Abstract
PURPOSE
Polymorphonuclear neutrophils (PMNs) play an important role in mediating the innate immune response after severe traumatic injury; however, the cellular proteome response to traumatic condition is still largely unknown.
EXPERIMENTAL DESIGN
We applied 2D-LC-MS/MS based shotgun proteomics to perform comparative proteome profiling of human PMNs from severe trauma patients and healthy controls.
RESULTS
A total of 197 out of ~2500 proteins (being identified with at least two peptides) were observed with significant abundance changes following the injury. The proteomics data were further compared with transcriptomics data for the same genes obtained from an independent patient cohort. The comparison showed that the protein abundance changes for the majority of proteins were consistent with the mRNA abundance changes in terms of directions of changes. Moreover, increased protein secretion was suggested as one of the mechanisms contributing to the observed discrepancy between protein and mRNA abundance changes. Functional analyses of the altered proteins showed that many of these proteins were involved in immune response, protein biosynthesis, protein transport, NRF2-mediated oxidative stress response, the ubiquitin-proteasome system, and apoptosis pathways.
CONCLUSIONS AND CLINICAL RELEVANCE
Our data suggest increased neutrophil activation and inhibited neutrophil apoptosis in response to trauma. The study not only reveals an overall picture of functional neutrophil response to trauma at the proteome level, but also provides a rich proteomics data resource of trauma-associated changes in the neutrophil that will be valuable for further studies of the functions of individual proteins in PMNs.
Keywords: human neutrophil, LC-MS/MS, Proteomics, Trauma, Genomics
Introduction
Polymorphonuclear neutrophils (PMNs) are the most abundant white blood cells that play an important role in innate immune response by providing the first line of defense against microbial threats[1–3]. Extensive investigations have been made to elucidate the biological function of PMNs in immune response. The most accepted hypothesis is that PMNs are activated by “damage-associated molecular patterns” (DAMPs)[4], such as pro-inflammatory cytokines, followed by the migrating to infected/injured tissues by chemotaxis, mediating the inflammatory functions through adhesion, rolling, firm adhesion, and transendothelial migration[1, 5, 6]. After entering the target tissue, PMNs mediate secondary tissue damage by activation of the NADPH oxidase enzyme complex[7], resulting in a burst of oxygen consumption, generation of excessive reactive oxygen species (ROS), and release of ROS and toxic enzymes[8]. The release of chemokines, cytokines, complex antibiotic arsenal and granule enzymes play as alarm signals that activate antigen presenting cells[9]. PMNs have been recognized as life-saving decision-makers that coach dendritic cells, monocytes, and lymphocytes, and help the organism to decide whether to initiate and maintain an immune response[10].
The activity of PMNs is directly linked to the immune response induced by trauma. As one of the leading causes of human death, trauma is usually associated with over-activation of innate immune responses followed by a subsequent immune-suppression, which leads to enhanced susceptibility to infection, sepsis, and multiple organ dysfunction syndrome (MODS)[5, 6, 11]. Following extensive basic science research, the roles of various inflammatory signal molecules, the innate immune systems, and its pattern recognition receptors have been recognized for initiating systemic inflammatory response syndrome (SIRS) following traumatic injury[11–13]; however, the complex underlying mechanisms for PMN response to trauma, SIRS and MODS development after trauma are still poorly understood.
Given the importance of PMNs, there has been an increasing interest in applying discovery-oriented genomics and proteomics approaches with the aims of elucidating the underlying signaling pathways of the complex human diseases[14–16]. For example, several studies that reported the application of genome-wide expression analyses to circulating blood leukocytes and tissue samples derived from trauma patients have gained insights into the pathways that underlie systemic inflammation in humans[14, 17]. Several studies have reported the profiling of the PMN proteome or subproteome by either two-dimensional gel electrophoresis or liquid chromatography-mass spectrometry (LC-MS) based approaches[18–22], allowing a large number of PMN proteins to be identified. In this study, we performed a comprehensive analysis of the trauma-associated alterations of the neutrophil proteome by comparing the PMNs isolated from the blood samples collected from five severe trauma patients around 4–7 days post injury, when a peak modified Marshall Score[23] was observed, and five healthy controls applying two-dimensional liquid chromatography separations coupled with tandem mass spectrometry (2D-LC-MS/MS). The results revealed 197 proteins showing significant changes in protein abundances following injury, including proteins associated with immune response pathways and functional categories relevant to neutrophil activation and cell survival such as EIF2 signaling, aminoacyl-tRNA biosynthesis, NRF2-mediated oxidative stress response, the ubiquitin-proteome system, and apoptosis.
Materials and Methods
Human neutrophil samples
Blood samples from 5 controls and 5 severe trauma patients were collected for the global proteomics analysis. All patients selected had no clinical evidence of infection and were of the same age group, gender, and ethnicity, and a brief summary of patient demographics is provided in Supplemental Table 1. All blood samples for the proteomics study were collected at peak modified Marshall score[23] (i.e., 4–7 days). Blood was collected with BD Vacutainer brand tubes with EDTA as anticoagulant (Becton Dickinson, NJ). Neutrophils were isolated from individual subjects immediately following blood collection by a ficol-dextran method as previously described[24, 25]. Cell pellets were washed with ice-cold phosphate buffered saline (PBS) and transferred to 1.5 ml Fisherbrand low-retention microcentrifuge tubes (Fisher Scientific) and were frozen at −80 °C. ~5–10 million cells were obtained for each subject. The purity of neutrophils was greater than 95% as determined by fluorescence activated cell sorter light scatter patterns.
Blood samples from an independent cohort of 10 controls and 101 severe trauma patients were obtained for the microarray analysis. A brief summary of patient demographics is provided in Supplemental Table 2. All blood samples were collected into EDTA Vacutainer collection tubes (Becton Dickinson) and run on the microfluidic device to generate mRNA and cell lysate samples for genomics and proteomics studies as described by Kotz et al.[26]. A portion of the samples (10 controls and 8 trauma subjects) was used for targeted validation for selected candidates identified from the global profiling. All experimental procedures were approved by the Institutional Review Boards of the University of Rochester (Rochester, NY), Pacific Northwest National Laboratory (Richland, WA), Massachusetts General Hospital (Boston, MA), and University of Florida College of Medicine (Gainesville, FL) in accordance with federal regulations.
Protein digestion and fractionation
PMN cell pellets isolated from individual subjects were lysed in 50% 2,2,2-trifluoroethanol (TFE) by sonication for 30 s in ice-water. Protein concentrations were measured by the BCA assay (Pierce, Rockford, IL) and ~200–300 µg protein was recovered for each sample. All protein samples were digested with a TFE-based protocol[27]. Digested peptide samples were pooled into two trauma pools and two control pools, respectively. ~300 µg total peptides were generated for each pooled sample for subsequent strong cation exchange (SCX) fractionation. Each pooled peptide sample was fractionated into 25 fractions similarly as previously described[28].
LC-MS/MS analysis
Each of the 25 SCX fractions was further analyzed using a fully automated custom-built capillary HPLC system coupled online with an LTQ ion trap mass spectrometer (ThermoFinnigan, San Jose, CA) using an in-house manufactured electro spray ionization interface. The reversed-phase capillary column was slurry packed using 3 µm Jupiter C18 particles (Phenomenex, Torrance, CA) in a 75 µm (inside diameter) × 65 cm fused silica capillary (Polymicro Technologies, Phoenix, AZ). The mobile phases consisted of A (0.2% acetic acid and 0.05% TFA in water) and B (0.1% TFA in 90% acetonitrile). An exponential gradient was employed during the separation, which started with 100% A gradually increased to 60% B over the course of 100 min. The instrument was operated in data-dependent mode with an m/z range of 400–2000. The 10 most abundant ions from the MS analysis were selected for MS/MS analysis using a normalized collision energy setting of 35%. A dynamic exclusion of 1 min was used to avoid repetitive analysis of the same abundant precursor ion. The heated capillary was maintained at 200 °C, and the ESI voltage was held at 2.2 kV.
Proteomics data analysis
LC-MS/MS spectra were analyzed by the SEQUEST algorithm against the human International Protein Index (IPI) database with a total of 51,252 protein entries (Version 3.19) with the decoy database searching option for assessing false discovery rate (FDR)[29]. The search parameters used were: 3 Da precursor ion mass tolerance, 1 Da fragment ion mass tolerance, and a maximum of three missed tryptic cleavages. Filtering criteria similar to those previously reported[30] were applied to limit the FDR at the unique peptide level to <1%. Identified proteins were grouped to a non-redundant protein groups using ProteinProphet software[31] and only one protein IPI reference number was used to represent each protein group. Only those proteins or protein groups with two or more unique peptide identifications were considered to be confident protein identifications.
