Abstract
Races may exist in humans in a cultural sense, but biological concepts of race are needed to access their reality in a non-species-specific manner and to see if cultural categories correspond to biological categories within humans. Modern biological concepts of race can be implemented objectively with molecular genetic data through hypothesis-testing. Genetic data sets are used to see if biological races exist in humans and in our closest evolutionary relative, the chimpanzee. Using the two most commonly used biological concepts of race, chimpanzees are indeed subdivided into races but humans are not. Adaptive traits, such as skin color, have frequently been used to define races in humans, but such adaptive traits reflect the underlying environmental factor to which they are adaptive and not overall genetic differentiation, and different adaptive traits define discordant groups. There are no objective criteria for choosing one adaptive trait over another to define race. As a consequence, adaptive traits do not define races in humans. Much of the recent scientific literature on human evolution portrays human populations as separate branches on an evolutionary tree. A tree-like structure among humans has been falsified whenever tested, so this practice is scientifically indefensible. It is also socially irresponsible as these pictorial representations of human evolution have more impact on the general public than nuanced phrases in the text of a scientific paper. Humans have much genetic diversity, but the vast majority of this diversity reflects individual uniqueness and not race.
Keywords: admixture, evolutionary lineage, gene flow, genetic differentiation, race, human evolution
1. The Biological Meaning of ‘Race’
Many human societies classify people into racial categories. These categories often have very real effects politically, socially, and economically. Even if race is culturally real, that does not mean that these societal racial categories are biologically meaningful. For example, individuals who classify themselves as “white” in Brazil are often considered “black” in the U.S.A., and many other countries use similar or identical racial terms in highly inconsistent fashions (Fish, 2002). This inconsistency is only reinforced when examined genetically. For example, Lao et al. (2010) assessed the geographical ancestry of self-declared “whites” and “blacks” in the United States by the use of a panel of geographically informative genetic markers. It is well known that the frequencies of alleles vary over geographical space in humans. Although the differences in allele frequencies are generally very modest for any one gene, it is possible with modern DNA technology to infer the geographical ancestry of individuals by scoring large numbers of genes. Using such geographically informative markers, self-identified “whites” from the United States are primarily of European ancestry, whereas U.S. “blacks” are primarily of African ancestry, with little overlap in the amount of African ancestry between self-classified U.S. “whites” and “blacks”. In contrast, Santos et al. (2009) did a similar genetic assessment of Brazilians who self-identified themselves as “whites”, “browns”, and “blacks” and found extensive overlap in the amount of African ancestry among all these “races”. Indeed, many Brazilian “whites” have more African ancestry than some U.S. “blacks”. Obviously, the culturally defined racial categories of “white” and “black” do not have the same genetic meanings in the United States and Brazil. The inconsistencies in the meaning of “race” across cultures and with genetic ancestry provide a compelling reason for a biological-based, culture-free definition of race. Another reason is that humans are the product of the same evolutionary processes that have led to all the other species on this planet. The subdivision of a species into groups or categories is not unique to our species. Since evolutionary biology deals with all life on this planet, biologists need a definition of race that is applicable to all species. A definition of “race” that is specific to one human culture at one point of time in its cultural history is inadequate for this purpose. Therefore, a universal, culture-free definition of race is required before the issue of the existence of races in humans (or any other species) can be addressed in a biological context.
The word “race” is not commonly used in the non-human biological literature. Evolutionary biologists have many words for subdivisions within a species (Templeton, 2006). At the lowest level are demes, local breeding populations. Demes have no connotation of being a major subdivision or type within a species. In human population genetics, even small ethnic groups or tribes are frequently subdivided into multiple demes, whereas “race” always refers to a much larger grouping. Another type of subdivision is “ecotype”, which refers to a group of individuals sharing one or more adaptations to a specific environment. Sometimes the defining environmental variable is widespread, so an ecotype can refer to a large geographical population. However, sometimes the environmental heterogeneity can exist on a small geographical scale. In such circumstances, a single local area with no significant genetic subdivision for almost all genes can contain more than one ecotype (e.g., Oberle & Schaal, 2011). Ecotypes are therefore not universally a major subdivision or type within a species, but sometimes merely a local polymorphism. Ecotypes cannot define “race” in a manner applicable to all species, and whether or not ecotypes can define human races will be addressed later. Of all the words used to describe subdivisions or subtypes within a species, the one that has been explicitly defined to indicate major geographical “races” or subdivisions is “subspecies” (Futuyma, 1986, pg. 107–109; Mayr, 1982, pg. 289). Because of this well-established usage in the evolutionary literature, “race” and “subspecies” will be regarded as synonyms from a biological perspective. In this manner, human “race” can be placed into a broader evolutionary context that is no longer species-specific or culturally dependent.
The question of the existence of human “races” now becomes the question of the existence of human subspecies. This question can be addressed in an objective manner using universal criteria. The Endangered Species Act of the USA mandates the protection of endangered vertebrate subspecies (Pennock & Dimmick, 1997). Accordingly, conservation biologists have developed operational definitions of race or subspecies that are applicable to all vertebrates, and two have been used extensively in the non-human literature. These two biological definitions of subspecies or “race” will be applied to humans and to our nearest evolutionary relative, the chimpanzee, in order to avoid an anthropocentric, culture-specific definition of race.
One definition regards races as geographically circumscribed populations within a species that have sharp boundaries that separate them from the remainder of the species (Smith, Chiszar, & Montanucci, 1997). In traditional taxonomic studies, the boundaries were defined by morphological differences, but now these boundaries are typically defined in terms of genetic differences that can be scored in an objective fashion in all species. Most demes or local populations within a species show some degree of genetic differentiation from other local populations, by having either some unique alleles or at least different frequencies of alleles. If every genetically distinguishable population were elevated to the status of race, then most species would have hundreds to tens of thousands of races, thereby making race nothing more than a synonym for a deme or local population. A race or subspecies requires a degree of genetic differentiation that is well above the level of genetic differences that exist among local populations. One commonly used threshold is that two populations with sharp boundaries are considered to be different races if 25% or more of the genetic variability that they collectively share is found as between population differences (Smith, et al., 1997). A common measure used to quantify the degree of differentiation is a statistic known as pairwise fst. The pairwise fst statistic in turn depends upon two measures of heterozygosity. The frequency with which two genes are different alleles given that they have been randomly drawn from the two populations pooled together is designated by Ht, the expected heterozygosity of the total population. Similarly, Hs is the average frequency with which two randomly drawn genes from the same subpopulation are different alleles. Then, fst=(Ht-Hs)/Ht. In many modern genetic studies, the degree of DNA sequence differences between the randomly drawn genes is quantified, often with the use of a model of mutation, instead of just determining if the two DNA sequences are the same or different. When this done, the analysis is called an Analysis of MOlecular VAriation (AMOVA), and various measures of population differentiation analogous to fst exist for different mutation models. Regardless of the specific measure, the degree of genetic differentiation can be quantified in an objective manner in any species. Hence, human races can indeed be studied with exactly the same criteria applied to non-human species. The main disadvantage of this definition is the arbitrariness of the threshold value of 25%, although it was chosen based on the observed amount of subdivision found within many species.
A second definition defines races as distinct evolutionary lineages within a species. An evolutionary lineage is a population of organisms characterized by a continuous line of descent such that the individuals in the population at any given time are connected by ancestor/descendent relationships. Because evolutionary lineages can often be nested together into a larger, more ancestral evolutionary lineage, the evolutionary lineages that are relevant for defining subspecies in conservation biology are the smallest population units that function as an evolutionary lineage within a species. The phylogenetic species concept elevates all evolutionary lineages to the status of species (Cracraft, 1989), but most species concepts allow for multiple lineages to exist within a species. For example, the cohesion species concept defines a species as an evolutionary lineage that maintains its cohesiveness over time because it is a reproductive community capable of exchanging gametes and/or an ecological community sharing a derived adaptation or adaptations needed for successful reproduction (Templeton, 1989, 2001). Two or more evolutionary lineages nested within an older lineage that are capable of exchanging gametes and/or share the same adaptations necessary for successful reproduction are considered lineages nested within a single cohesion species. The biological species concept only uses the criterion of gamete exchangeability and is a proper logical subset of the cohesion concept (Templeton, 1998b; Templeton, 2001). Hence, the biological species concept also allows multiple evolutionary lineages to exist within a species. The possibility of multiple evolutionary lineages within a species is commonly recognized in the area of conservation biology, and indeed the evolutionary lineage definition of race or subspecies has become the dominant definition in much of conservation and evolutionary biology, in large part because it is a natural historical population unit that emerges from modern phylogenetic theory and practice (Amato & Gatesy, 1994; Crandall, Binida-Emonds, Mace, & Wayne, 2000).
