Abstract
Mass spectrometric imaging (MSI) has rapidly increased its presence in the pharmaceutical sciences. While quantitative whole-body autoradiography and microautoradiography are the traditional techniques for molecular imaging of drug delivery and metabolism, MSI provides advantageous specificity that can distinguish the parent drug from metabolites and modified endogenous molecules. This review begins with the fundamentals of MSI sample preparation/ionization, and then moves on to both qualitative and quantitative applications with special emphasis on drug discovery and delivery. Cutting-edge investigations on sub-cellular imaging and endogenous signaling peptides are also highlighted, followed by perspectives on emerging technology and the path for MSI to become a routine analysis technique.
Keywords: ADME, spatial distribution, quantitative mass spectrometry, PK/PD, molecular imaging, pharmaceuticals
1. Introduction
For many biological systems, a sufficient characterization may require more than a catalog of the molecules that are present. It may also rely on their anatomical distribution patterns and relative spatial relationships. Researchers in drug discovery and delivery are also in need to spatially characterize and quantify a drug’s absorption, distribution, metabolism, and excretion (ADME). Quantitative whole-body autoradiography (QWBA) is among the most commonly used methods to do so [1]. It begins with dosing an animal specimen with a test drug labeled with radioactive beta-emitters such as 3H or 14C. After a desired amount of time, the specimen is euthanized, snap frozen, and sectioned [2]. Whole-body sections are then placed opposite to phosphor detectors and an image is produced within a few days to a few weeks. Radioactive standards are simultaneously imaged with the specimen to quantify the drug’s penetration. Microautoradiography (MARG) is a similar method that is employed when high spatial resolution images are needed, but is not often used for quantitative purposes [2].
Radiochemical imaging methods have proven to be an invaluable part of the mass balance study—that is, a quantitative account of a drug from ingestion through excretion [3]. But as reputable as they are, they possess fundamental shortcomings in answering questions about spatial relationships. The information in the QWBA or MARG images may rightfully allow an inductive leap to drug distribution, but the raw data itself only shows the detection of radiation. The 14C isotope could be from the drug, a metabolite, or even a derivatized endogenous molecule, but a radiochemical method would not be able to distinguish the difference.
Over the past decade, mass spectrometry (MS) imaging (MSI) has gained considerable interest from the pharmaceutical community. In contrast to QWBA and MARG, MS detects the actual molecules in the image based on their characteristic mass-to-charge ratios (m/z) and does not require the use of labeled compounds. Using MSI, one can distinctly detect and characterize drugs, metabolites, and endogenous drug-modified molecules in their native states within a biological matrix. Furthermore, MSI can be a label-free method. No prior knowledge about target analyte is required, allowing for discovery of unknowns. Additionally, radioactive materials always present potential biological hazard, even under carefully controlled working conditions.
The aim of this review is to highlight the potential and realized benefits of drug and metabolite MSI and identify its practical capabilities and common pitfalls. It is primarily written for readers mostly unfamiliar with MSI, but can also be useful for MSI investigators interested in the latest applications in drug delivery and discovery. Many pharmaceuticals have molecular weights less than or equal to 1000 Da and would be considered small molecules in MS analysis. Therefore, this review will also cover methods for endogenous small molecule MSI that could also be applied to drugs and metabolites.
Many excellent MSI reviews have been published [4-10], and the authors also recommend some informative radiochemical imaging reviews for comparison [1-3]. The following sections will begin with a very brief overview of fundamentals and will then heavily focus on applications in drug discovery and drug delivery. The applications will demonstrate what types of questions can be answered by MSI, and special attention will be paid to the efforts towards developing quantitative MSI. Because an accurate quantitative analysis is vital to ADME, absolute and relative quantification is one of the most sought-after methods in pharmaceutical MSI. Additionally, short sections will be devoted to subcellular MSI and MSI of neuropeptides, two other areas with relevance to pharmaceutical research. Finally, this review will conclude with an outlook on emerging technology and the challenges MSI must overcome to achieve routine analysis in the pharmaceutical sciences.
2. Mass Spectrometry Imaging Fundamentals
2.1. From Molecular Ions to Molecular Image
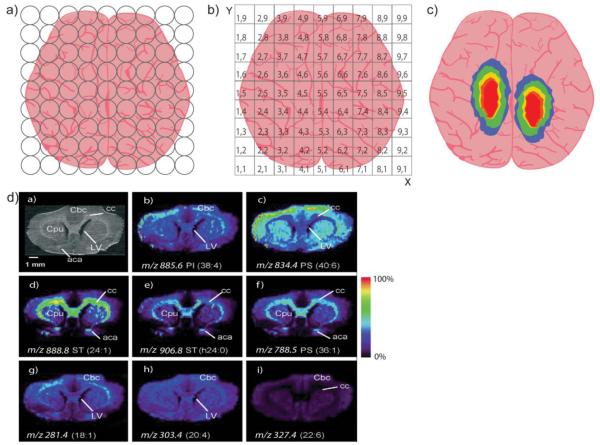
This section of the review will explain the basic MSI workflow and highlight the pertinent fundamentals. Scheme 1 is a graphical representation of an MSI experiment, from creating ions to forming an image. Typically, the first step of an ex vivo MSI experiment involves obtaining a specimen via snap-freezing and cryosectioning, followed by tissue section mounting (Scheme 1A). Spectra are collected from tissue in a raster pattern, creating a grid of points on the tissue where molecules have been ionized and detected according to their m/z. Once MS analysis is complete, each point is converted into a two-dimensional spatial coordinate (Scheme 1B). Finally, an image is constructed by displaying the intensity of a specific m/z at each coordinate (Scheme 1C). Real MSI results are shown in Scheme 1D with an optical image of a rat brain tissue followed by the mapped spatial distributions of eight m/z values belonging to different lipid species [11]. These images were acquired simultaneously, and one could theoretically acquire hundreds of molecular images in one experiment [12].
Scheme 1. Creating and Imaging Ions by Mass Spectrometry.
(a) After tissue is prepared, mass spectra are collected at points across the entire tissue. (b) Each sampled point is converted into spatial coordinates and (c) an image is created by displaying the intensity of a specific ion at each point. (d) Real images of 8 different lipids acquired simultaneously from a rat brain, reprinted with permission from [11].
The acquisition and construction of an image from ionized tissue compounds is often performed by one of two methods. In the first method, the intensity of an ion created directly from tissue is displayed at each sampled point. It is a simple and straightforward approach, but it is easy to imagine that a tissue slice may contain many isomeric or isobaric compounds whose m/z overlap. This is especially true with certain methods that use small molecules to assist in ionization, and these small molecules fall into the mass range of the analyte. Therefore, using tandem mass spectrometry (MS/MS) is a common way to increase confidence in compound identification and improve the dynamic range of MSI [13, 14]. Scheme 2 presents an overview of selected reaction monitoring (SRM), a common MS/MS technique employed for imaging, through an illustrative example of clozapine imaging. After tissue compounds enter the mass spectrometer, the ions with an m/z of (327 ± 2) are isolated during the initial mass analysis. Clozapine and its isobaric species are then fragmented, and a second mass analysis isolates the signature fragment of clozapine (m/z 270.08) and uses its intensity to create an image. The downside to SRM-MSI is that it is difficult to use in a non-targeted study. The parent m/z and fragment m/z must be known a priori.
Scheme 2. Selected Reaction Monitoring.
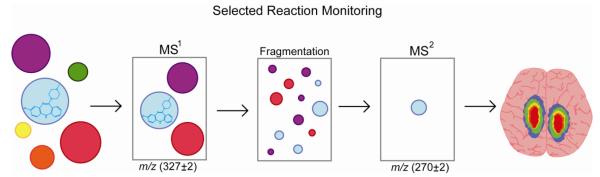
Clozapine (blue) and other compounds have been desorbed and ionized prior to mass analysis. In selected reaction monitoring (SRM), the first round of mass analysis removes all ions outside the mass window of 325 Da – 329 Da that allows isolation of precursor ions for subsequent fragmentation. The remaining ions are then fragmented. For the second round of mass analysis, ions outside of the 268 Da – 272 Da are removed, leaving only the signature fragment of clozapine (blue) to be imaged.