Relative protein abundance quantification was performed based on the spectral count data as recently described[32]. Briefly, after achieving the list of confidently identified peptides, all low scoring MS/MS spectra that match to this set of peptides were recovered for spectral counting quantification. All datasets were normalized based on the total spectral counts. A pseudo spectral count number of 0.5 was added to the 0 spectral counts to avoid taking logarithm to zero. A G test[33] was used to determine the statistical significance of the protein abundance difference[34, 35]. A threshold of five total spectral counts for either the trauma or the control conditions was applied to qualify for the statistical test, where 2060 proteins passed this spectral count threshold. The G value of each protein was calculated as
G =2 × (C × ln{C / [(C + T) / 2]} + T × ln{T/[(C + T) / 2]})
Where for a given protein, C is the total spectral count of controls, and T is the total spectral count of trauma condition. Proteins with p<0.05 (G value > 3.84, degree of freedom = 1) and consistent change in all individual comparisons were considered as significant abundance changes.
Microarray analysis
RNA extraction followed a modified commercial protocol (QIAGEN RNeasy Plus) yielding purified total RNA that was analyzed on an Agilent Bioanalyzer 2100 system. cDNA was synthesized with the Ovation Biotin RNA Amplification and Labeling System (NuGEN Technologies) from 20 ng of total RNA as starting material. The labeled cDNA was hybridized onto a custom-designed, 6.9 million–feature Affymetrix human exon-junction array as described by Xu et al.[36]. MAExpress[37] was applied to perform quantile normalization of the raw expression data and then EDGE[38] was used to perform time series analysis.
Targeted verification using selected reaction monitoring (SRM)
Targeted verification of selected proteins was performed by using SRM with a subset of the same cohort (10 healthy controls and 8 trauma patients with blood samples collected at both 4-day and 1-week post injury) that was used for the microarray analyses. Five proteins along two housekeeping proteins identified from shotgun proteomics were selected for targeted quantification (See Supplementary Table 6). The peptides and SRM transitions was selected and screened as previously described [39]. 10 final tryptic peptides of the seven proteins (one or two peptides per protein) were selected for label-free SRM quantification based on their unambiguous detection. At least 6 transitions of each peptide were monitored for confident identification and accurate peak assignment. The predicted collision energies from Skyline were used for all peptides.
All LC-SRM experiments were performed on a Waters nanoACQUITY UPLC system (Waters Corporation, Milford, MA) directly coupled to a Waters Xevo TQS instrument (Waters Corporation). Peptide separations were performed at a mobile phase flow rate of 400 nL/min using a BEH 1.7 µm C18 column (100 µm i.d. × 10 cm, Waters Corporation). The mobile phases consisted of 0.1% formic acid in water (A) and 0.1% formic acid in ACN (B). 4 µL of sample (0.25 µg/µl) was injected for each analysis using a binary gradient of 10–15% B in 3.5 min, 15–25% B in 21 min, 25–38.5% B in 11 min, 38.5–95% B in 1 min and 95% B for 8 min with a total of ~44.5 min. The inlet capillary of the mass spectrometer was maintained at 110 °C with an electrospray ionization voltage of 2.6 kV.
Datasets were analyzed by Skyline software (Version 1.4.0) [40]. The peak areas were calculated without any smoothing and the best transition of each peptide was used for relative quantification. In order to eliminate any variations from the amount of sample injections, actin B (ACTB) and glyceraldehyde-3-phosphate dehydrogenase (GADPH) were monitored as the internal reference for normalization. Relative abundances for each protein were calculated as the ratio against the average area of the reference peptides from these two proteins. All subsequent data analyses were performed in DAnTE [41], a statistical tool for quantitative analysis.
Results
Human neutrophil proteome coverage
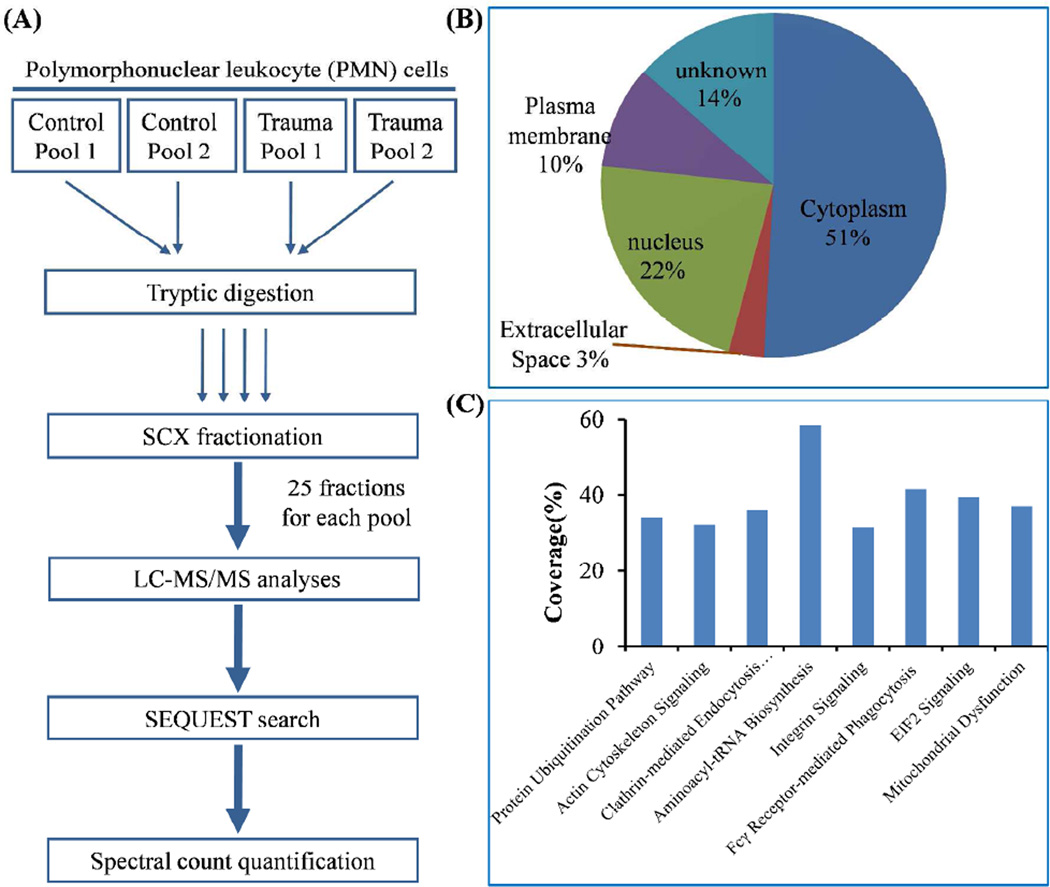
Five trauma patients and five healthy subjects matched by age, sex, and ethnicity were selected for comparative proteomics profiling of enriched PMNs in order to identify proteins with differential abundances in response to traumatic injury. The experimental workflow of comparative profiling using two-dimensional LC-MS/MS is illustrated in Figure 1A. Both trauma and control samples were combined into two pools with three subjects in one pool and two subjects in the other pool. All four pooled samples were individually digested by trypsin and fractionated into 25 fractions per pool by SCX and each fraction was analyzed by LC-MS/MS. On average, ~1,571,000 spectra were acquired and ~98,000 spectra were confidently identified as peptides. The extensive profiling resulted in confident identification of a total ~22,880 unique peptides with <1% FDR from all samples, which corresponded to a total of 2536 proteins identified by at least two unique peptides. Gene ontology analysis of the 2536 proteins revealed protein identifications from all major cellular compartments (Figure 1B). Good coverage of major cell signaling pathways was also observed within this dataset (Figure 1C). All peptides and proteins identified were listed in Supplemental Table 3 and 4.
Figure 1. Experimental workflow and observed proteome coverage of neutrophils.
(A) Workflow of comparative proteomic profiling. (B) Subcellular distribution of all neutrophil proteins identified by 2 or more peptides. (C) Proteome coverage of major canonical pathways.
