Abstract
Integral membrane proteins (IMPs) play a central role in cell communication with the environment. Their structures are essential for our understanding of the molecular mechanisms of signaling and for drug design, yet they remain badly underrepresented in the protein structure databank. Solution NMR is, aside from X-ray crystallography, the major tool in structural biology. Here we review recently reported solution NMR structures of polytopic IMPs and discuss the new approaches, which were developed in the course of these studies to overcome barriers in the field. Advances in cell-free protein expression, combinatorial isotope labeling, resonance assignment, and collection of structural data greatly accelerated IMP structure determination by solution NMR. In addition, novel membrane-mimicking media made possible determination of solution NMR structures of IMPs in a native-like lipid environment.
Introduction
Structural studies of integral membrane proteins (IMPs) show impressive progress with more than 100 new structures determined in the last two years (http://www.pdb.org, X-ray structures: http://blanco.biomol.uci.edu/mpstruc/listAll/list, NMR structures: http://www.drorlist.com/nmr/MPNMR.html). Advances in the production of isotope-labeled IMPs as well as the development of solution NMR-based techniques for structure analysis have resulted in multiple new NMR structures of IMPs [1**,2**,3**,4]. Recent introduction of a rotational alignment solid state NMR method [5] has allowed the determination of IMP structures in a lipid bilayer under physiological conditions [5,6*]. Concurrently with the advances in NMR techniques, improvements in IMP production and crystallization have contributed to the growing number of X-ray structures of medically relevant IMPs [7,8]. However, in spite of this progress and the fact that membrane proteins constitute almost 30% of the proteome [9], structures of IMPs represent less than 1% of known protein structures (http://www.pdb.org).
Of all deposited protein structures, only approximately 11% were determined by NMR spectroscopy (http://www.pdb.org). However, for the most difficult and desired targets, human IMPs, this percentage is much higher: thanks for recent improvements in solution and solid-state NMR methods almost half of 37 known unique structures of human IMPs were determined by NMR spectroscopy [3**,6*,10,11].
It has to be said that with a few exceptions [1**,2**,6*,11–15,16*,17], NMR spectroscopy deals with small IMPs. This is because a large size of a complex of a polytopic IMP with detergents or lipids, together with the internal mobility of the IMP's TM helical bundle, causes fast relaxation and strong non-uniform broadening of NMR resonances and, as a result, multiple problems with signal assignment, spectra analysis, and detection of long-range interactions. These limitations can only be overcome with new technologies. Some recent technological improvements in cell-free protein synthesis, selective isotope labeling, and systematic paramagnetic labeling and data analysis, have already made the determination of the structures of several polytopic α-helical IMPs possible [1**,2**,3**], but many more are needed.
In this review we discuss new IMP structures obtained by solution NMR [1**,2**,3**,4] together with new strategies combining advanced methods of IMP production and isotope labeling with significant improvements in data collection and analysis.
New solution NMR structures of IMPs
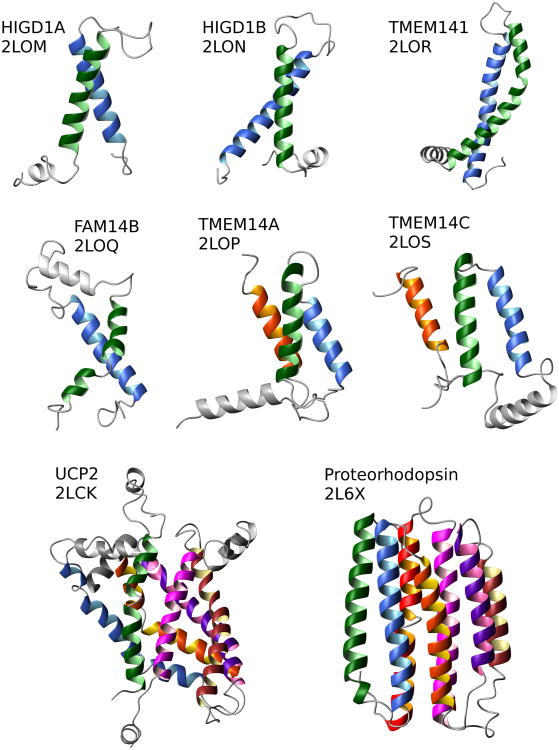
New solution NMR structures of full-length polytopic α-helical IMPs, mouse Mitochondrial uncoupling protein 2, UCP2 [1**], bacterial Proteorhodopsin [2**], and six human IMPs [3**] have recently been reported (Figure 1). Also, the structure of Pseudomonas aeruginosa β-barrel outer membrane protein H, OprH, was calculated based on solution NMR data [4]. A combination of solution and solid-state NMR methods was used in the determination of the pentameric structure of rabbit Phospholamban in lipids [18]. UCP2, OprH, and Phospholamban were produced in E. coli cells, while both Proteorhodopsin and the human IMPs were synthesized using a similar setup of an E. coli-based cell-free (CF) system [19] and assigned with the assistance of combinatorial labeling approaches [20,21]. For the studies of UCP2, Proteorhodopsin, and the human IMPs, a systematic collection of paramagnetic relaxation enhancement (PRE) data was instrumental in overcoming the shortage of long-distance NOE data. For UCP2 and for five human IMPs the PRE data were the sole sources of long-range distance constraints (Table).
Figure 1.
Recent new solution NMR structures of polytopic IMPs. The TM helices are colored in the following order starting from the N-terminus: green, blue, orange, dark violet, brown, magenta, and red. Non-TM helices are shown in gray. The PDB codes are indicated under the protein names. Figures are prepared using the Molmol program [65].
Table.