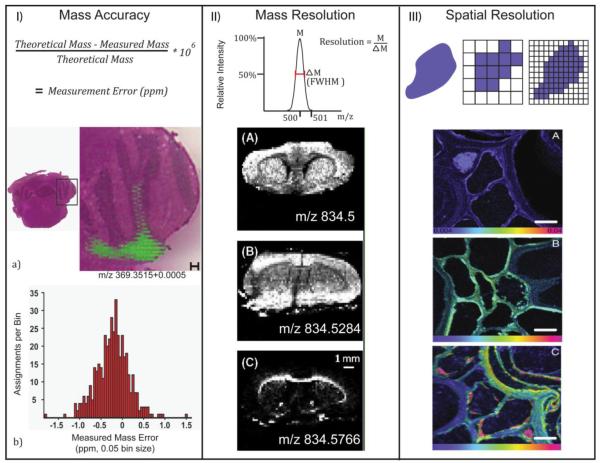
MSI has three primary figures of merit defined in Scheme 3. The first two are from MS: mass accuracy and mass resolution. Mass accuracy refers to the agreement of an ion’s detected mass to its theoretical mass and is often measured in parts-per-million (ppm) [15]. Scheme 3(I) shows an overlaid cholesterol image on mouse brain tissue constructed from ions detected within a window of 0.0005 Daltons (Da) of cholesterol’s theoretical mass, 369.3515 Da [16]. The mass window corresponds to an approximate mass accuracy of 1.4 ppm to reflect the histogram below.
Scheme 3. MSI Figures of Merit.
(I) An image of cholesterol at 1.4 ppm mass accuracy, reprinted with permission [16]. (II) Three lipid images requiring high mass resolution, reprinted with permission [17]. (III) A secondary ion mass spectrometry image of root cells with submicron spatial resolution, reprinted with permission [18].
Mass resolution, or resolving power, describes the minimum difference between two m/z that can be identified as unique ions [15]. One way to calculate mass spectral resolution is to divide a peak’s apex m/z by its full-width at half-maximum (FWHM). In Scheme 3(II), the images of three similar m/z values display very different spatial distributions in a mouse brain [17]. The top brain image is actually the summation of two isobaric lipids, whose unique distributions are shown below. Without mass resolution of at least 29000, they would falsely appear as a single peak and produce a false spatial distribution.
Spatial resolution is a term from microscopy imaging. It refers to the minimum distance between two objects in an image at which they can be distinctly discerned. In MSI, spatial resolution calculation is usually dependent on the type of ionization employed, but one can often use the distance between sampled points. If each pixel in an image represents the entire area of a sampled point, then the minimum distance that must exist between two objects is the center-to-center length between two sample areas. Scheme 3(III) shows time-resolved nitrogen enrichment in Triticum aestivum root cells. These secondary ion mass spectrometry images were acquired with less than 1 μm spatial resolution [18].
Mass accuracy, mass resolution, and spatial resolution are not the only figures of merit that exist for MSI, but they are the most cited figures throughout the literature. The amount of resolution and accuracy needed to produce a quality image depends upon the compounds being imaged and the complexity of the tissue. For example, a drug with an exact mass of 239.1077 Da needs 419 ppm mass accuracy and 2400 mass resolution to be discerned from isobaric molecules with mass differences of 0.1 Da or greater. To be discerned from mass differences of 0.001 Da, 4.2 ppm mass accuracy and nearly mass resolution at 240000 is required. Fourier transform (FT) mass spectrometers are known for parts-per-billion mass accuracy and mass resolution exceeding 1000000 [19, 20], but they can be very expensive and often require longer acquisition times to construct an ion image. Time-of-flight (TOF) instruments have resolving powers ranging from 10000 to 100000 and can routinely achieve less than 5 ppm mass accuracy [21]. Quadrupole and ion trap instruments are usually considered robust “low resolution” instruments with nominal mass resolving powers. However, triple-quadrupole-ion trap instruments have proven to be very powerful in MSI when used for SRM [22]. When the first and third quadrupoles are set to constantly monitor a single transition or a couple of transitions with very short dwell time, the mass spectrometer is effectively turned into an ion counter and can offer unparalleled sensitivity. If the third quadrupole is used as an ion trap, the experiment can alternate between SRM scans, full MS scans, and enhanced product ion scans that can detected many fragmentation transitions from a single ion. This approach can allow nearly simultaneous targeted and non-targeted imaging.
The power of SRM-MSI should not be underestimated. Even using an instrument with the best specified mass accuracy and mass resolution could result in ambiguity for compound identification in a complex sample. Some isobaric compounds might not be resolved even with the use of high resolution FT-MSI, but might be easily imaged and identified using sequence-specific fragmentation via SRM [23]. High mass resolution will do nothing to resolve isomeric compounds with identical m/z, but if the isomers show different fragmentation patterns, SRM-MSI would be able to resolve them. SRM-MSI is most easily performed on an ion trap or any hybrid mass spectrometer containing two mass analyzers and a fragmentation cell (e.g., triple quadrupole, quadrupole-TOF, and TOF-TOF).
While mass accuracy and mass resolution rely heavily on instrumentation, spatial resolution is more dependent on sample preparation and ionization. The following fundamentals section will focus on common methods for sample preparation and MS ionization. Many excellent MSI reviews have covered traditional and novel preparation protocols extensively, so we will only detail the most basic aspects and will refer the readers to cited works for more information.
2.2 Sample Preparation and Ionization
The sacrifice of an animal specimen marks beginning of rapid molecular breakdown. Although MSI of formalin-fixed paraffin-embedded (FFPE) tissue is possible [24], use of freshly extracted tissue immediately frozen at -80 degree Celsius is more common [25]. In a recent study, Sugiura et al. presented a series of acetylcholine images from mouse sagittal brain sections [26]. Even with as short as 1 minute between organ extraction and freezing, the images showed significant postmortem acetylcholine degradation. To mitigate this problem, in-situ freezing (ISF) was performed wherein the head of a deeply anesthetized living specimen was dipped into liquid nitrogen to freeze the brain simultaneously with specimen sacrifice. Other sacrifice and tissue preparation protocols exist to mitigate degradation, and they have shown to be especially useful in neuropeptide imaging. For a further discussion on these methods, refer to section 3.4.
Next, the tissue is ready to be cut into thin sections on cryostat. The thickness of sectioned tissue should be on the order of the organism’s cellular diameter, although other considerations such as sensitivity and ionization method of choice could also play a role. For example, if a particular tissue’s cells have an average diameter of 10-20 μm, the slices should be no greater than 10-20 μm thick [25].