Protein abundance alterations in response to trauma
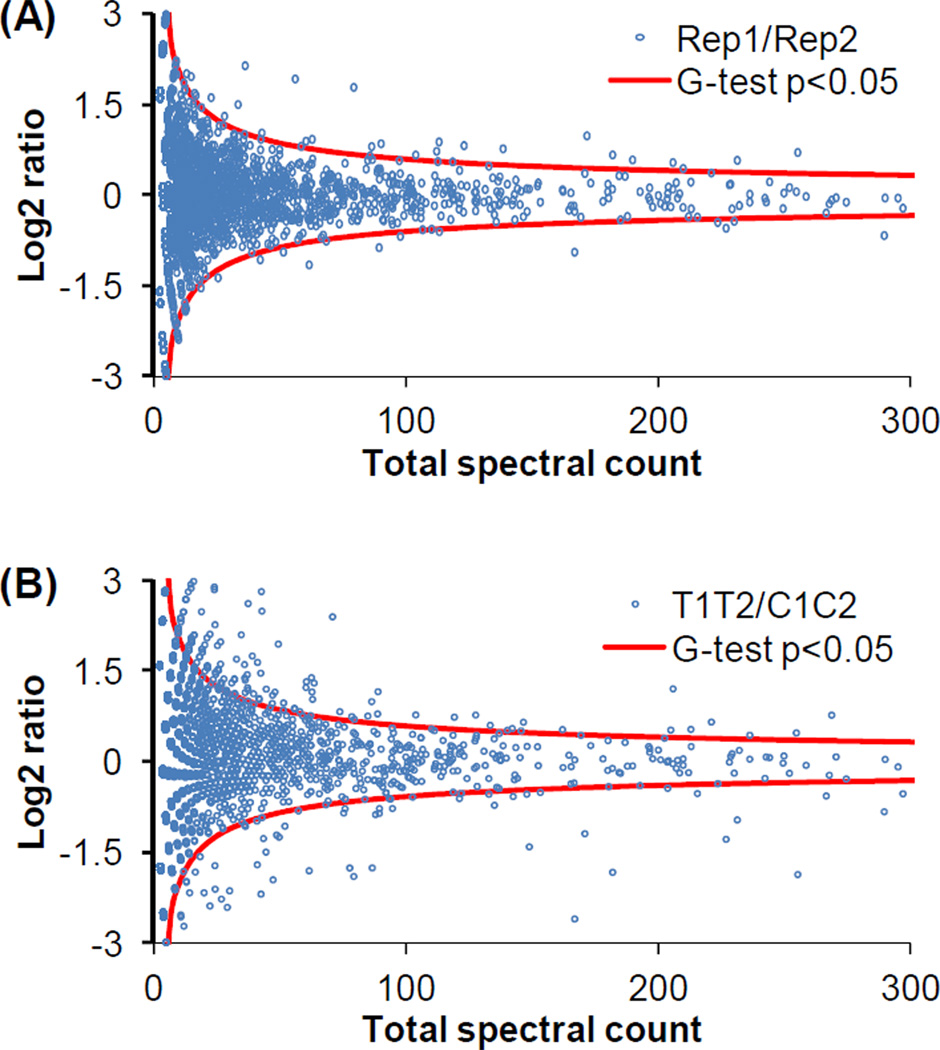
In order to identify protein abundance changes in response to trauma, spectral count[32, 42, 43] as the number of MS/MS spectra identifying a given protein was used as a semi-quantitative measure. A G test was further used to determine the statistical significance. As shown in Figure 2A, only a few outliers were identified after the significance analysis by G-test (p<0.05, G value >3.84, degree of freedom =1, see Supplemental table 5) in the comparison between replicate datasets. However, a relatively large number of proteins passed the significance test in the comparison between trauma datasets and control datasets (Figure 2B). Furthermore, consistent changes in each pair of comparisons between control and trauma conditions (i.e., control pool 1 vs trauma pool 1, control pool 1 vs trauma pool 2, control pool 2 vs trauma pool 1, control pool 2 vs trauma pool 2) with a minimum of 40% difference were required for final significant proteins. A total of 197 proteins passed all the filters. Among them, 144 proteins were up-regulated and 53 proteins were down-regulated following trauma.
Figure 2. Candidate protein selection by G-test.
(A) Log2 ratio distribution of the comparison between two replicates. The data from trauma and control conditions were combined from one pooled sample as one replicate. (B) Log2 ratio distribution of the comparison between datasets from trauma and controls. Both replicates were combined for each condition.
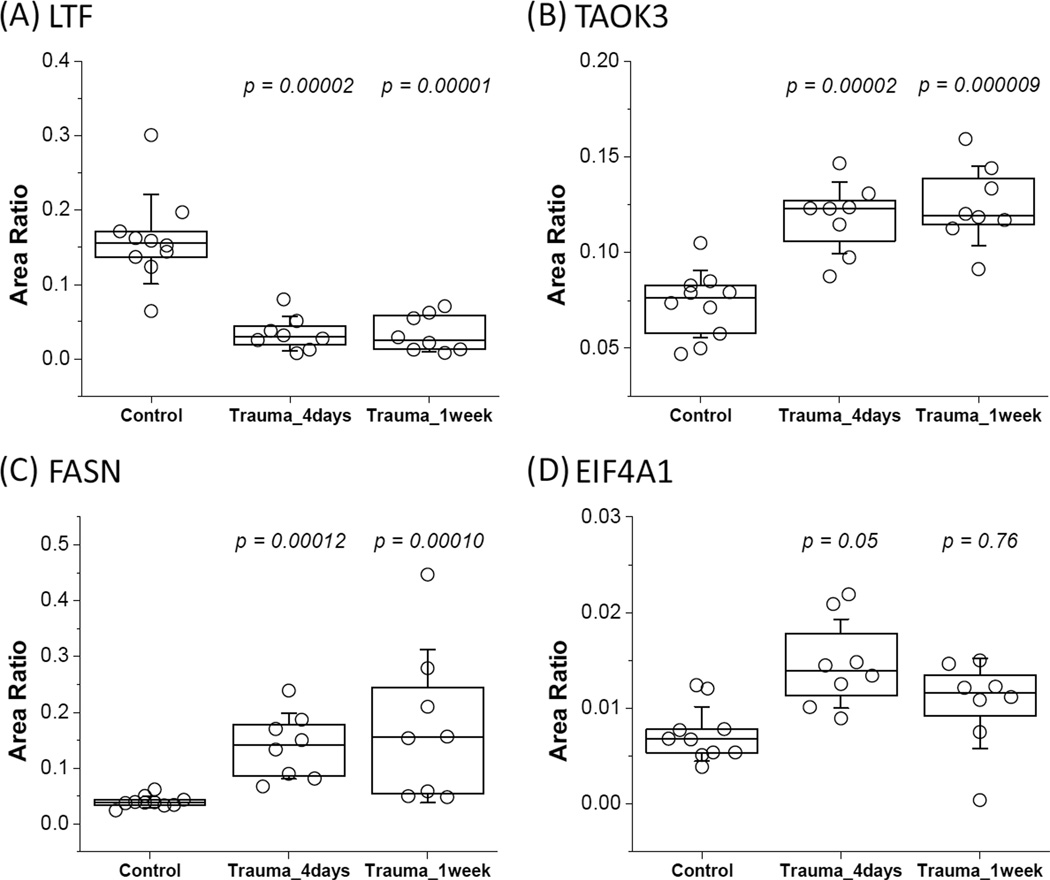
Five proteins, growth-inhibiting protein 12 (LTF), serine/threonine-protein kinase TAO3 (TAOK3), fatty acid synthase (FASN), eukaryotic initiation factor 4A-1 (EIF4A1), and caspase-1 (CASP1), were selected for validation by targeted SRM-based quantification using a cohort of 10 controls and 8 trauma subjects with blood collected at two time-points. As shown in Figure 3, the relative abundance changes of four proteins in response to injury were statistically significant. The observed directions of changes are in good agreement with the spectral count data from pooled samples. One of the proteins, caspase-1 (CASP1), was not confidently detected by 1D LC-SRM, presumably due to its low abundance.
Figure 3. Protein abundance changes between control and trauma conditions as quantified by LC-SRM.
(A) LTF, DLLFKDSAIGFSR (m/z: 490.3/620.8). (B)TAOK3, PTQSVQSQALHYR (m/z: 505.6/437.7) (C) FASN, TLLEGSGLESIISIIHSSLAEPR (m/z: 808.1/883.0) (D) EIF4A1, VLITTDLLAR (m/z: 557.8/789.4). The relative abundances were plotted from the best transition of the given peptide for each protein. Area ratios were calculated against the average area of reference peptides (GYSFTTTAER and QEYDESGPSIVHR of ACTB; GALQNIIPASTGAAK and LISWYDNEFGYSNR of GAPDH). Statistical p-values were calculated against the control group using the ANOVA test.