Summary of experimental approach and data used in determination of new IMP structures by solution NMR.
| Protein | Source/Expression systema | Mediab | MW, kDa/ Number of TMsc | Assignment method | experimental constraints | Calculation method | PDB | Ref | ||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| Long-range | ||||||||||
|
|
||||||||||
| NOE | PREd | RDC | ||||||||
| UCP2 | Mm/Ec | DPC | 33.0/6α | Sequential | 0 | 146/4 | 470 | MFR | 2LCK | [1**] |
| DMPC/CLe | XPLOR-NIH | |||||||||
| PR | gPB/CF | DH(7)PC | 26.6/7α | Combinatorial | 87 | 290/13 | 81 | CYANA | 2L6X | [2**] |
| HIGD1A | Hs/CF | LMPG | 10.2/2α | Combinatorial | 0 | 156/6 | 0 | CYANA | 2LOM | [3**] |
| HIGD1B | Hs/CF | LMPG | 11.2/2α | Combinatorial | 0 | 224/6 | 0 | CYANA | 2LON | [3**] |
| TMEM14A | Hs/CF | LMPG | 11.1/3α | Combinatorial | 18 | 0 | 0 | CYANA | 2LOO | [3**] |
| 0 | 334/7 | 2LOP | ||||||||
| TMEM14C | Hs/CF | LMPG | 11.6/3α | Combinatorial | 0 | 283/9 | 0 | CYANA | 2LOS | [3**] |
| TMEM141 | Hs/CF | LMPG | 11.8/2α | Sequential | 0 | 162/5 | 0 | CYANA | 2LOR | [3**] |
| FAM14B | Hs/CF | LMPG | 9.6/3α | Combinatorial | 0 | 195/8 | 0 | CYANA | 2LOQ | [3**] |
| OprH | Pa/Ec | DH(6)PC | 19.4/8β | Sequential | 95 | 0 | 0 | CNS | 2LHF | [4] |
| VDAC-1 | Hs/Ec | LDAO | 30.8/19β | Sequentialf | 272 | 0 | 0 | CYANA | 2K4T | [11] |
| DsbB | Ec/Ec | DPC | 20.1/4α | Sequential | 39 | 871/9 | 337 | XPLOR-NIH | 2K73 | [12] |
| DAGK | Ec/Ec | DPC | (13.1/3α) x3g | Sequential | 0 | (208/9) x3g | 67 x3g | XPLOR-NIH | 2KDC | [13] |
| pSRII | Np/Ec | DHPC | 25.4/7α | Sequential | 1536 | 0 | 0 | CNS | 2KSY | [14] |
|
| ||||||||||
| OmpXh | Ec/Ec | NLP | 16.4/8β | Sequential | 58 | 0 | 0 | XPLOR-NIH | 2M06 | [64**] |
Ec, E. coli; Mm, M. musculus; gPB, uncultured marine gamma proteobacterium EBAC31A08; Hs, H. sapiens; Pa, Pseudomonas aeruginosa; Np, Natronomonas pharaonis; CF, cell-free system.
DPC, dodecylphosphocholine; LDAO, lauryldimethylamine oxide; DMPC, 1,2-dimyristoyl-sn-glycero-3-phosphocholine; CL, cardiolipin; DH(7)PC, 1,2-diheptanoyl-sn-glycero-3-phosphocholine; DH(6)PC, 1,2-dihexanoyl-sn-glycero-3-phosphocholine; LMPG, 1-myristoyl-2-hydroxy-sn-glycero-3-[phospho-rac-(1-glycerol)]; NLP, Nanodisks (scaffold protein MSP1D1ΔH5, lipids 3:1 DMPC: 1,2-dimyristoyl-sn-glycero-3-phosphoglycerol).
a, number of TM helices; b – number of TM β-strands.
number of upper PRE distance restraints/number spin-labeled cysteine mutants.
DMPC and CL added in small quantities (2 and 1 mM, respectively).
sequential assignment with assistance of selective 13C1-labeling.
x3 indicate the corresponding number for DAGK monomer in the trimer structure.
the first solution NMR structure of IMP in Nanodisks.
In case of UCP2, a severe overlap of backbone (1H, 15N) resonances complicated the NOE data collection. Therefore, the secondary structure of UCP2 was determined by a molecular fragment replacement approach using residual dipolar coupling (RDC) data collected from a weakly aligned sample. In a calculation of the tertiary fold of UCP2, RDC and PRE data were used to define the orientation and spatial arrangement of the secondary structure elements respectively [1**].
A side chain assignment of Proteorhodopsin became possible with the use of the stereo-array isotope labeling (SAIL) technique [22]. This technique allowed extraction of essential long-range inter-helical distance constraints between methyl groups and tryptophan side chains using non-uniform sampled 4D [13C, 13C]-separated NOESY of the protein sample with selectively labeled amino acids. Nevertheless, the majority of long-range distance constraints used in Proteorhodopsin structure calculation were derived from the PRE data collected from 13 single-cysteine mutants [2**].
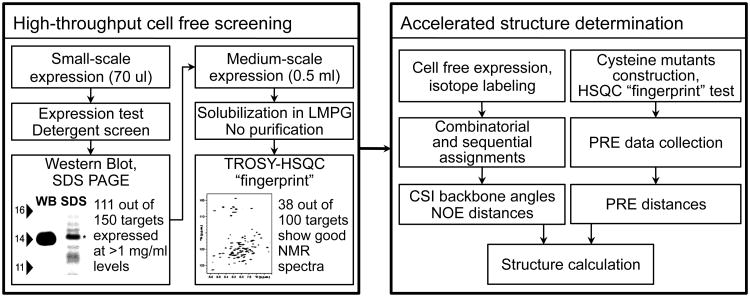
Kalmmt et al. [3**] selected six small polytopic human IMPs for structural analysis from a large number of human IMPs efficiently screened by the CF expression system. In 18 months they determined backbone structures of the selected human IMPs using the powerful combination of CF-assisted amino acid type (AAT) selective isotope labeling, fast NMR assignment using combinatorial dual labeling (CDL) approach [20], and fast PRE data collection for long-range distance constraints [3**] (Figure 2).
