Abstract
Increased hepatic lipid content is an early correlate of insulin resistance, and can be caused by nutrient-induced mTor activation. The latter increases basal Akt activity, leading to a self-perpetuating lipogenic cycle. We have previously shown that the developmental Notch pathway has metabolic functions in adult liver. Acute or chronic inhibition of Notch dampens hepatic glucose production and increases Akt tone, and might therefore be predicted to increase hepatic lipid content. Surprisingly, we show that constitutive liver-specific ablation of Notch signaling, or its acute inhibition with a decoy Notch1 receptor, prevents hepatosteatosis by blocking mTorc1. Conversely, Notch gain-of-function causes fatty liver through constitutive activation of mTorc1, an effect reversible by rapamycin treatment. We demonstrate that Notch signaling increases mTorc1 complex stability, augmenting mTorc1 function and Srebp1c-mediated lipogenesis. The data identify Notch as a therapeutically actionable branch point of metabolic signaling, where hepatic Akt activation can be uncoupled from steatosis.
Introduction
Metabolic diseases in their protean incarnations are likely to define health, public policy, and economics of the 21st century.1 Aside from surgical remediation, progress in their treatment with lifestyle or pharmacologic therapies has been disappointing.
Altered insulin signaling is often associated with excessive hepatic triglyceride content (hepatosteatosis), a correlate of hepatic failure, hepatocellular cancer and need for liver transplantation.2 Activation of the nutrient-sensing mTorc1 pathway, a substrate of insulin/Akt signaling,3 stimulates hepatic de novo lipogenesis,4 leading to hepatosteatosis. These parallel pathways allow the dissociation of insulin signaling in liver in obesity – FoxO1 action is unrestrained in the “insulin-resistant” state to stimulate gluconeogenesis and glycogenolysis, whereas higher plasma insulin levels accelerates flux through the preserved Akt/mTorc1 pathway, to simultaneously promote hepatic glucose production and hepatosteatosis.4 Thus, treatment of hepatocytes with rapamycin, an allosteric inhibitor of mTorc1, prevents insulin activation of the lipogenic transcription factor Srepb1c.4,5 Although interpretation of in vivo rodent studies, and clinical experience in rapamycin-treated patients, is clouded by their effects to disrupt insulin signaling in other tissues and possible effects on mTorc2 function, mice with disruptions in hepatic mTorc1 signaling have offered insight into its role in regulation of glucose and lipid metabolism.6–9 For instance, liver-specific knockout of the mTorc1-defining component Raptor protects from diet-induced hepatic steatosis, likely due to reduced lipogenesis.10 Interestingly, hepatocyte-specific knockout of Tsc1, a native mTor inhibitor, protects from diet-induced fatty liver due to mTorc1-independent effects on Insig2a, an Akt-dependent regulator of Srebp1c function, suggesting that the Akt and mTorc1 pathways intersect at multiple levels to integrate insulin and nutrient signals in the liver.11
The bifurcation of the insulin signaling pathways after Akt – to FoxO1 for glucose production, and to mTor/Srebp1c for lipogenesis – raises the question of whether these pathways have additional inputs. Notch signaling is critical for cell type specification and lineage restriction.12 Cell surface-tethered ligands (Jagged and Delta-like) bind Notch receptors on neighboring cells, resulting in a series of cleavage events that culminate in γ-secretase-dependent liberation of the Notch intracellular domain (NICD).13 NICD translocates to the nucleus, where it binds to and co-activates the transcriptional effector Rbp-Jk, promoting expression of the Hairy enhancer of split (Hes) and Hes-related (Hey) family of genes.14 Homozygous null alleles of components of this signaling pathway result in embryonic lethality, demonstrating their importance to normal development.15–17 Importantly, Notch signaling is therapeutically accessible, and inhibitors are in advanced clinical development for cancer.18
The homeostatic functions of Notch in the adult animal have received less attention, except in neoplastic processes.19 We have shown that liver Notch signaling is regulated in response to metabolic stimuli, and that Notch1 increases hepatic glucose production by co-activating FoxO1 at the Glucose-6-phosphatase promoter.20 Conversely, liver-specific deletion of Rbp-Jk (L-Rbpj mice), or γ-secretase inhibitor (GSI) treatment improves glucose tolerance, and reduces hepatocyte glucose production.20 Interestingly, previous studies demonstrated that Notch1 can activate mTorc1 in leukemic cells, whereas GSIs decrease mTorc1 activity in breast cancer.21,22 Thus, we hypothesized that hepatic Notch could modulate the coordinate actions of insulin on gluconeogenesis (via FoxO1) and lipogenesis (via mTorc1). We describe here that inhibition of hepatic Notch protects from obesity-induced fatty liver, likely through decreased de novo lipogenesis. Conversely, constitutive hepatic Notch signaling stabilizes and activates mTorc1, leading to increased lipogenesis and fatty liver. We show that Notch-mediated hepatic steatosis is rapamycin-sensitive, whereas Notch-induced glucose tolerance is mTor-independent. These results establish Notch as a unique pharmacological target in liver, whose inhibition can prevent the twin abnormalities of hepatic insulin resistance – excessive glucose production as well as fatty liver – by virtue of its ability to uncouple Akt from mTor.