Comparison between protein abundance and gene expression changes
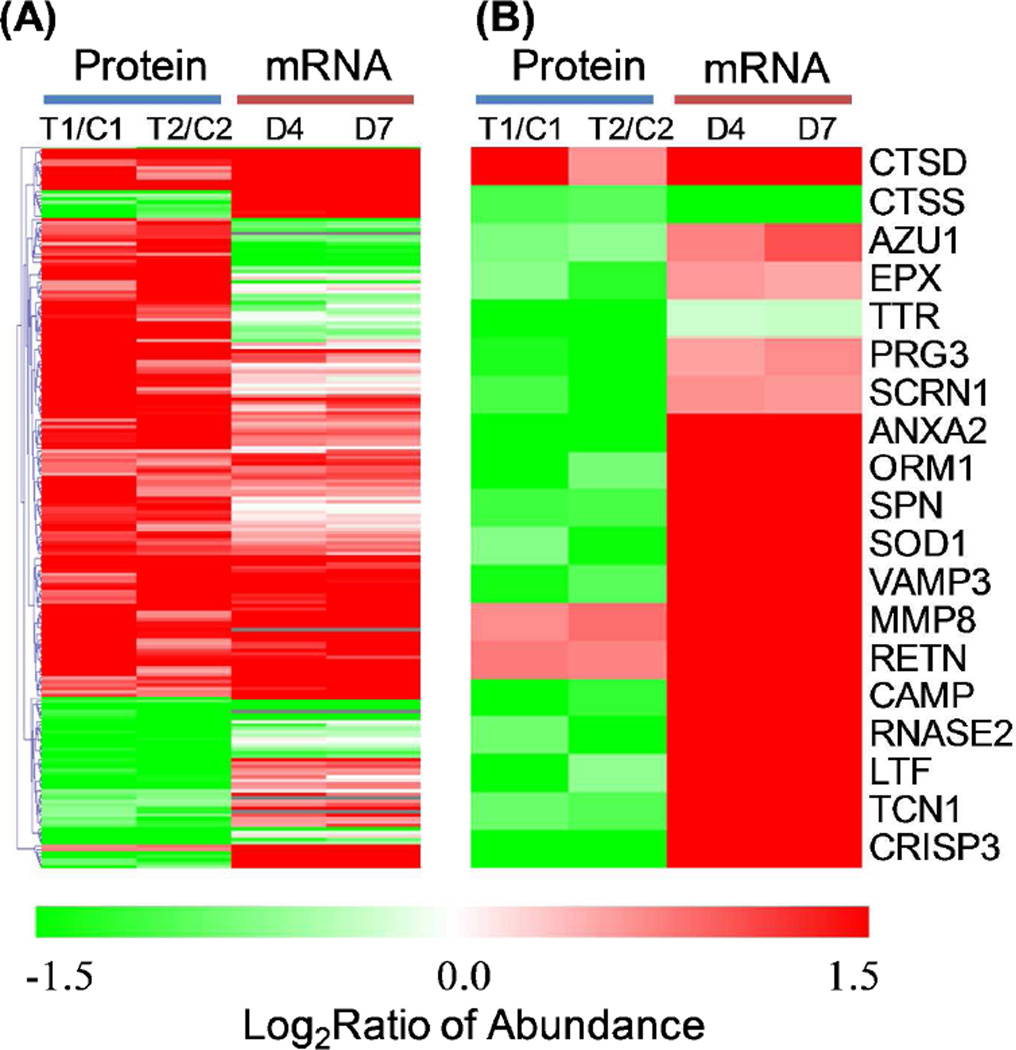
The observed protein abundance changes were further compared to gene expression data from an independent cohort of patients (101 trauma patients and 10 controls) where genome wide expression analyses were performed on enriched PMNs from controls and trauma patients at time points ranging from half day to 28 days post injury. A subset of 174 out of 197 protein candidates from proteomics study were observed with significant changes in their gene expression levels following trauma (FDR<0.01). We selected the gene expression changes observed for the 4- and 7-day time points post-injury for comparing to protein abundance changes for the set of 197 proteins since the samples for proteomics were collected at similar time periods following injury. As shown in Figure 4A, the directions of abundance changes (up or down) for ~67% proteins are consistent with those observed changes in mRNA abundance levels.
Figure 4. Comparison between protein abundance and gene expression changes.
(A) Heatmap displaying the protein level and mRNA level changes of 197 proteins. (B) Heatmap displaying selected secretory proteins. T1 and T2 represent trauma pool 1 and pool 2, respectively. C1 and C2 represent control pool 1 and pool 2, respectively. Color scale is based on the log2 (abundance ratio) for trauma condition dividing by the control. Protein abundance data are displayed in two biological replicates and mRNA abundance data are displayed with 4-day and 7-day time point. Note that the ± 1.5 log2ratio color scale is chosen to highlight all changes. Changes for many proteins are greater than shown on the color scale and their exact changes are listed in supplemental tables.
Considering that PMNs are known to secrete multiple anti-microbial products[10], we next examined the concordance between protein abundance and gene expression changes for genes that were known to produce secretory proteins. Figure 4B shows the patterns of protein abundance and gene expression changes for 19 proteins that were reported as potential secretory proteins [19, 22, 44]. A number of microbicidal products are known to secrete from PMN granules through a process called degranulation during inflammation [45], which includes leukosialin (gene symbol:SPN), cathepsins (CTSD, CTSS), antibacterial protein Fall-39 (CAMP), growth inhibiting protein 12 (LTF), and azurocidin (AZU1), all of which play critical roles in bacterial clearance [46, 47]. As shown, most of these genes were observed to have a significant increase in their mRNA levels following injury; however, the abundances for most proteins were decreased significantly in the trauma conditions. The observation with significant decreased intracellular protein abundances supports that neutrophils have increased secretion to the extracellular milieu following degranulation[45] in response to the inflammatory conditions. Therefore, these data illustrate the value of an integrated genomics and proteomics data to gain insights into the dynamics of individual gene products by revealing increased protein secretion under trauma conditions.
Functional Analyses of trauma-responsive proteins
The functions and associated pathways of trauma-associated proteins were analyzed based on the Ingenuity Pathway Analysis (IPA) knowledge base, gene ontology information, and relevant literature. A number of functional categories were revealed to be associated with this set of trauma-responsive proteins, including several known immune response associated pathways, protein biosynthesis, protein transport, NRF2-mediated oxidative stress response, the ubiquitin-proteasome pathway, and apoptosis (see Supplemental Table 5 for full listed functional annotations).
Known immune response associated pathways
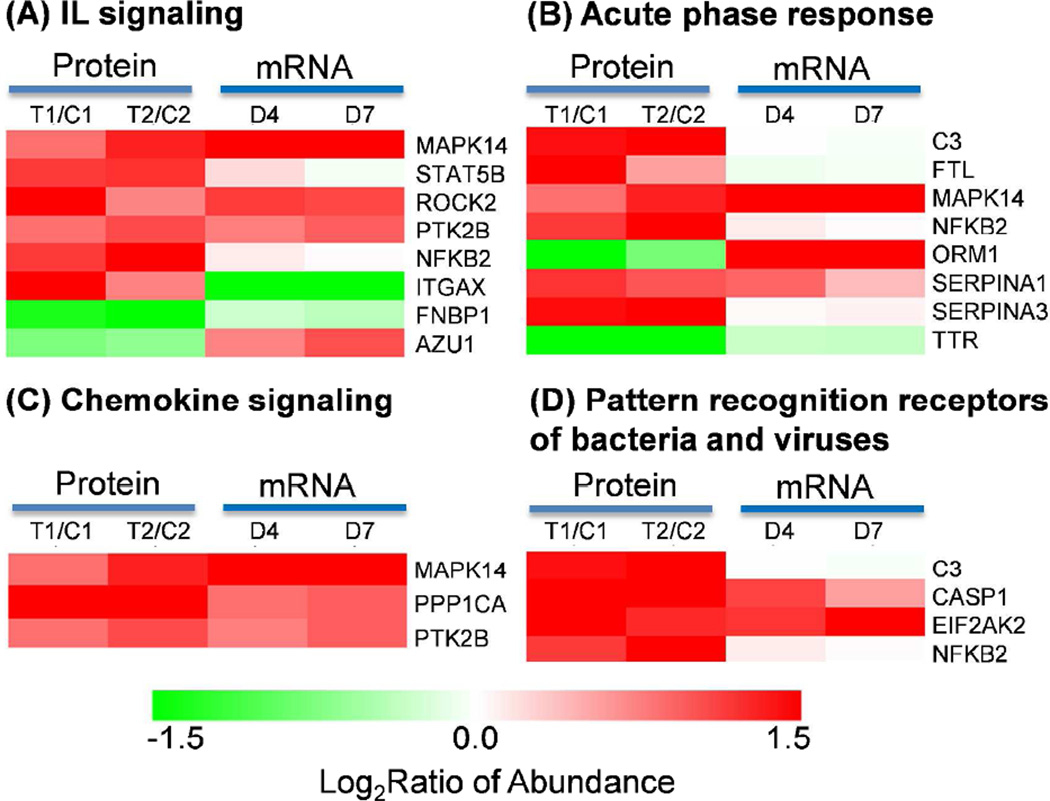
Significant up-regulations were observed for proteins involved in several known immune response associated pathways, as shown in Figure 5. Pro-inflammatory cytokines such as interleukins (IL), pattern recognition receptors, and acute phase response proteins were previously reported to be associated with early activation of PMNs [48] and trauma injury [11–13, 49, 50]. Chemokine signaling pathway is associated with the migration of PMNs to target tissues. All these pathways appear to be significantly up-regulated in response to trauma (Figure 5). The discrepancy of protein abundance data and mRNA data for azurocidin 1 (AZU1) and orosomucoid 1(ORM1) is potentially related to the increased protein secretion as described previously. The up-regulation of complement component 3 (C3) and integrin alpha X (ITGAX) suggest an increased potential for the adhesion of PMNs to endothelial cells and accumulation of immune cells at the injured tissue [51–53].
