Abstract
The nuclear receptor Rev-erb-α modulates hepatic lipid and glucose metabolism, adipogenesis and the inflammatory response in macrophages. We show here that Rev-erb-α is highly expressed in oxidative skeletal muscle and plays a role in mitochondrial biogenesis and oxidative function, in gain- and loss-of function studies. Rev-erb-α-deficiency in skeletal muscle leads to reduced mitochondrial content and oxidative function, resulting in compromised exercise capacity. This phenotype was recapitulated in isolated fibers and in muscle cells upon Rev-erbα knock-down, while Rev-erb-α over-expression increased the number of mitochondria with improved respiratory capacity. Rev-erb-α-deficiency resulted in deactivation of the Stk11–Ampk–Sirt1–Ppargc1-α signaling pathway, whereas autophagy was up-regulated, resulting in both impaired mitochondrial biogenesis and increased clearance. Muscle over-expression or pharmacological activation of Rev-erb-α increased respiration and exercise capacity. This study identifies Rev-erb-α as a pharmacological target which improves muscle oxidative function by modulating gene networks controlling mitochondrial number and function.
Keywords: Rev-erb-α, skeletal muscle, oxidative capacity, mitochondrial biogenesis, autophagy
Skeletal muscle contractility is important for locomotion and posture, activities performed by different myofiber types with distinct contractile and metabolic properties. Mitochondria serve a critical function in the maintenance of skeletal myofiber homeostasis and to adjust energy production to the demand. They do so by the oxidation of glucose-derived pyruvate and the β-oxidation of fatty acids generating an electrochemical proton gradient through the respiratory complexes of the electron transport chain, which can be used to drive the phosphorylation of ADP to ATP, a reaction called oxidative phosphorylation (OXPHOS). What determines mitochondrial content and function is not completely understood.
Nuclear receptors and their cofactors regulate metabolism in response to environmental signals and trigger homeostatic responses by coordinately regulating transcriptional networks. Previous studies have established that nuclear receptors, such as the Peroxisome Proliferator-Activated Receptor (Ppar)-β/δ, estrogen-related receptor-α and γ, along with co-regulators such as Ppar-γ coactivator-1 (Ppargc1)-α and Ppargc1-β, as well as the nuclear receptor co-repressor 1, among others, control muscle physiology by modulating mitochondrial biogenesis and function, fiber type determination and switch, as well as muscle vascularization 1–5. Ppargc1-α is a master driver of mitochondrial biogenesis and its over-expression in skeletal muscle resulted in increased mitochondrial number and function 1, whereas skeletal muscle Ppargc1-α-deficiency led to a reduced number of mitochondria and a marked reduction of muscle oxidative capacity 6, 7.
The nuclear receptor Rev-erb-α is expressed in tissues such as liver and adipose tissue 8, where it modulates lipid, bile acid and glucose metabolism 8–13. In addition, Rev-erb-α controls adipogenesis 14, 15 as well as the macrophage inflammatory response16. Rev-erb-α interacts with the nuclear receptor co-repressor 1 and chromatin modifiers, such as the histone deacetylase 3, to form a complex repressing target gene transcription 17. Remarkably, a Rev-erb-α/Ppargc1-α cross-talk pathway regulates heme synthesis in hepatic cells 18, 19. Whether Rev-erb-α interacts with Ppargc1-α in muscle and whether Reverb-α controls skeletal muscle oxidative capacity has not been investigated yet.
Here we report that Rev-erb-α-deficient mice display severely reduced running capacity due to compromised skeletal muscle oxidative capacity. This was accompanied by both reduced mitochondrial function and reduced mitochondria content associated with lower mitochondrial biogenesis. We further show that Rev-erb-α modulates Ppargc1-α expression and function by modulating adenosine monophosphate-activated protein kinase (Ampk)/sirtuin (Sirt)-1 signaling. Additionally, Rev-erb-α regulates the autophagy process by repressing genes involved in autophagosome formation and lysosomal degradation. These observations were reproduced in muscle cells upon Rev-erb-α silencing, while Rev-erb-α over-expression induced the opposite phenotype (i.e. improved mitochondrial respiration, up-regulation of mitochondrial biogenesis genes and repression of autophagy genes). Over-expression or pharmacological activation of Rev-erb-α profoundly improved running capacity and mitochondrial respiration in mice. Rev-erb-α thus emerges as a novel regulator of skeletal muscle oxidative capacity.
Results
Rev-erb-α−/− mice display lower exercise capacity
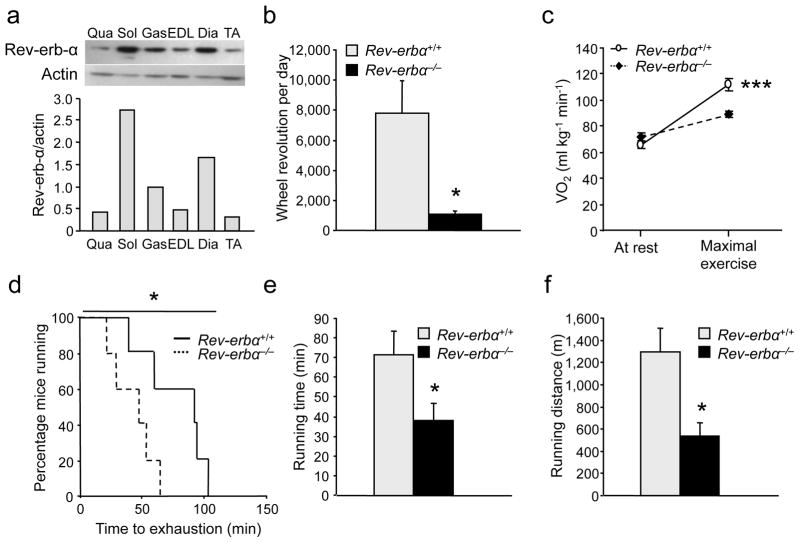
Rev-erb-α expression is significantly higher in oxidative compared to more glycolytic muscles (Fig. 1a). Notably, its expression is higher in soleus and gastrocnemius muscle upon exercise training (Fig. S1a). Thus we explored whether Rev-erb-α plays a role in skeletal muscle oxidative capacity and exercise capacity. Rev-erbα–deficient (Rev-erbα−/−) mice exhibited markedly reduced spontaneous locomotor activity in a free wheel exercise compared to wild-type littermates (Fig. 1b). We next assessed basal (VO2b) and maximal (VO2max) oxygen consumption, reflecting aerobic capacity, by submitting Rev-erbα−/− mice to a forced, progressive treadmill exercise. VO2b measured at rest was not significantly different between the two genotypes, whereas VO2max measured at exhaustion was significantly lower in Rev-erbα−/− mice (Fig. 1c), resulting in a >60% reduced aerobic capacity during exercise. In a standard endurance exercise test performed at 70% of their respective VO2max, 50% of Rev-erbα−/− mice, compared to only 20% of the wild-type mice, stopped running within 50 min indicative of their inability to sustain a long-lasting exercise (Fig. 1d). In this setting, Rev-erbα−/− mice ran for a significantly shorter time and distance than their wild-type littermates (Fig. 1e,f).
Figure 1. Rev-erbα-deficient mice display reduced voluntary activity and exercise performance.
(a) Western blot analysis of mouse muscles; actin was used as control. Qua, quadriceps; Sol, soleus; Gas, gastrocnemius; EDL, extensor digitorum longus; Dia, diaphragm; TA, tibialis anterior (n = 7 per group). (b) Wheel voluntary activity of Rev-erbα−/− mice compared to Rev-erbα+/+ mice (n = 5 per genotype). (c) Basal (at rest) and maximal aerobic capacity (VO2) during a progressive treadmill test to exhaustion in an enclosed single lane treadmill (n = 9–10 per genotype). (d) Endurance capacity of Rev-erbα−/− mice compared to Rev-erbα+/+ mice (n = 6 per genotype). (e–f) Running time (e) and distance (f) until exhaustion in endurance exercise (n = 6 per genotype). Results are expressed as means ± sem; *P < 0.05, ***P < 0.001 by unpaired t-test (a–c,e–f) and by log-rank Mantel cox (d).