Results
Liver Notch activity peaks after prolonged fasting and in late refeeding
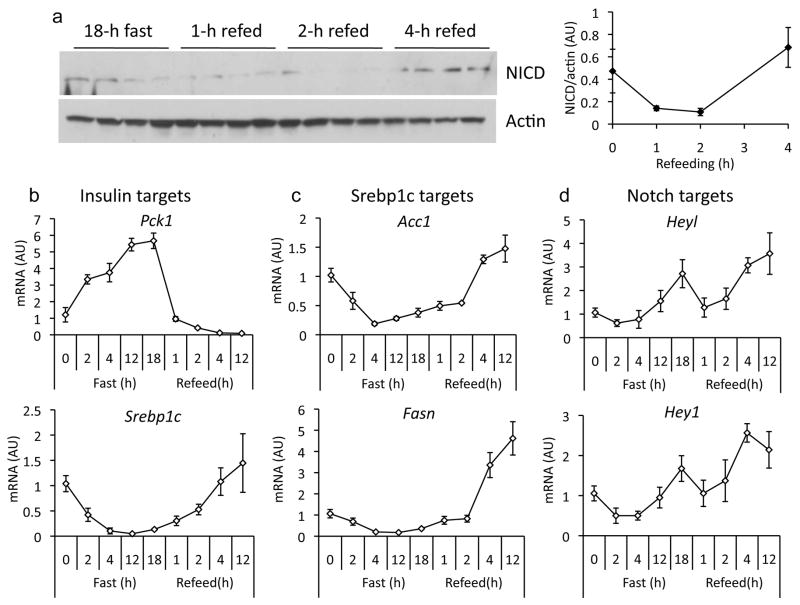
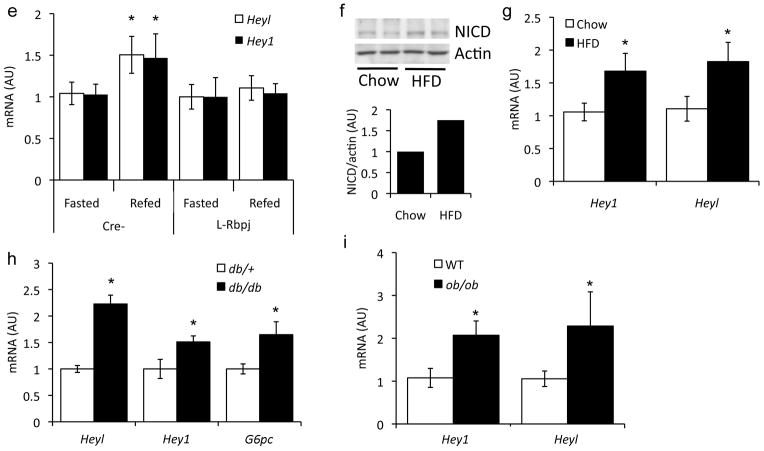
Notch1 activation in liver, as reflected by cleavage at Val1744 and increased expression of Notch targets, increases with fasting.20 In early refeeding (0–2 h), Notch1 cleavage and target gene expression declined, followed by a second peak of Notch activation at later time points (4–12 h) (Fig. 1a and Supplementary Fig. 1). Notably, Notch activation during fasting coincides with increased gluconeogenic gene expression, while the second peak coincides with expression of Srebp1c and its targets (Fatty acid synthase, Fasn; and Acetyl-CoA-carboxylase, Acc1) (Fig. 1b–d), as well as activation of mTor (not shown). This induction was expectedly absent in livers from mice lacking hepatocyte Rbp-Jk (L-Rbpj) (Fig. 1e),20 confirming that classical Notch activation is affected by the nutritional state. Livers from mice fed a high-fat diet (HFD) also showed greater Notch activation than chow-fed littermates (Fig. 1f, g), as did hepatocytes and livers from leptin-signaling deficient mice as compared to controls (Fig. 1h, i), suggesting a cell-autonomous dysregulation of Notch signaling in obesity and fatty liver.
Figure 1.
Regulation of hepatic Notch activity. (a) Western blot and quantitation of cleaved Notch1 receptor (NICD) in livers from fasted and refed 9-week-old, chow-fed C57/Bl6 mice (n=4/group). Expression of insulin (b), Srebp1c (c) and Notch (d) targets in livers from fasted and refed 9-week-old, chow-fed C57/Bl6 mice (n=5/group). (e) Regulation of Notch targets in 16-week-old L-Rbpj and control (Creminus;) mice, fasted for 16-h or fasted for 16-h followed by 4-h refeeding (n=6/group). Fasted values are set arbitrarily at 1 for both groups. *p < 0.05 vs. fasted mice. (f) Western blot of cleaved Notch1 and (g) Notch target gene expression in livers from fasted, 16-week old chow-fed or high-fat diet (HFD)-fed mice (n=12/group). *p < 0.05 vs. chow-fed mice. (h) Notch target expression in livers from db/db or control (db/+) mice (n=5/group), or (i) in hepatocytes from ob/ob or control (WT) mice, all sacrificed in the ad libitum state (triplicate wells, representative of 2 individual experiments). *p < 0.05 vs. db/+ or WT mice. Data show means ± SEM.
Liver-specific Rbp-Jk deletion protects from diet-induced steatosis
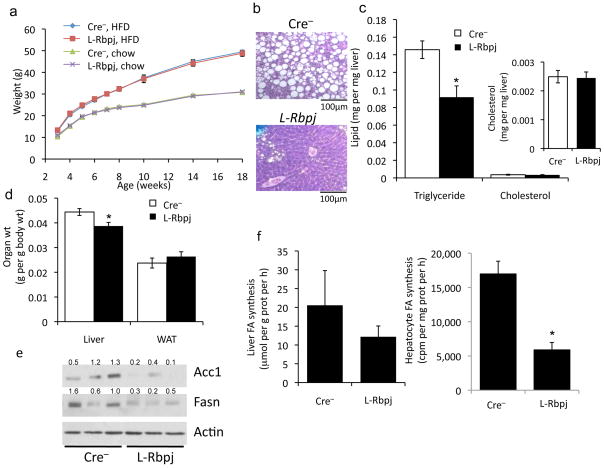
As whole-body disruption results in embryonic lethality,16 we generated liver-specific Rbp-Jk knockout (L-Rbpj) mice, in which hepatocyte Rbp-Jk was deleted post-natally,20 with full recombination by 6–12 weeks of age.23 We have previously shown that L-Rbpj mice are protected from obesity-induced insulin resistance.20 Given the interaction between Rbp-Jk and FoxO1,24 we hypothesized that L-Rbpj mice would have similarly increased hepatic triglyceride as mice lacking liver FoxOs.25,26 Notably, despite unchanged body weight, L- Rbpj mice showed lower HFD-induced hepatic steatosis (Fig. 2a, b), due to a 30–50% reduction in hepatic triglycerides (Fig. 2c). L-Rbpj livers were smaller, without changes in adiposity (Fig. 2d) or serum lipids (Supplementary Fig. 2), as compared to Cre− control mice. Moreover, Rbp-Jk knockout prevented steatosis in mice lacking hepatic FoxO1 (Supplementary Fig. 3a),25 suggesting that Notch regulates hepatic lipid deposition independent of its known co-activation of FoxO1 targets.20
Figure 2.