Figure 5. Known pathways involved in activation of neutrophils.
(A) IL signaling. (B) Acute phase response. (C) Chemokine signaling. (D) Pattern recognition receptors of bacteria and viruses.
Protein biosynthesis
Figure. 6A shows significant up-regulation in both protein abundances and gene expression for a set of proteins that are known to be involved in pathways associated with protein biosynthesis. Most of these proteins are associated with eukaryotic translation initiation factor (EIF) 2 signaling and aminoacyl-tRNA biosynthesis pathways. EIF2 signaling and aminoacyl-tRNA biosynthesis pathways play essential roles in the initiation and translational phases of protein biosynthesis, respectively. The concordant up-regulation in both mRNA and protein abundance levels for multiple EIFs, tRNA synthases, as well as 40S and 60S ribosomal proteins provides solid evidence that protein biosynthesis is activated in PMNs under trauma conditions. These genomics and proteomics observations of increased protein synthesis are supportive for the observed increased protein secretion (Figure 4B) and it is also in good agreement with previous reports on activated PMNs [54, 55].
Figure 6. Specific protein functional categories.
(A) Protein biosynthesis. (B) Protein transport. (C) Nrf2-mediated oxidative stress response. (D) Ubiquitin-proteasome system. (E) Apoptosis.
Protein transport
Increased protein abundances and mRNA abundances were observed for the majority proteins and genes that were known to be involved in the regulation of protein transport (Figure 6B). These proteins include several coatomer subunits, exocyst complex component, nuclear pore complex proteins, small GTPases, and several other proteins. Coatomer is known as a large protein complex that coats membrane-bound transport vesicles and potentially plays a role in forward transport from the endoplasmic reticulum to the Golgi apparatus and through the Golgi apparatus [56, 57]. The observed up-regulation of three coatomer subunits (COPA, COPB, COPG), and exocyst complex component 4 suggests enhanced intracellular vesicle trafficking under trauma conditions.
NRF2-mediated oxidative stress response
Besides the activation of protein synthesis and transport, we also observed trauma-induced up-regulation in protein and mRNA abundances for most of the proteins associated with the NRF2-mediated oxidative stress response pathway (Figure 6C), suggesting activation in the oxidative stress response. NRF2-mediated oxidative stress response pathway is essential for cellular defense response to oxidative stress by regulating a battery of detoxifying enzymes and antioxidant enzymes. NRF2 was previously reported to be associated with oxidative regulation of lipopolysaccharide (LPS) induced innate immune response in PMNs and the activation of NRF2-mediated oxidative response pathway led to a protective role from the LPS induced inflammatory response and mortality [58]. Moreover, it has been recently demonstrated that Cullin-3 directly interacts with oxidative stress sensor keap1 to regulate NRF2 turnover [59]. A number of chaperone and stress response proteins known to be activated by NRF2 were also observed with increased protein abundances in response to injury. These chaperone and stress response proteins include heat shock protein group (DNAJB11, DNAJA1, DNAJC1), stress-induced phosphoprotein 1 (STIP1), and Fk506-binding protein 4 and 5 (FKBP4 and FKBP5). Other proteins observed in the pathway include antioxidant protein Ferritin (FTL and FTH1) and signaling protein MAPK14.
Ubiquitin-proteasome pathway
The up-regulation in both protein abundance and gene expression for several 26S proteasome regulatory subunits and several members of ubiquitin-ligase complex was observed (Figure 6D), suggesting an increase in the activity of the ubiquitin-proteasome system (UPS). Such an increase in the UPS is common in response to cellular stresses such as trauma injury and infection. Our observation of the up-regulation of several heat shock proteins such as DNAJB11, DNAJA1, and DNAJC1 (Figure 6C) also supports the activation of the UPS since heat shock proteins were also implicated in the increase of UPS activities [60]. The importance of the proteasome activity in the nuclear factor kappa beta (NFκβ) activation, which inhibits neutrophil apoptosis in severe trauma, has also been reported [61, 62].
Apoptosis
Neutrophil numbers are known to be significantly increased in trauma patients with MODS compared to healthy controls and an inhibition of apoptosis or delayed apoptosis in severe trauma is anticipated [17, 62]. A number of proteins in our dataset have been previously reported as potentially involved in apoptosis and their protein abundance and gene expression patterns are shown in Figure 6E. The observed protein abundance increase of NFκβ suggests an increase in its activity, which is consistent with previously reported NFκβ dependent inhibition of neutrophil apoptosis [62]. Increased protein abundances in response to trauma were observed for most of these apoptosis associated proteins and many of these proteins have been previously reported as anti-apoptotic. These potential anti-apoptotic proteins include caspase-1 (CASP1) [63], cyclin-dependent kinase (CDK2) [64], heat shock proteins (HSPD1, HYOU1) [65], protein kinase B (PTK2B) [66], fatty acid synthase (FASN) [67], peptidylprolyl isomerase D (PPID) [68], HECT, UBA and WWE domain-containing protein 1 (HUWE1) [69], nucleoside diphosphate kinase A (NME1) [70], and serine/threonine-protein kinases SRPK1 [71] and TAOK3 [72]. The anti-apoptotic implications of most of these proteins have not been reported in PMNs, thus representing potential novel regulators for PMN apoptosis. Only a few proteins were explicitly reported for their anti-apoptotic roles in neutrophils. For example, caspase-1, one of the most up-regulated proteins in our data, was also reported as up-regulated for providing an inhibition of apoptosis of inflammatory neutrophils through activation of IL-1β [63]. The inhibition of cyclin-dependent kinase (CDK) was shown to induce neutrophil apoptosis [64]. It was also reported that protein kinase B (PTK2B) was involved in the inhibition of neutrophil apoptosis via the signaling through PI-3-kinase and downstream pathways [66].
In addition to the up-regulation of potential anti-apoptotic proteins, we have also observed a significant down-regulation of several potential pro-apoptotic proteins, including arachidonate 15-lipoxygenase (ALOX15) [73], thymosin beta-10 (TMSB10) [74], and 40S ribosomal protein s3a (RPS3A) [75]. Arachidonate lipoxygenases have been reported as essential regulators of cell survival and apoptosis [76] and inhibition of ALOX15 expression has been shown to prevent cancer cell apoptosis [73]. A dramatic down-regulation (~10-fold) in both protein abundance and gene expression for ALOX15 was observed in PMNs in response to trauma, implying that ALOX15 may be a novel important regulator for neutrophil apoptosis. Taken together, the observation of upregulation of many anti-apoptotic proteins and down-regulation of pro-apoptotic proteins provides solid evidence of inhibited or delayed apoptosis in PMNs under trauma conditions.
Other novel proteins
Besides the above described functional categories, many proteins were also reported with their implications in other functional categories such as inflammatory response, infectious disorder, anti-viral response, cell adhesion, and lipid metabolism. However, many proteins still lack prior knowledge of their functions despite their observed significant abundance changes induced by trauma. Many of these proteins may represent novel candidates worthy of further functional studies. For instance, protein WDR40A, a WD repeat-containing protein that interacts with the COP9 signalosome, was observed with significant down-regulation in mRNA and protein expression, suggesting functional relevance of this protein in trauma-induced response; however, few studies have been undertaken on this protein to date[77]. SAM-domain protein (SAMSN1) is another novel protein with unknown functions while its protein abundance and gene expression levels are observed with significant up-regulation in response to trauma.
