting with target substrates, binding to Cas3 and blocking nuclease activity or preventing Cas3 recruitment to the target-bound surveillance complex (Fig. 1).
Ecological Implications of CRISPR-System Suppression
Viruses are obligate intracellular parasites that all compete for common cellular resources. These resources are limited, so many phages encode superinfection exclusion (Sie) systems that block subsequent infection by the same or related phages.1 These systems allow a founding phage to shut the door on subsequent infections and, thus, selfishly preserve the cellular resources for the replication of its own genome. For phages that maintain a lysogenic lifestyle, this relationship can serve the interests of both the virus and the host by protecting the host from superinfection by related phage. However, the viral suppressors described by Bondy-Denomy et al. effectively prop the door open by disarming the immune system and allowing for superinfections by specific phages, while simultaneously attenuating the plaquing efficiency of others.33 This strategy raises some interesting questions about the evolutionary benefit of a CRISPR suppression mechanism that disarms the immune system in ways that render the cell susceptible to superinfection by some phages, while inhibiting infection of others. This complex relationship may contribute to the selective pressures between different viruses, which, like their hosts, are also competing for common resources.
Cease Fire?
It is unlikely viruses and their hosts will lay down their arms. In fact, the recent discovery of these VSCs may offer an explanation for the extreme diversity of CRISPR and cas genes.35-37 The suppressors identified by Bondy-Denomy et al. only block the immune system in PA14 (a type I-F immune system), while they have no effect on the immune system in E. coli K12 (a type I-E CRISPR system). The type I immune systems are all anticipated to function via similar mechanisms, but the phylogenetic and mechanistic distinction between these different immune systems is highlighted by viral suppressors that distinguish between these systems. In fact, the remarkable diversity of CRISPRs and cas genes may be a direct reflection of the selective pressures imposed on these immune systems by phages. We anticipate that viral suppressors have evolved to target-specific elements in each of the different CRISPR-systems, and that understanding the mechanisms of suppression will reveal unforeseen aspects of CRISPR biology. The discovery of these suppressors is evidence of an ongoing arms race that is being waged at the interface between adaptive immunity and viral persistence.
The Rise of Antibiotic Resistance and the Return of Phage Therapy
The evolutionary arms race between phages and their bacterial hosts may have medically relevant implications. Bacteriophages were originally discovered by Ernest Hanbury Hankin in the late 1800s, and the potential of these bacterial antagonists in treating human disease was almost immediately recognized.38 In 1919, Felix d’Herelle first demonstrated that phages could be used to cure patients suffering from dysentery, and in the years that followed, phages were used with remarkable success to treat a range of bacterial diseases (reviewed by Keen, E.C.39). However, the early success of phage therapy coincided with Aexander Flemming’s discovery of Penicillin, and antibiotics became medicine’s new wonder drugs. Antibiotics were rapidly commercialized, mass-produced and overused, contributing to the widespread proliferation of antibiotic resistance in bacteria.
As the “chemical shield” of our antibiotic repertoire becomes increasingly fractured, the medical field is again considering the power of phages for treating bacterial infections. The anti-CRISPR proteins described by Bondy-Denomy et al. are likely to be the first of many VSCs with diverse modes of action and these suppressors may prove to be valuable in augmenting phage therapy by enabling targeted suppression of bacterial immune systems.
Acknowledgments
I am grateful to the members of my lab for critical reading and thoughtful feedback on this manuscript. Our research is supported by the National Institutes of Health (GM 103500), the M.J. Murdock Charitable Trust and the Montana State University Agricultural Experimental Station.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/23591
References
- 1.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–27. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 2.Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. 2011;45:273–97. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 3.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–90. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu Rev Genet. 2012;46:311–39. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- 5.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–8. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 6.van der Oost J, Brouns SJ. RNAi: prokaryotes get in on the act. Cell. 2009;139:863–5. doi: 10.1016/j.cell.2009.11.018. [DOI] [PubMed] [Google Scholar]
- 7.Meyerson NR, Sawyer SL. Two-stepping through time: mammals and viruses. Trends Microbiol. 2011;19:286–94. doi: 10.1016/j.tim.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Valen L. A new evolutionary law. Evol Theory. 1973;1:1–30. [Google Scholar]
- 9.Matranga C, Zamore PD. Small silencing RNAs. Curr Biol. 2007;17:R789–93. doi: 10.1016/j.cub.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 10.Obbard DJ, Gordon KHJ, Buck AH, Jiggins FM. The evolution of RNAi as a defence against viruses and transposable elements. Philos Trans R Soc Lond B Biol Sci. 2009;364:99–115. doi: 10.1098/rstb.2008.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waterhouse PM, Wang MB, Lough T. Gene silencing as an adaptive defence against viruses. Nature. 2001;411:834–42. doi: 10.1038/35081168. [DOI] [PubMed] [Google Scholar]
- 12.van Rij RP, Andino R. The silent treatment: RNAi as a defense against virus infection in mammals. Trends Biotechnol. 2006;24:186–93. doi: 10.1016/j.tibtech.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 14.Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–8. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 15.Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–12. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 16.Burgyán J, Havelda Z. Viral suppressors of RNA silencing. Trends Plant Sci. 2011;16:265–72. doi: 10.1016/j.tplants.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–26. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Q, Wang X, Ding SW. Viral suppressors of RNA-based viral immunity: host targets. Cell Host Microbe. 2010;8:12–5. doi: 10.1016/j.chom.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–56. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lintner NG, Kerou M, Brumfield SK, Graham S, Liu H, Naismith JH, et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE) J Biol Chem. 2011;286:21643–56. doi: 10.1074/jbc.M111.238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJ, van der Oost J, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–9. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci USA. 2011;108:10092–7. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell. 2012;45:303–13. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cady KC, Bondy-Denomy J, Heussler GE, Davidson AR, O’Toole GA. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J Bacteriol. 2012;194:5728–38. doi: 10.1128/JB.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garneau JE, Dupuis MÈ, Villion M, Romero DA, Barrangou R, Boyaval P, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 28.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci USA. 2011;108:10098–103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–50. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- 30.Levin BR. Nasty viruses, costly plasmids, population dynamics, and the conditions for establishing and maintaining CRISPR-mediated adaptive immunity in bacteria. PLoS Genet. 2010;6:e1001171. doi: 10.1371/journal.pgen.1001171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinberger AD, Sun CL, Pluciński MM, Denef VJ, Thomas BC, Horvath P, et al. Persisting viral sequences shape microbial CRISPR-based immunity. PLoS Comput Biol. 2012;8:e1002475. doi: 10.1371/journal.pcbi.1002475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, Pasić L, Thingstad TF, Rohwer F, et al. Explaining microbial population genomics through phage predation. Nat Rev Microbiol. 2009;7:828–36. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- 33.Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2012 doi: 10.1038/nature11723. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009;34:401–7. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 35.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–77. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hankin EH. L'action bactericide des eaux de la Jumna et du Gange sur le vibrion du cholera. Ann Inst Pasteur (Paris) 1896;10:511–23. [Google Scholar]
- 39.Keen EC. Phage therapy: concept to cure. Front Microbiol. 2012;3:238. doi: 10.3389/fmicb.2012.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lakatos L, Csorba T, Pantaleo V, Chapman EJ, Carrington JC, Liu YP, et al. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006;25:2768–80. doi: 10.1038/sj.emboj.7601164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin H, Zhu JK. A viral suppressor protein inhibits host RNA silencing by hooking up with Argonautes. Genes Dev. 2010;24:853–6. doi: 10.1101/gad.1927310. [DOI] [PMC free article] [PubMed] [Google Scholar]