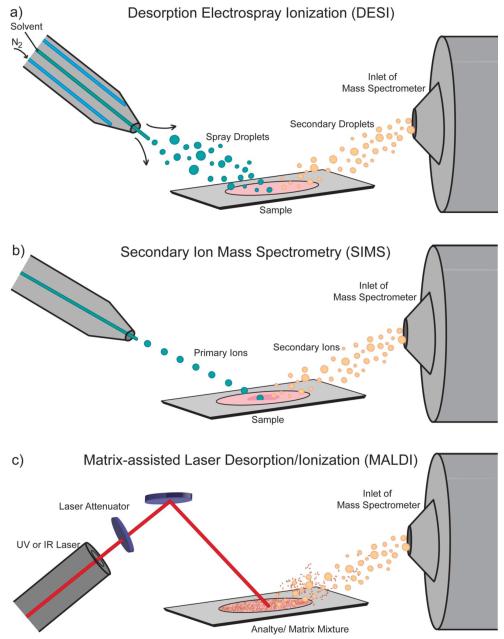
After tissue freezing and sectioning, sample preparation is largely dependent on the ionization source. Scheme 4 provides a visual overview of the three most commonly used sources: secondary ion mass spectrometry (SIMS), matrix-assisted laser desorption/ionization (MALDI), and desorption electrospray ionization (DESI). SIMS is the oldest of the three and was first used for imaging in 1962 by Castaing and Slodzian [27]. The mechanism of ionization is beyond the scope of this review [28], but essentially ions are formed when a focused primary ion beam hits sample and sputters off ionized tissue compounds into the mass spectrometer. SIMS is often limited to analyzing molecules under 1000 Da without significant sensitivity loss or unwanted fragmentation [9]. However, the different types of ion beams can be used to increase the intact ion yield of larger or more labile compounds [16, 29, 30]. SIMS imaging does not require special preparation after sectioning and mounting the tissue, but there are some optional methods that improve the imaging results. Coating the tissue with gold, silver, small organic acids, or the use of gold nanoparticle as substrates on the surface of the sample holder or inserted into the sample can improve the ionization of intact molecules larger than 1000 Da [31-37].
Scheme 4. MSI Ionization Sources.
The three most common sources for MSI: (a) Desorption electrospray ionization which uses a stream of solvent ions to desorb and ionize analyte molecules, (b) secondary ion mass spectrometry which uses a beam of ions from an ion gun to sputter analyte ions off the sample, and (c) matrix-assisted laser desorption/ionization which uses laser irradiation of a matrix-coated sample for desorption and ionization.
SIMS is able to reliably achieve less than 1 μm spatial resolution [38], making it the method of choice for subcellular pharmaceutical investigations. The high lateral spatial resolution and utility SIMS can provide is seen in Scheme 3 (III). If such high spatial resolution is not required, MALDI MSI is a versatile alternative that has been employed for imaging drugs and metabolites [39], lipids [40], peptides [41], and proteins [42]. In MALDI MSI, the tissue is coated with a thin layer of matrix and irradiated with a laser beam. The matrix absorbs much of the energy from the incident laser and provides a very “soft” ionization for analyte compounds [43]. Although MALDI is most commonly performed at high vacuum pressures, it can also be utilized at atmospheric pressure (AP-MALDI) for an even softer process [44]. The mechanism for ion formation is still an active area of study [45, 46], but beyond the scope of this review.
Matrix selection and application is a crucial step for MALDI MSI. It has a great effect on sensitivity, spatial resolution, and selective analyte ionization [25]. Matrices must absorb light at the laser wavelength and must not react with tissue-bound analyte. Imaging with an infrared laser (IR-MALDI-MSI) allows one to use water as a matrix due to high absorptivity in the mid-IR range [47]. Water can also be added for IR laser ablation by freezing the tissue to form a thin layer of frost, as shown by Muddiman and coworkers [48]. More commonly, however, an ultraviolet laser is used (UV-MALDI-MSI) with small, conjugated organic acid matrices. For low-molecular weight pharmaceuticals, CHCA is generally considered the matrix of choice, though far from exclusive use [25]. For example, Jackson et al. showed the utility of a gold nanoparticle matrix to cationize neutral cerebrosides and improve their detection among abundant, positively charged phosphatidylcholines [49]. In a targeted MSI experiment, different matrices can be tested on a standard of the target analyte to determine the optimal choice of matrix.
Generally, matrix application methods are optimized to produce a homogenous coating of small crystals. Inhomogeneities can create local ionization biases that may cause extensive signal suppression [50, 51]. Solvent-based methods prepare matrix dissolved in solution at or near the point of saturation followed by tissue deposition via nebulizers and airbrushes [52] or automated systems [53-55]. Solvent-free methods were developed to circumvent spatial delocalization of soluble analytes from excessive amounts of solvent during matrix application [13], and a myriad of preparation protocols have been developed [56-59]. Solvent-free methods work well for most molecules, but have a noticeable sensitivity drop-off for larger analytes [57].
Until 2006, SIMS and MALDI were the two main pathways for MSI. It was not long after Takats et al. introduced DESI [60] that its imaging capabilities were realized [11]. DESI is a form of ambient ionization method where ions are analyzed from their native matrices with little or no sample preparation [61]. In a DESI ion source, solvent is sprayed through a high-voltage needle at AP and directly onto a sample. Molecules from the sample are ionized and desorbed into a heated capillary leading to a mass spectrometer. Ions are formed from either an electrospray ionization (ESI) mechanism [62] or heterogeneous charge transfer [63, 64]. DESI imaging is mostly used for lipids [11] and small molecules [65].
There is virtually no sample preparation needed for DESI aside from sectioning and mounting the tissue. Additives can be added to the solvent spray to increase selectivity and sensitivity for certain analytes, so-called reactive DESI [66], and has been used by Wu et al. to image cholesterol [67]. Cholesterol is difficult to ionize because of its low proton affinity and low acidity, but the addition of betaine aldehyde to the DESI spray derivatized cholesterol’s OH-group with a permanently charged hemiacetal, thus promoting ionization and detection.
Ionization and sample preparation methods should be selected based on what compound classes and tissue locations are intended for the image. SIMS is best used for ionizing small molecules and sensitive enough to spatially characterize sub-cellular amounts of analyte. However, maintaining such spatial resolution with compounds larger than 1000 Da is difficult. Additionally, commercially available SIMS instruments do not have MS/MS capabilities as MALDI or DESI instruments equipped with hybrid TOF or quadrupole-based mass analyzers, thereby limiting SIMS-SRM-MSI. Nonetheless, SIMS has been shown to make use of post-source decay (PSD) for SRM-MSI with the appropriate analysis software [68]. MALDI is capable of imaging molecules small and large, but often the spatial resolution is limited to 10-30 μm. Ambient DESI is a viable option for high-throughput analysis of pharmaceuticals with minimal tissue adulteration. DESI commonly produces spatial resolutions of 100 μm or greater, but a resolution of 35 μm was reported in a recent study [69].
3. Mass Spectrometry Imaging Applications
3.1 Qualitative MSI
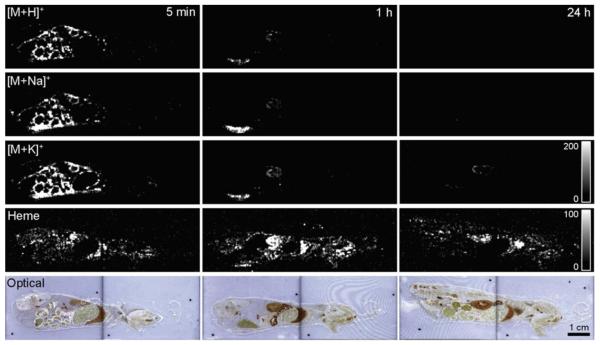
In the initial stages of modern MSI, the focus was largely on characterizing spatial distribution. A study by Stoeckli et al. 2007 on β-peptides gives a basic demonstration of its unique qualitative and semi-quantitative capabilities [70]. In this study, mice were intravenously dosed with peptide solutions, three with β-peptides and a fourth with an α-peptide. The β- peptide dosed mice were sequentially sacrificed after periods of 5 minutes, 1 hour, and 24 hours before being frozen and sectioned. Figure 1 shows the MALDI-MSI of each mouse’s β-peptide spatial distribution. These images can provide some qualitative information about ADME. The peptide is clearly absorbed and distributed throughout several organs, and it stays intact until it reaches the kidney where it is excreted. Additionally, the presence of β-peptide signal after 24 hours provides kinetic information, and the absence of the α-peptide signal after 1 hour indicates the greater metabolic stability of β-peptides, as expected. Of course, Stoeckli stated that similar results, with the added benefit of absolute quantitation, could have been achieved by QWBA. However, MSI provided a quicker image acquisition without the need of costly radioactive labels.