Many processes can create an evolutionary lineage. For example, hybridization can create a new lineage by either having the hybrid state stabilized (often through polyploidy) or having a stable recombinant type emerge (Templeton, 1981). This mode for the origin of new lineages is common in plants, but rare in vertebrates (Templeton, 1981). In terrestrial vertebrates, evolutionary lineages are commonly created within a species when an ancestral population is split into two or more subpopulations, often by some sort of geographical barrier, such that there is no or extremely limited genetic interchange after the split (Crandall, et al., 2000). Recall that lineages are defined in terms of ancestor/descendent relationships. DNA is the molecule that is passed on from ancestors to descendents, so genetic surveys provide a direct means of identifying lineages. The primary genetic impact of the establishment of a new evolutionary lineage is that the lineage accumulates genetic differences from the remaining descendents of the ancestral population with increasing time since the split. Immediately after the split, the subpopulations would share most ancestral polymorphisms (gene loci with more than one allele) and would therefore be difficult to diagnose as separate lineages. With increasing time since the split, genetic divergence accumulates and diagnosing the separate lineages becomes easier. A split into separate lineages also means that the genetic differences among the races would define an evolutionary tree analogous to an evolutionary tree of species. Statistical methods exist for testing the null hypothesis that the genetic variation within a species has a tree-like structure, and other statistics test the null hypothesis that the entire sample defines a single evolutionary lineage (Templeton, 1998b, 1999; Templeton, 2001). Therefore, just as with the fst definition, the lineage definition of race can be implemented in an objective fashion using uniform criteria, thereby avoiding an anthropocentric or cultural definition of race.
It is critical to note that genetic differentiation alone is insufficient to define a subspecies or race under either of these definitions of race. Both definitions require that genetic differentiation exists across sharp boundaries and not as gradual changes, with the boundaries reflecting the historical splits. These sharp boundaries are typically geographic, but not always. For example, even non-genetic behavioral differences, such as learned song dialects in birds or linguistic boundaries in humans, can serve as the basis for a sharp genetic boundary when these non-genetic traits are associated with evolutionary history. The fst definition in addition requires that the genetic differentiation across the geographical boundary exceeds a quantitative threshold, and the evolutionary lineage definition requires that the genetic differentiation fits a tree-like evolutionary structure. Hence, genetic differentiation is necessary but not sufficient to infer a race. Human populations certainly show genetic differences across geographical space, but this does not necessarily mean that races exist in humans.
2. To Tree Or Not To Tree; That Is The Question
Genetic differentiation will arise among a set of populations when they have been split historically into isolated lineages, but genetic differentiation can also arise from recurrent but restricted genetic interchange or gene flow among the populations with no historical splits (Templeton, 2006). For example, restricted gene flow can occur when most dispersal is limited to nearby local populations. Because genes are passed on from generation to generation, a new allele can still spread throughout a species’ range over multiple generations by using nearby populations as “stepping-stones” to reach more distant populations. Stepping stone models yield a pattern of genetic differentiation known as isolation-by-distance in which the degree of genetic differentiation between two populations increases with increasing geographical distance between them. Geographical distance is often not a straight-line distance, but rather a distance measure than can incorporate known dispersal barriers, such as oceans for a terrestrial species. It is also possible to include non-geographic factors, such as song dialect or language, into a distance measure. All the populations within a species could be in an interconnected network of stepping-stones such that there are no sharp genetic boundaries separating populations. Instead, genetic differentiation exists in a gradual, clinal pattern across the appropriate distance measure. Therefore, genetic differentiation alone does not imply a tree-like structure, which should always be regarded as a hypothesis to be tested and not assumed (Smouse, 1998). There are multiple ways of testing the fit of the genetic data to an evolutionary tree (called “treeness”).
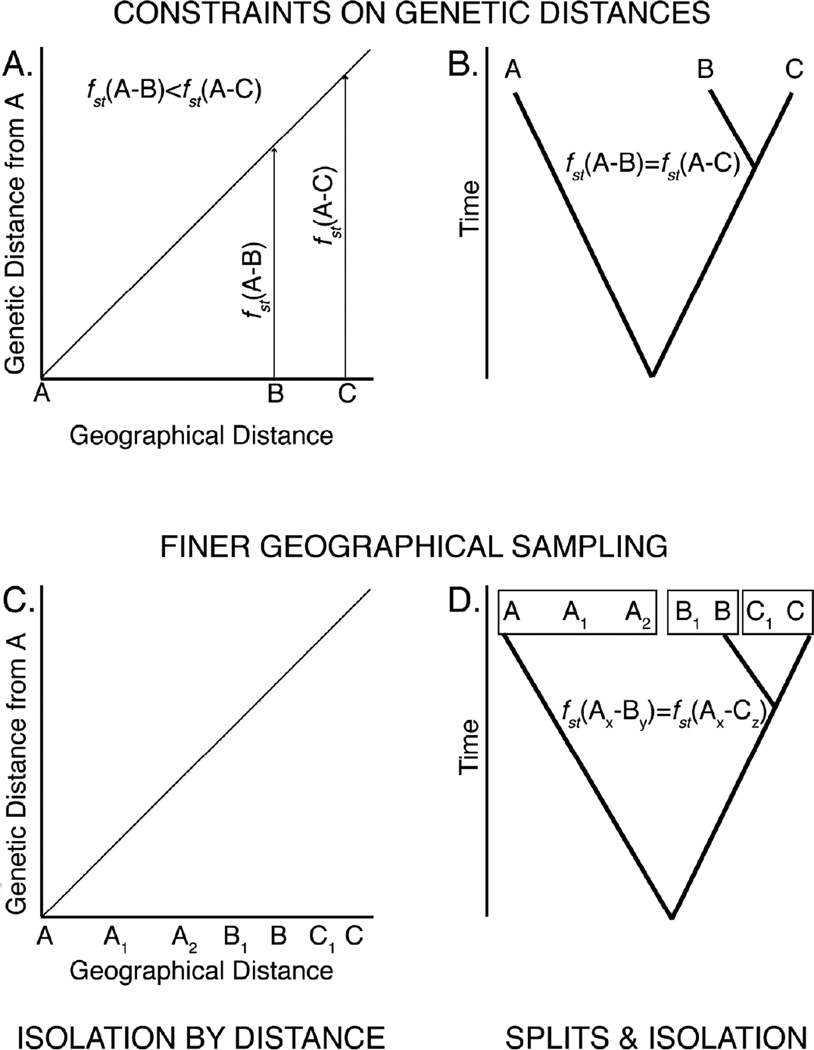
One method of testing for treeness is based on the constraints imposed on patterns of genetic differentiation under a tree-like structure of splits, as shown in Figure 1. Figure 1 shows three hypothetical populations (A, B, and C) such that B and C are the closest geographical pair, but with A closer to B than to C (Figure 1.A). Under isolation-by-distance, the genetic distance (measured, say, by the fst value between a pair of populations) should increase with increasing geographical distance. In particular, Figure 1.A shows a line that represents the pairwise fst between population A and any other population at a given geographical distance from A. As can be seen in Figure 1.A, the fst between A and B is less than the fst between A and C under isolation by distance. In contrast, Figure 1.B shows populations A, B and C as representing separate evolutionary lineages (races) such that A split from the common ancestral population of B and C in the past, followed by a more recent split between populations B and C. This results in an evolutionary tree of populations such that genetic distance between any two populations should be proportional to the time at which they split from a common ancestral population in the tree. In this hypothetical case, the genetic distances between populations A and B and between populations A and C should be the same since they both involve a split from the same ancestral population. Hence, the pattern of genetic distances and the constraints they should obey differ for evolutionary trees versus isolation-by-distance. The constraints imposed by an evolutionary tree (treeness) provide a basis for quantitatively testing the fit of the data to the tree hypothesis. The cophenetic correlation measures how well the observed genetic distances fit the predicted genetic distances from an evolutionary tree model and provides a heuristic goodness of fit to treeness (Rohlf, 1993). Long and Kittles (2003) provide an update to a log-likelihood ratio test of the null hypothesis of treeness first proposed by Cavalli-Sforza and Piazza (1975). Either or both of these tests can be applied to a genetic data set to make an objective assessment of whether or not an evolutionary tree is an appropriate model for the populations found within a species.