Rev-erb-α controls muscle mitochondrial content and function
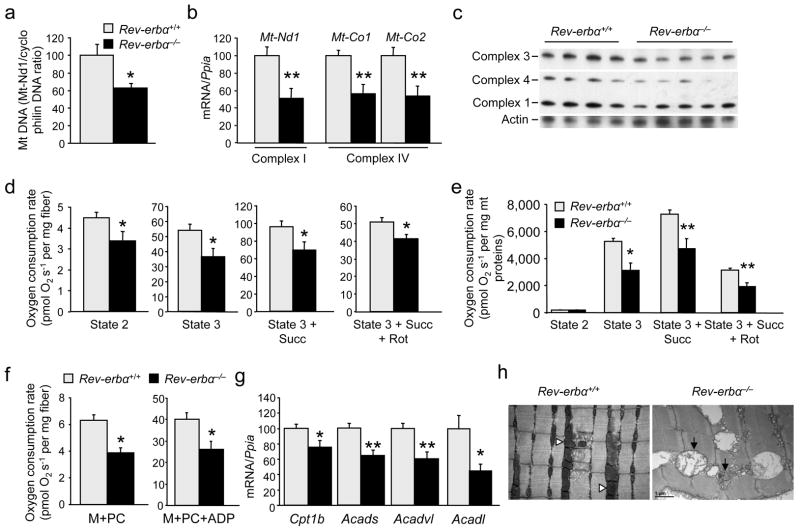
Mitochondrial DNA content was ~40% lower in skeletal muscle from Rev-erbα−/− mice compared to wild-type littermates (Fig. 2a), suggesting a regulatory role of Rev-erb-α on skeletal muscle mitochondrial content. Accordingly, expression of genes encoding subunits of the mitochondrial electron transport respiratory chain, such as NADH dehydrogenase 1, a subunit of complex I, and cytochrome c oxidase -1 and 2, two subunits of complex IV, was lower in soleus and quadriceps muscles from Rev-erbα−/− mice (Fig. 2b, S1b). In addition, protein relative amounts of the mitochondrial oxidative phosphorylation complexes were lower in Rev-erbα−/− compared to Rev-erbα+/+ muscle (Fig. 2c, S1c). Moreover, glutamate-malate-stimulated (state 2) respiration was lower in isolated fibers from Rev-erbα−/− vs. Reverbα+/+ muscle, in line with the reduced mitochondrial content (Fig. 2d). Mitochondrial oxidative capacity was impaired in Rev-erbα-deficient muscle as attested by a marked reduction of ADP-driven glutamate-malate (state 3) respiration in saponin-permeabilized fibers isolated from Rev-erbα−/− compared to wild-type mice (Fig. 2d). The lower respiration upon addition of succinate and rotenone demonstrates a reduced capacity of the entire chain rather than a deficiency in a specific complex. This was further underscored by the significantly lower respiration when comparing equal amounts of isolated mitochondria (Fig. 2e), illustrating not only a decreased mitochondrial content, but also lower respiratory chain function of isolated Rev-erbα−/− compared to Rev-erbα+/+ mitochondria.
Figure 2. Rev-erb-α modulates mitochondria content and function.
(a) Mitochondrial DNA (mtDNA) content (n = 6 per genotype). (b) RT-qPCR analysis of mitochondrial respiratory chain subunit genes in soleus muscle (n = 7 per genotype). (c) Western blot analysis of mitochondrial complexes in soleus from Rev-erbα−/− and Rev-erbα+/+ mice (n = 4–5 per genotype). (d) Mitochondrial respiration in permeabilized soleus fiber from Rev-erbα−/− and Rev-erbα+/+ mice (Succ, succinate; Rot, rotenone) (n = 7 per genotype). (e) Mitochondrial respiration from isolated muscle mitochondria (n = 5 per genotype). (f) Mitochondrial fatty acid β-oxidation-dependent respiration in isolated permeabilized soleus fibers from Rev-erbα−/− and Rev-erbα+/+ mice (n = 6 per genotype). (g) Skeletal muscle (soleus) expression of genes encoding proteins involved in fatty acid oxidation (n = 7 per genotype). (h) Electron microscopy analysis of Rev-erbα−/− and Rev-erbα+/+ muscle. Black arrows: swollen, less dense mitochondria; white arrowheads: normal mitochondria. Representative pictures from n = 5 mice per genotype. Results are expressed as means ± sem; *P < 0.05 and **P < 0.01, by unpaired t-test.
Although gene expression analysis of fiber type markers suggested a switch toward a more glycolytic profile (tropomyosin 3, a marker of oxidative type 1 fiber, myosin heavy chain IIa and IIx, markers of mostly oxidative fibers, are lower in Rev-erbα-deficient soleus and quadriceps muscles, (data not shown), specific type 1, type2a and 2b immunostaining did not reveal any significant changes between the two genotypes (Fig. S1d). In addition, Myoglobin gene expression and vascularization, assessed by CD31 immunostaining on soleus muscle sections, were similar between the two genotypes (Fig. S5d,e).
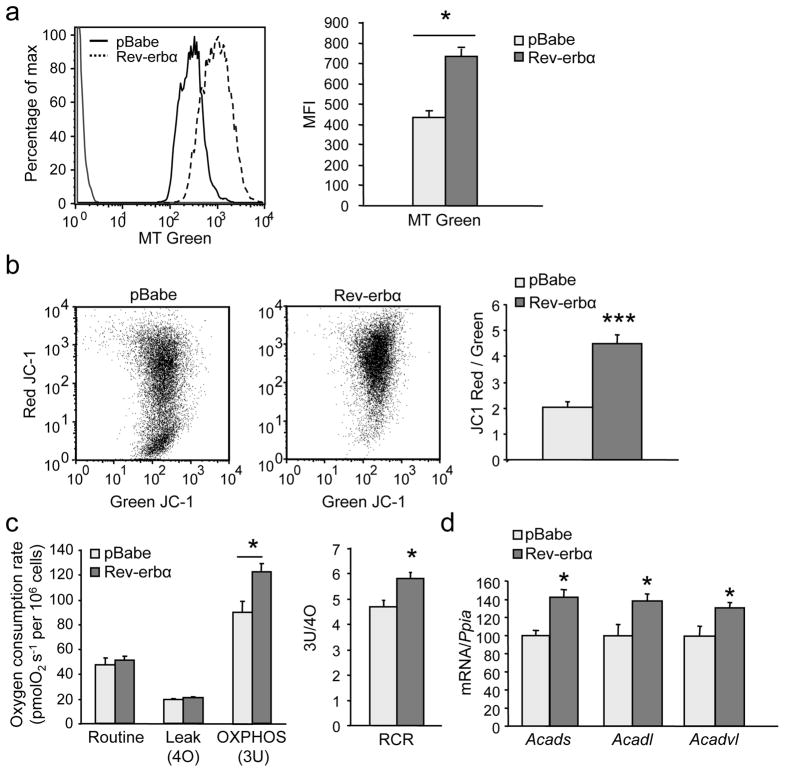
To study whether Rev-erb-α regulates mitochondrial function in a cell-autonomous manner, C2C12 muscle cells were stably infected with a Rev-erb-α coding retrovirus. MitoTracker green staining, a marker of mitochondria content, was induced upon Rev-erb-α over-expression in differentiated C2C12 myotubes (Fig. 3a). Notably, the JC–1 red/green ratio, an indicator of membrane potential, was also increased by Rev-erb-α over-expression in C2C12 cells, suggesting increased mitochondrial activity (Fig. 3b). Rev-erb-α expression enhanced the maximal respiratory capacity due to enhanced coupled ATP-producing oxidative phosphorylation as illustrated by higher state 3U respiration and respiratory control ratio (RCR, OXPHOS/Leak ratio), whereas the uncoupling proton leakage remained unchanged (Fig. 3c). Conversely, Rev-erbα silencing substantially reduced mitochondrial respiration (Fig. S2a) as well as the quantity of total (green Mitotracker) and functional (red Mitotracker) mitochondria (Fig. S2b) in differentiated C2C12 cells. Analysis of mitochondrial fatty acid oxidation in permeabilized fibers of Rev-erbα-deficient muscle revealed compromised oxygen consumption in the presence of palmitoyl-L-carnitine/malate alone and in presence of ADP (Fig. 2f). In parallel, the expression of genes encoding enzymes of fatty acid β–oxidation, notably carnitine palmitoyltransferase1B (Cpt1b), (very) long chain (Acadvl, Acadl) and short chain (Acads) acyl-CoA dehydrogenases, was lower in Rev-erbα−/− vs Reverbα+/+ skeletal muscle (Fig. 2g, S3a), whereas Rev-erb-α retrovirus infected C2C12 cells displayed a mirror phenotype (i.e. higher Acadvl, Acadl and Acads acyl-CoA dehydrogenase expression) compared to control-infected pBabe cells (Fig. 3d). These results indicate that Rev-erb-α modulates fatty acid β–oxidation-driven generation of reducing equivalents to feed in the electron transport chain in vitro and ex vivo in skeletal muscle. Together these results demonstrate that Rev-erb-α regulates muscle cell mitochondrial content and function.