Figure 2.
Flowchart of high-throughput cell free expression screening and cell-free assisted accelerated structure determination.
Production and isotope labeling of IMPs
E. coli expression system is often a system of choice in IMP structural studies because of its low cost, flexibility [23,24,25*], and already developed labeling approaches [26]. However, it is no panaceum for overexpression of eukaryotic IMPs in prokaryotic cells due to substantial differences between protein assembly machineries [23]. Finally, prokaryotic cells cannot provide posttranslational protein modifications, which are detrimental for folding and function of many eukaryotic proteins.
As an alternative to the E. coli expression system, expression in yeast, insect, or mammalian cells has become a common source of eukaryotic IMPs for X-ray studies. An overexpression of stable and functional uniformly 13C, 15N-labeled Rhodopsin in Pichia pastoris using minimal defined medium was recently reported [27]. New media, based on yeast autolysates, were recently developed for reduced-cost production of isotope-labeled IMPs in insect and mammalian cells [28,29].
Cell-free (CF) expression systems, based on E. coli, wheat germ, or insect cell extracts have proven effective in overcoming many limitations inherent in in vivo expression of different proteins, including IMPs [20,30,31]. A presence of membrane mimetics in a CF reaction mixture can support direct expression of solubilized IMPs [30,32,33,34*]. Alternatively, IMPs can be expressed in the absence of a hydrophobic milieu, precipitate, and be subsequently solubilized in a mild detergent [20,30]. Since the target protein is the only labeled protein produced in a CF reaction, a purification of an NMR sample can be minimized or even eliminated for precipitant IMPs [20]. A high yield of the IMPs, above 1 mg per 1 ml of reaction mixture, and a small volume of CF expression chamber (20-60 ml for preparative scale expression, which is 20-100 times smaller than for typical in vivo expression) keep the costs of isotope labeling reasonably low even for of triple-labeled samples [20]. Excellent reproducibility of the CF system has enabled expression screening in small volumes (below 100 ul) of 134 targets from E. coli inner membrane [35] and 150 human integral membrane proteins [3**] (Figure 2). Customizable CF systems have excellent potential for further development, which may lead to post-translational modifications of expressed proteins. For example, establishing the right redox potential and the right amount of catalytic enzymes resulted in successful CF expression of natively-folded disulfide-rich mammalian proteins [36*].
Selective labeling of proteins with stable isotopes, as well as selective back-protonation or “unlabeling” of deuterated proteins, is extremely useful in NMR studies of IMPs characterized by overcrowded spectra and fast NMR signal relaxation. Selective labeling with 13C and 15N and unlabeling with 1H are aimed at obtaining NMR-visible atoms or group of atoms on an “invisible” background, in other words, at selective NMR visualization.
An E. coli expression system allows relatively inexpensive NMR-visualization of specific groups by using amino acid precursors in minimal media [26]. However, complex amino acid metabolism in E. coli causes cross-labeling and label dilution in AAT-selective labeling. In prototrophic E. coli strains AAT-labeling can be done only with histidine, lysine, methionine, and alanine [37]. To amend this problem, Lin et al. [38] developed a library of 20 auxotroph E. coli mutant strains for selective isotope labeling of other amino acids and their combinations. On the other hand, selective AAT-labeling in insect and mammalian cells is quite efficient due to very little, compared with bacterial systems, cross-labeling problems.
The best approach to ATT-selective labeling, however, comes from CF systems, which are characterized by very limited amino acid metabolism and small volume of reaction, and, as a result, provide efficient and low cost labeling. The limited scrambling and dilution of isotope labels can be almost eliminated by an inhibition of specific enzymes in a CF reaction [39–41]. Several approaches for AAT-selective labeling in CF systems were introduced to accelerate the resonance assignment [20,21,42]. Substantially reduced amino acid metabolism and small volume of reaction make SAIL-labeled amino acids affordable [22]. Reckel et al. extensively used SAIL approach [22] to reduce signal overlap, perform stereospecific assignment, and obtain long-range distance constraints for the calculation of Proteorhodopsin structure [2**]. Recently, Loscha and Otting [43*] described a way of partial stereospecific 2H-labeling of glycine in CF reaction, which simplifies the process of stereospecific assignment of glycine's Hα protons.
Additionally, it is noteworthy that Fluorine labeling of IMPs is also under continuous development. A covalent modification of cysteines with 19F-labeled compound was used to describe conformational changes in β2-adrenergic receptor [44*]. 19F-labels incorporated into DAGK with unnatural amino acids were used to evaluate the protein dynamics in DPC micelles and in the native E. coli membrane [45,46].
Accelerated NMR analysis of MPs
Fast assignment based on selective labeling
The classical sequential implementation of NMR resonance assignment is particularly laborious for α-helical IMPs because of signal overlap and very fast relaxation, which hampers many experiments designed for sequential assignment [47]. An alternative assignment method utilizes selective dual 15N- and 13C-labeling and detection of heteronuclear 13C-15N spin coupling through a peptide bond in proteins [48]. The alternative method generated several combinatorial labeling approaches which use different subsets of 13C- and/or 15N-labeled amino acids in each sample in order to reduce the number of samples required for the assignment as compared to one-by-one selective labeling [49]. As many as 70% of the backbone 1HN, 15NH, and 13CO resonances, which are evenly distributed throughout the sequence, can be unambiguously assigned using 6-8 combinatorial labeled samples [20,21,49]. Additionally, such combinatorial assignment defines the type of amino acid for all cross peaks in an [1H-15N]-HSQC spectrum.