Figure 1.
Distinguishing between population trees arising from splits and isolation versus recurrent gene flow with isolation by distance. In all cases, the symbol “ fst(X–Y)” indicates the genetic distance between populations X and Y. Part A graphs the expected relationship between a genetic distance from a reference population (population “A”) to other populations (“B” and “C”) as a function of their geographical distance. Part B indicates the genetic distance between two populations as the sum of the branch lengths that interconnect them in an evolutionary tree. Parts C and D show how finer geographical sampling affects these relationships.
Another method for testing for a tree-like structure is based upon finer geographical sampling (Figure 1.C and 1.D). As more sites are sampled under an isolation-by-distance model, the geographically intermediate populations should also have intermediate genetic distances (Figure 1.C). In contrast, when the populations are grouped into a smaller number of evolutionary lineages, genetic distances among populations within a lineage should be relatively small, although they may show an isolation-by-distance pattern within the geographical range occupied by a particular lineage. However, the genetic distances are expected to show a large, sudden increase when crossing the geographical boundary between two lineages. For example, all the “A” lineage populations in Figure 1.D (A, A1, and A2) should show the same genetic distance to any of the “B” lineage populations (B and B1), in great contrast to the isolation by distance model in which A2 and B should have a smaller genetic distance (because they are geographically close in Figure 1.C) than the genetic distance between A and A2, which are geographically farther apart.
A more direct method for testing for evolutionary lineages within a species is multi-locus nested clade phylogeographic analysis (NCPA). This technique uses genetic data only from those regions of the genome that show little to no recombination. Mitochondrial DNA and most Y-chromosome DNA fall into this category, as well as much nuclear DNA because recombination in mammals and other organisms is concentrated into hotspots. The evolutionary history of how mutations are accumulated over time and space is written most clearly in regions with little or no recombination, so this subset of the genome is particularly informative about past evolutionary events and processes. In such regions, haplotypes can be constructed that specify a specific genetic state for every polymorphic site within the region of little to no recombination. These haplotypes in turn can then be used to estimate an evolutionary tree of haplotypes that indicates how mutational changes were accumulated in this genomic region over time. These haplotype trees are not population trees. Indeed, in a species that is completely panmictic throughout its range, there are no subpopulations at all and therefore a population tree cannot even exist. Nevertheless, such a species will have haplotype trees for every genomic region that has no recombination. However, splits followed by isolation can have a large effect on the haplotype tree and the spatial distribution of different branches of the haplotype tree (Templeton, Routman, & Phillips, 1995). NCPA is a statistical method of extracting phylogeographic inferences from haplotype trees that can be coupled with a maximum likelihood hypothesis testing framework (A.R. Templeton, 2004). The inference criteria have been validated by the use of 150 positive controls (cases in which prior information exists about past events) and found to be accurate with a false positive rate of 4% per tested clade (branch of the haplotype tree) per locus when the nominal level of significance is set at 5% (A. R. Templeton, 2004). A strong vindication of NCPA inferences specifically for human evolution has emerged from studies on ancient DNA. Most models of human evolution have anatomically modern humans arising in Africa, and then spreading out to the rest of the world. Because the genus Homo was already in Eurasia at the time of this out-of-Africa expansion, the question arises as to how the expanding modern population interacted with the pre-existing Eurasian populations. NCPA was the only method of phylogeographic analysis to reject the null hypothesis of no admixture with strong statistical significance (Templeton, 2002; Templeton, 2005); that is, there was genetic interchange among these populations at a low level (the mostly out-of-Africa hypothesis). This rejection of no admixture occurred at a time during which most genetic analyses were interpreted as supporting the out-of-Africa replacement hypothesis, including studies that assigned a low probability to any hypothesis with even a slight amount of admixture (Fagundes et al., 2007). With advances in molecular genetics, it became possible to directly test for admixture by examining DNA extracted from human fossils. Two fossil Eurasian populations have been examined, and both indicate low levels of admixture (Green et al., 2010; Reich et al., 2010), thereby supporting the mostly-out-of-Africa hypothesis and rejecting replacement. No other method of phylogeographic inference has been subjected to such a thorough validation through its performance with actual data sets.
Computer simulation also validates the accuracy of NCPA inferences. Knowles and Maddison (2002) simulated a situation of splits followed by isolation that occurred in large populations over a short time scale; a situation that leads to much retention of ancestral polymorphism and lineage sorting which in turn can make many methods of inference perform poorly. Indeed, Knowles and Maddison (2002) reported that a coalescent simulation approach performed poorly with these simulated data. NCPA utilizes local contrasts of branches (clades) within the haplotype tree and not the overall haplotype tree topology. As a consequence, NCPA can deal well with the difficulties caused by ancestral polymorphism, lineage sorting, and even some gene flow or introgression after the initial split of populations (Templeton, 2009a). It is therefore not surprising that when multi-locus (ML) NCPA was applied to the difficult situation simulated by Knowles and Maddison (2002) that the splits and their temporal sequence were identified with 100% accuracy with no false positives (Templeton, 2009b).
ML-NCPA can be used to identify and test for evolutionary lineages within a species in both a positive and negative sense. First, ML-NCPA can infer past population fragmentation events; that is, cases in which a geographically contiguous subset of the species splits off from the remainder of the species and behaves primarily as an isolated lineage or race after the split, although some limited gene flow or introgression could still occur. Such races are strong inferences that arise from rejecting the null hypothesis of no fragmentation. ML-NCPA can identify such lineages within a species before they define monophyletic groups in a species tree and even when there is some limited gene flow and introgression after the split (see the example of lineages of African elephants in Templeton, 2009a). Moreover, NCPA can identify populations that behaved as isolated lineages in the past even though they are undergoing current admixture (Templeton, 2001). Thus, “races” under the positive evolutionary lineage criteria of ML-NCPA need not be monophyletic groups nor totally genetically isolated. Moreover, computer simulations reveal that the positive criteria of inferring evolutionary lineages via multi-locus NCPA have extremely low rates of false positives, varying from 0.00% to 0.21% for 5% nominal levels under various simulated scenarios (Panchal & Beaumont, 2010). ML-NCPA can also be used to test the null hypothesis that populations from two geographical regions have experienced no gene flow in a specified time period (Templeton, 2009a). In this case, rejection of the null hypothesis indicates that separate evolutionary lineages do not exist. Panchal and Beaumont claimed that their simulations reveal that this inference is subject to a high false positive rate “because there is no stipulation that the inferences should be concordant across time” (Panchal & Beaumont, 2010, pg. 418). However, likelihood ratio tests for concordance across a time interval for gene flow inferences are part of ML-NCPA (Templeton, 2009a, equation 2; A.R. Templeton, 2004, equation 12), so this conclusion of Panchal and Beaumont is due to a fundamental misrepresentation of ML-NCPA. Whenever Panchal and Beaumont actually implemented ML-NCPA, the false positive rates were always below the nominal rate. Thus, ML-NCPA provides an appropriate statistical framework for testing for the existence of races as evolutionary lineages within a species in both a positive and negative sense.
As shown in this section, the question of to tree or not to tree can be tested in a statistically objective fashion with a variety of techniques and molecular genetic data. This allows an objective assessment of races as evolutionary lineages in any species.