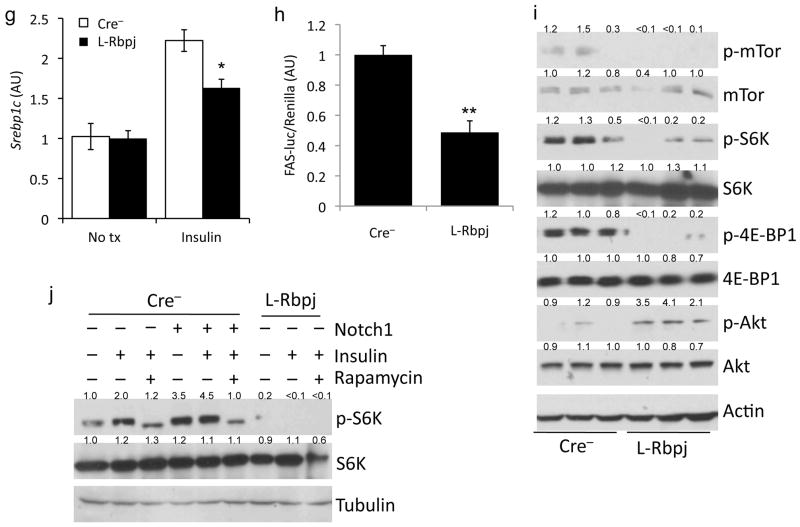
Lower hepatic triglycerides in HFD-fed L-Rbpj mice. (a) Body weight of Cre− and L-Rbpj male mice (n=6–8/group) on standard chow, or HFD started at weaning. (b) H&E staining from age- and weight-matched L-Rbpj and Cre− mice on HFD. (c) Hepatic lipids (inset shows expanded graph for cholesterol), and (d) liver and epididymal white adipose tissue (WAT) weight in mice fed HFD from weaning and sacrificed at 20 weeks after a 16-h fast (n=8/group). (e) Hepatic lipogenic protein expression and (f, left) de novo lipogenesis in livers from 20-week-old, HFD-fed mice sacrificed after a 16-h fast, followed by 6-h refeeding (n=7/group). (f, right) De novo lipogenesis in hepatocytes from chow-fed L-Rbpj and Cre− 16-week-old mice (triplicate wells, representative of 2 individual experiments). (g) Basal and insulin-stimulated Srebp1c expression and (h) Fasn-luciferase activity in primary hepatocytes from chow-fed, 16-week-old Cre− and L-Rbpj littermates, transferred to serum-free medium for 16-h, followed by addition of 10nM insulin for 6-h prior to lysis (triplicate wells, representative of 2 individual experiments). (i) Western blot analysis of Akt and mTor signaling in livers from HFD-fed mice sacrificed after a 5-h fast. (j) Western blots from hepatocytes isolated from L-Rbpj and Cre− 16-week-old mice, transduced with control or Notch1-IC adenovirus, treated with 10nM insulin and/or 25nM rapamycin for 4-h. Protein expression normalized to either actin or tubulin. *p < 0.05, **p < 0.01, ***p < 0.001 vs. Cre− mice or hepatocytes. Data show means ± SEM.
L-Rbpj mice show reduced de novo lipogenesis
We evaluated cell-autonomous and non-autonomous pathways that regulate hepatic triglyceride accumulation.2,27 VLDL secretion was unaltered in L-Rbpj mice (Supplementary Fig. 3b), as were plasma triglyceride levels after olive oil gavage (Supplementary Fig. 3c). Liver expression of fatty acid oxidation enzymes Acox and Cpt1a, serum ketones and β-oxidation of exogenous fatty acids in primary hepatocytes were similarly unchanged (Supplementary Fig. 3d–f). Next, we studied lipogenesis - L-Rbpj livers showed lower Fasn and Acc1 expression (Fig. 2e), leading to less fatty acid synthesis (Fig. 2f). In L-Rbpj-derived primary hepatocytes, we found impaired insulin-dependent Srebp1c expression and activity, as assessed by lower expression of Fasn promoter-driven luciferase containing a consensus Srebp1c binding site28 (Fig. 2g, h). Alternative lipogenic pathways, including PPARγ signaling, were unaltered in L-Rbpj mice (Supplemental Fig. 3g).29 We observed a similar protection from insulin resistance associated with lower hepatic triglycerides following short-term HFD feeding (Supplemental Fig. 4). These data indicate that blocking hepatic Notch reduces hepatic triglyceride, likely due to impaired Srebp1c-mediated lipogenesis.
Reduced mTorc1 signaling in L-Rbpj mice
We next studied pathways that converge on Srebp1c, insulin/Akt and nutrient/mTor.4 L-Rbpj livers show higher insulin sensitivity, with higher Akt phosphorylation at the Pdk1 site, T308.20 Conversely, we noted repressed mTorc1 signaling, as indicated by lower phosphorylation of mTor and mTorc1 targets, p70 S6 kinase and 4E-BP1, after either 5h or 16h fasting as compared to control animals (Fig. 2i and not shown).30–32 To determine if this effect was cell-autonomous, we isolated primary hepatocytes from Cre− and L-Rbpj mice, and found that while Akt phosphorylation was higher (not shown), basal and insulin-stimulated p70 S6k phosphorylation were repressed (Fig. 2j). These data suggest that Notch is required for maximal hepatocyte mTorc1 activity.