Discussion
The complexity of human diseases presents a significant opportunity for applying high throughput technologies such as genome-wide gene expression profiling and proteomics to investigate the underlying mechanisms of human diseases. In this work we demonstrate a comparative proteomics profiling of PMNs isolated from human trauma patients for revealing functional changes of immune cells induced by traumatic injury. In response to injury, we expect that there is somewhat of an overlap of changes induced by tissue injury and infection, which may be difficult to distinguish. To minimize the potential impact of infection, we have selected all the patients for this proteomics study without clinical evidence of infection. Although an occult infection could not be ruled out, none of these patients developed any evidence of infection during their entire clinical course.
Despite being the most abundant leukocyte and the first line of defense against intruding microorganisms, the importance of PMNs in the innate immune response has not been fully understood until recently [1, 10]. Our work represents the first global comparative proteomics study of human patient PMNs with the aim to identify functional changes of PMNs in response to trauma. The study revealed significant neutrophil proteome response to traumatic injury with the abundances of 197 proteins significantly changed.
The overall good agreement between an independently acquired gene expression dataset and proteomics data provides a degree of support on the quality of this comparative proteomics data. Overall, the directions of abundance changes (up or down) for ~67% proteins are consistent with those observed changes in mRNA abundance levels. More interestingly, the mRNA and protein abundance changes are nearly in perfect agreement for some specific functional categories such as protein biosynthesis and oxidative stress response as shown in Figure 5 and 6, supporting the high confidence of the observed proteomic changes. Validation of selected proteins using targeted quantification in individual samples (Figure 3) further supports the data quality.
This study also provides clear evidence that increased protein secretion to extracellular milieu is one of the major mechanisms for discrepancy observed between mRNA and protein abundance changes. Given the fact that increased protein secretion cannot be measured by whole cell proteomics alone without the measurement of blood protein concentrations, the concurrent observation of significantly increased gene expression and decreased intracellular protein abundances for secreted proteins represents an interesting and important example of the value for integrating proteomics and transcriptomics measurements.
The observation of imperfect correlation between protein and mRNA results is also in good agreement with previous investigations that also reported a rather poor correlation between mRNA and protein abundance changes [78, 79]. These results suggest that the control mechanisms for regulating the mRNA and protein abundances are different. Protein abundances are known to be controlled by many post-transcriptional mechanisms. Therefore, it is not surprising that we did not observe significant protein abundance changes for many genes with significant changes in mRNA and vice versa.
Based on the functional implications of the observed significant proteins, the neutrophil response to trauma can be well consolidated into two main aspects: (1) increased neutrophil activation and (2) improved neutrophil survival. The neutrophil activation is represented by the observed increase in known immune response associated pathways (Figure 5), protein synthesis pathways including the EIF2 signaling and aminoacyl-tRNA biosynthesis pathways (Figure 6A), and an increase in vesicular protein transport (Figure 6B). Moreover, there are several well-known neutrophil activation markers such as high affinity immunoglobulin gamma Fc receptor I (CD64) and complement receptor type 1 (CD35) [80, 81]. While CD64 and CD35 were not detected in our proteomics data presumably due to their low abundances, the up-regulation in gene expression [26] was observed for both CD64 and CD35 with 3.4 and 2.0-fold increases, respectively, further supporting neutrophil activation post traumatic injury.
The increase in protein synthesis and vesicular protein transport supports the notion that secretory granule proteins are produced and secreted in a faster rate following neutrophil activation in response to trauma. The increased protein secretion through degranulation in response to inflammation is well known[45], and this is further supported by the observation of a decrease in intracellular protein abundances for these proteins (Figure 4B). The observed significant increase in gene expression for known secretory proteins along with general activation of protein synthesis represents the cells’ compensatory mechanism to keep up the need for protein secretion. These observations fall well in line with the prior knowledge about the central role of granules and their associated proteins in antimicrobial functions of PMNs in providing a first line of defense against microorganisms [10].
The improved neutrophil survival is supported by the observation of the activation of NRF2-mediated oxidative stress response (Figure 6C), and the observation of upregulation in many anti-apoptotic proteins and down-regulation of pro-apoptotic proteins (Figure 6E). The integration of this pathway information strongly suggest that neutrophil apoptosis is inhibited or delayed under the trauma conditions being investigated, which is also in good agreement with recent studies on neutrophil apoptosis [61, 62].
While the functional implications of many of these proteins were reported in other cell types and disease conditions, the functions of most of proteins have not yet been reported in human PMNs. We anticipate that many of these proteins represent novel regulatory factors for neutrophil activation and survival. For example, as described in the Results section, the observation of significant down-regulation of ALOX15 suggests that it can be a novel regulator for neutrophil apoptosis [76]. The significant up-regulation of fatty-acid synthase (FASN) suggests a role of fatty acids and lipid metabolism in neutrophil activation.
In summary, this comparative proteomics profiling of human PMNs revealed the overall functional changes of PMNs in response to trauma injury. While the observed proteome alterations generally correlate with the increased activation and survival of neutrophils, the functions of many of the altered proteins have not yet been reported for PMNs or inflammatory diseases. This dataset of trauma-associated neutrophil protein alterations provides a rich resource for further studies of the functions of individual proteins in PMNs and some of these proteins may even be potential candidate markers for predicting disease outcomes.
Supplementary Material
Statement of Clinical Relevance.
Polymorphonuclear neutrophils (PMNs) play an important role in mediating the innate immune response after severe traumatic injury. In order to gain understanding of the underlying molecular mechanisms of PMNs in response to trauma, it is necessary to study the cellular proteome response to the traumatic condition. Our study performed the first comparative proteome profiling of human PMNs from severe trauma patients and healthy controls by applying 2D-LC-MS/MS-based shotgun proteomics. Our observed proteome changes revealed increased neutrophil activation and inhibited neutrophil apoptosis in response to trauma. The study not only reveals an overall picture of functional neutrophil response to trauma at the proteome level, but also provides a rich proteomics data resource of trauma-associated changes in the neutrophils that may be valuable for further studies of the functions of individual proteins in PMNs and some of these proteins may even be potential candidate markers for predicting disease outcomes.
ACKNOWLEDGEMENTS
Portions of this research were supported by NIH grants U54 GM-62119-02 (to R.G.T) and T32 GM-008256 (to R.G.T), RR18522 (To R.D.S), DP2OD006668 (to. W.J.Q.) and EMSL (Environmental Molecular Science Laboratory). EMSL is a national scientific user facility sponsored by the U.S. Department of Energy (DOE) Office of Biological and Environmental Research on the Pacific Northwest National Laboratory (PNNL) campus in Richland, Washington. PNNL is operated by Battelle for the DOE under contract DE-AC05-76RLO-1830.