3. Do Races Exist in Chimpanzees?
3.1. Do Races Exist in Chimpanzees Using fst Thresholds?
To avoid the emotional issues associated with race in humans, the threshold and lineage definitions of race will first be applied to our closest evolutionary relative, the chimpanzee. The common chimpanzee (Pan troglodytes) has been traditionally subdivided into five races or subspecies on the basis of morphological differences: P .t. verus in the Upper Guinea region of western Africa, P. t. ellioti in the Gulf of Guinea region (southern Nigeria and western Cameroon), P. t. troglodytes in central Africa, P. t. schweinfurthii in the western part of equatorial Africa (mostly southern Cameroon), and P. t. marungensis in central and eastern equatorial Africa. Gonder et al. (2011) surveyed chimpanzees throughout their range and found that sharp genetic differences separate the Upper Guinea and Gulf of Guinea populations from each other and from all other populations, but with less sharp genetic boundaries between the equatorial African populations. Table 1 shows the pairwise AMOVA results for these population contrasts. The Upper Guinea and Gulf of Guinea populations are above the 25% threshold for contrasts with each other and with all other chimpanzee populations. However, the three regions sampled in equatorial Africa are all well below the 25% threshold used for the recognition of subspecies. Hence, there are three races or subspecies of common chimpanzees using the threshold criterion: P .t. verus in the Upper Guinea region, P. t. ellioti in the Gulf of Guinea region, and the chimpanzee populations from equatorial Africa, which includes three of the traditional morphological subspecies.
Table 1.
Genetic differentiation among populations of chimpanzees as measured by Rst, a measure related to fst but that incorporates a mutational model for microsatellites (modified from Gonder et al. 2011).
| Upper Guinea | Gulf of Guinea | Southern Cameroon | Central Africa | |
|---|---|---|---|---|
| Gulf of Guinea | 0.41 | |||
| Southern Cameroon | 0.43 | 0.25 | ||
| Central Africa | 0.46 | 0.27 | 0.07 | |
| Eastern Africa | 0.44 | 0.28 | 0.05 | 0.03 |
3.2. Do Races Exist in Chimpanzees As Evolutionary Lineages?
Gonder et al (2011) also used the chimpanzee genetic data to estimate an evolutionary tree of populations. The resulting tree has the Upper Guinea population splitting off first, followed by the Gulf of Guinea population, and then splits among the equatorial Africa populations. This tree predicts that the Upper Guinea population should be equally distant from all the other populations, and Table 1 shows that this prediction is supported when the error in estimating the distances is taken into account. This tree also predicts that the Gulf of Guinea population should be equally distance from all the equatorial African populations, but that this distance should be smaller (less time since the split) than the distances involving the Upper Guinea population. Table 1 shows that this prediction is also supported. However, Gonder et al (2011) also showed that the genetic distances among the three equatorial African populations follow an isolation-by-distance pattern on an east-west axis and do not define a tree-like structure. These three populations are therefore collapsed into a single lineage. Hence, chimpanzees do show a partial tree-like structure of genetic differentiation with three lineages: Upper Guinea, Gulf of Guinea, and the combined equatorial African populations. Hence, races do exist in chimpanzees under the lineage definition, and they correspond exactly to the same three races defined by the quantitative threshold definition of race.
4. Do Biological Races Exist in Humans?
4.1. Genetic Differentiation Among Human Populations
Do human races exist using the same criteria applied to chimpanzees? Rosenberg et al. (2002) performed a genetic survey of 52 human populations. They used a computer program to sort individuals or portions of their genomes into five groups, and discovered that the genetic ancestry of most individuals was inferred to come from just one group. Moreover, the five groups corresponded to 1) sub-Saharan Africans; 2) Europeans, Near & Middle Easterners, and Central Asians; 3) East Asians; 4) Pacific populations; and 5) Amerindians. This paper was the most widely cited article from the journal Science in 2002, and many of these citations claimed that this paper supported the idea that races were biologically meaningful in humans (e.g., Burchard et al., 2003). However, Rosenberg et al. (2002) were more cautious. When they increased the number of groups beyond five, they also obtained an excellent classification into smaller, more regional groups. Hence, they were showing that with enough genetic markers, it is possible to discriminate most local human populations from one another. Recall that genetic differentiation alone does not necessarily mean that any of these groups are races.
4.2. Do Races Exist in Humans Using fst Thresholds?
Assuming for now that the five major geographical groups are the meaningful populations, do these groups satisfy the quantitative threshold definition of race? Table 2 shows the AMOVA results for these five human groups, along with a comparable analysis of the three races of chimpanzees that satisfy both the threshold and lineage definitions of race. Table 2 shows how the genetic variation is partitioned into differences among individuals within the same local population, differences between local populations within the same “race”, and between “races”. Table 2 confirms the reality of race in chimpanzees using the threshold definition, as 30.1% of the genetic variation is found in the among-race component, a result expected from the pairwise analysis shown in Table 1. In contrast to chimpanzees, the five major “races” of humans account for only 4.3% of human genetic variation – well below the 25% threshold. The genetic variation in our species is overwhelmingly variation among individuals (93.2%).
Table 2.
AMOVA of genetic variation in chimpanzees (data from Gonder et al. 2011) and from humans (data from Rosenberg et al. 2002).
| Genetic Variance Components |
|||||
|---|---|---|---|---|---|
| Species | Number of “Races” |
Number of Populations |
Among Individuals Within Populations |
Among Populations Within Races |
Among Races |
| Chimpanzees | 3 | 5 | 64.2% | 5.7% | 30.1% |
| Humans | 5 | 52 | 93.2% | 2.5% | 4.3% |
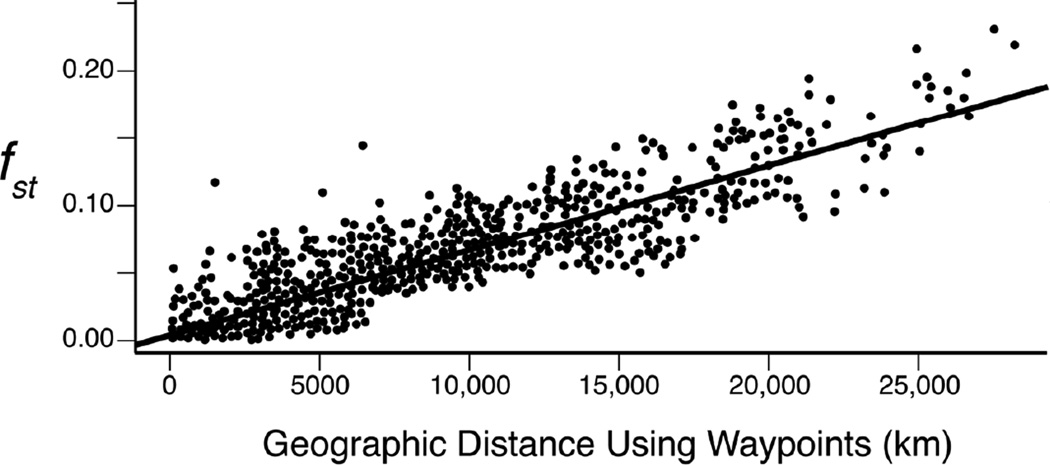
The threshold definition also requires sharp genetic boundaries between the “races.” Figure 2 shows a plot of the pairwise fst values of humans as a function of geographical distance (Ramachandran et al., 2005). As can be seen, the pairwise fst values increase smoothly with increasing geographical distance (in this case based on waypoints to minimize travel across oceans and seas). There are no indications of the discontinuities expected when sharp geographical boundaries of genetic differentiation exist. A more detailed analysis reveals that the spatial patterns of human genetic variation are explained well by a series of long-range migrations and population founder events coupled with gene flow with isolation-by-distance (Hunley, Healy, & Long, 2009). The gene flow arising from long-range migrations and isolation-by-distance has obscured any sharp boundaries that may have temporarily existed after the founder events (Figure 2) as well as has reduced the quantitative amount of genetic differentiation. Consequently, neither aspect of the threshold definition is satisfied; there are no sharp boundaries separating human populations, and the degree of genetic differentiation among human groups, even at the continental level, is extremely low. Using the threshold definition, there are no races in humans.