Acute Notch inhibition protects from insulin resistance and fatty liver
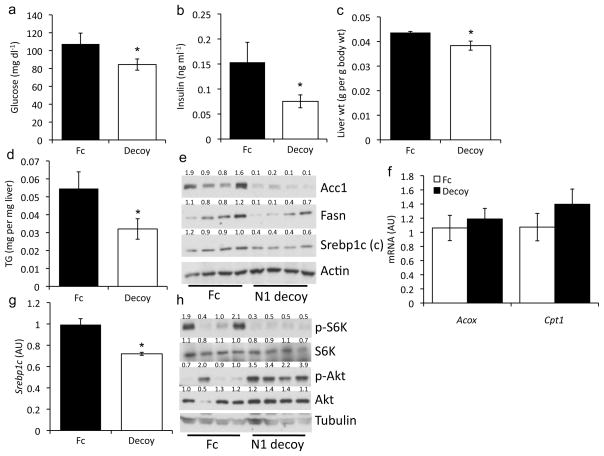
To exclude the possibility of a developmental phenotype in L-Rbpj mice, we tested if acute inhibition of Notch signaling can similarly protect from diet-induced fatty liver and reduce mTorc1 function. We transduced adult mice with a “decoy” Notch1 receptor that encodes only the extracellular domain33,34 and acts in a dominant-negative manner by sequestering endogenous ligand. Adenovirus-driven Notch1 decoy is preferentially expressed in the liver, and is poorly secreted into the circulation (not shown). Consistent with results from L-Rbpj mice, Notch decoy administration to HFD-fed mice lowered glucose and insulin levels (Fig. 3a, b), as well as liver weight and triglyceride content (Fig. 3c, d), without affecting body or adipose weight (Supplementary Fig. 5a, b), as compared to Cre− control mice. Notch decoy inhibited Srebp1c cleavage, and Fasn and Acc1 expression (Fig. 3e) but did not affect fatty acid oxidation or Ppary expression (Fig. 3f) or serum lipids (Supplementary Fig. 5c, d). Notch decoy-transduced primary hepatocytes similarly showed lower Srebp1c expression (Fig. 3g), but no change in Ppary or its targets (Supplementary Fig. 5e). Livers from Notch decoy-transduced mice demonstrated higher pAkt-T308, but lower pS6k-S389 (Fig. 3h). In sum, similar to L-Rbpj mice, acute reduction in hepatic Notch signaling increases insulin sensitivity, while simultaneously lowering mTorc1-mediated Srebp1c activity and hepatic triglycerides.
Figure 3.
Notch decoy increases insulin sensitivity and decreases hepatic lipid content. (a) Glucose, (b) insulin, (c) liver weight, and (d) hepatic triglyceride 14-d after delivery of Notch1 decoy or Fc control adenovirus in HFD-fed mice, fasted for 16-h (n=6/group). (e) Western blots of liver proteins from HFD-fed mice transduced with Notch decoy or Fc adenovirus and fasted for 16-h. (f) Expression of fatty acid oxidation genes in livers from fasted mice transduced with Notch decoy (n=6/group). (g) Srebp1c levels in primary mouse hepatocytes transduced with Notch decoy or Fc adenovirus (triplicate wells, representative of 2 individual experiments). (h) Western blots of liver protein from HFD-fed mice, sacrificed after a 16-h fast, transduced with Notch decoy or Fc adenovirus. Protein expression normalized to either Actin or Tubulin. Mice were 12-week-old male C57Bl/6, unless otherwise indicated. *p < 0.05 vs. Fc. Data show means ± SEM.
Hepatic Notch1 induces mTorc1 signaling and fatty liver
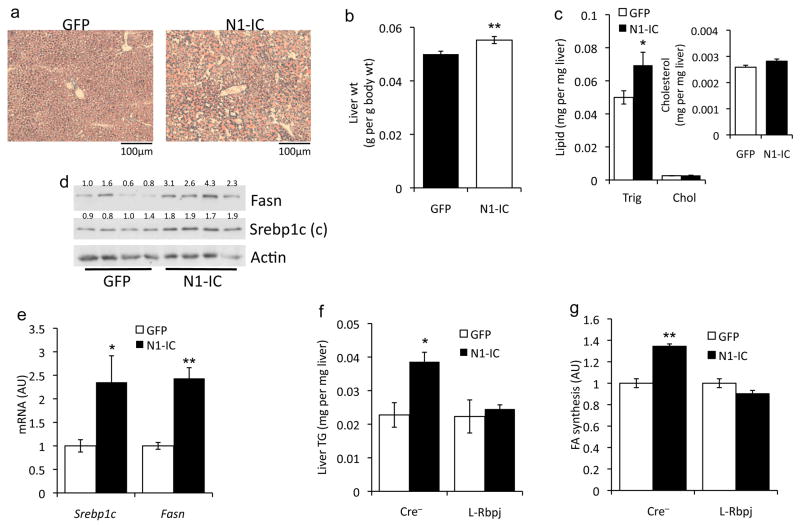
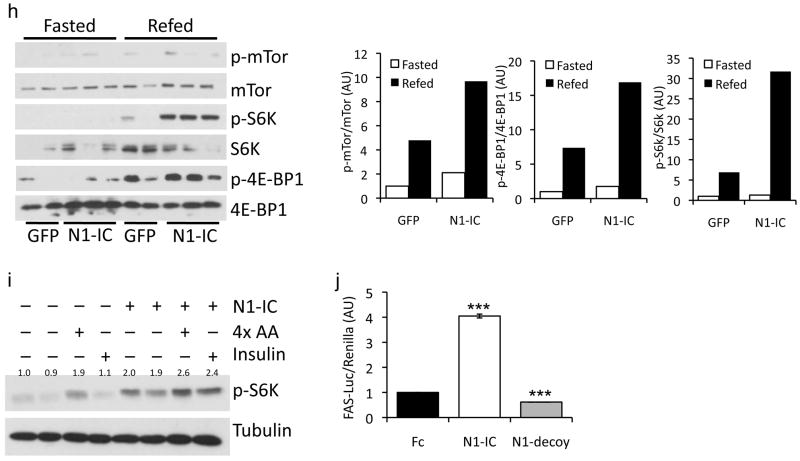
Our loss-of-function studies suggest that Notch signaling is permissive for mTorc1 activation and diet-induced steatosis. We thus tested whether Notch gain-of-function would be sufficient to induce fatty liver in vivo. Chow-fed mice transduced with adenovirus encoding constitutively active Notch1 (N1-IC) showed higher liver weight and triglyceride (Fig. 4a–c), without concomitant changes in body weight or composition (not shown). N1-IC–transduced livers demonstrated higher Srebp1c cleavage, resulting in increased expression of Srebp1c and Fasn (Fig. 4d, e). Consequently, primary hepatocytes transduced with N1-IC showed greater lipogenesis (Supplementary Fig. 6a). Importantly, N1-IC expression failed to alter lipogenic gene expression, fatty acid synthesis or hepatic triglycerides in L-Rbpj mice and hepatocytes (Fig. 4f, g and Supplementary Fig. 6b), suggesting that Notch-induced lipogenesis requires Rbp-Jk, similar its activation of hepatic glucose production.20
Figure 4.