Figure 1. Whole-body MSI of Drug Metabolism.
The distribution of a β-peptide in mice after 5 minutes, 1 hour, and 24 hours. The first three rows show the β-peptide distribution imaged from its three most common ion types. The fourth row shows the image of heme as a molecular reference, and the fifth row shows an optical image of the specimen for a physical reference. Reprinted with permission from [70].
MSI with qualitative emphasis still provides great utility. It can give relative pharmacokinetic (PK) and pharmacodynamic (PD) information, as well as allow the identification of novel metabolites that could help elucidate metabolic mechanisms. Enthaler et al. recently employed MALDI-MSI in a sophisticated method to probe ex vivo compound penetration in human and porcine skin [71]. Although the model compound was a cosmetic, Nile Red, the exact same protocols could be applied to cutaneous drug delivery. A problem with MALDI-MSI of skin samples was adhesion of skin to ITO-coated glass slides. The group attempted to improve adhesion by pre-treating the slides with coronal discharge and modify its surface to have stronger interactions with the proteins in skin. This method allowed them to image skin samples after a Nile Red treatment and a common penetration enhancer, dimethyl sulfoxide, for 24 hours. The 30 μm spatially resolved images showed that Nile Red penetrated past the upper stratum corneum layer and into the epidermis, but did not reach the dermis. Additionally, endogenous cholesterol sulfate, a molecule that regulates protective barrier formation in skin, was also imaged and showed distributions mainly in the epidermis. This example presents a remarkable potential for MSI to not only characterize a drug’s delivery and penetration, but to also identify endogenous molecular distributions that may explain the observed absorption properties.
Although QWBA enables a similar quantitative analysis of radio-labeled Nile Red penetration, MARG would be required to achieve a spatial resolution at 30 μm and limited quantitation can be achieved. Nile Red is amenable to characterization by fluorescence microscopy and Enthaler even used the technique to validate the MSI. However, fluorescent microscopy would not be a label-free alternative for molecules that do not fluoresce.
The utility of pharmaceutical MSI goes beyond direct imaging of tissue. Kreye et al. used MSI to probe the dynamics of controlled theophylline release from lipid implants [72]. Though no actual tissue was imaged, the 4 mm-long cylindrical implants were embedded in gelatin, frozen, cryosectioned, and then sprayed with matrix, nearly analogous to the preparation of tissue. Images of the initial theophylline distribution throughout the implant were created using radial and longitudinal cross-sections. Both views showed macroscopic homogeneity with steep gradients at the micrometer level, to which Kreye attributed to the initial particles of lipid and drug the implants were fabricated from. Very useful information about the mechanism of release was also obtained from radial cross-section images of implants after 0, 3, and 14-day exposures to release media. It is clearly shown that the release is not homogeneous, but starts from the edges of the implant.
Stoeckli, Enthaler, and Kreye performed MSI studies that were able to obtain important information, although they could have been similarly acquired with QWBA. In contrast, Drexler et al. used MSI where QWBA would be less suited for the study of phototoxicity [73]. Phototoxicity assays must identify photoreactivity of a target molecule. Radiochemical methods cannot distinguish a drug from its metabolite and therefore are less applicable to such assays. Drexler used QWBA to first determine if an orally administered proprietary drug would accumulate in tissues in the eye. The radiograms showed a significant presence of the drug in retina where it could possibly react with incident ultraviolet and visible light. MSI of eye tissue indicated that the parent drug was present but could not identify any photochemical derivatizations, therefore providing a negative result for phototoxicity.
Drexler’s phototoxicity assay via MSI may present a clear advantage over QWBA in this particular case, although QWBA offers desired quantitative imaging capability. The next section will discuss quantitative MSI, one of the most innovative and fast-moving area in the field.
3.2 Quantitative MSI
The inherently heterogeneous nature of ionization makes accurate quantitation in MSI challenging. The intensity of an MS signal is not only related to an analyte’s concentration, but to its ionization efficiency and environmental extractability as well. Heeren et al. discussed this issue at length in a critical insight article [74]. A simple MSI experiment was presented wherein three aqueous protein digests; ubiquitin, cytochrome C, and bovine serum albumin; were spotted and mixed at equal concentrations. The spots were imaged by MALDI-MSI. Spots containing only ubiquitin and cytochrome C in the mixture were able to display peptides from both proteins, but whenever the mixture contained albumin, only the albumin peptides were detected. As evident in this example, certain molecules are preferentially ionized and can “steal” signal from co-localized species.
The complex tissue environment could make quantitation difficult. Luxembourg et al. showed that variations in tissue salt concentration and histological features will create heterogeneous matrix crystallization [51]. The variations in crystals may cause different degrees of tissue desorption and recovery of the analyte, and they may also result in different degrees of fragmentation during desorption and ionization. The end result is a signal that is more reflective of the tissue histology and microenvironment than of the analyte concentration.
MSI will require accurate means of quantification if it is ever to become an acceptable alternative to QWBA. ADME characterization of drugs seeks to answer a question of mass balance, and simply imaging the relative locations cannot give a satisfactory answer. The absolute amount must be known throughout the distribution to provide true pharmacokinetic and pharmacodynamics data, and such knowledge can also identify unaccounted for drug that just may not show up in an image.
In 2010, Hattori et al. investigated the metabolism of ATP in ischemic penumbra mice models [75]. Because the study required quantitative imaging that could clearly distinguish ATP from its metabolites, they had to find a way to merge MSI with another quantitative method. The solution was to extract a contralateral brain slice and perform quantitative analysis with capillary electrophoresis (CE)-ESI-MS/MS. The following equation was used to calculate concentrations of molecules in the image:
The tissue concentration of a specific area (Ci) is equal to ratio of the maximum intensity of that area (Ii) and the median maximum intensities from the contralateral hemisphere (I) multiplied by the whole tissue concentration (C’) determined by CE-ESI-MS/MS. Hattori et al. validated their method by comparison to previous spatial metabolic results from a radiochemical method [76]. Although Hattori’s approach proved powerful and accurate, the difficulties of coupling CE to ESI-MS/MS lead others to an analogous liquid chromatography (LC) approach developed by Koeniger et al. [77]
Under certain circumstances, quantitative MSI can be performed by creating an on-tissue calibration curve. Figure 2 from Nilsson et al. shows the distribution of tiotropium in rat lung tissue [78]. The adjacent tissue is a piece of control lung upon which known concentrations of tiotropium were spotted and imaged. If the tissue was fairly morphologically homogeneous, this could account for the universal response and extractability of the tiotropium. The calibration curves show the linearity of normalized intensities from the on-tissue calibration compared with LC-MS/MS quantitation of dosed lung tissue. The two curves are in strong agreement and provide a proof-of-principle for label-free on-tissue quantitative MSI.
Figure 2. On-Tissue Calibration Curve.
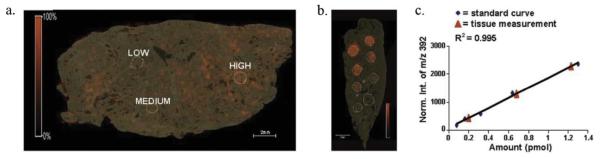
(a) Absolute quantitation of tiotropium on lung tissue shows areas that contain 0.20 pmol (low), 0.65 pmol (medium), and 1.25 pmol (high). (b) Signals from standard spotted on tissue were matched (c) with normalized signals from the LC-ESI-MS calibration. The red triangles show the linear response of the on-tissue MALDI standards and the blue diamonds show the linear response of corresponding LC-ESI-MS standards. Reprinted with permission from [78].