Figure 2.
Isolation-by-distance in human populations. The x-axis is the geographical distance between two populations, as measured through waypoints that minimize travel over oceans. The y-axis is the pairwise fst between two populations. Modified from Ramachandran et al. (2005). Copyright (2005) National Academy of Sciences, U.S.A.
4.3. Do Races Exist in Humans As Evolutionary Lineages?
Turning to the lineage definition, Figure 2 is consistent with an isolation-by-distance pattern and not a tree-like structure. Hence, using the same criteria for rejecting the racial status of the traditionally named subspecies of chimpanzees from equatorial Africa, the existence of human races as evolutionary lineages is similarly rejected. This rejection is quantified with the cophenetic correlation. The cophenetic correlations for various data sets that have been used to portray human population trees vary from 0.45 to 0.79 (Templeton, 1998a). A tree-like structure of genetic differentiation requires a cophentic correlation greater than 0.9, and any value less than 0.8 is regarded as a poor fit (Rohlf, 1993), so all of these datasets reject the hypothesis of an evolutionary tree of human populations. Similarly, the log-likelihood ratio test for treeness results in a strong rejection of the null hypothesis of treeness for human populations (Long & Kittles, 2003).
Increased geographical sampling further undermines the idea of separate lineages. The geographical sampling of Rosenberg et al. (2002) was coarse. It is now known that the computer program STRUCTURE used in these studies generates well-differentiated populations as an artifact of coarse sampling from any species characterized by isolation-by-distance (Frantz, Cellina, Krier, Schley, & Burke, 2009; Safner, Miller, McRae, Fortin, & Manel, 2011). As pointed out earlier, Figure 2 reveals that human genetic distances fit an isolation-by-distance model. Consequently, it is not surprising that when Behar et al. (2010) sampled Old World populations more finely and used the computer program STRUCTURE, most individuals showed significant genetic inputs from two or more populations, indicating that most human individuals have mixed ancestries and do not belong to a “pure” group. The “races” so apparent to many who cited Rosenberg et al. (2002) simply disappeared with better sampling. These results and Figure 2 further falsify the hypothesis that humans are subdivided into evolutionary lineages.
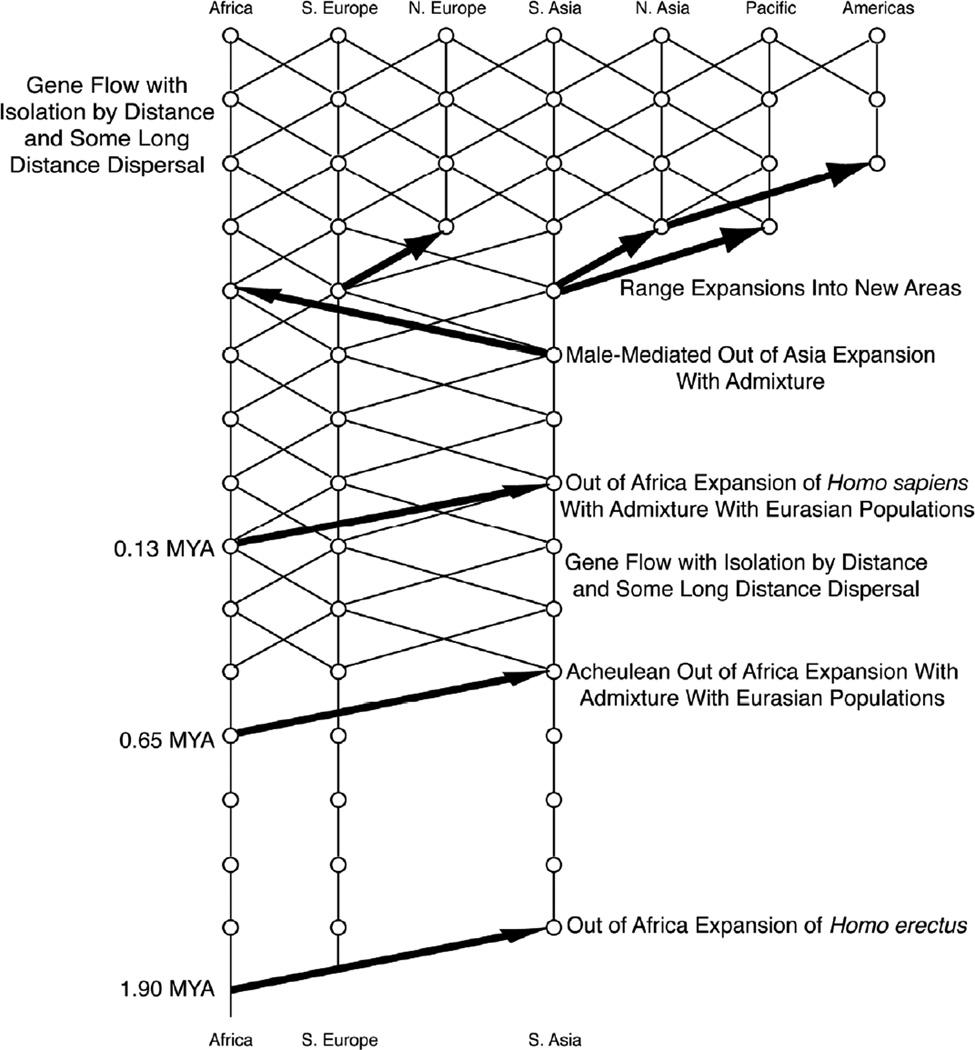
Multi-locus nested clade phylogeographic analysis (ML-NCPA) also undermines the hypothesis of human races as lineages. Figure 3 shows the inferences from ML-NCPA about human evolution based on 25 regions of the human genome (Templeton, 2005, 2007) with some modifications due to a new test for testing the null hypothesis of no gene flow between two regions over a specified time interval in the past (Templeton, 2009a). The oldest inferred event is an out-of-Africa range expansion into Eurasia that is genetically dated to about 1.9 million years ago – the same time that the fossil evidence indicates that Homo erectus spread out of Africa into Eurasia during a major wet period in the Sahara. There were seven inferences of gene flow constrained by isolation-by-distance involving both African and Eurasian populations that were dated between the first out-of-Africa expansion at 1.9 Ma and 650,000 years ago, but applying the likelihood ratio test of the null hypothesis of isolation (no gene flow) between Africa and Eurasia given in Templeton (2009a) yields a log-likelihood ratio statistic of 11.86 with 7 degrees of freedom, which is not significant at the 5% level. Recurrent gene flow with isolation by distance between Africa and Eurasia is therefore suggested but not cross-validated for this early part of the Pleistocene, so no cross lines indicating gene flow are depicted in Figure 3 between the major geographical lineages of humans established by the first out-of-Africa expansion during the early Pleistocene.
Figure 3.
Significant inferences about human evolution from multi-locus, nested-clade phylogeographic analysis. Geographical location is indicated on the x-axis, and time on the y-axis, with the bottom of the figure corresponding to two million years ago. Vertical lines indicate genetic descent over time, and diagonal lines indicate gene flow across space and time. Thick arrows indicate statistically significant population range expansions, with the base of the arrow indicating the geographical origin of the expanding population. Lines of descent are not broken because the population range expansion events were accompanied by statistically significant admixture when they involved expansion into previously inhabited areas. Modified from Templeton (2005).