ABBREVIATIONS
- PMN
Polymorphonuclear neutrophils
- ROS
Reactive oxygen species
- MODS
Multiple organ dysfunction syndrome (MODS)
- SIRS
Systemic inflammatory response syndrome (SIRS)
- LC-MS
Liquid chromatography-mass spectrometry (LC-MS)
- SCX
Strong Cation Exchange
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest
References
- 1.Borregaard N. Neutrophils, from marrow to microbes. Immunity. 2010;33:657–670. doi: 10.1016/j.immuni.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 2.Muller I, Munder M, Kropf P, Hansch GM. Polymorphonuclear neutrophils and T lymphocytes: strange bedfellows or brothers in arms? Trends Immunol. 2009;30:522–530. doi: 10.1016/j.it.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi ME. DAMPs, PAMPs and alarmins: all we need to know about danger. J Leukoc Biol. 2007;81:1–5. doi: 10.1189/jlb.0306164. [DOI] [PubMed] [Google Scholar]
- 5.Stahel PF, Smith WR, Moore EE. Role of biological modifiers regulating the immune response after trauma. Injury. 2007;38:1409–1422. doi: 10.1016/j.injury.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 6.Hietbrink F, Koenderman L, Rijkers G, Leenen L. Trauma: the role of the innate immune system. World J Emerg Surg. 2006;1:15. doi: 10.1186/1749-7922-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy O. Antibiotic proteins of polymorphonuclear leukocytes. Eur J Haematol. 1996;56:263–277. doi: 10.1111/j.1600-0609.1996.tb00714.x. [DOI] [PubMed] [Google Scholar]
- 8.Nauseef WM. Assembly of the phagocyte NADPH oxidase. Histochem Cell Biol. 2004;122:277–291. doi: 10.1007/s00418-004-0679-8. [DOI] [PubMed] [Google Scholar]
- 9.Matzinger P. The danger model: a renewed sense of self. Science. 2002;296:301–305. doi: 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
- 10.Nathan C. Neutrophils and immunity: challenges and opportunities. Nat Rev Immunol. 2006;6:173–182. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- 11.Matsuda N, Hattori Y. Systemic inflammatory response syndrome (SIRS): molecular pathophysiology and gene therapy. J Pharmacol Sci. 2006;101:189–198. doi: 10.1254/jphs.crj06010x. [DOI] [PubMed] [Google Scholar]
- 12.Calfee CS, Matthay MA. Clinical immunology: Culprits with evolutionary ties. Nature. 2010;464:41–42. doi: 10.1038/464041a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Q, Raoof M, Chen Y, Sumi Y, et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calvano SE, Xiao W, Richards DR, Felciano RM, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 15.Cobb JP, Mindrinos MN, Miller-Graziano C, Calvano SE, et al. Application of genome-wide expression analysis to human health and disease. Proc Natl Acad Sci U S A. 2005;102:4801–4806. doi: 10.1073/pnas.0409768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cobb JP, O’Keefe GE. Injury research in the genomic era. Lancet. 2004;363:2076–2083. doi: 10.1016/S0140-6736(04)16460-X. [DOI] [PubMed] [Google Scholar]
- 17.Laudanski K, Miller-Graziano C, Xiao W, Mindrinos MN, et al. Cell-specific expression and pathway analyses reveal alterations in trauma-related human T cell and monocyte pathways. Proc Natl Acad Sci U S A. 2006;103:15564–15569. doi: 10.1073/pnas.0607028103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu P, Crawford M, Way M, Godovac-Zimmermann J, et al. Subproteome analysis of the neutrophil cytoskeleton. Proteomics. 2009;9:2037–2049. doi: 10.1002/pmic.200800674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomazella GG, da Silva I, Laure HJ, Rosa JC, et al. Proteomic analysis of total cellular proteins of human neutrophils. Proteome Sci. 2009;7:32. doi: 10.1186/1477-5956-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasper B, Thole HH, Patterson SD, Welte K. Cytosolic proteins from neutrophilic granulocytes: a comparison between patients with severe chronic neutropenia and healthy donors. Electrophoresis. 1997;18:142–149. doi: 10.1002/elps.1150180126. [DOI] [PubMed] [Google Scholar]
- 21.Lominadze G, Ward RA, Klein JB, McLeish KR. Proteomic analysis of human neutrophils. Methods Mol Biol. 2006;332:343–356. doi: 10.1385/1-59745-048-0:343. [DOI] [PubMed] [Google Scholar]
- 22.Lominadze G, Powell DW, Luerman GC, Link AJ, et al. Proteomic analysis of human neutrophil granules. Mol Cell Proteomics. 2005;4:1503–1521. doi: 10.1074/mcp.M500143-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Marshall JC, Cook DJ, Christou NV, Bernard GR, et al. Multiple organ dysfunction score: a reliable descriptor of a complex clinical outcome. Crit Care Med. 1995;23:1638–1652. doi: 10.1097/00003246-199510000-00007. [DOI] [PubMed] [Google Scholar]
- 24.Sheth K, Friel J, Nolan B, Bankey P. Inhibition of p38 mitogen activated protein kinase increases lipopolysaccharide induced inhibition of apoptosis in neutrophils by activating extracellular signal-regulated kinase. Surgery. 2001;130:242–248. doi: 10.1067/msy.2001.115902. [DOI] [PubMed] [Google Scholar]
- 25.Bankey PE, Banerjee S, Zucchiatti A, De M, et al. Cytokine induced expression of programmed death ligands in human neutrophils. Immunol Lett. 2010;129:100–107. doi: 10.1016/j.imlet.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kotz KT, Xiao W, Miller-Graziano C, Qian WJ, et al. Clinical microfluidics for neutrophil genomics and proteomics. Nat Med. 2010;16:1042–1047. doi: 10.1038/nm.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Qian WJ, Mottaz HM, Clauss TR, et al. Development and evaluation of a micro- and nanoscale proteomic sample preparation method. J Proteome Res. 2005;4:2397–2403. doi: 10.1021/pr050160f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Zhou JY, Chin MH, Schepmoes AA, et al. Region-specific protein abundance changes in the brain of MPTP-induced Parkinson's disease mouse model. J Proteome Res. 2010;9:1496–1509. doi: 10.1021/pr901024z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qian WJ, Liu T, Monroe ME, Strittmatter EF, et al. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. Journal of proteome research. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 30.Zhou JY, Schepmoes AA, Zhang X, Moore RJ, et al. Improved LC-MS/MS spectral counting statistics by recovering low-scoring spectra matched to confidently identified peptide sequences. Journal of proteome research. 2010;9:5698–5704. doi: 10.1021/pr100508p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 32.Zhou JY, Schepmoes AA, Zhang X, Moore RJ, et al. Improved LC-MS/MS spectral counting statistics by recovering low-scoring spectra matched to confidently identified peptide sequences. J Proteome Res. 2010;9:5698–5704. doi: 10.1021/pr100508p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sokal RR, Rohlf FJ. Biometry: the principles and practice of statistics in biological research. 3rd edition. New York: W. H. Freeman and Co.; 1995. p. 887. [Google Scholar]
- 34.Zhou JY, Afjehi-Sadat L, Asress S, Duong DM, et al. Galectin-3 is a candidate biomarker for amyotrophic lateral sclerosis: discovery by a proteomics approach. J Proteome Res. 2010;9:5133–5141. doi: 10.1021/pr100409r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang B, VerBerkmoes NC, Langston MA, Uberbacher E, et al. Detecting differential and correlated protein expression in label-free shotgun proteomics. J Proteome Res. 2006;5:2909–2918. doi: 10.1021/pr0600273. [DOI] [PubMed] [Google Scholar]
- 36.Xu W, Seok J, Mindrinos MN, Schweitzer AC, et al. Human transcriptome array for high-throughput clinical studies. Proc Natl Acad Sci U S A. 2011;108:3707–3712. doi: 10.1073/pnas.1019753108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irizarry RA, Bolstad BM, Collin F, Cope LM, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic acids research. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leek JT, Monsen E, Dabney AR, Storey JD. EDGE: extraction and analysis of differential gene expression. Bioinformatics. 2006;22:507–508. doi: 10.1093/bioinformatics/btk005. [DOI] [PubMed] [Google Scholar]
- 39.Kim JS, Fillmore TL, Liu T, Robinson E, et al. 18O–labeled proteome reference as global internal standards for targeted quantification by selected reaction monitoring-mass spectrometry. Molecular & cellular proteomics : MCP. 2011;10:M110 007302. doi: 10.1074/mcp.M110.007302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacLean B, Tomazela DM, Shulman N, Chambers M, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polpitiya AD, Qian WJ, Jaitly N, Petyuk VA, et al. DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics. 2008;24:1556–1558. doi: 10.1093/bioinformatics/btn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 43.Qian WJ, Jacobs JM, Camp DG, 2nd, Monroe ME, et al. Comparative proteome analyses of human plasma following in vivo lipopolysaccharide administration using multidimensional separations coupled with tandem mass spectrometry. Proteomics. 2005;5:572–584. doi: 10.1002/pmic.200400942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Souza Castro M, de Sa NM, Gadelha RP, de Sousa MV, et al. Proteome analysis of resting human neutrophils. Protein Pept Lett. 2006;13:481–487. doi: 10.2174/092986606776819529. [DOI] [PubMed] [Google Scholar]