Two approaches, which were specifically developed for NMR analysis of IMPs, combinatorial dual-labeling (CDL) strategy [20] (Figure 3) and UPLABEL algorithm [21], calculate the optimal labeling scheme depending on the sequence of the protein. Such customized schemes balance the number of samples required for the assignment and the complexity of spectra for each labeled sample. Starting from the results of this assignment, the standard sequential assignment of IMPs was tremendously accelerated yielding an assignment for >90% of backbone resonances in a few weeks [3**,20]. This was also possible because combinatorial dual 15N- and 13C-labeling schemes use two short and most sensitive heteronuclear NMR experiments, [1H-15N]-HSQC and HN(CO) (Figure 4). An ability to obtain backbone assignment by utilizing simple 2D heteronuclear NMR experiments gives rise to the idea of de novo backbone structure determination, which is based on fast assignment and structural constraints derived from 2D [1H-15N]-HSQC-type experiments.
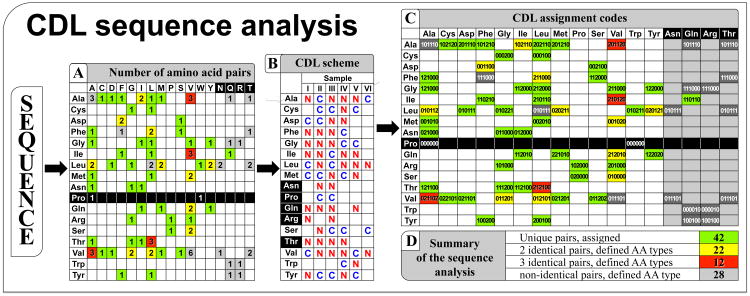
Figure 3.
Protein sequence analysis using combinatorial dual-labeling strategy [20] for accelerated assignment. From a distribution of amino acids in a sequence, represented as a matrix of amino acid pairs (A), MCCL program (http://sbl.salk.edu/combipro) calculates the combinatorial dual-labeling scheme (B) for maximal possible assignment with available amino acids (E). Each type of the amino acid pairs (A) in a protein sequence receives an assignment tag (C) according to the labeling pattern of the peptide bond between the two amino acids in the pair in each sample (0 – no 15N labeling of the peptide bond; 1 – 15N-only; 2 – 13C and 15N labeling).
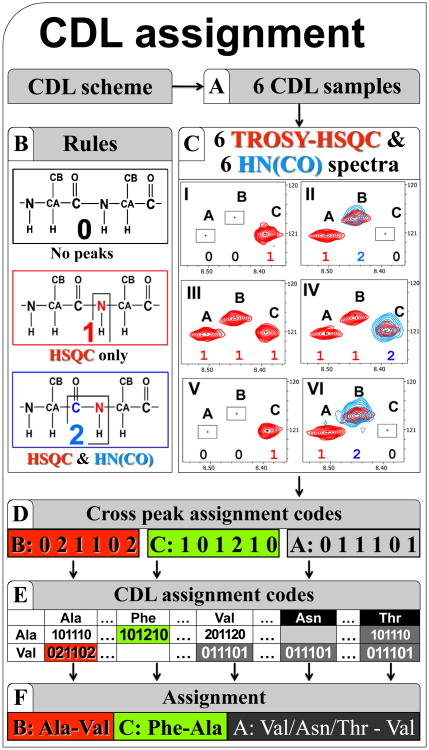
Figure 4.
Accelerated assignment using the CDL strategy [21]. (A) The CDL samples are expressed simultaneously in the CF system according to the CDL scheme and solubilized in the same buffer. The 1H-15N-TROSY-HSQC and HN(CO) spectra are acquired for each CDL sample. Panel (C) shows superposition of a fragment of 1H-15N-HSQC and HN(CO) spectra for each CDL sample. Analysis of the spectra reveals the assignment code (D) for the cross peak in each sample (according to the rules (B): 0 – absent in both spectra; 1 – present only in HSQC; 2 – present in both HSQC and HNCO). Each of 1H-15N cross-peaks is assigned by matching the tag of codes derived from the spectra (D) with the code pre-calculated using the CDL scheme (E).
Fast data collection for structural restraints
The chemical shift index (CSI), a byproduct of NMR resonances assignment, gives very useful information about protein secondary structure [50**] and, complimented with distance constraints between the secondary structure elements, allows determination of the protein's 3D fold. The paramagnetism-based data, PRE and pseudocontact shifts, can be obtained for backbone atoms using 2D [1H-15N]-HSQC-type experiments and converted into distance constraints up to 40 Å. The determination of many recent IMP structures [1**,2**,3**,12,13,20,51,52] (summarized in Table) relied on the PRE data, obtained using sulfhydryl-specific spin-label [53]. During last decade the PRE data became a major source of long-range constraints for membrane proteins deficient in NOE-derived long-range distance constraints.
The sulfhydryl-specific spin-labeling [53] requires a design of a number of target protein mutants with a single accessible cysteine at strategic positions. This is a crucial step in the PRE experiment because the positions and the number of the spin labels in TM domains determine the completeness of the distance constraint set, which is a very important factor for the success of structure calculation [54*,55*]. A flexibility of the region containing a mutated cysteine substantially increases an error in calculated distance constraints [3**,13]. Several works describe important details of the spin labeling technique for structural studies of IMPs [3**,12,54*,55*,56,57].
Structure calculation based on PRE distance constraints
The PRE effect is usually quantified based on the drop in the intensity of the cross-peak in 2D [1H-15N]-HSQC spectra recorded for the diamagnetic and paramagnetic samples [58] or as a difference in transverse relaxation rates measured in two-time-points experiments for paramagnetic and diamagnetic samples [59]. The direct distance calculation [1**,2**,3**,12,51] and the qualitative analysis of PRE-induced drop of cross- peak intensities [13,20,60] were used to derive the distance constraints between HN protons and the unpaired electron of the spin label. The constraints cover distances in the range between 12 and 25 Å, depending on the size of IMP-detergent complex and the protein's internal mobility [58].