The next major event shown in Figure 3 is a second population expansion out of Africa into Eurasia around 650,000 years ago, corresponding to the spread of the Acheulean tool culture out of Africa into Eurasia during the second major Saharan wet period of the Pleistocene. The null hypothesis of no admixture between the expanding population and the Eurasian populations is rejected with a probability level of 0.035. Hence, the Acheulean expansion was marked by genetic interchange between African and Eurasian populations, further weakening the hypothesis of isolated Pleistocene lineages of humans. Statistically significant (p ≤ 0.13) recurrent gene flow with isolation by distance is inferred between African and Eurasian populations until a third major expansion of humans out-of-Africa into Eurasia occurred around 130,000 years ago, the time of the last major Saharan wet period. Accordingly, a trellis structure indicating gene flow rather than isolated lineages is depicted after the Acheulean expansion in Figure 3. The fossil record indicates that modern humans began expanding out of sub-Saharan Africa at 130,000 years ago (Vanhaeren et al., 2006) and reached China no later than 110,000 to 100,000 years ago (Jin et al., 2009; Liu et al., 2010). The null hypothesis of no admixture is overwhelmingly rejected for this expansion event with a probability level less than 10−17. As noted earlier, the inference of admixture has since been directly supported by studies on fossil DNA of archaic Eurasian populations (Green, et al., 2010; Reich, et al., 2010).
Following the expansion with admixture of modern humans from Africa, there have been additional expansions, mostly into areas not formerly occupied by humans (Figure 3). Wherever humans lived, gene flow was soon established, mostly limited by isolation-by-distance but with some long-distance dispersal in more recent times. These inferences of long distance range expansions followed by gene flow mostly constrained by isolation by distance have been subsequently supported by extensive computer simulations (Hunley, et al., 2009). On a time scale of tens of thousands of years (the temporal resolution of the ML-NCPA studies), there is not one statistically significant inference of splitting during the last 1.9 million years. Hence, the null hypothesis of a single human lineage is not rejected, so there is no evidence for lineage races in humans. Furthermore, ML-NCPA strongly rejects the null hypotheses of no gene flow and no admixture under the null hypothesis that isolated lineages did exist, so there is strong evidence against lineage races in humans. Hence, there are no races in humans under the lineage definition.
5. Are Human Races Defined By Adaptive Traits?
Races, when they exist, occupy a subset of the geographical range of their species. Sometimes environmental factors vary over the geographical range of the species, and some of these environmental factors induce natural selection that results in local adaptation. Hence, when races exist, they sometimes display local adaptations to the environment associated with their geographical sub-range that are not adaptive in the remainder of the species’ geographical range. In such cases, these geographic subpopulations also represent an ecotype. As stated earlier, the ecotype concept can be and has been applied to many different types of populations and even individuals within a local area, but at least in some circumstances in some species an ecotype and subspecies or race can refer to the same populations. This reasoning leads to the idea that local adaptations can sometimes be biological markers of racial status in humans; that is, human races are ecotypes (Pigliucci & Kaplan, 2003). However, human ecotypes do not correspond to races under either subspecies definition. Even the advocates of the ecotype race concept acknowledge that the same adaptation can arise independently in different parts of the species’ range (Pigliucci & Kaplan, 2003). Hence, ecotypes do not in general correspond to evolutionary lineages, and specifically human ecotypes cannot correspond to evolutionary lineages in humans since the hypothesis of multiple evolutionary lineages within the human species is rejected, as noted earlier. Moreover, variation in environmental factors can induce natural selection that results in local adaptations even in species that are not genetically subdivided at all (Templeton, 2006); that is, the ecotypes are only genetically differentiated at the gene loci under selection and show little to no genetic differentiation over the remainder of the genome. In these cases, the geographic distributions of the local adaptations reflect the geography of environmental factors and not boundaries of overall genetic differentiation. Hence, the ecotype concept in general does not correspond to populations demarcated by sharp boundaries of genetic differentiation that exceed some threshold. This is also certainly the case in humans because there are no human populations with sharp genetic boundaries that exceed the thresholds used in the non-human literature.
One solution to these problems of using ecotypes as races in humans is to simply abandon the subspecies concept of race entirely and define human races solely through ecotype status (Pigliucci & Kaplan, 2003). This solution does not solve the main problem that the subspecies concept of race addressed: avoiding cultural biases in a definition of human race. All species, humans included, adapt to many environmental factors, not just one. Frequently, different adaptive traits display discordant geographical distributions because the underlying environmental factors have discordant geographical distributions. As a result, one will get different ecotypes for different adaptations. This is not a problem for how the ecotype concept is used in the general evolutionary literature, but it raises a critical problem for implementing the ecotype concept as a definition of race in humans. Depending upon which adaptive trait is chosen, one will get very different “races”. So which adaptive traits should be used and which should be ignored? Evolutionary biology provides no objective way of addressing the question of choice of adaptive traits, so the ecotype concept of race in humans is yet another subjective, culturally sensitive concept of “race.”
Skin color is historically the locally adaptive trait most commonly considered by European cultures as a “racial trait” in humans. Skin color is an adaptation to the amount of ultraviolet (uv) radiation in the environment: dark skins are adaptive in high uv environments in order to protect from radiation damage that can kill and burn cells and damage DNA if not protected by melanin, and light skins are adaptive in low uv environments in order to make sufficient vitamin D, which requires uv (Hochberg & Templeton, 2010; Jablonski & Chaplin, 2010). The geographical distribution of skin color follows the environmental factor of uv intensity. Skin color differences do not reflect overall genetic divergence. For example, the native peoples with the darkest skins live in tropical Africa and Melanesia. The dark skins of Africans and Melanesians are adaptive to the high uv found in these areas. Because Africans and Melanesians live on opposite sides of the world, they are more highly genetically differentiated than many other human populations (Figure 2) despite their similar skin colors. Europeans, who are geographically intermediate between Africa and Melanesia, are likewise intermediate at the molecular genetic level between Africans and Melanesians, even though Europeans have light skins that are adapted to the low uv environment of Europe. Skin color differences in humans are not a reliable indicator of overall genetic differentiation or evolutionary history. Moreover, skin color varies continuously among humans in a clinal fashion rather than categorical ecotypes (Relethford, 2009). Hence, there is a compelling biological reason to exclude skin color as the racially-defining adaptive trait under the ecotype concept of race.
Another adaptive trait in humans is resistance to malaria, which is widespread in African populations. However, malaria is also common in some areas outside of Africa, and malarial resistance is found in many European and Asian populations as well. Indeed, one of the alleles underlying malarial resistance, the sickle-cell allele, has its highest frequency in certain populations on the Arabian Peninsula and in India despite frequently being regarded as a disease of “blacks”. Each adaptive trait in humans has its own geographical distribution that reflects the distribution of the underlying environmental factor for which it is adaptive. The discordance in the distributions of adaptive traits in humans means that different adaptive traits will define different ecotypes/”races”. No guidance, other than cultural preference, is given for choosing which adaptive trait should define the ecotypes that are regarded as races, and which ecotypes should not be regarded as races. Hence, equating ecotypes to races, even if limited just to humans, does not yield an objective, culture-free definition of race.
6. A Trellis or a Tree?
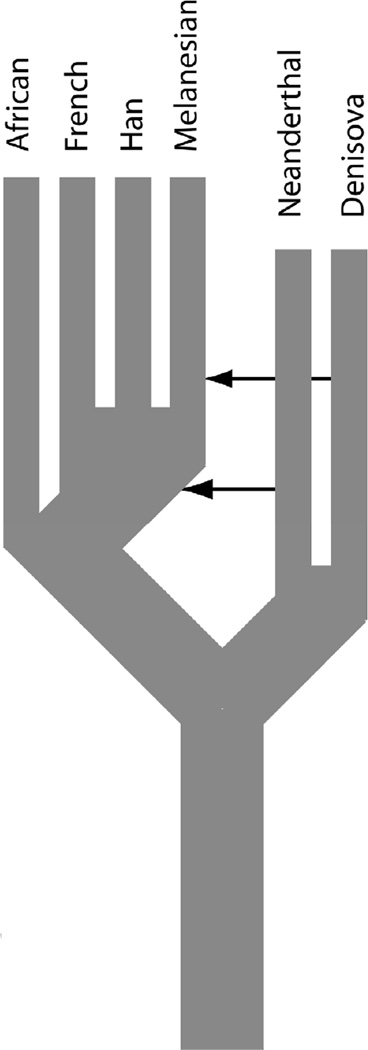
The imagery of recent human evolution is dominated by evolutionary trees of human populations. Human populations are shown again and again as separate branches on an evolutionary tree, related to other human populations by splits that occurred at specific times in the past. Even papers that document genetic interchange among human populations, such as the recent papers on admixture with archaic populations (Green, et al., 2010; Reich, et al., 2010), place human populations on an evolutionary tree with only weak arrows indicating isolated events of admixture that minimally violate an otherwise tree-like structure (see Figure 4, adapted from Reich, et al., 2010). In particular, as is typical of the human population genetic literature, Africans are portrayed in Figure 4 as having “split” from the rest of humanity a long time ago with not one episode of genetic interchange being portrayed since that ancient “population separation” (Reich, et al., 2010, pg. 1058).