Figure 3. Rev-erb-α modulates mitochondrial respiration.
(a) Mitochondria content in Rev-erb-α-expressing (Rev-erbα) or control (pBabe) retrovirus infected C2C12 cells by flow cytometry analysis of green mitotracker (MT) staining (Left, representative histogram; Right, mean fluorescence intensity (MFI)) (n = 6 per condition). (b) Functional mitochondria content by flow cytometry analysis of JC–1 (Left) and JC1red/JC1green (i.e. high/low mitochondrial membrane potential) MFI ratio (Right) (n = 6 per condition). (c) Oxygen consumption in Rev-erbα and control pBabe cells (n = 5 per condition). (d) RT-qPCR analysis of fatty acid oxidation gene expression in differentiated C2C12 cells infected with Rev-erbα or control (pBabe) retrovirus (n = 6 per group). Results are expressed as means ± sem; *P < 0.05, *** P < 0.001 by unpaired t-test.
Electron microscopy analysis on muscle sections revealed a slight misalignment of Z lines, the presence of vacuolated fibers, and the presence of abnormal, swollen and less dense mitochondria in muscle sections from Rev-erbα−/− mice, compared to wild-type littermates (Fig. 2h, S4), revealing a severe skeletal muscle phenotype and mitochondrial dysfunction of Rev-erbα−/− mice. The repair process, assessed by the presence of centro-cellular nuclei (Fig. S5a shows that the nuclei are localized at the periphery of the fibers, testifying the absence of regeneration) and the absence of change in the expression of Pax7 and Myf5 (Fig. S5b), was not different between Rev-erbα−/− mice and wild-type littermates. In line, immunofluorescent staining of Pax7 indicates its localization at the boundary of the myofiber (Fig. S5c), implying the presence of quiescent satellite cells located on the border of myofibers, which are similar in the two genotypes.
Rev-erb-α increases skeletal muscle mitochondrial biogenesis
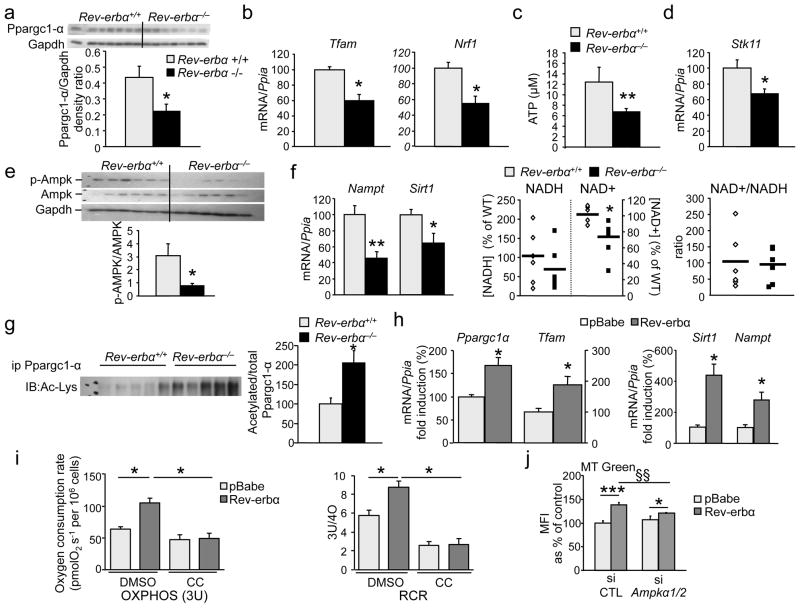
mRNA (−56%) and protein (−50%) concentrations of Ppargc1-α, which plays a pivotal role in mitochondrial biogenesis, were lower in soleus and quadriceps muscle from Reverbα−/− mice (Fig. 4a, S3b, S3d). Consistently, the expression of mitochondrial transcription factor A (Tfam) and nuclear respiratory factor-1, genes involved in mitochondrial biogenesis, was lower in Rev-erbα−/− mice (Fig. 4b). By contrast, Rev-erb-α over-expression increased Ppargc1α and Tfam in C2C12 cells (Fig. 4h).
Figure 4. Rev-erb-α modulates mitochondrial biogenesis by interfering with Ampk-Sirt1-Ppargc1-α signaling.
(a–g) Soleus Ppargc1-α protein relative amounts (a), mRNA concentrations of Tfam and Nuclear respiratory factor 1 (Nrf1) (b), cellular ATP concentrations (c), Stk11 mRNA concentrations (d), western blot analysis of Ampk phosphorylation (e), Nampt and Sirt1 mRNA concentrations (f), NAD+ and NADH cellular concentrations, (g) acetylated Ppargc1-α protein relative amounts in Rev-erbα−/− and Rev-erbα+/+ soleus muscle (n = 7 per genotype). (h) Ppargc1α, Tfam, Sirt1 and Nampt gene expression in Rev-erb-α (Rev-erbα) vs. control (pBabe) retrovirus infected C2C12 cells (n = 6 per group). (i) OXPHOS (3U) mitochondrial respiration and RCR in absence or presence of the Ampk phosphorylation inhibitor C compound (CC) in C2C12 cells infected with Rev-erb-α or control (pBabe) retrovirus. (n = 6 per condition) (j) Mitochondria content in Rev-erb-α-expressing (Rev-erbα) or control (pBabe) retrovirus infected C2C12 cells transfected with a siRNA to reduce Ampkα1 and Ampkα2 expression or a scramble control siRNA: green Mitotracker mean fluorescence intensity (MFI) (n = 3 per condition). Results are expressed as means ± sem *P < 0.05, **P < 0.01, and ***P < 0.001, §§P < 0.01by unpaired t-test.