Activation of hepatic Notch increases mTor, lipogenic genes, and steatosis in chow-fed mice. (a) Oil-Red-O staining, (b) weight, and (c) lipid content in livers of 16-h fasted mice (inset shows expanded graph for cholesterol) 7-d after adenoviral delivery of GFP (control) or constitutively active Notch1 (N1-IC) (n=6/group). (d) Liver Western blots and (e) gene expression analysis in mice transduced with GFP or N1-IC adenovirus, sacrificed after a 16-h fast followed by 2-h refeeding. (n=6/group). (f) Hepatic triglyceride content 7-d after GFP or N1-IC transduction in 16-h fasted, 24-week-old, chow-fed Cre− and L-Rbpj mice. (g) De novo lipogenesis in hepatocytes isolated from L-Rbpj and Cre− mice after transduction with GFP (arbitrarily set to a value of 1) or N1-IC and incubation with 10nM insulin (triplicate wells, representative of 2 individual experiments). (h) Western blot and quantitation of bands from livers of mice transduced with GFP or N1-IC, fasted for 16-h, or refed for 2-h. (i) Western blot from FAO hepatoma cells transduced with Fc (−) or N1-IC (Notch1), incubated in serum-free and amino acid-free medium for 4-h, followed by treatment with 10nM insulin or 4× amino acid mixture for 4-h. (j) Fasn-luciferase assays in FAO hepatoma cells transduced with N1-IC, N1-decoy or Fc (control) adenovirus and treated with 10nM insulin. *p < 0.05, **p < 0.01, ***p < 0.001 vs. Fc or GFP. Protein expression normalized to either Actin or Tubulin. Mice were 8-week-old C57Bl/6 males, unless otherwise indicated. Data show means ± SEM.
Notch-induced lipogenic gene expression paralleled higher hepatic mTorc1 activity in fasted and (more markedly) refed animals (Fig. 4h), consistent with enhanced physiologic regulation of mTorc1. In hepatoma cells and mouse primary hepatocytes, activation of mTorc1 signaling by insulin and amino acids was potentiated by N1-IC (Fig. 4i), resulting in Srebp1c cleavage and activation (Fig. 4j and Supplemental Fig. 6c). These data suggest that Notch modulates, but does not override endogenous mTor regulation, in a cell-autonomous manner.
Inhibition of mTor prevents Notch-induced fatty liver
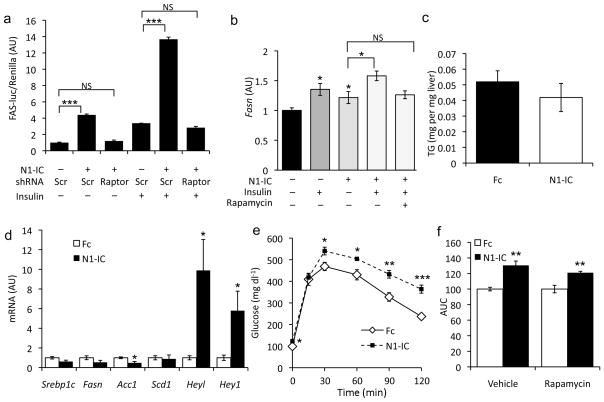
To test the hypothesis that Notch induction of lipogenic gene expression and fatty liver requires mTorc1 signaling, we co-transfected hepatoma cells with Fasn-luciferase and shRNA to Raptor,35 the defining component of the mTorc1 complex, then transduced cells with N1-IC. Notch-induction of Fasn-luciferase activity was potentiated by insulin, but reversed by Raptor knockdown or rapamycin treatment (Fig. 5a and Supplementary Fig. 7). Similarly, Notch induction of endogenous Fasn in primary hepatocytes was augmented by insulin, and suppressed by rapamycin (Fig. 5b), suggesting that N1-IC-induced Fasn expression is mTorc1-dependent.
Figure 5.
mTor inhibition prevents Notch-induced fatty liver. (a) Fasn-luciferase in FAO hepatoma cells transfected with either scrambled (Scr) or Raptor shRNA, transduced with either Fc (−) or N1-IC (Notch) adenovirus, serum-starved overnight and subsequently treated for 6-h with 10nM insulin. (b) Gene expression in primary hepatocytes after transduction with GFP (−) or N1-IC (Notch) adenovirus, followed by incubation with 10nM insulin and/or 25nM rapamycin (triplicate wells, representative of 2 individual experiments). (c) Hepatic triglyceride content and (d) gene expression in rapamycin-treated Fc or N1-IC-transduced mice, sacrificed after a 16-h fast followed by 6-h refeeding. (e) Glucose tolerance test and (f) AUC from GTT in mice transduced with Fc (arbitrarily set to a value of 1 for both treatments) or N1-IC, injected daily with rapamycin or vehicle. Mice were 10-week-old, short-term (3 weeks) HFD-fed C57Bl/6 males. *p < 0.05, **p < 0.01 and ***p < 0.001 vs. Fc-transduced cells or mice. Data show means ± SEM.