Figure 4.
A population tree of humans with arrows indicating admixture from archaic human populations in the past. Modified from Reich et al. (2010).
Contrast Figure 4 to Figure 3, which also depicts recent human evolution. All aspects of Figure 3 are supported by explicit hypothesis testing and statistically significant inferences. Indeed, as shown in this paper, our evolutionary history has been dominated by gene flow and admixture that unifies humanity into a single evolutionary lineage, as shown by the trellis structure and arrows of expansion that overlay upon, not replace, earlier populations. This finding does not mean that all human populations are genetically identical. Past founder events, isolation-by-distance, and other restrictions on gene flow ensure that human populations are genetically differentiated from one another, and local adaptation ensures that some of these differences reflect adaptive evolution to the environmental heterogeneity that our globally distributed species experiences. However, most of our genetic variation exists as differences among individuals, with between population differences being very small. In every case in which treeness has been tested for human populations, it has been rejected. In contrast, the evolutionary trees found throughout the human genetic literature, such as that portrayed in Figure 4, are simply invoked. There is no hypothesis testing, even though treeness or multiple lineages are testable hypotheses. Simply invoking conclusions without testing them is scientifically indefensible; yet, that is the norm for population trees in much of the human evolution literature.
Many of the papers that portray human population trees caution in the text that the populations are not truly genetically isolated, but this makes the tree portrayal even less defensible as the authors are knowingly portraying human evolution in a false fashion. Moreover, it is socially irresponsible. Scientific papers on human genetics and evolution often attract much attention in the popular media, and it is the pictures and figures from these papers that are primarily transmitted to the general public and not nuanced text. That is certainly the case for Figure 4, which has appeared in various versions in many newspapers and websites (e.g., http://ufomaniacs.blogspot.com/2010/12/ancient-humansthe-denisovansinterbred.html). The message of these figures is both that there was some admixture with archaic human populations in the past and also that Africans have been separated from the rest of humanity for a long period of time with no genetic interchange. Africans in these figures are clearly presented as a distinct lineage of human evolution; that is, a separate race. This pictorial conclusion has been definitively falsified as indicated in this paper, so figures such as Figure 4 mislead, not educate, the public about our scientific knowledge of human evolution and race.
Scientists should take seriously what their work communicates to the general public. If they applied the most straightforward concept of science, the idea that hypotheses should be tested whenever possible, then human evolutionary trees such as Figure 4 would disappear and would be replaced by trellises that emphasize the genetic interconnections among all humans on this planet. Humans are an amazingly diverse species, but this diversity is not due to a finite number of subtypes or races. Rather, the vast majority of human genetic diversity reflects local adaptations and, most of all, our individual uniqueness.
Acknowledgements
I wish to thank Quayshawn Spencer for organizing a symposium on race, his encouragement and patience in translating my symposium address into this paper, and his many insightful critiques of an earlier draft. This work is supported by NIH grant P50-GM65509.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amato G, Gatesy J. PCR assays of variable nucleotide sites for identification of conservation units. In: Schierwater B, Streit B, Wagner GP, DeSalle R, editors. Molecular Ecology and Evolution: Approaches and Applications. Basel: Birkhäuser Verlag; 1994. pp. 215–226. [DOI] [PubMed] [Google Scholar]
- Behar Doron M, Yunusbayev Bayazit, Metspalu Mait, Metspalu Ene, Rosset Saharon, Parik Juri, Villems Richard. The genome-wide structure of the Jewish people. [10.1038/nature09103] Nature. 2010;466(7303):238–242. doi: 10.1038/nature09103. doi: http://www.nature.com/nature/journal/v466/n7303/abs/nature09103.html#supplementary-information. [DOI] [PubMed] [Google Scholar]
- Burchard EG, Ziv E, Coyle N, Gomez SL, Tang H, Karter AJ, Risch N. The importance of race and ethnic background in biomedical research and clinical practice. New England Journal Of Medicine. 2003;348(12):1170–1175. doi: 10.1056/NEJMsb025007. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Piazza A. Analysis of evolution: evolutionary rates, independence, and treeness. Theoretical Population Biology. 1975;37:3–25. doi: 10.1016/0040-5809(75)90029-5. [DOI] [PubMed] [Google Scholar]
- Cracraft J. Species as entities of biological theory. In: Ruse M, editor. What the Philosophy of Biology Is. Dordrecht, Netherlands: Kluwer Academic Publishers; 1989. pp. 31–52. [Google Scholar]
- Crandall KA, Binida-Emonds ORP, Mace GM, Wayne RK. Considering evolutionary processes in conservation biology. Trends In Evolution and Ecology. 2000;15:290–295. doi: 10.1016/s0169-5347(00)01876-0. [DOI] [PubMed] [Google Scholar]
- Fagundes Nelson JR, Ray Nicolas, Beaumont Mark, Neuenschwander Samuel, Salzano Francisco M, Bonatto Sandro L, Excoffier Laurent. Statistical evaluation of alternative models of human evolution. Proceedings of the National Academy of Sciences. 2007;104(45):17614–17619. doi: 10.1073/pnas.0708280104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish JM. The myth of race. In: Fish JM, editor. Race and Intelligence: Separating Science From Myth. Mahwah, New Jersey: Lawrence Erlbaum Associates; 2002. pp. 113–141. [Google Scholar]
- Frantz AC, Cellina S, Krier A, Schley L, Burke T. Using spatial Bayesian methods to determine the genetic structure of a continuously distributed population: clusters or isolation by distance? Journal of Applied Ecology. 2009;46(2):493–505. [Google Scholar]
- Futuyma DJ. Evolutionary Biology. 2nd ed. Sunderland, Massachusetts: Sinauer Associates, Inc; 1986. [Google Scholar]
- Gonder Mary Katherine, Locatelli Sabrina, Ghobrial Lora, Mitchell Matthew W, Kujawski Joseph T, Lankester Felix J, Tishkoff Sarah A. Evidence from Cameroon reveals differences in the genetic structure and histories of chimpanzee populations. Proceedings of the National Academy of Sciences. 2011;108(12):4766–4771. doi: 10.1073/pnas.1015422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green Richard E, Krause Johannes, Briggs Adrian W, Maricic Tomislav, Stenzel Udo, Kircher Martin, Paabo Svante. A Draft Sequence of the Neandertal Genome. Science. 2010;328(5979):710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Z, Templeton AR. Evolutionary perspective in skin color, vitamin D and its receptor. HORMONES. 2010;9(4):307–311. doi: 10.14310/horm.2002.1281. [DOI] [PubMed] [Google Scholar]
- Hunley Keith L, Healy Meghan E, Long Jeffrey C. The global pattern of gene identity variation reveals a history of long-range migrations, bottlenecks, and local mate exchange: Implications for biological race. American Journal of Physical Anthropology. 2009;139(1):35–46. doi: 10.1002/ajpa.20932. [DOI] [PubMed] [Google Scholar]