It should be noted, that the precision of the PRE-based constraints obtained for the spin-labeled protein is usually lower than the precision of the constraints derived from NOEs (discussed in [58,59]). In the calculation of distance constraints the errors, which came from the estimation of a correlation time and the measurements of linewidths and intensities, usually do not exceed 15% if the ratio of cross-peak intensities in paramagnetic and diamagnetic samples is between 0.15 and 0.80. However, while the dynamics of the spin-labeled side chain is assumed to be restricted [57,61], uncertainty resulting from protein internal dynamics can substantially increase the error for the distances calculated from averaged PRE data [62]. All factors combined contribute to the relatively large error of ±2…5 Å for PRE-based distance constraints [3**,56]. In spite of the large error, carefully obtained, extensive sets of long-range PRE-based distance constraints were shown to be sufficient for the calculation of the backbone structure of α-helical and β-barrel IMPs [1**,3**,54*,56] (Table 1). The availability of additional constraints (long-range NOEs, RDCs, etc.) considerably improves the quality of structures including details of side chain packing, and allows determination of the structure of the extramembrane loops [2**,12].
New membrane-mimicking media
Solubilization of IMPs in a membrane-mimicking environment is a very important step in solution NMR studies. Substantial efforts had been devoted to develop Nanodisks, new membrane mimetic media, which are closest artificial media to the natural membrane phospholipid bilayer environment and suitable for solution NMR studies of IMPs [63].
The major advantage of Nanodisks is the absence of detergents, which may affect the structure of the extramembrane regions of an IMP or disrupt interactions with a ligand. In a recent study, Hagn et al. [64**] designed several novel Nanodisks media with reduced sizes. Bacteriorhodopsin and outer membrane protein X, OmpX, incorporated into these Nanodisks show ∼30% reduced apparent correlation time compared with the time measured in original Nanodisks media. The reduced-size Nanodisks environment combined with a high level of deuteration of both the protein and the lipids allowed resonance assignment and structure determination of OmpX [64**].
Conclusions
Current progress in structural studies of IMPs by NMR spectroscopy relies on synergetic approaches, which merge innovation in protein synthesis, sample preparation, data acquisition and analysis, and structure determination methods. However, future development of isotope labeling methods, especially those for proteins expressed in eukaryotic cells, and of methods for protein post-translational modifications in CF systems, is necessary for IMPs studies. Additionally, the advancement in the design of new membrane mimicking media capable of preserving the structures of both TM and extramembrane regions of IMPs and suitable for solution NMR continues to be an essential task. The current status of acquired knowledge and implementation of innovative methods allow a favorable and optimistic projection regarding the impact which solution NMR studies will have on the structural biology field in the coming years.
Highlights.
Solution NMR studies of polytopic integral membrane proteins
Fast structure determination of mammalian integral membrane proteins
Cell-free synthesis and combinatorial labeling accelerate structural studies
Paramagnetic relaxation enhancement became a major source of structural data
Solution NMR structure of membrane protein in lipid nanoparticles is feasible
Acknowledgments
The authors thank W. Kwiatkowski for useful discussions and careful reading of the manuscript. This work has been partly supported by the National Institute of Health grants GM098630 and GM095623 and by IFEZ (joint Center for Biosciences).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1**.Berardi MJ, Shih WM, Harrison SC, Chou JJ. Mitochondrial uncoupling protein 2 structure determined by NMR molecular fragment searching. Nature. 2011;476:109–113. doi: 10.1038/nature10257. The authors use molecular fragment replacement to define the local and secondary structures of UCP2 that best fit RDC data. The relative orientation of the secondary structural elements and their spatial arrangement in the 3D fold were determined from RDC and PRE constraints respectfully. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2**.Reckel S, Gottstein D, Stehle J, Löhr F, Verhoefen MK, Takeda M, Silvers R, Kainosho M, Glaubitz C, Wachtveitl J, et al. Solution NMR Structure of Proteorhodopsin. Angewandte Chemie International Edition. 2011;50:11942–11946. doi: 10.1002/anie.201105648. Determination of the structure of CF expressed Proteorhodopsin in DHPC micelles. The assignment and collection of long-range NOEs assisted with CF selective isotope labeling. The structure with well-defined TM domain and extramembrane regions calculated using NOE, PRE, and RDC constraints. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3**.Klammt C, Maslennikov I, Bayrhuber M, Eichmann C, Vajpai N, Chiu EJ, Blain KY, Esquivies L, Kwon JH, Balana B, et al. Facile backbone structure determination of human membrane proteins by NMR spectroscopy. Nat Methods. 2012;9:834–839. doi: 10.1038/nmeth.2033. Combination of efficient CF expression and selective isotope labeling for fast assignment, data collection and analysis. Backbone structures of six human IMPs determined by fast analysis of PRE and CSI data. Efficient application of CF system for screening of NMR quality of >100 human IMPs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edrington TC, Kintz E, Goldberg JB, Tamm LK. Structural Basis for the Interaction of Lipopolysaccharide with Outer Membrane Protein H (OprH) from Pseudomonas aeruginosa. J Biol Chem. 2011;286:39211–39223. doi: 10.1074/jbc.M111.280933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das BB, Nothnagel HJ, Lu GJ, Son WS, Tian Y, Marassi FM, Opella SJ. Structure determination of a membrane protein in proteoliposomes. J Am Chem Soc. 2012;134:2047–2056. doi: 10.1021/ja209464f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6*.Park SH, Das BB, Casagrande F, Tian Y, Nothnagel HJ, Chu M, Kiefer H, Maier K, De Angelis AA, Marassi FM, et al. Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature. 2012 doi: 10.1038/nature11580. The first 3D structure of human GPCR determined by NMR and the first structure of GPCR determined under physiologically relevant conditions, a phospholipid bilayer. The rotational alignment solid-state NMR method combines an oriented-sample and magic-angle spinning NMR techniques. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherezov V. Lipidic cubic phase technologies for membrane protein structural studies. Current Opinion in Structural Biology. 2011;21:559–566. doi: 10.1016/j.sbi.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steyaert J, Kobilka BK. Nanobody stabilization of G protein-coupled receptor conformational states. Current Opinion in Structural Biology. 2011;21:567–572. doi: 10.1016/j.sbi.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallin E, Heijne GV. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Call ME, Wucherpfennig KW, Chou JJ. The structural basis for intramembrane assembly of an activating immunoreceptor complex. Nat Immunol. 2010;11:1023–1029. doi: 10.1038/ni.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G. Solution Structure of the Integral Human Membrane Protein VDAC-1 in Detergent Micelles. Science. 2008;321:1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y, Cierpicki T, Jimenez RHF, Lukasik SM, Ellena JF, Cafiso DS, Kadokura H, Beckwith J, Bushweller JH. NMR Solution Structure of the Integral Membrane Enzyme DsbB: Functional Insights into DsbB-Catalyzed Disulfide Bond Formation. Molecular Cell. 2008;31:896–908. doi: 10.1016/j.molcel.2008.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Horn WD, Kim HJ, Ellis CD, Hadziselimovic A, Sulistijo ES, Karra MD, Tian C, Sonnichsen FD, Sanders CR. Solution Nuclear Magnetic Resonance Structure of Membrane-Integral Diacylglycerol Kinase. Science. 2009;324:1726–1729. doi: 10.1126/science.1171716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gautier A, Mott HR, Bostock MJ, Kirkpatrick JP, Nietlispach D. Structure determination of the seven-helix transmembrane receptor sensory rhodopsin II by solution NMR spectroscopy. Nat Struct Mol Biol. 2010;17:768–774. doi: 10.1038/nsmb.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bayrhuber M, Meins T, Habeck M, Becker S, Giller K, Villinger S, Vonrhein C, Griesinger C, Zweckstetter M, Zeth K. Structure of the human voltage- dependent anion channel. Proceedings of the National Academy of Sciences; 2008; pp. 15370–15375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16*.Shahid SA, Bardiaux B, Franks WT, Krabben L, Habeck M, van Rossum BJ, Linke D. Membrane-protein structure determination by solid-state NMR spectroscopy of microcrystals. Nature Methods. 2012;9:1212–1217. doi: 10.1038/nmeth.2248. The authors determine the structure of 30 kDa trimeric autotransporter adhesin using the sample derived from crystallization trials that yielded only poorly diffracting microcrystals. A good example of solid-state MAS NMR spectroscopy as an emerging method for IMP structural biology. [DOI] [PubMed] [Google Scholar]
- 17.Baker KA, Tzitzilonis C, Kwiatkowski W, Choe S, Riek R. Conformational dynamics of the KcsA potassium channel governs gating properties. Nature Structural & Molecular Biology. 2007;14:1089–1095. doi: 10.1038/nsmb1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verardi R, Shi L, Traaseth NJ, Walsh N, Veglia G. Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method. PNAS. 2011;108:9101–9106. doi: 10.1073/pnas.1016535108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reckel S, Sobhanifar S, Durst F, Löhr F, Shirokov VA, Dötsch V, Bernhard F. Strategies for the cell-free expression of membrane proteins. Methods Mol Biol. 2010;607:187–212. doi: 10.1007/978-1-60327-331-2_16. [DOI] [PubMed] [Google Scholar]
- 20.Maslennikov I, Klammt C, Hwang E, Kefala G, Okamura M, Esquivies L, Mörs K, Glaubitz C, Kwiatkowski W, Jeon YH, et al. Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis. Proc Natl Acad Sci U S A. 2010;107:10902–10907. doi: 10.1073/pnas.1001656107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hefke F, Bagaria A, Reckel S, Ullrich SJ, Dötsch V, Glaubitz C, Güntert P. Optimization of amino acid type-specific 13C and 15N labeling for the backbone assignment of membrane proteins by solution- and solid-state NMR with the UPLABEL algorithm. J Biomol NMR. 2011;49:75–84. doi: 10.1007/s10858-010-9462-4. [DOI] [PubMed] [Google Scholar]
- 22.Kainosho M, Torizawa T, Iwashita Y, Terauchi T, Mei Ono A, Güntert P. Optimalisotope labelling for NMR protein structure determinations. Nature. 2006;440:52–57. doi: 10.1038/nature04525. [DOI] [PubMed] [Google Scholar]
- 23.Freigassner M, Pichler H, Glieder A. Tuning microbial hosts for membrane protein production. Microbial Cell Factories. 2009;8:69. doi: 10.1186/1475-2859-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlegel S, Löfblom J, Lee C, Hjelm A, Klepsch M, Strous M, Drew D, Slotboom DJ, De Gier JW. Optimizing membrane protein overexpression in the Escherichia coli strain Lemo21(DE3) J Mol Biol. 2012;423:648–659. doi: 10.1016/j.jmb.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 25*.Vaiphei ST, Tang Y, Montelione GT, Inouye M. The use of the condensed single protein production system for isotope-labeled outer membrane proteins, OmpA and OmpX in E. coli. Mol Biotechnol. 2011;47:205–210. doi: 10.1007/s12033-010-9330-1. The adaptation of the condensed single protein production system for IMP expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruschak AM, Kay LE. Methyl groups as probes of supra-molecular structure, dynamics and function. Journal of Biomolecular NMR. 2010;46:75–87. doi: 10.1007/s10858-009-9376-1. [DOI] [PubMed] [Google Scholar]