The Nilsson study is impressive but only applicable to compounds that do not suffer from signal suppression at any place on a piece of tissue. The recent push in quantitative MSI is to add isotopically labeled internal standards (IS) to the tissue. Unlike radiochemical labels, stable isotopes such as 13C, 15N, 18O, or 2H are used. Pirman and Yost demonstrated the effectiveness of this method with the absolute quantification of endogenous acetyl-L-carnitine (AC) in pig brain [79]. A glass microscope slide was homogenously spotted with d3-AC and sectioned pig brain was subsequently mounted on top. The image was made from the ratio of signature MS/MS fragment ions from the endogenous compound and the isotope-encoded internal standard. It is assumed that co-localized isotopologues will have identical ionization efficiencies, extractabilities, and MS/MS fragmentation behaviors, therefore allowing direct quantitative comparison between a target analyte’s MS intensity in tissue sample and the heavy standard’s MS intensity.
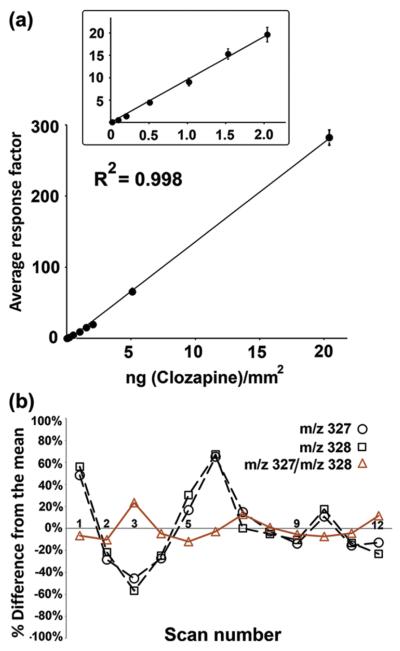
Vismeh et al. used DESI MSI with similar quantitative methods for absolute quantification of clozapine distributions [80]. Rats were dosed with clozapine before sacrifice and a deuterated standard was pipetted on top of the sectioned tissue. Figure 3 shows the calibration curve of the clozapine ratios on tissue. In addition to the high degree of linearity in calibration curve, it is interesting to note the superior stability of the ratio of intensities compared to the stability of clozapine or IS alone.
Figure 3. Absolute Quantitation with Internal Standards.
(a) The black circles show the linearly increasing response of the intensity ratio between clozapine (m/z 327) and d-clozapine (m/z 328) at standard concentrations spotted on tissue. (b) When scanning across a constant concentration of 0.3 ng/mm2, the MS signal is more stable with the clozapine/d-clozapine ratio than with either molecule alone. Reprinted with permission from [80].
The preliminary quantitative results from the last couple of years have created a lot of excitement in the field. There is a great potential that MSI will one day become a common tool for mass balance and ADME assays. Further refinement and biological application validation remain an active area of research.
3.3 MSI At and Below the Cellular Scale
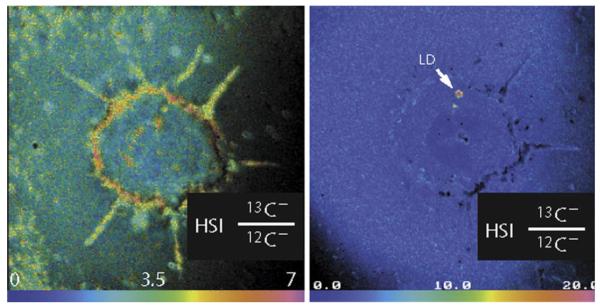
Molecular imaging at the cellular scale can offer critical details on the mechanisms, dynamics, and kinetics of drug delivery. The increased resolution brings the experiment from identifying drugs that interact with specific organs to drugs that interact with specific cell types. High-performance MSI can even elucidate specific organelles or cellular regions that are crucial to a drug’s delivery. Figure 4 shows SIMS images of a single cultured adipocyte at 33 nm spatial resolution [81]. Cells were incubated with 13C-labeled oleic acid to investigate how free fatty acid was utilized and metabolized. Localization of the oleic acid will result in an increase of 13C relative to 12C, and so an image of the 13C / 12C ratio will identify all areas where it has been integrated. The first image shows that a significant proportion of oleic acid localized in the lipid-rich cell membrane. A second image at a different depth of the same cell shows a very high concentration of oleic acid as part of a lipid droplet inside of the cell.
Figure 4. Cellular Fatty Acid Metabolism.
(a) A single adipocyte cell shows incorporation of 13C into the cell membrane after incubation with 13C-labeled oleic acid, as well as incorporation into (b) intracellular lipid droplets. Reprinted with permission from [81].
Sensitivity becomes increasingly important in sub-cellular MSI. With less total material in each sample point, fewer ions will be created. Ion microscopy is a technique that aids sensitivity by using detectors that are sensitive to an ion’s 2-D position [82]. As such, the spatial resolution becomes independent of sampling size. Chandra et al. used SIMS microscopy to make quantitative images of neutron-capture therapy drug delivery in cultured human glioblastoma cells [83]. 10B-labeled p-boronophenylalanine-fructose (BPA-F) and 11B-labeled sodium borocaptate (BSH) were introduced into the culture medium in two separate experiments, one with separate introduction of each drug and one with simultaneous introduction. The overall goal of the experiment was to determine whether there was a synergistic effect on the delivery of either drug during co-administration. The boron drugs seemed to homogenously penetrate all parts of the cell except the perinuclear cytoplasm, and the authors concluded that BSH and BPA-F show only additive boron delivery when co-administered to the cell media.
Altelaar et al. published a protocol paper that describes two approaches for MALDI MSI of cellular dimensions [84]. Ion microscopy is one such approach, with the other approach being the scanning microprobe MALDI (SMALDI). Altelaar demonstrates that both methods can attain high-resolution images of whole rat brain tissue, but SMALDI may require unfavorably high laser fluence. SMALDI involves using very small laser beam diameters and distances between sampling points, about 0.7 μm and 0.25 μm respectively [85]. Astonishingly, even though SMALDI ablates such little material into the mass spectrometer, peptide standards have been detected at attomole and zeptomole levels in a single 1 μm × 1 μm pixel. An investigation by Bouschen et al. used SMALDI on human carcinoma cells and peptide mixtures to produce images with an effective spatial resolution of 2 μm [86].
Very recently, the use of alternative laser ablation geometry has demonstrated the ability for subcellular MALDI imaging without a microprobe or ion microscopy. As shown in Scheme 4C, the incident MALDI irradiation commonly strikes the front side of the sample holder in what is known as reflection geometry (RG). If the sample holder is made of transparent material that will not absorb the laser wavelength, the incident radiation can instead strike from the back at 1 angle from the sample in what is known as transmission geometry (TG). TG-MSI was first used by Richards et al. to image sulfatide lipids [87] and fragile gangliosides [88]. Although the images were acquired at 10 – 20 μm spatial resolution, single shot ablation in TG allowed images of the entire mouse brain to be created in less than an hour, a substantial reduction of imaging time compared to the use of conventional RG mode. Zavalin et al. published a proof-of-principle experiment for TG-MALDI-MSI of single cells [89]. Lipids are shown localized in the cell membranes of HEK-293 cells in images that were acquired with a 1 μm laser diameter and 1.5 μm center-to-center sampling points. Although no exogenous compounds were imaged at subcellular resolution, there is no reason to suspect it would not be possible.