As a functional reflection of impaired mitochondrial electron transport chain activity, ATP concentrations were severely reduced in Rev-erbα-deficient muscle (Fig 4c). Ampk, a ‘fuel gauge’ activated by liver kinase B1 (Lkb1 also known as serine threonine/kinase 11 or Stk11) when the AMP/ATP ratio increases, induces Sirt1-dependent deacetylation of Ppargc1-α as well as the expression of nicotinamide phosphoribosyltransferase (Nampt), the rate-limiting enzyme in the synthesis of the Sirt1 cofactor NAD+, thus impacting on mitochondrial and lipid oxidation genes 20. Of note, Stk11 gene expression (Fig. 4d, S3d), Ampk phosphorylation (Fig. 4e) and activity illustrated by the p–Acac/Acac (Acetyl-coenzyme A carboxylase) ratio (Fig. S3c), as well as Nampt gene expression (Fig. 4f) were lower in Rev-erbα-deficient compared to Rev-erbα+/+ muscle. Concentrations of NAD+ and NADH, the reducing equivalent produced by fatty acid oxidation, were lower in skeletal muscle from Rev-erbα−/− mice compared to their littermates (Fig. 4f). Moreover, a significantly lower Sirt1 expression was observed in skeletal muscle from Rev-erbα−/− compared to Rev-erbα+/+ mice (Fig. 4f, S3d), and blunted SIRT1 activity was attested by increased Ppargc1-α acetylation (Fig. 4g). Conversely, Sirt1 and Nampt expression was induced in Rev-erb-α over-expressing C2C12 cells (Fig. 4h), accompanied by significantly improved mitochondrial respiration, an effect which was fully blocked by the addition of the C compound, an inhibitor of Ampk phosphorylation (Fig. 4i). Consistently, siRNA knock-down of Ampkα1 and Ampkα2 prevented the increase in mitochondrial content upon Rev-erb-α over-expression (Fig. 4j), whereas Ampk activation by AICAR increased mitochondrial number in shRev-erbα cells, but not to the same extent as in control cells (Fig. S2c). Together these data indicate that Rev-erb-α regulates skeletal muscle mitochondria biogenesis through modulation of the Stk11–Ampk–Sirt1–Ppargc1-α pathway.
Rev-erbα-deficiency induces skeletal muscle autophagy
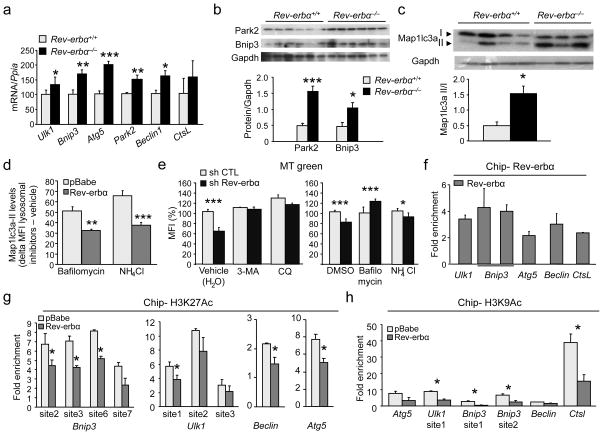
We next explored whether Rev-erb-α also modulates mitochondria degradation. Autophagy is a process mediating the selective clearance of cytoplasmic components, such as damaged mitochondria, which could otherwise become deleterious. Autophagy is a self-digestion process occurring via the formation of a vesicle (nucleation) expanding to become an autophagosome which engulfs cellular components and directs them to the lysosome for degradation. Expression of the genes encoding Ulk1, a protein of the initiation complex implicated in vesicle formation, Beclin1, a protein of the nucleation complex, as well as autophagy related gene-5 and Bnip3, which are responsible for vesicle elongation and autophagosome formation, and the lysosomal enzymes cathepsin L and ATPase6v1b2 were higher in skeletal muscle of Rev-erbα−/− compared to Rev-erbα+/+ mice (Fig. 5a, 5b), and lower in Rev-erb-α infected C2C12 cells vs. control cells (Fig. S6). Rev-erb-α also regulated gene and protein expression of Parkin (also known as Park2), a protein specifically involved in mitochondrial clearance 21 (Fig. 5b, S6). The functional increase of autophagy was illustrated by the maturation of microtubule-associated protein 1 light chain 3 alpha (Map1lc3a, also known as LC3) Map1lc3a–I into its lipidated form Map1lc3a–II, a marker of ongoing autophagy associated with the autophagosome membrane (Fig. 5c). Moreover, treatment with the lysosome inhibitors bafilomycin and NH4Cl which block the fusion of the lysosome with mature autophagosomes which subsequently accumulate, increased Map1lc3a–II levels to a significantly lesser extent in Rev-erb-α over-expressing C2C12 cells compared to treated control (pBabe) cells (Fig. 5d). This indicates that autophagy flux is decreased upon Rev-erb-α over-expression. Conversely, the number of mitochondria was lower upon Rev-erbα silencing in C2C12 cells and addition of the autophagy blockers 3-methyl adenine and chloroquine or the lysosome inhibitors bafilomycin and NH4Cl prevented this decrease (Fig 5e), suggesting that Rev-erbα deficiency results in an increased autophagy flux, thus contributing to the lower mitochondria number.
Figure 5. Rev-erb-α modulates skeletal muscle autophagy.
(a) RT-qPCR analysis of autophagy gene expression in soleus from Rev-erbα−/− and Reverbα+/+ littermates. (n = 7 per genotype). (b) Western blot analysis of soleus muscle Park2 and Bnip3 proteins in Rev-erbα−/− mice compared to Rev-erbα+/+ littermates. (c) Western blot analysis of Map1lc3a–I and Map1lc3a–II protein in soleus from Rev-erbα−/− and Rev-erbα+/+ mice (n = 3–4 per genotype). (d) Flow cytometry delta (treated – vehicle) mean fluorescence intensity (MFI) of Map1lc3a–II protein levels in C2C12 cells infected with Rev-erbα (Reverbα) vs control (pBabe) retrovirus and treated with lysosome inhibitors (50 nM bafilomycin or 25 mM NH4Cl) or their respective vehicle after 16 h serum deprivation (n = 6 per condition). (e) Mitochondria content in shRev-erbα and control (shCTL) cells treated or not with 5 mM 3-methyl adenine (3MA) or 50 μM chloroquine (CQ) or 100 nM bafilomycin or 25 mM NH4Cl: specific green mitotracker (MT) flow cytometry MFI (n = 6 per condition). (f) Rev-erb-α binding to regulatory regions of the indicated autophagy genes measured by ChIP-qPCR (n = 3 independent experiments). (g–h) ChIP-qPCR examining the Rev-erb-α binding regions from (e) for changes in H3K27 and H3K9 acetylation (n = 3 independent experiments). (f–h) ChiP experiments were conducted on Rev-erb-α over-expressing (Rev-erbα) and control (pBabe) C2C12 cells. Results are expressed as means ± sem *P < 0.05, **P < 0.01 and ***P < 0.001, by unpaired t-test.
To gain further mechanistic insight, the distribution of Rev-erb-α binding sites in the vicinity of autophagy genes, assessed using chromatin immunoprecipitation (ChIP)-sequencing data17, was examined and overlaid with epigenetic marks obtained in C2C12 myotubes 22, reasoning that intense transcriptional activity at these marks may increase the likelihood of Rev-erb-α binding to these locations. Primers for ChIP-qPCR targeting these regions were then designed to assess Rev-erb-α binding and changes in acetylation marks in Rev-erb-α-expressing C2C12 cells. An enrichment of Rev-erb-α binding in the regulatory regions of several autophagy genes was observed (Fig 5f), which was associated with decreased H3K27 (Fig 5g) and H3K9 (Fig 5h) acetylation on these sites.