Based on these data, we hypothesized that higher lipogenic gene expression and fatty liver seen in mice transduced with N1-IC adenovirus would be ameliorated by rapamycin treatment. Indeed, Notch-mediated hepatic steatosis was completely reversed by rapamycin treatment (Fig. 5c). The effect of rapamycin was specific to Notch induction of lipogenic genes, as Heyl and Hey1 were unaffected (Fig. 5d). Similarly, although rapamycin induced mild glucose intolerance (not shown),6 N1-IC-transduced mice showed further exacerbation (Fig. 5e, f). These data show that Notch-induced hepatic steatosis, but not -hyperglycemia, is prevented by mTor inhibition.
Notch increases mTorc1 complex stability
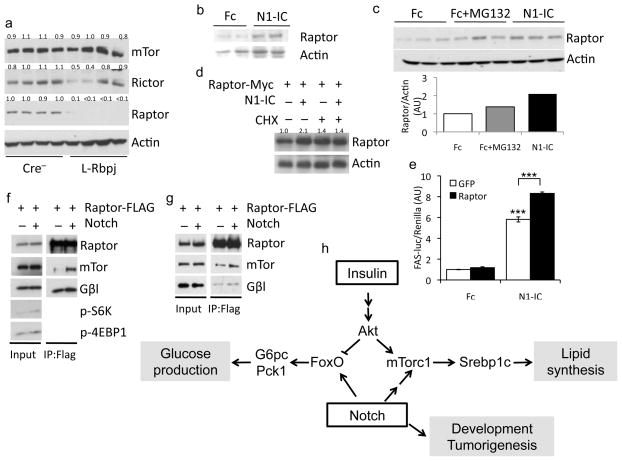
To study the mechanism of altered Notch-induced mTorc1 activation, we examined mTor complex levels in L-Rbpj mouse liver. We found unchanged levels of the shared mTorc1/mTorc2 components, mTor and Gβl, and of the mTorc2-specific component Rictor, but a surprising reduction in the levels of Raptor protein (Fig. 6a), independent of changes in Raptor mRNA (not shown), suggesting that the effects of Rbp-Jk deficiency on Raptor are post-transcriptional. Conversely, mice transduced with N1-IC adenovirus demonstrated higher liver Raptor protein (Fig. 6b). We found a similar increase of endogenous Raptor protein in hepatoma cells (Fig. 6c) or primary hepatocytes (not shown) transduced with N1-IC, without changes in Raptor mRNA (Supplementary Fig. 8a). Transient transfection of Raptor cDNA in primary hepatocytes showed a similar effect, demonstrating that the action of Notch is independent of locus effects (Supplementary Fig. 8b). Notably, the effect of N1-IC was not recapitulated by proteosomal inhibition by MG132 (Fig. 6c), but was reversed by treatment of hepatocytes with the protein synthesis inhibitor, cycloheximide (Fig. 6d).
Figure 6.
Notch induces mTorc1 complex stability. (a) Western blots of liver proteins from 5h-fasted L-Rbpj and control mice (mTor and Actin blots reproduced from Figure 2i). (b) Western blot of liver protein from chow-fed, 12-week-old C57Bl/6 male mice transduced with Fc or N1-IC, sacrificed at day 7, after overnight fasting. (c) Western blots from FAO hepatoma cells transduced with either Fc or N1-IC, with or without treatment with MG132 for 4-h. (d) Western blots of primary hepatocytes, transfected with Raptor cDNA, then transduced with Fc or N1-IC and treated for 2-h with cycloheximide. (e) Fasn-luciferase activity in FAO hepatoma cells transfected with Fasn-luciferase reporter, co-transduced with Fc or N1-IC and GFP or Raptor. 24-h after transduction, hepatoma cells were transferred to serum-free medium for 16-h, then treated with 10 nM insulin for 6-h prior to lysis. ***p<0.001 vs. Fc. Data show means ± SEM. (f) Western blots of HEK 293 cells or (g) primary hepatocytes transfected with Raptor-FLAG, followed by transduction with GFP or N1-IC and immunoprecipitation with anti-FLAG antibody. Protein expression normalized to either actin or tubulin. (h) Model of Notch effects on hepatic glucose and lipid metabolism.
Raptor overexpression was insufficient to induce Fasn-luciferase whereas co-expression of N1-IC and Raptor produced a synergistic effect (Fig. 6e), consistent with previous work that Raptor overexpression per se does not increase mTorc1 function.34,35 Likewise, overexpression of Raptor was insufficient to activate mTorc1 in either primary hepatocytes or HEK 293 cells (not shown). We concluded that Notch induction of Raptor levels parallels, but does not cause increased mTorc1 activation, and hypothesized that increased Raptor levels are secondary to higher mTorc1 complex stability. Indeed, we found that Notch overexpression increased association among mTorc1 components in HEK 293 cells (Fig. 6f), regardless of whether Raptor (Supplemental Fig. 8c) or mTor (Supplemental Fig. 8d) was immunoprecipitated. Similar mTorc1 stabilization was observed in FAO hepatoma cells (Supplemental Fig. 8e) and mouse primary hepatocytes (Fig. 6g). Finally, Notch-stabilized mTorc1 complexes were resistant to increasing concentrations of CHAPS detergent known to disrupt mTor-Raptor interaction (Supplementary Fig. 8f).36–38 In sum, these data indicate that the Notch stabilizes and activates mTorc1, resulting in increased de novo lipogenesis and fatty liver.