3.4 Neuropeptide Imaging: Mapping Endogenous Signaling Molecules and Drug Delivery
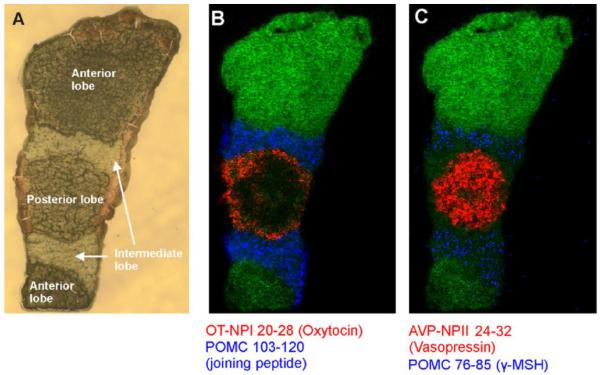
Organisms use neuropeptides and other signaling peptides to regulate a great variety of physiological processes. These endogenous molecules can be templates for synthetic drug development and delivery, and peptide-based pharmaceuticals have been of great interest for decades. Neuropeptides and structural analogs can also be of use for drug discovery and delivery as there is great interest in peptide-based pharmaceuticals. Neuropeptides and other signaling peptides are commonly studied by MS with workflows that often include MSI at the whole tissue scale all the way down to single neurons [90-92]. But even at the tissue level, high spatial resolution is important. Tissues rich in neuropeptides, such as the pituitary gland, can have dimensions of just a few millimeters. Geunther et al. obtained astonishing AP-MALDI-MS images of neuropeptides in a mouse pituitary [93]. Their results suggest that new MALDI-MSI technology is leading the method to a mature level that can probe very specific tissue-peptide interactions. Figure 5 shows an optical image of a pituitary gland followed by molecular images of four peptides. The pituitary gland was measured at dimensions of 3 mm by 1 mm. With 5 μm spatial resolution, oxytocin and vasopressin were primarily localized in the posterior lobe while a joining peptide and γ-MSH were localized in the intermediate lobe.
Figure 5. Neuropeptide MSI of a Mouse Pituitary Gland.
(a) An optical image of a pituitary shows that the localization of (b) oxytocin (red), joining peptide (blue), (c) vasopressin (red), and γ-MSH (blue) can be segregated to specific lobes. Reprinted with permission [93].
In a study performed by Hui et al., a neuropeptide workflow included MALDI-MSI localization to aid the functional study of a novel tachykinin neuropeptide [94]. Brains from Callinectes sapidus were imaged to show the distributions and expression levels of two peptides, CalsTRP and CabTRP Ia, between fed and unfed animals. Both peptides showed consistent upregulation and co-localization in fed animals, with CabTRP Ia at a higher intensity than CalsTRP. The combined evidence of the co-regulation, co-localization, and nearly identical amino acid sequences led the investigators to posit that both neuropeptides may originate from the same preprotachykinin. Additionally, the higher intensity of CabTRP Ia may indicate which sequence from the preprotachykinin is preferentially expressed, although the authors concede it may also be a result of CalsTRP having a higher post-mortem turnover.
Post-mortem protease activity is a significant problem for neuropeptide imaging. In fact, early MS neuropeptide studies suggested that more than 90% of detected peptides were non-active fragments from protein degradation [95]. On-tissue protease deactivation has been successfully achieved by raising the tissue temperature to a level that deactivates most enzymes. Microwave irradiation, either applied as a focused beam for animal sacrifice [96] or to tissues post-sacrifice in a microwave oven [97], has dramatically increased the detected post-mortem levels of neuropeptides and small molecules such as cyclic-AMP and arachidonic acid. Alternatively, the commercially available Denator AB that uses a combination of pressure and heat to quickly denature enzymes immediately following extraction [98]. MALDI-MSI of heat-treated tissues by Goodwin et al. shows that the Denator heating at 95 C significantly reduced protein turnover [99]. However, the stabilization process did cause detrimental damage to tissue morphology which would complicate MSI. The authors conclude that heat-treated tissue in MSI would be best utilized as complementary sample source for MSI from intact structures of non-treated tissue. Microwave irradiation may also cause inconsistencies detrimental to imaging such as the differential heating and enzyme inactivation of certain areas in a tissue [96].
4. Emerging Technology, Perspectives, and Conclusions
After nearly a decade and a half of extensive research efforts, MSI is finally approaching a stage of more widespread applications. Qualitative molecular imaging is now a rather straight-forward process, and even more novel approaches are being developed, such as three-dimensional MSI that simultaneously maps lateral distribution and depth penetration within tissues [100, 101]. However, MSI has not reached universal acceptance in pharmaceutical science due to remaining challenges that must be overcome.
The importance of robust and accurate quantitation for drug and metabolite MSI cannot be underemphasized. The proof-of-principle quantitative MSI has certainly been demonstrated. It is possible under some circumstances to detect and localize drugs with similar sensitivity to QWBA, and may even surpass radiochemical quantitation at cellular and subcellular scales. However, QWBA excels in its reproducibility and robustness. If a radio-labeled compound is present, it will produce a signal. MSI compounds, on the other hand, may be present but undetectable due to matrix ionization suppression, extractability, or analyte degradation. Using a stable isotope-labeled IS can account for suppression and extractability and is likely well-adopted in future quantitative MSI. More methods to improve reproducibility may also emerge, and biggest factor may just be giving MSI time to be employed in novel biological applications.
The sheer complexity of biological tissues creates challenges for MSI. Front-end separation would have the biggest impact on dynamic range, and some recently developed liquid extraction (LE) methods have made it possible to separate compounds from tissue microsections using LC [102, 103]. Small liquid junctions can make contact with the surface of the tissue and collect local molecules to be separated by LC, effectively decoupling analyte extraction from ionization. Although some LE methods have shown spatial resolution near 30 μm [104], most of them produce localization which spatial resolution well in excess of 100 μm.
A possibility that will not affect ionization suppression but that can deconvolute MSI spectra is to employ ion mobility (IM) separation, which separates ions in the gas phase based on shape and size [105]. IM-MSI can differentiate structural metabolite isomers [106] and separate isobaric molecular classes like lipids and peptides prior to MS analysis [49, 59, 107]. On-tissue derivatization techniques can help to bring compound signals out of the tissue background noise, as shown by Manier et al. when imaging isoniazid, an anti-tuberculosis drug [108]. The SRM transitions displayed by a neat sample of isoniazid were observed from isobaric compounds in control tissue. However, when tissue-bound isoniazid reacted with glass slides precoated with trans-cinnamaldehyde, the resulting drug derivative had SRM transitions unique to dosed tissue.
With a single MS image containing 5,000 to 50,000 spectra, statistical analyses of MSI presents its own significant challenges [109]. Pixel-to-pixel variation, an unavoidable MSI phenomenon, is chief among them. Pre-processing methods aimed at de-noising spectra or clustering pixels can help mitigate the variations, but a sound physical model of the origin of variation would certainly lead to more effective algorithms [110]. Additionally, Thiele et al. have demonstrated that performing in vivo MRI before MSI can help match features in the MS image to anatomical structures in the tissue, especially in the construction of a 3-D MS image [111], highlighting advantages of multi-modality imaging approaches.
McDonnell et al. published a very insightful review that addresses perhaps the most important issue regarding the spread of MSI technology [112]. The overall reproducibility of MSI can be greatly improved by increasing the accessibility of the method and creating well-defined standardized practices. As stated by McDonnell, participants at a 2009 MSI training course revealed that matrix deposition protocols varied greatly even when the same commercial deposition device was being used. European countries have since established an MSI network, COST Action BM1104 that aims to create best-practice guidelines and method standards, as well as provide resources and hands-on training. If other communities are quick to follow this example, the time before universal acceptance of MSI by pharmaceutical science will be drastically reduced.