- Jablonski Nina G, Chaplin George. Human skin pigmentation as an adaptation to UV radiation. Proceedings of the National Academy of Sciences. 2010;107(Supplement 2):8962–8968. doi: 10.1073/pnas.0914628107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin ChangZhu, Pan WenShi, Zhang YingQi, Cai YanJun, Xu QinQi, Tang ZhiLu, Cheng Hai. The Homo sapiens Cave hominin site of Mulan Mountain, Jiangzhou District, Chongzuo, Guangxi with emphasis on its age. Chinese Science Bulletin. 2009;54(21):3848–3856. [Google Scholar]
- Knowles LLacey, Maddison Wayne P. Statistical phylogeography. Molecular Ecology. 2002;11:2623–2635. doi: 10.1046/j.1365-294x.2002.01637.x. [DOI] [PubMed] [Google Scholar]
- Lao O, Vallone PM, Coble MD, Diegoli TM, van Oven M, van der Gaag KJ, Kayser M. Evaluating Self-declared Ancestry of US Americans With Autosomal, Y-chromosomal and Mitochondrial DNA. Human Mutation. 2010;31(12):E1875–E1893. doi: 10.1002/humu.21366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Wu, Jin Chang-Zhu, Zhang Ying-Qi, Cai Yan-Jun, Xing Song, Wu Xiu-Jie, Wu Xin-Zhi. Human remains from Zhirendong, South China, and modern human emergence in East Asia. Proceedings of the National Academy of Sciences. 2003;107(45):19201–19206. doi: 10.1073/pnas.1014386107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Kittles RA. Human genetic diversity and the nonexistence of biological races. HUMAN BIOLOGY. 2003;75:449–471. doi: 10.1353/hub.2003.0058. [DOI] [PubMed] [Google Scholar]
- Mayr E. The Growth of Biological Thought: Diversity, Evolution, and Inheritance. Cambridge, Massachusetts: The Belknap Press of Harvard University Press; 1982. [Google Scholar]
- Oberle Brad, Schaal Barbara A. Responses to historical climate change identify contemporary threats to diversity in Dodecatheon. Proceedings of the National Academy of Sciences. 2011;108(14):5655–5660. doi: 10.1073/pnas.1012302108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchal Mahesh, Beaumont Mark A. Evaluating Nested Clade Phylogeographic Analysis under Models of Restricted Gene Flow. Systematic Biology. 2010;59(4):415–432. doi: 10.1093/sysbio/syq022. [DOI] [PubMed] [Google Scholar]
- Pennock DS, Dimmick WW. Critique of the evolutionarily significant unit as a definition for distinct population segments under the US Endangered Species Act. Conservation Biology. 1997;11(3):611–619. [Google Scholar]
- Pigliucci M, Kaplan J. On the concept of biological race and its applicability to humans. Philosophy of Science. 2003;70(5):1161–1172. [Google Scholar]
- Ramachandran Sohini, Deshpande Omkar, Roseman Charles C, Rosenberg Noah A, Feldman Marcus W, Cavalli-Sforza LLuca. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. PNAS. 2005;102(44):15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich David, Green Richard E, Kircher Martin, Krause Johannes, Patterson Nick, Durand Eric Y, Paabo Svante. Genetic history of an archaic hominin group from Denisova Cave in Siberia. [10.1038/nature09710] Nature. 2010;468(7327):1053–1060. doi: 10.1038/nature09710. doi: http://www.nature.com/nature/journal/v468/n7327/abs/nature09710.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relethford JH. Race and global patterns of phenotypic variation. American Journal of Physical Anthropology. 2009;139(1):16–22. doi: 10.1002/ajpa.20900. [DOI] [PubMed] [Google Scholar]
- Rohlf FJ. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System, Version 1.80. Setauket, NY: Exeter Software; 1993. [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic structure of human populations. Science. 2002;298(5602):2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Safner Toni, Miller Mark P, McRae Brad H, Fortin Marie-Josée, Manel Stéphanie. Comparison of Bayesian Clustering and Edge Detection Methods for Inferring Boundaries in Landscape Genetics. International Journal of Molecular Sciences. 2011;12(2):865–889. doi: 10.3390/ijms12020865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos RV, Fry PH, Monteiro S, Maio MC, Rodrigues JC, Bastos-Rodrigues L, Pena SDJ. Color, Race, and Genomic Ancestry in Brazil Dialogues between Anthropology and Genetics. Current Anthropology. 2009;50(6):787–819. doi: 10.1086/644532. [DOI] [PubMed] [Google Scholar]
- Smith HM, Chiszar D, Montanucci RR. Subspecies and classification. Herpetological Review. 1997;28:13–16. [Google Scholar]
- Smouse PE. To tree or not to tree. Molecular Ecology. 1998;7:399–412. [Google Scholar]
- Templeton AR. Human races: A genetic and evolutionary perspective. American Anthropologist. 1998a;100(3):632–650. [Google Scholar]
- Templeton AR. Species and speciation: geography, population structure, ecology, and gene trees. In: Howard DJ, Berlocher SH, editors. Endless Forms: Species and Speciation. Oxford: Oxford University Press; 1998b. pp. 32–43. [Google Scholar]
- Templeton AR. Using gene trees to infer species from testable null hypothesis: cohesion species in the Spalax ehrenbergi complex. In: Wasser SP, editor. Evolutionary Theory and Processes: Modern Perspectives, Papers in Honour of Eviatar Nevo. Dordrecht: Kluwer Academic Publishers; 1999. pp. 171–192. [Google Scholar]
- Templeton AR. Out of Africa again and again. [10.1038/416045a] Nature. 2002;416(6876):45–51. doi: 10.1038/416045a. doi: http://www.nature.com/nature/journal/v416/n6876/suppinfo/416045a_S1.html. [DOI] [PubMed] [Google Scholar]
- Templeton AR. Statistical phylogeography: methods of evaluating and minimizing inference errors. Molecular Ecology. 2004;13(4):789–809. doi: 10.1046/j.1365-294x.2003.02041.x. [DOI] [PubMed] [Google Scholar]
- Templeton AR. Population Genetics and Microevolutionary Theory. Hoboken, New Jersey: John Wiley & Sons; 2006. [Google Scholar]
- Templeton AR. Statistical hypothesis testing in intraspecific phylogeography: nested clade phylogeographical analysis vs. approximate Bayesian computation. Molecular Ecology. 2009a;18(2):319–331. doi: 10.1111/j.1365-294X.2008.04026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR. Why does a method that fails continue to be used: the answer. Evolution. 2009b;63(4):807–812. doi: 10.1111/j.1558-5646.2008.00600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR. Mechanisms of speciation -- a population genetic approach. Ann. Rev. Ecol. Syst. 1981;12:23–48. [Google Scholar]
- Templeton AR. The meaning of species and speciation: A genetic perspective. In: Otte D, Endler JA, editors. Speciation and its Consequences. Sunderland, Massachusetts: Sinauer; 1989. pp. 3–27. [Google Scholar]
- Templeton AR. Using phylogeographic analyses of gene trees to test species status and processes. Molecular Ecology. 2001;10:779–791. doi: 10.1046/j.1365-294x.2001.01199.x. [DOI] [PubMed] [Google Scholar]
- Templeton AR. A maximum likelihood framework for cross validation of phylogeographic hypotheses. In: Wasser SP, editor. Evolutionary Theory and Processes: Modern Horizons. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2004. pp. 209–230. [Google Scholar]
- Templeton AR. Haplotype trees and modern human origins. Yearbook of Physical Anthropology. 2005;48:33–59. doi: 10.1002/ajpa.20351. [DOI] [PubMed] [Google Scholar]
- Templeton AR. Population biology and population genetics of Pleistocene Hominins. In: Henke W, Tattersall I, editors. Handbook of Palaeoanthropology. Vol. 3. Berlin: Springer-Verlag; 2007. pp. 1825–1859. [Google Scholar]
- Templeton AR, Routman E, Phillips C. Separating population structure from population history: a cladistic analysis of the geographical distribution of mitochondrial DNA haplotypes in the Tiger Salamander, Ambystoma tigrinum . Genetics. 1995;140:767–782. doi: 10.1093/genetics/140.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaeren Marian, d Errico Francesco, Stringer Chris, James Sarah L, Todd Jonathan A, Mienis Henk K. Middle Paleolithic Shell Beads in Israel and Algeria. Science. 2006;312(5781):1785–1788. doi: 10.1126/science.1128139. [DOI] [PubMed] [Google Scholar]