Discussion
The role of developmental pathways in metabolic homeostasis of adult tissues is only beginning to be appreciated.39 We have shown that genetic or pharmacologic inhibition of Notch protects from diet-induced glucose intolerance, without effects on body weight or adiposity, in a FoxO1-dependent manner.20 In this work, we demonstrate a similar protection from fatty liver with inhibition of hepatic Notch signaling. This is unexpected, as inhibition of hepatic FoxO1 is associated with increased hepatic lipid deposition, 25,26,40,41 an increasingly recognized effect of shifting hepatic carbon flux from glucose to lipid production.42 In this regard, it appears that chronic (L-Rbpj mice) or acute Notch inhibition (Notch decoy), achieves the long-sought goal of decreasing hepatic glucose production without compensatory increases in hepatic lipid content. Interestingly, GSIs induce fatty liver, but do so in a Notch-independent fashion (U.B.P., manuscript in preparation), consistent with the idea that substrates of γ-secretase include Notch-unrelated pathways, and restricting the repertoire of therapeutically viable Notch inhibitors that can be pursued for treatment of metabolic disease. Nonetheless, the many potential benefits of Notch inhibition, which include amelioration of atherosclerosis,43 provide in our opinion a strong rationale to pursue Notch inhibition as a treatment of the metabolic syndrome.44
The identification of Notch as a regulator of carbon flux towards hepatic glucose or lipid production (Fig. 6h) is a conceptual advance, as is the surprising finding that a molecular pathway thought to be specialized toward differentiation is regulated by physiologic (fasting/re-feeding), as well as pathologic (insulin resistance) metabolic cues in hepatocytes. We hypothesize that in the overfed and insulin-resistant state, Notch signaling is inappropriately activated, and reprises its developmental interactions with FoxO1 and mTorc1. The mechanisms underlying nutritional activation of hepatic Notch require further clarification. For example, it should be determined whether Notch activation in the hepatocyte requires input from neighboring hepatocytes or other resident liver cells (endothelial, stellate, Kupffer, etc.). Similarly, which of the five Notch ligands drives signaling in response to nutrients is unknown, and the possibility that different ligands signal in different metabolic states to direct carbon flux or drive differentiation is teleologically attractive.
Besides the further validation of hepatic Notch as a therapeutic target, our data demonstrate a physiologic, and potentially pharmacologic, means of regulating mTorc1 activity and lipogenesis. Previous studies have indicated that tight control of hepatic mTorc1 signaling is critical for hepatic lipid metabolism.10,11 The tandem, but not necessarily related, findings of mTorc1 stabilization and activation by Notch deserve further study. Since the identification of Raptor as the mTorc1-regulatory subunit, it has been known that the mTor-Raptor association is sensitive to detergent concentrations;38 subsequent reports have confirmed this finding and identified potential post-translational modifications on Raptor,36,37,45 but none have been shown to mediate mTor-Raptor interaction. How Notch induces mTorc1 stability, but the demonstration that Raptor levels are decreased in L-Rbpj mice and that cycloheximide prevents Notch-induced stabilization indicates that a transcriptional target(s) of Notch regulates complex stability.
In summary, Notch antagonism uncouples Akt from mTor activation, suggesting that Notch antagonists from oncology and neuroscience46,47 may be repurposed to treat fatty liver and diabetes. Furthermore, as Notch-mediated mTorc1 activation does not appear to be cell type-specific, modulators of mTorc1 processing and degradation may represent a therapeutic avenue to block mTorc1 activity without the metabolic liabilities of current mTor inhibitors.6
Supplementary Material
Acknowledgments
Supported by NIH grants DK093604 (UBP), DK57539 (DA), HL062454 (JK), and DK63608 (Columbia Diabetes Research Center). We thank D. Conlon, C. Eng, I. Goldberg, R. Haeusler and I. Tabas, as well as members of the Accili, Kitajewski and Ginsberg laboratories, for insightful discussion of the data. We acknowledge excellent technical support from A. Flete, T. Kolar and J. Lee, as well as plasmids from D. Sabatini and B. Spiegelman.
Footnotes
Competing financial interest statement
The Authors declare that they have no competing financial interest in the work described.
Author contributions
U.B.P. designed and performed experiments, analyzed data, and wrote the manuscript. L.Q. and T.K. designed and performed experiments and analyzed data. J.K. and D.A. designed the studies, analyzed the data and wrote the manuscript.
References
- 1.Wang YC, McPherson K, Marsh T, Gortmaker SL, Brown M. Health and economic burden of the projected obesity trends in the USA and the UK. Lancet. 2011;378:815–825. doi: 10.1016/S0140-6736(11)60814-3. [DOI] [PubMed] [Google Scholar]
- 2.Savage DB, Semple RK. Recent insights into fatty liver, metabolic dyslipidaemia and their links to insulin resistance. Current opinion in lipidology. 2010;21:329–336. doi: 10.1097/MOL.0b013e32833b7782. [DOI] [PubMed] [Google Scholar]
- 3.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes & development. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 4.Li S, Brown MS, Goldstein JL. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3441–3446. doi: 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nature reviews Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 6.Blattler SM, et al. Yin Yang 1 deficiency in skeletal muscle protects against rapamycin-induced diabetic-like symptoms through activation of insulin/IGF signaling. Cell metabolism. 2012;15:505–517. doi: 10.1016/j.cmet.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamming DW, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335:1638–1643. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Houde VP, et al. Chronic rapamycin treatment causes glucose intolerance and hyperlipidemia by upregulating hepatic gluconeogenesis and impairing lipid deposition in adipose tissue. Diabetes. 2010;59:1338–1348. doi: 10.2337/db09-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howell JJ, Manning BD. mTOR couples cellular nutrient sensing to organismal metabolic homeostasis. Trends in endocrinology and metabolism: TEM. 2011;22:94–102. doi: 10.1016/j.tem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson TR, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yecies JL, et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell metabolism. 2011;14:21–32. doi: 10.1016/j.cmet.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolos V, Grego-Bessa J, de la Pompa JL. Notch signaling in development and cancer. Endocrine reviews. 2007;28:339–363. doi: 10.1210/er.2006-0046. [DOI] [PubMed] [Google Scholar]