- 27.Fan Y, Shi L, Ladizhansky V, Brown LS. Uniform isotope labeling of a eukaryotic seven-transmembrane helical protein in yeast enables high-resolution solid-state NMR studies in the lipid environment. J Biomol NMR. 2011;49:151–161. doi: 10.1007/s10858-011-9473-9. [DOI] [PubMed] [Google Scholar]
- 28.Egorova-Zachernyuk TA, Bosman GJCGM, DeGrip WJ, Shvets VI. Stable isotope labelling of human histamine receptor H1R: Prospects for structure-based drug design. Dokl Biochem Biophys. 2010;433:164–167. doi: 10.1134/S160767291004006X. [DOI] [PubMed] [Google Scholar]
- 29.Egorova-Zachernyuk TA, Bosman GJCGM, Degrip WJ. Uniform stable-isotope labeling in mammalian cells: formulation of a cost-effective culture medium. Appl Microbiol Biotechnol. 2011;89:397–406. doi: 10.1007/s00253-010-2896-5. [DOI] [PubMed] [Google Scholar]
- 30.Sobhanifar S, Reckel S, Junge F, Schwarz D, Kai L, Karbyshev M, Löhr F, Bernhard F, Dötsch V. Cell-free expression and stable isotope labelling strategies for membrane proteins. J Biomol NMR. 2010;46:33–43. doi: 10.1007/s10858-009-9364-5. [DOI] [PubMed] [Google Scholar]
- 31.Madono M, Sawasaki T, Morishita R, Endo Y. Wheat germ cell-free protein production system for post-genomic research. New Biotechnology. 2011;28:211–217. doi: 10.1016/j.nbt.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 32.Klammt C, Perrin MH, Maslennikov I, Renault L, Krupa M, Kwiatkowski W, Stahlberg H, Vale W, Choe S. Polymer-based cell-free expression of ligand- binding family B G-protein coupled receptors without detergents. Protein Sci. 2011;20:1030–1041. doi: 10.1002/pro.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajesh S, Knowles T, Overduin M. Production of membrane proteins without cells or detergents. New Biotechnology. 2011;28:250–254. doi: 10.1016/j.nbt.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 34*.Bazzacco P, Billon-Denis E, Sharma KS, Catoire LJ, Mary S, Le Bon C, Point E, Banères JL, Durand G, Zito F, et al. Nonionic homopolymeric amphipols: application to membrane protein folding, cell-free synthesis, and solution nuclear magnetic resonance. Biochemistry. 2012;51:1416–1430. doi: 10.1021/bi201862v. The authors use the new synthetic nonionic polymer for refolding of functional GPCR from inclusion bodies. The nonionic polymer is also compatible with the CF expression system. [DOI] [PubMed] [Google Scholar]
- 35.Schwarz D, Daley D, Beckhaus T, Dötsch V, Bernhard F. Cell-free expression profiling of E. coli inner membrane proteins. Proteomics. 2010;10:1762–1779. doi: 10.1002/pmic.200900485. [DOI] [PubMed] [Google Scholar]
- 36*.Michel E, Wüthrich K. Cell-free expression of disulfide-containing eukaryotic proteins for structural biology. FEBS Journal. 2012;279:3176–3184. doi: 10.1111/j.1742-4658.2012.08697.x. An optimization of redox potential and amount of disulfide bond catalytic enzymes in the reaction mixture for preparative scale expression of natively folded mammalian proteins in the E. coli-based CF system. [DOI] [PubMed] [Google Scholar]
- 37.O'Grady C, Rempel BL, Sokaribo A, Nokhrin S, Dmitriev OY. One-step amino acid selective isotope labeling of proteins in prototrophic Escherichia coli strains. Anal Biochem. 2012;426:126–128. doi: 10.1016/j.ab.2012.04.019. [DOI] [PubMed] [Google Scholar]
- 38.Lin MT, Sperling LJ, Frericks Schmidt HL, Tang M, Samoilova RI, Kumasaka T, Iwasaki T, Dikanov SA, Rienstra CM, Gennis RB. A rapid and robust method for selective isotope labeling of proteins. Methods. 2011;55:370–378. doi: 10.1016/j.ymeth.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yokoyama J, Matsuda T, Koshiba S, Tochio N, Kigawa T. A practical method for cell-free protein synthesis to avoid stable isotope scrambling and dilution. Anal Biochem. 2011;411:223–229. doi: 10.1016/j.ab.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 40.Su XC, Loh CT, Qi R, Otting G. Suppression of isotope scrambling in cell-free protein synthesis by broadband inhibition of PLP enzymes for selective 15N- labelling and production of perdeuterated proteins in H2O. J Biomol NMR. 2011;50:35–42. doi: 10.1007/s10858-011-9477-5. [DOI] [PubMed] [Google Scholar]
- 41.Tonelli M, Singarapu KK, Makino S, Sahu SC, Matsubara Y, Endo Y, Kainosho M, Markley JL. Hydrogen exchange during cell-free incorporation of deuterated amino acids and an approach to its inhibition. J Biomol NMR. 2011;51:467–476. doi: 10.1007/s10858-011-9575-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Löhr F, Reckel S, Karbyshev M, Connolly PJ, Abdul-Manan N, Bernhard F, Moore JM, Dötsch V. Combinatorial triple-selective labeling as a tool to assist membrane protein backbone resonance assignment. J Biomol NMR. 2012;52:197–210. doi: 10.1007/s10858-012-9601-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43*.Loscha KV, Otting G. Biosynthetically directed (2)H labelling for stereospecific resonance assignments of glycine methylene groups. J Biomol NMR. 2012 doi: 10.1007/s10858-012-9690-x. The biosynthetic fractional labeling approach uses enzyme naturally presented in E coli-based CF system to convert serine into stereospecifically 2H-labeled glycine. [DOI] [PubMed] [Google Scholar]