Rev-erb-α activation improves muscle mitochondrial function
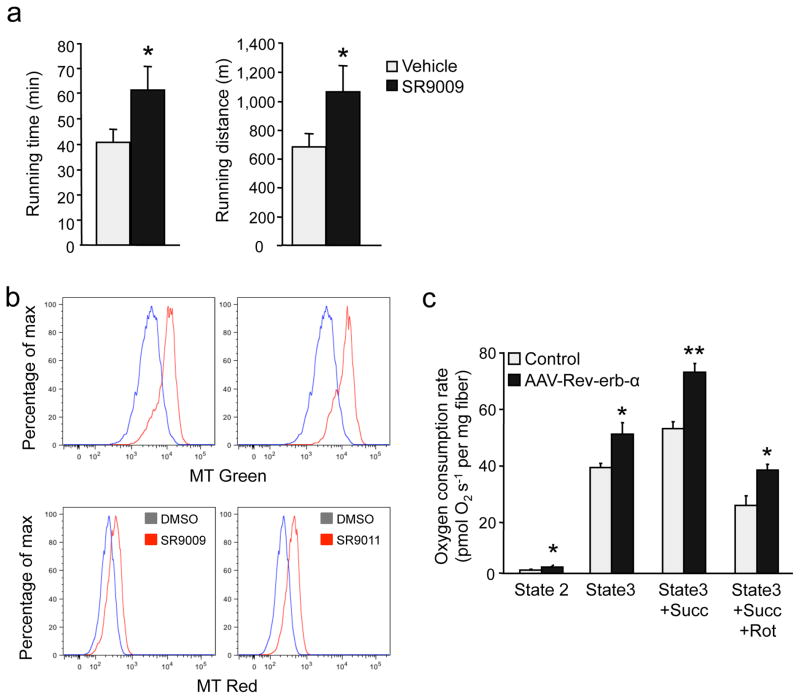
Next, we determined the effect of pharmacological activation of Rev-erb-α, by treating mice with the synthetic ligand SR9009 12, on exercise capacity. Notably, in an endurance exercise test, mice treated with SR9009 ran significantly longer both in time and distance than vehicle-treated mice (Fig. 6a). Moreover, incubation of C2C12 cells with two different agonists increased the number of total (Mitotracker Green) and active (Mitotracker red) mitochondria (Fig. 6b). Finally, skeletal muscle-specific Rev-erb-α over-expression in mice significantly improved mitochondrial function as attested by a significant increase in glutamate-malate-stimulated and ADP-driven respiration in absence and presence of succinate of permeabilized fibers (Fig. 6c). Together these data indicate that enhancement of Rev-erb-α expression and/or activity exerts beneficial direct effects on skeletal muscle to improve mitochondrial respiration and exercise capacity.
Figure 6. Rev-erb-α over-expression or pharmacological activity improves mitochondrial function and exercise capacity.
(a) Running distance and time until exhaustion in endurance exercise in mice treated with SR9009 (100 mpk for 30 days) (n = 6 per group). (b) Mitochondria content in C2C12 cells treated with SR9009 (5 μM), SR9011 (5 μM) or vehicle: representative bargraph of flow cytometry green mitotracker (upper panel) and red mitotracker (bottom panel) staining (n = 4 per condition). (c) Mitochondrial respiration in permeabilized fiber isolated from tibialis anterior muscle from mice intra-muscularly injected with a Rev-erb-α expressing AAV vector (n = 4 per group). Results are expressed as means ± sem; *P < 0.05, **P < 0.01 by unpaired t-test.
Discussion
Results from both gain- and loss-of-function experiments identify Rev-erb-α as a physiological regulator of muscle mitochondrial content and oxidative function. This is supported by the following observations. First, Rev-erb-α is preferentially expressed in more oxidative muscles, such as soleus muscle, and Rev-erbα-deficient mice display profoundly altered exercise capacity associated with a marked decrease of mitochondrial content and function, the presence of swollen mitochondria and abundant vacuoles within the fibers. Similar alterations were observed upon knock-down of Rev-erbα in differentiated C2C12 myotubes (lower mitochondrial content and impaired mitochondrial function). By contrast, pharmacological activation or skeletal muscle-specific Rev-erb-α over-expression in vivo and in vitro in C2C12 cells results in the opposite phenotype, highlighting that Rev-erb-α exerts a direct action on skeletal muscle cells. Second, our data indicate that Rev-erb-α controls mitochondrial biogenesis and respiration through the Stk11–Ampk–Sirt1–Ppargc1–α signaling pathway. Finally, we show that skeletal muscle Rev-erb-α regulates several genes involved in different steps of the autophagy process, including genes more specifically dedicated to the elimination of damaged mitochondria. Together these data support the concept that Rev-erb-α acts through a two-pronged mechanism implicating both biogenesis of new mitochondria and clearance of defective mitochondria.
The regulation of the Stk11–Ampk–Sirt1–Ppargc1-α pathway by Rev-erb-α provides a molecular mechanism whereby Rev-erb-α controls mitochondrial biogenesis and function. Ampk and Sirt1 are crucial in the metabolic flexibility that allows skeletal muscle to switch to lipid oxidation during fasting and exercise. Deficient Ampk activity in Rev-erbα−/− mice is associated with impaired Sirt1-mediated Ppargc1-α deacetylation and ensuing attenuated induction of mitochondrial fatty acid oxidation gene expression and reduced exercise capacity 20, 23. Likewise, Ppargc1-α acetylation is higher and its expression diminishes in Rev-erbα-deficiency, which may contribute to the reduced number of mitochondria, lower expression of transcription factors important for mitochondrial function, such as Tfam, and altered electron transport chain activity. In addition, expression of genes encoding fatty acid β–oxidation enzymes as well as palmitate-induced mitochondrial respiration is compromised in the absence of Rev-erb-α. This is further supported by defective Ampk activation, reduced NAD+ concentrations together with markedly altered Sirt1 and Nampt expression, testifying a marked de-activation of this pathway. Moreover, the observation that siRNA or C compound-mediated inhibition of Ampk activity blocks the stimulatory effect of Rev-erb-α over-expression on mitochondrial respiration, demonstrates that Rev-erb-α-mediated improvement of mitochondrial respiration requires active Ampk. Of note, Ampk phosphorylation is not induced even though ATP concentrations are low in Rev-erbα-deficient mice. Binding of AMP increases the ability of other kinases to phosphorylate and activate the Ampk-α subunit. Stk11 is a major upstream kinase expressed in skeletal muscle that phosphorylates and activates Ampk, and Stk11-deficiency results in blunted Ampk activation, decreases Ppargc1α expression together with reduced exercise capacity as well as compromised mitochondrial activity 24, 25, a phenotype similar to the one observed in the Rev-erbα-deficient mice. Our results also indicate that mitochondria are damaged and mitochondrial oxygen consumption is reduced in Rev-erbα−/− muscle, whereas skeletal muscle-specific Rev-erb-α over-expression improves mitochondrial respiration. Moreover, treatment of mice with a synthetic Rev-erb-α agonist increases oxygen consumption 12. Notably, and in contrast to other transcriptional regulators, such as Ppar-β/δ 2 and Ppargc1-α1, 6, Rev-erb-α modulates skeletal muscle oxidative capacity without inducing a fiber type switch, suggesting a disconnection between both phenomena upon Rev-erbα-deficiency. Together these data support a prominent role for Rev-erb-α in skeletal muscle mitochondria function.
Autophagy is a dynamic self-digestion process that ensures the selective clearance of damaged organelles or aggregated macromolecules, a particularly important mechanism in differentiated cells such as myotubes 26–28. Our data demonstrate that Rev-erb-α exerts a tight control on this pathway by regulating several genes involved in vesicle nucleation and expansion, autophagosome formation as well as genes encoding lysosomal enzymatic activities. The conversion of Map1lc3a–I to Map1lc3a–II and its lipidation is a hallmark of autophagy 29. The maturation into Map1lc3a–II was elevated in the absence of Rev-erb-α, whereas autophagy flux is decreased by Rev-erbα over-expression. Of note, Rev-erb-α represses genes triggering mitophagy, such as Park2, a cytosolic E3 ubiquitin ligase that translocates to depolarized mitochondria to induce mitophagy, thereby maintaining a pool of functioning mitochondria and limiting oxidative damage 21. Likewise, Rev-erb-α down-regulates Ulk1, which not only participates in the initiation complex, but may also play an important role in triggering mitophagy 30. Ulk1–deficiency indeed results in accumulation of defective mitochondria in mature red blood cells 31. Finally, we demonstrate that autophagy blockade by autophagy or lysosome inhibitors reverses the effect of Rev-erbα knock-down on total mitochondria number. Autophagy is also a survival mechanism by which cells produce energy in times of nutrient paucity. One of the most potent inducers of autophagy is Ampk, which directly phosphorylates Ulk1 and, simultaneously, turns off mTORC1 signaling, an inhibitor of autophagy 26, 28, 32. However, neither Ampk (which is down-regulated in the absence of Rev-erb-α) nor Mtor, whose signaling is not regulated by Rev-erb-α (not shown), are likely to mediate Rev-erb-α’s action on autophagy. Another potent autophagy inducer, Sirt1 33, is also down-regulated in the absence of Rev-erbα, again excluding that Sirt1 plays a role in the observed phenotype. Together these data indicate that Rev-erb-α by-passes these regulators and represses autophagy genes directly likely by binding to the DNA in the regulatory regions of these genes. Indeed, our ChIP-qPCR data confirmed Rev-erb-α binding enrichment in the regulatory regions of autophagy genes. This is accompanied by a modification of epigenetic marks at these sites as indicated by decreased H3K27 and H3K9 acetylation, consistent with the repressive role of Rev-erb-α on gene transcription.