- 13.Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Developmental cell. 2009;16:633–647. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 14.Dufraine J, Funahashi Y, Kitajewski J. Notch signaling regulates tumor angiogenesis by diverse mechanisms. Oncogene. 2008;27:5132–5137. doi: 10.1038/onc.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swiatek PJ, Lindsell CE, del Amo FF, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes & development. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- 16.Oka C, et al. Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development. 1995;121:3291–3301. doi: 10.1242/dev.121.10.3291. [DOI] [PubMed] [Google Scholar]
- 17.Shen J, et al. Skeletal and CNS defects in Presenilin-1-deficient mice. Cell. 1997;89:629–639. doi: 10.1016/s0092-8674(00)80244-5. [DOI] [PubMed] [Google Scholar]
- 18.Rizzo P, et al. Rational targeting of Notch signaling in cancer. Oncogene. 2008;27:5124–5131. doi: 10.1038/onc.2008.226. [DOI] [PubMed] [Google Scholar]
- 19.Weinmaster G, Kopan R. A garden of Notch-ly delights. Development. 2006;133:3277–3282. doi: 10.1242/dev.02515. [DOI] [PubMed] [Google Scholar]
- 20.Pajvani UB, et al. Inhibition of Notch signaling ameliorates insulin resistance in a FoxO1-dependent manner. Nature medicine. 2011;17:961–967. doi: 10.1038/nm.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan SM, Weng AP, Tibshirani R, Aster JC, Utz PJ. Notch signals positively regulate activity of the mTOR pathway in T-cell acute lymphoblastic leukemia. Blood. 2007;110:278–286. doi: 10.1182/blood-2006-08-039883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Efferson CL, et al. Downregulation of Notch pathway by a gamma-secretase inhibitor attenuates AKT/mammalian target of rapamycin signaling and glucose uptake in an ERBB2 transgenic breast cancer model. Cancer research. 2010;70:2476–2484. doi: 10.1158/0008-5472.CAN-09-3114. [DOI] [PubMed] [Google Scholar]
- 23.Postic C, Magnuson MA. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 24.Kitamura T, et al. A Foxo/Notch pathway controls myogenic differentiation and fiber type specification. The Journal of clinical investigation. 2007;117:2477–2485. doi: 10.1172/JCI32054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haeusler RA, Pratt-Hyatt M, Welch CL, Klaassen CD, Accili D. Impaired generation of 12-hydroxylated bile acids links hepatic insulin signaling with dyslipidemia. Cell metabolism. 2012;15:65–74. doi: 10.1016/j.cmet.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tao R, et al. Hepatic FoxOs regulate lipid metabolism via modulation of expression of the nicotinamide phosphoribosyltransferase gene. The Journal of biological chemistry. 2011;286:14681–14690. doi: 10.1074/jbc.M110.201061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Postic C, Girard J. Contribution of de novo fatty acid synthesis to hepatic steatosis and insulin resistance: lessons from genetically engineered mice. The Journal of clinical investigation. 2008;118:829–838. doi: 10.1172/JCI34275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim JB, Wright HM, Wright M, Spiegelman BM. ADD1/SREBP1 activates PPARgamma through the production of endogenous ligand. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:4333–4337. doi: 10.1073/pnas.95.8.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang YL, et al. Aberrant hepatic expression of PPARgamma2 stimulates hepatic lipogenesis in a mouse model of obesity, insulin resistance, dyslipidemia, and hepatic steatosis. The Journal of biological chemistry. 2006;281:37603–37615. doi: 10.1074/jbc.M604709200. [DOI] [PubMed] [Google Scholar]
- 30.Gingras AC, et al. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes & development. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiang GG, Abraham RT. Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. The Journal of biological chemistry. 2005;280:25485–25490. doi: 10.1074/jbc.M501707200. [DOI] [PubMed] [Google Scholar]
- 32.Weng QP, et al. Regulation of the p70 S6 kinase by phosphorylation in vivo. Analysis using site-specific anti-phosphopeptide antibodies. The Journal of biological chemistry. 1998;273:16621–16629. doi: 10.1074/jbc.273.26.16621. [DOI] [PubMed] [Google Scholar]
- 33.Funahashi Y, et al. A notch1 ectodomain construct inhibits endothelial notch signaling, tumor growth, and angiogenesis. Cancer research. 2008;68:4727–4735. doi: 10.1158/0008-5472.CAN-07-6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Funahashi Y, et al. Notch modulates VEGF action in endothelial cells by inducing Matrix Metalloprotease activity. Vascular cell. 2011;3:2. doi: 10.1186/2045-824X-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterson TR, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foster KG, et al. Regulation of mTOR complex 1 (mTORC1) by raptor Ser863 and multisite phosphorylation. The Journal of biological chemistry. 2010;285:80–94. doi: 10.1074/jbc.M109.029637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaizuka T, et al. Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. The Journal of biological chemistry. 2010;285:20109–20116. doi: 10.1074/jbc.M110.121699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim DH, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 39.Liu H, et al. Wnt signaling regulates hepatic metabolism. Science signaling. 2011;4:ra6. doi: 10.1126/scisignal.2001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsumoto M, Han S, Kitamura T, Accili D. Dual role of transcription factor FoxO1 in controlling hepatic insulin sensitivity and lipid metabolism. The Journal of clinical investigation. 2006;116:2464–2472. doi: 10.1172/JCI27047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haeusler RA, Kaestner KH, Accili D. FoxOs function synergistically to promote glucose production. The Journal of biological chemistry. 2010;285:35245–35248. doi: 10.1074/jbc.C110.175851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun Z, et al. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nature medicine. 2012;18:934–942. doi: 10.1038/nm.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fukuda D, et al. Notch ligand Delta-like 4 blockade attenuates atherosclerosis and metabolic disorders. Proceedings of the National Academy of Sciences of the United States of America; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim-Muller JY, Accili D. Cell biology. Selective insulin sensitizers. Science. 2011;331:1529–1531. doi: 10.1126/science.1204504. [DOI] [PubMed] [Google Scholar]
- 45.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Molecular cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noguera-Troise I, et al. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature. 2006;444:1032–1037. doi: 10.1038/nature05355. [DOI] [PubMed] [Google Scholar]
- 47.Wu Y, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.