Acknowledgements
Preparation of this manuscript was supported in part by National Science Foundation (CHE-0957784) and National Institutes of Health through grant 1R01DK071801. C.L. acknowledges an NIH-supported Chemistry Biology Interface Training Program Predoctoral Fellowship (grant number T32-GM008505). L.L. acknowledges an H. I. Romnes Faculty Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Solon EG. Use of Radioactive Compounds and Autoradiography to Determine Drug Tissue Distribution. Chem Res Toxicol. 2012;25:543–555. doi: 10.1021/tx200509f. [DOI] [PubMed] [Google Scholar]
- [2].Solon EG, Schweitzer A, Stoeckli M, Prideaux B. Autoradiography, MALDI-MS, and SIMS-MS Imaging in Pharmaceutical Discovery and Development. Aaps J. 2010;12:11–26. doi: 10.1208/s12248-009-9158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Roffey SJ, Obach RS, Gedge JI, Smith DA. What is the objective of the mass balance study? A retrospective analysis of data in animal and human excretion studies employing radiolabeled drugs. Drug Metab Rev. 2007;39:17–43. doi: 10.1080/03602530600952172. [DOI] [PubMed] [Google Scholar]
- [4].Greer T, Sturm R, Li LJ. Mass spectrometry imaging for drugs and metabolites. J Proteomics. 2011;74:2617–2631. doi: 10.1016/j.jprot.2011.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Seeley EH, Caprioli RM. MALDI imaging mass spectrometry of human tissue: method challenges and clinical perspectives. Trends Biotechnol. 2011;29:136–143. doi: 10.1016/j.tibtech.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chaurand P. Imaging mass spectrometry of thin tissue sections: a decade of collective efforts. J Proteomics. 2012 doi: 10.1016/j.jprot.2012.04.005. [DOI] [PubMed] [Google Scholar]
- [7].McDonnell LA, Heeren RM. Imaging mass spectrometry. Mass spectrometry reviews. 2007;26:606–643. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- [8].Hsieh Y, Chen J, Korfmacher WA. Mapping pharmaceuticals in tissues using MALDI imaging mass spectrometry. Journal of Pharmacological and Toxicological Methods. 2007;55:193–200. doi: 10.1016/j.vascn.2006.06.004. [DOI] [PubMed] [Google Scholar]
- [9].van Hove ERA, Smith DF, Heeren RMA. A concise review of mass spectrometry imaging. J Chromatogr A. 2010;1217:3946–3954. doi: 10.1016/j.chroma.2010.01.033. [DOI] [PubMed] [Google Scholar]
- [10].Castellino S, Groseclose MR, Wagner D. MALDI imaging mass spectrometry: bridging biology and chemistry in drug development. Bioanalysis. 2011;3:2427–2441. doi: 10.4155/bio.11.232. [DOI] [PubMed] [Google Scholar]
- [11].Wiseman JM, Ifa DR, Song QY, Cooks RG. Tissue imaging at atmospheric pressure using desorption electrospray ionization (DESI) mass spectrometry. Angew Chem Int Edit. 2006;45:7188–7192. doi: 10.1002/anie.200602449. [DOI] [PubMed] [Google Scholar]
- [12].Seeley EH, Caprioli RM. Molecular imaging of proteins in tissues by mass spectrometry. P Natl Acad Sci USA. 2008;105:18126–18131. doi: 10.1073/pnas.0801374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Troendle FJ, Reddick CD, Yost RA. Detection of pharmaceutical compounds in tissue by matrix-assisted laser desorption/ionization and laser desorption/chemical ionization tandem mass spectrometry with a quadrupole ion trap. J Am Soc Mass Spectr. 1999;10:1315–1321. [Google Scholar]
- [14].Rohner TC, Staab D, Stoeckli M. MALDI mass spectrometric imaging of biological tissue sections. Mech Ageing Dev. 2005;126:177–185. doi: 10.1016/j.mad.2004.09.032. [DOI] [PubMed] [Google Scholar]
- [15].Gross JH. Mass Spectrometry. Springer; Berlin: 2004. [Google Scholar]
- [16].Smith DF, Robinson EW, Tolmachev AV, Heeren RM, Pasa-Tolic L. C60 secondary ion Fourier transform ion cyclotron resonance mass spectrometry. Analytical chemistry. 2011;83:9552–9556. doi: 10.1021/ac2023348. [DOI] [PubMed] [Google Scholar]
- [17].Manicke NE, Dill AL, Ifa DR, Cooks RG. High-resolution tissue imaging on an orbitrap mass spectrometer by desorption electrospray ionization mass spectrometry. J Mass Spectrom. 2010;45:223–226. doi: 10.1002/jms.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Clode PL, Kilburn MR, Jones DL, Stockdale EA, Cliff JB, Herrmann AM, Murphy DV. In Situ Mapping of Nutrient Uptake in the Rhizosphere Using Nanoscale Secondary Ion Mass Spectrometry. Plant Physiol. 2009;151:1751–1757. doi: 10.1104/pp.109.141499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Marshall AG, Hendrickson CL, Jackson GS. Fourier transform ion cyclotron resonance mass spectrometry: A primer. Mass spectrometry reviews. 1998;17:1–35. doi: 10.1002/(SICI)1098-2787(1998)17:1<1::AID-MAS1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- [20].Hu QZ, Noll RJ, Li HY, Makarov A, Hardman M, Cooks RG. The Orbitrap: a new mass spectrometer. J Mass Spectrom. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- [21].Glauser G, Veyrat N, Rochat B, Wolfender JL, Turlings TC. Ultra-high pressure liquid chromatography-mass spectrometry for plant metabolomics: A systematic comparison of high-resolution quadrupole-time-of-flight and single stage Orbitrap mass spectrometers. J Chromatogr A. 2012 doi: 10.1016/j.chroma.2012.12.009. [DOI] [PubMed] [Google Scholar]
- [22].Hopfgartner G, Varesio E, Stoeckli M. Matrix-assisted laser desorption/ionization mass spectrometric imaging of complete rat sections using a triple quadrupole linear ion trap. Rapid Commun Mass Sp. 2009;23:733–736. doi: 10.1002/rcm.3934. [DOI] [PubMed] [Google Scholar]
- [23].Stoeckli M, Staab D, Schweitzer A. Compound and metabolite distribution measured by MALDI mass spectrometric imaging in whole-body tissue sections. Int J Mass Spectrom. 2007;260:195–202. [Google Scholar]
- [24].Casadonte R, Caprioli RM. Proteomic analysis of formalin-fixed paraffin-embedded tissue by MALDI imaging mass spectrometry. Nat Protoc. 2011;6:1695–1709. doi: 10.1038/nprot.2011.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Schwartz SA, Reyzer ML, Caprioli RM. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J Mass Spectrom. 2003;38:699–708. doi: 10.1002/jms.505. [DOI] [PubMed] [Google Scholar]
- [26].Sugiura Y, Zaima N, Setou M, Ito S, Yao I. Visualization of acetylcholine distribution in central nervous system tissue sections by tandem imaging mass spectrometry. Anal Bioanal Chem. 2012;403:1851–1861. doi: 10.1007/s00216-012-5988-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Castaing R, Slodzian G. Microanalyse par émission ionique secondaire. J. Microsc. 1962;1:395–410. [Google Scholar]
- [28].Pachuta SJ, Cooks RG. Mechanisms in Molecular Sims. Chem Rev. 1987;87:647–669. [Google Scholar]
- [29].Touboul D, Halgand F, Brunelle A, Kersting R, Tallarek E, Hagenhoff B, Laprevote O. Tissue molecular ion imaging by gold cluster ion bombardment. Analytical chemistry. 2004;76:1550–1559. doi: 10.1021/ac035243z. [DOI] [PubMed] [Google Scholar]
- [30].Ostrowski SG, Szakal C, Kozole J, Roddy TP, Xu JY, Ewing AG, Winograd N. Secondary ion MS imaging of lipids in picoliter vials with a buckminsterfullerene ion source. Analytical chemistry. 2005;77:6190–6196. doi: 10.1021/ac0508189. [DOI] [PubMed] [Google Scholar]