Rev-erb-α is a component of the biological clock, which allows synchronisation of internal rhythms to daily environmental cues 34. Skeletal muscle displays circadian rhythmicity of gene expression 35, 36 and the clock components Clock and Brain and muscle ARNT-like-1 have been shown to participate in the maintenance of skeletal muscle function 37. mRNA concentrations of the clock genes are altered in skeletal muscle from Rev-erbα−/− mice (Fig. S7a) and Rev-erb-α over-expression in synchronized C2C12 cells leads to an altered circadian expression pattern of Ppargc1α (Fig. S7b). Pharmacological activation of Rev-erb-α alters the circadian rhythms of Cpt1b and Ppargc1α in skeletal muscle 12. Consequently, in addition to a direct action of Rev-erb-α on autophagy genes and mitochondrial biogenesis and function, disturbed circadian rhythmicity due to Rev-erbα deletion or over-expression may contribute to the observed phenotype.
In conclusion, Rev-erb-α is a major physiological regulator of mitochondrial content and oxidative function. Pharmacological activation of Rev-erb-α may be a promising approach for the treatment of skeletal muscle diseases with compromised exercise capacity.
ONLINE METHODS
Animal studies
We used Rev-erbα-deficient mice39 and their wild-type littermates (Rev-erbα+/+ mice). Mice were housed in a 12 h/12 h light/dark cycle and allowed ad libitum access to food and water. They received a regular chow diet. Mice were sacrificed by cervical dislocation and skeletal muscles were snap-frozen in liquid nitrogen and stored at −80 °C until RNA and protein isolation and histological staining. Rev-erbα-deficiency in mice was verified by qPCR analysis of skeletal muscle gene expression of Rev-erbα and its known target gene Bmal1 (fig. S8a).
Rev-erb-α over-expression: we cloned the coding sequence of Rev-erb-α in front of a MCK promoter allowing muscle-specific expression and introduced in a 1/2 AAV vector (Sirion Biotech, Germany). We injected 2.1 × 1010 infectious particles intra-muscularly in the tibialis anterior muscle using the tibialis anterior muscle of the contra-lateral limb as control. Pharmacological activation: SR9009 was administered i.p. at 100 mpk for 30 days as previously described 12.
Maximal exercise stress test and endurance capacity
Two days before the experiment, mice were acclimatized to a single lane treadmill by performing a 10 m min−1 run. The day of the exercise stress test, mice were placed into the treadmill enclosed in a metabolic chamber connected to an oxygen (O2) sensor (Oxymax, Columbus Instruments, Columbus, OH). We measured basal oxygen consumption (VO2b) after a 30 min resting period; then, mice were encouraged to run on the treadmill at 10 m min−1 and 0% incline. Every 3 min, the treadmill speed was incremented by 4 m min−1 until the mice reached exhaustion in order to determine their maximal VO2 (VO2max). The speed at which VO2max was obtained was considered as the maximal running speed (Vmax). One week later, we tested the mice for their endurance capacity. After a 6 min run at 30% of their maximal running speed, the treadmill speed was set at 70% of the VO2max. The experiment was stopped once mice stayed for 5 continuous seconds on the electrical grid. Time to exhaustion and total running distance were determined.
Daily wheel running activity
To record daily running wheel behavior, we placed the animals into individual cages containing running wheels for 3 weeks (Campden, Phymep, Paris). Wheel revolution was recorded daily during the last two weeks and averaged.
Training
We examined Rev-erb-α expression in control (untrained) and exercised mice. Mice were trained for 8 weeks, 5 days per week, 1 to 2 h per day starting at a speed of 8 m min−1 up to 16 m min−1 the last 4 weeks.
Animal care and use were performed according to approved institutional guidelines and all experimental procedures were approved by the relevant ethic committees (Comité d’éthique en expérimentation animale Nord-Pas-De-Calais and The Scripps Research Institute Florida Campus Institutional Animal Care and Use Committee). We used male mice for all experiments.
Cell culture
We cultured C2C12 myoblasts in Dulbecco modified Eagle’s medium (DMEM; 4.5 g l−1 D-Glucose; Gibco) supplemented with 10% fetal bovine serum and 1% gentamycin. Myogenic differentiation into myotubes was induced when cells reached 80% confluency by adding DMEM supplemented with 2% horse serum (HS) and 1% gentamycin at 37 °C in a humidified incubator under 5% CO2 during 2 to 5 days. When indicated, we added SR9009 or SR9011 (5 μM) to the medium one day prior to differentiation and for the length of the differentiation period (8 days).
Serum shock
We assessed circadian gene expression patterns in cells after synchronization by a 2-h horse serum shock on near confluent cells 40. After 2 h, the medium was changed to 2% serum-containing medium as above.
Retroviral production and infection
We cultured Phoenix Eco cells (Orbigen) in DMEM containing 10% fetal bovine serum, and 1% gentamycin at 37 °C under standard culture conditions. To generate cell lines constitutively over-expressing Rev-erb-α, we inserted the coding sequence of human Reverbα in the pBabe retrovirus plasmid (Addgene) using the BamHI-SalII sites to generate pBabe-Rev-erb-α (Rev-erbα). We used the empty vector (pBabe) as control. To down-regulate Rev-erbα expression, we inserted the shRev-erbα sequence in psilencer 5.1-U6 Retro (Ambion) using the BamH1 and HindIII sites. Phoenix cells (100,000 per cm2) were transfected with the pBabe plasmid constructs (20 μg) using the cationic lipid RPR 120535B as previously described41. C2C12 cells were infected with the supernatant from pBabe or pBabe-Rev-erbα and shControl (shCTL) or shRev-erbα, and puromycin-resistant infected cells were used within 15 days after infection. We verified Rev-erbα over-expression by RT-qPCR on Rev-erb-α C2C12 cells compared to pBabe control cells (Fig. S8b). We verified Rev-erb-α nuclear localization by western blot analysis (Fig. S8c), and we verified its repressive transcriptional activity by transient transfection on a consensus RevDR2 site in Rev-erbα vs pBabe C2C12 cells and compared to non-infected C2C12 cells after co-transfection of a pSG5Rev-erbα expression plasmid (Fig. S8d). Rev-erbα down-regulation was verified by qPCR analysis of Rev-erbα mRNA on shRev-erbα C2C12 cells vs. control cells (Fig. S8e).
Determination of cellular respiration
C2C12 cells (1 × 106 cells ml−1) suspended in cell culture media (RPMI + 10% FCS) were placed into the chambers of the O2K oxygraph, operating at 37 °C. Routine respiration (R) was measured 20 min later. Then, 2 μg ml−1 oligomycin was injected into the chambers to obtain the oligomycin-inhibited leak rate of respiration (4o, Leak, ‘L’), ie. uncoupled respiration. Finally, pulses of FCCP (1 μM) were added into the chambers until maximal oxygen consumption was reached (3u, OXPHOS, ‘P’). It represents the maximal respiratory capacity. The respiratory control ratio (RCR), i.e. the coupled oxidative phosphorylation, is expressed as 3u/4o ratio.