- 44**.Liu JJ, Horst R, Katritch V, Stevens RC, Wüthrich K. Biased Signaling Pathways in β2-Adrenergic Receptor Characterized by 19F-NMR. Science. 2012;335:1106–1110. doi: 10.1126/science.1215802. Using the 19F labels, attached to TM helices of β2Adrenergic Receptor, the authors detect differences in the receptor structural response depending on the nature of a bound ligand. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi P, Wang H, Xi Z, Shi C, Xiong Y, Tian C. Site-specific 19F NMR chemical shift and side chain relaxation analysis of a membrane protein labeled with an unnatural amino acid. Protein Sci. 2011;20:224–228. doi: 10.1002/pro.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi P, Li D, Chen H, Xiong Y, Wang Y, Tian C. In situ 19F NMR studies of an E. coli membrane protein. Protein Sci. 2012;21:596–600. doi: 10.1002/pro.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim HJ, Howell SC, Van Horn WD, Jeon YH, Sanders CR. Recent advances in the application of solution NMR spectroscopy to multi-span integral membrane proteins. Progress in nuclear magnetic resonance spectroscopy. 2009;55:335. doi: 10.1016/j.pnmrs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kainosho M, Tsuji T. Assignment of the three methionyl carbonyl carbon resonances in Streptomyces subtilisin inhibitor by a carbon-13 and nitrogen-15 double-labeling technique.A new strategy for structural studies of proteins in solution. Biochemistry. 1982;21:6273–6279. doi: 10.1021/bi00267a036. [DOI] [PubMed] [Google Scholar]
- 49.Parker MJ, Aulton-Jones M, Hounslow AM, Craven CJ. A Combinatorial Selective Labeling Method for the Assignment of Backbone Amide NMR Resonances. Journal of the American Chemical Society. 2004;126:5020–5021. doi: 10.1021/ja039601r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50**.Wishart DS. Interpreting protein chemical shift data. Progress in Nuclear Magnetic Resonance Spectroscopy. 2011;58:62–87. doi: 10.1016/j.pnmrs.2010.07.004. Comprehensive review on chemical shift data in biomolecular NMR. [DOI] [PubMed] [Google Scholar]
- 51.Page RC, Lee S, Moore JD, Opella SJ, Cross TA. Backbone structure of a small helical integral membrane protein: A unique structural characterization. Protein Science. 2009;18:134–146. doi: 10.1002/pro.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rumpel S, Becker S, Zweckstetter M. High-resolution structure determination of the CylR2 homodimer using paramagnetic relaxation enhancement and structure-based prediction of molecular alignment. Journal of Biomolecular NMR. 2007;40:1–13. doi: 10.1007/s10858-007-9204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Altenbach C, Flitsch SL, Khorana HG, Hubbell WL. Structural studies on transmembrane proteins. 2. Spin labeling of bacteriorhodopsin mutants at unique cysteines. Biochemistry. 1989;28:7806–7812. doi: 10.1021/bi00445a042. [DOI] [PubMed] [Google Scholar]
- 54*.Gottstein D, Reckel S, Dötsch V, Güntert P. Requirements on paramagnetic relaxation enhancement data for membrane protein structure determination by NMR. Structure. 2012;20:1019–1027. doi: 10.1016/j.str.2012.03.010. The authors perform a systematic study with simulated NMR data to define general requirements for spin labeling of helical TM domains for accurate calculation of IMP structures using PRE constraints. [DOI] [PubMed] [Google Scholar]
- 55*.Chen H, Ji F, Olman V, Mobley CK, Liu Y, Zhou Y, Bushweller JH, Prestegard JH, Xu Y. Optimal Mutation Sites for PRE Data Collection and Membrane Protein Structure Prediction. Structure. 2011;19:484–495. doi: 10.1016/j.str.2011.02.002. The authors describe a method for calculation of an optimal placement of minimal number of spin labels for correct determination of topology of a helical TM bundle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liang B, Bushweller JH, Tamm LK. Site-Directed Parallel Spin-Labeling and Paramagnetic Relaxation Enhancement in Structure Determination of Membrane Proteins by Solution NMR Spectroscopy. Journal of the American Chemical Society. 2006;128:4389–4397. doi: 10.1021/ja0574825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kroncke BM, Horanyi PS, Columbus L. Structural origins of nitroxide side chain dynamics on membrane protein α-helical sites. Biochemistry. 2010;49:10045–10060. doi: 10.1021/bi101148w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Battiste JL, Wagner G. Utilization of Site-Directed Spin Labeling and High-Resolution Heteronuclear Nuclear Magnetic Resonance for Global Fold Determination of Large Proteins with Limited Nuclear Overhauser Effect Data †. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 59.Iwahara J, Tang C, Marius Clore G. Practical aspects of 1H transverseparamagnetic relaxation enhancement measurements on macromolecules. Journal of Magnetic Resonance. 2007;184:185–195. doi: 10.1016/j.jmr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roosild TP, Greenwald J, Vega M, Castronovo S, Riek R, Choe S. NMR structureof Mistic, a membrane-integrating protein for membrane protein expression. Science. 2005;307:1317–1321. doi: 10.1126/science.1106392. [DOI] [PubMed] [Google Scholar]
- 61.Warshaviak DT, Serbulea L, Houk KN, Hubbell WL. Conformational Analysis of aNitroxide Side Chain in an α-Helix with Density Functional Theory. The Journal of Physical Chemistry B. 2011;115:397–405. doi: 10.1021/jp108871m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iwahara J, Schwieters CD, Clore GM. Ensemble Approach for NMR Structure Refinement against 1H Paramagnetic Relaxation Enhancement Data Arising from a Flexible Paramagnetic Group Attached to a Macromolecule. Journal ofthe American Chemical Society. 2004;126:5879–5896. doi: 10.1021/ja031580d. [DOI] [PubMed] [Google Scholar]
- 63.Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG. Methods in Enzymology. Elsevier; 2009. Chapter 11 Reconstitution of Membrane Proteins in Phospholipid Bilayer Nanodiscs; pp. 211–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64**.Hagn F, Etzkorn M, Raschle T, Wagner G. Optimized Phospholipid Bilayer Nanodiscs Facilitate High-Resolution Structure Determination of Membrane Proteins. J Am Chem Soc. 2013 doi: 10.1021/ja310901f. The reduced-size Nanodisks allow determination of the first solution NMR structure of IMP in a near-native lipid environment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]