- 45.Faurschou M, Borregaard N. Neutrophil granules and secretory vesicles in inflammation. Microbes and infection / Institut Pasteur. 2003;5:1317–1327. doi: 10.1016/j.micinf.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 46.Zanetti M. Cathelicidins, multifunctional peptides of the innate immunity. J Leukoc Biol. 2004;75:39–48. doi: 10.1189/jlb.0403147. [DOI] [PubMed] [Google Scholar]
- 47.Sorensen O, Arnljots K, Cowland JB, Bainton DF, Borregaard N. The human antibacterial cathelicidin, hCAP-18, is synthesized in myelocytes and metamyelocytes and localized to specific granules in neutrophils. Blood. 1997;90:2796–2803. [PubMed] [Google Scholar]
- 48.Leung BP, Culshaw S, Gracie JA, Hunter D, et al. A role for IL-18 in neutrophil activation. J Immunol. 2001;167:2879–2886. doi: 10.4049/jimmunol.167.5.2879. [DOI] [PubMed] [Google Scholar]
- 49.Felderhoff-Mueser U, Schmidt OI, Oberholzer A, Buhrer C, Stahel PF. IL-18: a key player in neuroinflammation and neurodegeneration? Trends Neurosci. 2005;28:487–493. doi: 10.1016/j.tins.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 50.Pancewicz SA, Kondrusik M, Zajkowska J, Grygorczuk S. [Concentrations of pro-inflammatory cytokines IFN-gamma, IL-6, IL-12 and IL-15 in serum and cerebrospinal fluid in patients with neuroborreliosis undergoing antibiotic treatment] Pol Merkur Lekarski. 2007;22:275–279. [PubMed] [Google Scholar]
- 51.Li Z. The alphaMbeta2 integrin and its role in neutrophil function. Cell Res. 1999;9:171–178. doi: 10.1038/sj.cr.7290015. [DOI] [PubMed] [Google Scholar]
- 52.Gower RM, Wu H, Foster GA, Devaraj S, et al. CD11c/CD18 expression is upregulated on blood monocytes during hypertriglyceridemia and enhances adhesion to vascular cell adhesion molecule-1. Arterioscler Thromb Vasc Biol. 2011;31:160–166. doi: 10.1161/ATVBAHA.110.215434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Languino LR, Plescia J, Duperray A, Brian AA, et al. Fibrinogen mediates leukocyte adhesion to vascular endothelium through an ICAM-1-dependent pathway. Cell. 1993;73:1423–1434. doi: 10.1016/0092-8674(93)90367-y. [DOI] [PubMed] [Google Scholar]
- 54.Humphreys JM, Hughes V, Edwards SW. Stimulation of protein synthesis in human neutrophils by gamma-interferon. Biochem Pharmacol. 1989;38:1241–1246. doi: 10.1016/0006-2952(89)90329-8. [DOI] [PubMed] [Google Scholar]
- 55.Hughes V, Humphreys JM, Edwards SW. Protein synthesis is activated in primed neutrophils: a possible role in inflammation. Biosci Rep. 1987;7:881–890. doi: 10.1007/BF01119479. [DOI] [PubMed] [Google Scholar]
- 56.Waters MG, Serafini T, Rothman JE. 'Coatomer': a cytosolic protein complex containing subunits of non-clathrin-coated Golgi transport vesicles. Nature. 1991;349:248–251. doi: 10.1038/349248a0. [DOI] [PubMed] [Google Scholar]
- 57.Cosson P, Letourneur F. Coatomer (COPI)-coated vesicles: role in intracellular transport and protein sorting. Curr Opin Cell Biol. 1997;9:484–487. doi: 10.1016/s0955-0674(97)80023-3. [DOI] [PubMed] [Google Scholar]
- 58.Thimmulappa RK, Scollick C, Traore K, Yates M, et al. Nrf2-dependent protection from LPS induced inflammatory response and mortality by CDDO-Imidazolide. Biochem Biophys Res Commun. 2006;351:883–889. doi: 10.1016/j.bbrc.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park SH, Bolender N, Eisele F, Kostova Z, et al. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol Biol Cell. 2007;18:153–165. doi: 10.1091/mbc.E06-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nolan B, Kim R, Duffy A, Sheth K, et al. Inhibited neutrophil apoptosis: proteasome dependent NF-kappaB translocation is required for TRAF-1 synthesis. Shock. 2000;14:290–294. doi: 10.1097/00024382-200014030-00008. [DOI] [PubMed] [Google Scholar]
- 62.Nolan B, Collette H, Baker S, Duffy A, et al. Inhibition of neutrophil apoptosis after severe trauma is NFkappabeta dependent. J Trauma. 2000;48:599–604. doi: 10.1097/00005373-200004000-00004. discussion 604–595. [DOI] [PubMed] [Google Scholar]
- 63.Watson RW, Rotstein OD, Parodo J, Bitar R, Marshall JC. The IL-1 beta-converting enzyme (caspase-1) inhibits apoptosis of inflammatory neutrophils through activation of IL-1 beta. Journal of immunology. 1998;161:957–962. [PubMed] [Google Scholar]
- 64.Rossi AG, Sawatzky DA, Walker A, Ward C, et al. Cyclin-dependent kinase inhibitors enhance the resolution of inflammation by promoting inflammatory cell apoptosis. Nat Med. 2006;12:1056–1064. doi: 10.1038/nm1468. [DOI] [PubMed] [Google Scholar]
- 65.Sanson M, Auge N, Vindis C, Muller C, et al. Oxidized low-density lipoproteins trigger endoplasmic reticulum stress in vascular cells: prevention by oxygen-regulated protein 150 expression. Circ Res. 2009;104:328–336. doi: 10.1161/CIRCRESAHA.108.183749. [DOI] [PubMed] [Google Scholar]
- 66.Webb PR, Wang KQ, Scheel-Toellner D, Pongracz J, et al. Regulation of neutrophil apoptosis: a role for protein kinase C and phosphatidylinositol-3-kinase. Apoptosis. 2000;5:451–458. doi: 10.1023/a:1009601220552. [DOI] [PubMed] [Google Scholar]
- 67.Menendez JA, Mehmi I, Atlas E, Colomer R, Lupu R. Novel signaling molecules implicated in tumor-associated fatty acid synthase-dependent breast cancer cell proliferation and survival: Role of exogenous dietary fatty acids, p53-p21WAF1/CIP1, ERK1/2 MAPK, p27KIP1, BRCA1, and NF-kappaB. Int J Oncol. 2004;24:591–608. [PubMed] [Google Scholar]
- 68.Schubert A, Grimm S. Cyclophilin D, a component of the permeability transitionpore, is an apoptosis repressor. Cancer Res. 2004;64:85–93. doi: 10.1158/0008-5472.can-03-0476. [DOI] [PubMed] [Google Scholar]
- 69.Adhikary S, Marinoni F, Hock A, Hulleman E, et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 70.Seong HA, Jung H, Ha H. NM23-H1 tumor suppressor physically interacts with serine-threonine kinase receptor-associated protein, a transforming growth factor-beta (TGF-beta) receptor-interacting protein, and negatively regulates TGF-beta signaling. J Biol Chem. 2007;282:12075–12096. doi: 10.1074/jbc.M609832200. [DOI] [PubMed] [Google Scholar]
- 71.Hayes GM, Carrigan PE, Miller LJ. Serine-arginine protein kinase 1 overexpression is associated with tumorigenic imbalance in mitogen-activated protein kinase pathways in breast, colonic, and pancreatic carcinomas. Cancer Res. 2007;67:2072–2080. doi: 10.1158/0008-5472.CAN-06-2969. [DOI] [PubMed] [Google Scholar]
- 72.MacKeigan JP, Murphy LO, Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat Cell Biol. 2005;7:591–600. doi: 10.1038/ncb1258. [DOI] [PubMed] [Google Scholar]
- 73.Shureiqi I, Xu X, Chen D, Lotan R, et al. Nonsteroidal anti-inflammatory drugs induce apoptosis in esophageal cancer cells by restoring 15-lipoxygenase-1 expression. Cancer Res. 2001;61:4879–4884. [PubMed] [Google Scholar]
- 74.Lee SH, Zhang W, Choi JJ, Cho YS, et al. Overexpression of the thymosin beta-10 gene in human ovarian cancer cells disrupts F-actin stress fiber and leads to apoptosis. Oncogene. 2001;20:6700–6706. doi: 10.1038/sj.onc.1204683. [DOI] [PubMed] [Google Scholar]
- 75.Hu ZB, Minden MD, McCulloch EA, Stahl J. Regulation of drug sensitivity by ribosomal protein S3a. Blood. 2000;95:1047–1055. [PubMed] [Google Scholar]
- 76.Tang DG, Chen YQ, Honn KV. Arachidonate lipoxygenases as essential regulators of cell survival and apoptosis. Proc Natl Acad Sci U S A. 1996;93:5241–5246. doi: 10.1073/pnas.93.11.5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 78.Griffin TJ, Gygi SP, Ideker T, Rist B, et al. Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol Cell Proteomics. 2002;1:323–333. doi: 10.1074/mcp.m200001-mcp200. [DOI] [PubMed] [Google Scholar]
- 79.de Godoy LM, Olsen JV, Cox J, Nielsen ML, et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 80.Mann BS, Chung KF. Blood neutrophil activation markers in severe asthma: lack of inhibition by prednisolone therapy. Respir Res. 2006;7:59. doi: 10.1186/1465-9921-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Costantini C, Micheletti A, Calzetti F, Perbellini O, et al. Neutrophil activation and survival are modulated by interaction with NK cells. International immunology. 2010;22:827–838. doi: 10.1093/intimm/dxq434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.