Oxygen consumption on permeabilized soleus fibers
After cervical dislocation, soleus or tibialis anterior muscles were excised and placed into a Petri dish containing ice-cold biopsy preservation solution (BIOPS) and permeabilized fibers were prepared as previously described 42. We placed fibers (3 to 6 mg wet weight) into the O2K oxygraph chambers (Oroboros Instruments, Innsbruck, Austria). In the first chamber, we added glutamate (10 mM) and malate (2 mM) to obtain state 2 respiration. Then, we injected 2.5 mM ADP into the chamber to measure ADP-coupled oxygen consumption (state 3). Further addition of succinate (10 mM) allowed estimation of the entire OXPHOS capacity. Finally, complex I and III were inhibited with rotenone (0.5 μM) and antimycin A (2.5 μM), respectively. In the second chamber, we added palmitoyl-carnitine (5 or 20 μM) and malate (2 mM). After stabilization, coupled respiration was obtained with 2.5 mM ADP. Addition of exogenous cytochrome c (10 μM) was done in all experiments in order to test external membrane integrity. Experiments were performed at 25 °C.
Muscle mitochondria isolation
We cut muscles into small pieces and placed them into a Trypsin-EDTA solution for 15 min at 4 °C. Samples were rinsed with mitochondrial isolation buffer (in mM: sucrose 300, TES 5, EGTA 0.2, pH 7.2) and homogenized with a glass tissue grinder. After centrifugation at 800 g for 7 min, supernatant was collected and spun twice at 8,800 g. The final mitochondrial pellet was suspended into respiration medium Mitomed2 and mitochondria (150 μg) were placed into the oxygraph chambers. Substrates and protocols were as described for permeabilized fiber respiration.
Mitochondria quantity and function Quantification of mtDNA copy number
We isolated mitochondrial DNA (mtDNA) from murine muscles after digestion with Proteinase K (100 μg ml−1) by phenol/chloroform extraction. Relative amounts of nuclear DNA and mtDNA were determined by quantitative real-time PCR. We selected NADH dehydrogenase 1 gene for quantification of mtDNA and ppia (also known as cyclophilin) for nuclear DNA quantification.
Mitotracker-FACS quantification
Rev-erb-α over-expressing, shRev-erbα cells and their respective control cells were washed with PBS, trypsinized and incubated at 37 °C for 20 min with 100 nM MitoTracker Green FM and Red FM dyes (Molecular Probes). Mitotracker Green probe preferentially accumulates in mitochondria, allowing estimation of mitochondrial quantity. Mitotracker Red probe is a red-fluorescent dye that stains mitochondria in living cells and its accumulation is dependent on the membrane potential (Molecular Probes). Samples were washed 3 times in PBS and subjected to flow cytometric analysis on a FACSCalibur apparatus (Becton Dickinson, San Jose, CA).
ATP assay
We measured ATP concentrations using a commercially available kit (EnzyLight™ ATP Assay Kit, Bioassay Systems, Hayward, CA, USA) following the manufacturer’s instructions.
NAD assay
We measured NAD+ and NADH concentrations using a commercially available kit (Biovision Research Products, Mountain View, CA, USA) following the manufacturer’s instructions.
Quantitative RT-QPCR
We isolated total RNA from tissues by guanidinium thiocyanate/phenol/chloroform extraction43 and from C2C12 cells using the Trizol reagent (Invitrogen). Isolated RNA was reverse transcribed into cDNA using commercially available reagents (Superscript II kit; Applied Bioscience). We performed quantitative PCR (qPCR) with the Brilliant III SYBR Green QPCR Master Mix (Agilent) and specific primers following manufacturer’s instructions and using a mx3005 apparatus (Agilent). Gene expression was normalized to cyclophillin and expressed as indicated in the legends to figures. A list of specific primers is available in Supplementary Table 1.
Antibodies and immunoblotting
Protein extracted from muscle biopsies or cells were separated by SDS-polyacrylamide gel electrophoresis and we performed immunoblot analyses according to standard procedures. Proteins were transferred onto PVDF membranes and immunoblot analyses were carried out using antibodies directed against p-AMPK (Thr172)(1:1,000, Cell Signaling technology 2535) and AMPK (1:1,000, Cell Signaling technology 2532), Ppargc1-α (1:500, Santa-cruz biotechnology, sc-13067), Gapdh (1:500, Santa-cruz biotechnology, sc-25778), Actin (1:1,000, Santa-cruz biotechnology sc-1616), Bnip3 and Map1lc3a (1:1,000, Abcam38621 and Abcam51520, respectively), Park2 (1:1,000, Millipore ab9244), Acac (1:1,000, Cell Signalling 3662), p-Acac (S79) (1:1,000, Cell Signalling 3661), OX-PHOS (1:1,000, MitoSciences InVitrogen, iv 458099), Rev-erb-α (1:1,000, Perseus Proteomix PP-A8740A-00) following the manufacturer’s instructions.
Ppargc1-α acetylation assay
We analyzed Ppargc1-α acetylation in quadriceps muscle by immunoprecipitation of Ppargc1-α from 500 μg of protein extract with anti-Ppargc1-α antibody (Santa Cruz, sc-13067, 2 μg per sample) followed by western blot analysis using an acetyl-lysine antibody (1:2,000, Cell Signaling 9441) antibody.
Electron microscopy analysis of muscle sections
We examined ultrastructural muscle morphology using transmission electron microscopy. To this end, muscle tissue blocks were fixed in 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4). Post-fixation was performed in 1% OsO4 in 0.1 M cacodylate buffer (pH 7.4) supplemented with 1.5% K4[Fe(CN)6|. Subsequently samples were dehydrated and embedded in epon. Ultrathin sections were examined using a Philips CM100 electron microscope.
Immunofluorescence assay
Frozen cross sections of muscle were fixed with acetone and incubated with antibodies directed to CD31 (1:100, BD Biosciences, BD557355), Pax7 (1:20, Developmental Studies Hybridoma Bank, PAX7), fiber types (1:25, Developmental Studies Hybridoma Bank, A4.840), and laminin (1:50, Sigma L9393). After incubation with the appropriate fluorescent-labeled secondary antibodies (Goat anti-rabbit IgG AlexaFluor 555 for CD31, Goat anti-mouse IgG1 AlexaFluor488 for Pax7, Goat anti-rabbit IgG AlexaFluor350 for laminin and Goat Anti-mouse IgM AlexaFlur488 for A4.840, all from InVitrogen/Moleculr Probes) and DAPI-stain (30 nM, Invitrogen) for nuclei, sections were mounted in Mowiol. Images were captured with a Nikon E800 fluorescence microscope 44.
Flow cytometry Map1lc3a analysis
Rev-erb-α over-expressing and control cells were seeded in 6 well-plates and serum-starved in presence or in absence of Bafilomycin (50 nM) or NH4Cl (25 mM) overnight at 37 °C. Before staining with a Map1lc3a antibody (1:50, MBL International M152-3 clone 4E12), we fixed the cells with 4% paraformaldehyde for 15 min at room temperature, washed them in PBS and then permeabilized them with digitonin (100 μg ml−1) for 15 min at room temperature. Digitonin permeabilization allowed the extraction of the cytosolic form of Map1lc3a protein (Map1lc3a–I) and the specific detection of the Map1lc3a–membrane-associated fraction (Map1lc3a–II), which correlates with the number of autophagosomes 45. Cells were finally incubated with alexa-488 conjugated anti-mouse IgG for 15 min (1:200, invitrogen, A11001). We acquired sample signals using FACSCalibur (Becton Dickinson, San Jose, CA) and analyzed them using FlowJo software (Tree Star).
Chromatin Immunoprecipitation (ChIP)
We performed ChIP-qPCR experiments as previously described 38 with minor changes as follows. Differentiated C2C12 cells infected with the Rev-erb-α or control (pBabe) retrovirus were cross-linked in 1% formaldehyde for 10min at room temperature. Nuclear extracts were prepared and the chromatin was sonicated for 15 min using the Bioruptor (Diagenode). Chromatins were immunoprecipitated over-night at 4 °C with either a Rev-erbα antibody (Cell Signaling Technology, 2124), a H3K9Ac antibody (Millipore, 17-658), or a H3K27Ac antibody (AbCam, ab4729), all at 3 μg per sample, and then incubated for 4 h at 4 °C with protein A and protein G Dynal magnetic beads (Dynal Biotech, Norway) and tRNA (10 μg ml−1). Cross-linking was reversed overnight at 65 °C. DNA was purified using the QIAquick PCR purification kit (Qiagen) and was analyzed by quantitative PCR using the Brilliant III SYBR Green QPCR Master Mix (Agilent) using specific primers (Supplementary Table 2).
Statistical Analysis
Values are means ± sem of the indicated number of measurements. Statistical significance was determined using two-tailed unpaired Student’s t test with a significance level of 0.05 or Log-rank Mantel cox test.
Supplementary Material
Acknowledgments
This research was supported by a Marie Curie International Reintegration Grant ‘IRG’ (FP7) (to H.D.), the European Commission (FP7) consortium Eurhythdia (to B.S.), Région Nord Pas-de-Calais/FEDER (to B.S.), a CPER ‘starting grant’ (to H.D.), the ‘European Genomic Institute for Diabetes’ (EGID, ANR-10-LABX-46) (to B.S.), an unrestricted ITMO/Astra Zeneca grant (to B.S.), a joint Société Francophone du Diabète (SFD)/MSD research fellowship (to H.D.), a Research Grant from the European Foundation for the Study of Diabetes (EFSD) (to H.D.), National Institutes of Health Grants (MH093429 and DK080201) (to T.B.), a National Research Service Award (DK088499) (to L.A.S.) and a VICI Research Grant for innovative research from the Netherlands Organization for Scientific Research (Grant 918.96.618) (to P.S.). BS is a member of the Institut Universitaire de France.
Footnotes
Competing financial interests:
The authors declare no competing financial interests.
Author contributions
E.W., Y.S., B.S. and H.D. were responsible for the study design, data analysis and interpretation and co-wrote the manuscript. E.W., Y.S., L.S., C.D., S.L., C.P., S.D., G.S. and R.N. performed the experiments and data analysis. J.E., P.L., M.K.C.H., P.S., T.B. were involved in data analysis. Y.S. and T.M.K. were involved in Rev-erbα ligand chemistry. All authors have read and approved the final manuscript.
References
- 1.Lin J, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 2.Schuler M, et al. PGC1alpha expression is controlled in skeletal muscles by PPARbeta, whose ablation results in fiber-type switching, obesity, and type 2 diabetes. Cell Metab. 2006;4:407–414. doi: 10.1016/j.cmet.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Narkar VA, et al. Exercise and PGC-1alpha-independent synchronization of type I muscle metabolism and vasculature by ERRgamma. Cell Metab. 2011;13:283–293. doi: 10.1016/j.cmet.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zechner C, et al. Total skeletal muscle PGC-1 deficiency uncouples mitochondrial derangements from fiber type determination and insulin sensitivity. Cell Metab. 2010;12:633–642. doi: 10.1016/j.cmet.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamamoto H, et al. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell. 2011;147:827–839. doi: 10.1016/j.cell.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Handschin C, et al. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1alpha muscle-specific knock-out animals. J Biol Chem. 2007;282:30014–30021. doi: 10.1074/jbc.M704817200. [DOI] [PubMed] [Google Scholar]
- 7.Leone TC, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duez H, Staels B. Nuclear receptors linking circadian rhythms and cardiometabolic control. Arterioscler Thromb Vasc Biol. 2010;30:1529–1534. doi: 10.1161/ATVBAHA.110.209098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duez H, et al. Regulation of bile acid synthesis by the nuclear receptor Rev-erbalpha. Gastroenterology. 2008;135:689–698. doi: 10.1053/j.gastro.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 10.Yin L, et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 11.Cho H, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solt LA, et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012;485:62–68. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bugge A, et al. Rev-erbalpha and Rev-erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontaine C, et al. The orphan nuclear receptor Rev-Erbalpha is a peroxisome proliferator-activated receptor (PPAR) gamma target gene and promotes PPARgamma-induced adipocyte differentiation. J Biol Chem. 2003;278:37672–37680. doi: 10.1074/jbc.M304664200. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Lazar MA. Bifunctional role of Rev-erbalpha in adipocyte differentiation. Mol Cell Biol. 2008;28:2213–2220. doi: 10.1128/MCB.01608-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fontaine C, et al. The nuclear receptor Rev-erbalpha is a liver X receptor (LXR) target gene driving a negative feedback loop on select LXR-induced pathways in human macrophages. Mol Endocrinol. 2008;22:1797–1811. doi: 10.1210/me.2007-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng D, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu N, Yin L, Hanniman EA, Joshi S, Lazar MA. Negative feedback maintenance of heme homeostasis by its receptor, Rev-erbalpha. Genes Dev. 2009;23:2201–2209. doi: 10.1101/gad.1825809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Estall JL, et al. PGC-1{alpha} negatively regulates hepatic FGF21 expression by modulating the heme/Rev-Erb{alpha} axis. Proc Natl Acad Sci U S A. 2009;106:22510–22515. doi: 10.1073/pnas.0912533106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canto C, et al. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab. 2010;11:213–219. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asp P, et al. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc Natl Acad Sci U S A. 2011;108:E149–E158. doi: 10.1073/pnas.1102223108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujii N, et al. Role of AMP-activated protein kinase in exercise capacity, whole body glucose homeostasis, and glucose transport in skeletal muscle -insight from analysis of a transgenic mouse model- Diabetes Res Clin Pract. 2007;77 (Suppl 1):S92–S98. doi: 10.1016/j.diabres.2007.01.040. [DOI] [PubMed] [Google Scholar]
- 24.Koh HJ, et al. Skeletal muscle-selective knockout of LKB1 increases insulin sensitivity, improves glucose homeostasis, and decreases TRB3. Mol Cell Biol. 2006;26:8217–8227. doi: 10.1128/MCB.00979-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomson DM, et al. Skeletal muscle dysfunction in muscle-specific LKB1 knockout mice. J Appl Physiol. 2010;108:1775–1785. doi: 10.1152/japplphysiol.01293.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joo JH, et al. Hsp90-Cdc37 chaperone complex regulates Ulk1- and Atg13-mediated mitophagy. Mol Cell. 2011;43:572–585. doi: 10.1016/j.molcel.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kundu M, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egan DF, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee IH, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bass J. Circadian topology of metabolism. Nature. 2012;491:348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 35.McCarthy JJ, et al. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol Genomics. 2007;31:86–95. doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller BH, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci U S A. 2007;104:3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrews JL, et al. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc Natl Acad Sci U S A. 2010;107:19090–19095. doi: 10.1073/pnas.1014523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eeckhoute J, Lupien M, Brown M. Combining chromatin immunoprecipitation and oligonucleotide tiling arrays (ChIP-Chip) for functional genomic studies. Methods Mol Biol. 2009;556:155–164. doi: 10.1007/978-1-60327-192-9_11. [DOI] [PubMed] [Google Scholar]
- 39.Duez H, et al. Regulation of bile acid synthesis by the nuclear receptor Rev-erbalpha. Gastroenterology. 2008;135:689–698. doi: 10.1053/j.gastro.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 40.Nagoshi E, et al. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 41.Raspe E, et al. Identification of Rev-erbalpha as a physiological repressor of apoC-III gene transcription. J Lipid Res. 2002;43:2172–2179. doi: 10.1194/jlr.m200386-jlr200. [DOI] [PubMed] [Google Scholar]
- 42.Aragones J, et al. Deficency or inhibition of oxygen sensor Phd1 induces hypoxia tolerance by reprogramming basal metabolism. Nat Genet. 2008;40:170–180. doi: 10.1038/ng.2007.62. [DOI] [PubMed] [Google Scholar]
- 43.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 44.Delhaas T, et al. Steep increase in myonuclear domain size during infancy. Anat Rec. 2013;296:193–197. doi: 10.1002/ar.22631. [DOI] [PubMed] [Google Scholar]
- 45.Kaminskyy V, et al. A quantitative assay for the monitoring of autophagosome accumulation in different phases of the cell cycle. Autophagy. 2011;7:83–90. doi: 10.4161/auto.7.1.13893. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.