- [31].Keune K, Boon JJ. Enhancement of the static SIMS secondary ion yields of lipid moieties by ultrathin gold coating of aged oil paint surfaces. Surf Interface Anal. 2004;36:1620–1628. [Google Scholar]
- [32].Nygren H, Malmberg P, Kriegeskotte C, Arlinghaus HF. Bioimaging TOF-SIMS: localization of cholesterol in rat kidney sections. Febs Lett. 2004;566:291–293. doi: 10.1016/j.febslet.2004.04.052. [DOI] [PubMed] [Google Scholar]
- [33].Kim YP, Oh E, Shon HK, Moon DW, Lee TG, Kim HS. Gold nanoparticle-enhanced secondary ion mass spectrometry and its bio-applications. Appl Surf Sci. 2008;255:1064–1067. [Google Scholar]
- [34].Delcorte A, Bour J, Aubriet F, Muller JF, Bertrand P. Sample metallization for performance improvement in desorption/ionization of kilodalton molecules: Quantitative evaluation, imaging secondary ion MS, and laser ablation. Analytical chemistry. 2003;75:6875–6885. doi: 10.1021/ac0302105. [DOI] [PubMed] [Google Scholar]
- [35].Wu KJ, Odom RW. Matrix-enhanced secondary ion mass spectrometry: A method for molecular analysis of solid surfaces. Analytical chemistry. 1996;68:873–882. doi: 10.1021/ac950717i. [DOI] [PubMed] [Google Scholar]
- [36].Szymczak W, Wittmaack K. Effect of water treatment on analyte and matrix ion yields in matrix-assisted time-of-flight secondary ion mass spectrometry: the case of insulin in and on hydroxycinnamic acid. Rapid Commun Mass Sp. 2002;16:2025–2033. doi: 10.1002/rcm.821. [DOI] [PubMed] [Google Scholar]
- [37].Adriaensen L, Vangaever F, Lenaerts J, Gijbels R. Matrix-enhanced secondary ion mass spectrometry: the influence of MALDI matrices on molecular ion yields of thin organic films. Rapid Commun Mass Sp. 2005;19:1017–1024. [Google Scholar]
- [38].ToF SIMS: Surface analysis by mass spectrometry. IM Publications and Surface Spectra Limited; Chichester, Manchester: 2001. [Google Scholar]
- [39].Shanta SR, Kim Y, Kim YH, Kim KP. Application of MALDI Tissue Imaging of Drugs and Metabolites: A New Frontier for Molecular Histology. Biomol Ther. 2011;19:149–154. [Google Scholar]
- [40].Goto-Inoue N, Hayasaka T, Zaima N, Setou M. Imaging mass spectrometry for lipidomics. Bba-Mol Cell Biol L. 2011;1811:961–969. doi: 10.1016/j.bbalip.2011.03.004. [DOI] [PubMed] [Google Scholar]
- [41].Chen RB, Li LJ. Mass spectral imaging and profiling of neuropeptides at the organ and cellular domains. Anal Bioanal Chem. 2010;397:3185–3193. doi: 10.1007/s00216-010-3723-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chaurand P, Norris JL, Cornett DS, Mobley JA, Caprioli RM. New developments in profiling and imaging of proteins from tissue sections by MALDI mass spectrometry. J Proteome Res. 2006;5:2889–2900. doi: 10.1021/pr060346u. [DOI] [PubMed] [Google Scholar]
- [43].Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y, Yoshida T, Matsuo T. Protein and polymer analyses up to m/z 100000 by laser ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 1988;2:151–153. [Google Scholar]
- [44].Laiko VV, Baldwin MA, Burlingame AL. Atmospheric pressure matrix-assisted laser desorption/ionization mass spectrometry. Anal Chem. 2000;72:652–657. doi: 10.1021/ac990998k. [DOI] [PubMed] [Google Scholar]
- [45].Karas M, Kruger R. Ion formation in MALDI: The cluster ionization mechanism. Chem Rev. 2003;103:427–439. doi: 10.1021/cr010376a. [DOI] [PubMed] [Google Scholar]
- [46].Knochenmuss R, Zenobi R. MALDI ionization: The role of in-plume processes. Chem Rev. 2003;103:441–452. doi: 10.1021/cr0103773. [DOI] [PubMed] [Google Scholar]
- [47].Vertes A, Nemes P, Shrestha B, Barton AA, Chen ZY, Li Y. Molecular imaging by Mid-IR laser ablation mass spectrometry. Appl Phys a-Mater. 2008;93:885–891. [Google Scholar]
- [48].Robichaud G, Barry JA, Garrard KP, Muddiman DC. Infrared Matrix-Assisted Laser Desorption Electrospray Ionization (IR-MALDESI) Imaging Source Coupled to a FT-ICR Mass Spectrometer. J Am Soc Mass Spectrom. 2012 doi: 10.1007/s13361-012-0505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Jackson SN, Ugarov M, Egan T, Post JD, Langlais D, Schultz J. Albert, Woods AS. MALDI-ion mobility-TOFMS imaging of lipids in rat brain tissue. J Mass Spectrom. 2007;42:1093–1098. doi: 10.1002/jms.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Knochenmuss R, Karbach V, Wiesli U, Breuker K, Zenobi R. The matrix suppression effect in matrix-assisted laser desorption/ionization: Application to negative ions and further characteristics. Rapid Commun Mass Sp. 1998;12:529–534. [Google Scholar]
- [51].Luxembourg SL, McDonnell LA, Duursma MC, Guo XH, Heeren RMA. Effect of local matrix crystal variations in matrix-assisted ionization techniques for mass spectrometry. Analytical chemistry. 2003;75:2333–2341. doi: 10.1021/ac026434p. [DOI] [PubMed] [Google Scholar]
- [52].Wang HYJ, Post SNJJ, Woods AS. A minimalist approach to MALDI imaging of glycerophospholipids and sphingolipids in rat brain sections. Int J Mass Spectrom. 2008;278:143–149. doi: 10.1016/j.ijms.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Baluya DL, Garrett TJ, Yost RA. Automated MALDI matrix deposition method with inkjet printing for imaging mass spectrometry. Analytical chemistry. 2007;79:6862–6867. doi: 10.1021/ac070958d. [DOI] [PubMed] [Google Scholar]
- [54].Yamada Y, Hidefumi K, Shion H, Oshikata M, Haramaki Y. Distribution of chloroquine in ocular tissue of pigmented rat using matrix-assisted laser desorption/ionization imaging quadrupole time-of-flight tandem mass spectrometry. Rapid Commun Mass Sp. 2011;25:1600–1608. doi: 10.1002/rcm.5021. [DOI] [PubMed] [Google Scholar]
- [55].Schuerenberg M, Luebbert C, Deininger SO, Ketterlinus R, Suckau D. MALDI tissue imaging: mass spectrometric localization of biomarkers in tissue slices. Nat Methods. 2007;4:iii–iv. [Google Scholar]
- [56].Grove KJ, Frappier SL, Caprioli RM. Matrix pre-coated MALDI MS targets for small molecule imaging in tissues. J Am Soc Mass Spectrom. 2011;22:192–195. doi: 10.1007/s13361-010-0013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hankin JA, Barkley RM, Murphy RC. Sublimation as a method of matrix application for mass spectrometric imaging. J Am Soc Mass Spectr. 2007;18:1646–1652. doi: 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Puolitaival SM, Burnum KE, Cornett DS, Caprioli RM. Solvent-free matrix dry-coating for MALDI Imaging of phospholipids. J Am Soc Mass Spectr. 2008;19:882–886. doi: 10.1016/j.jasms.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Trimpin S, Herath TN, Inutan ED, Wager-Miller J, Kowalski P, Claude E, Walker JM, Mackie K. Automated Solvent-Free Matrix Deposition for Tissue Imaging by Mass Spectrometry. Analytical chemistry. 2010;82:359–367. doi: 10.1021/ac902065u. [DOI] [PubMed] [Google Scholar]
- [60].Takats Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306:471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- [61].Alberici RM, Simas RC, Sanvido GB, Romao W, Lalli PM, Benassi M, Cunha IBS, Eberlin MN. Ambient mass spectrometry: bringing MS into the “real world”. Anal Bioanal Chem. 2010;398:265–294. doi: 10.1007/s00216-010-3808-3. [DOI] [PubMed] [Google Scholar]
- [62].Dole M, Mack LL, Hines RL. Molecular Beams of Macroions. J Chem Phys. 1968;49:2240. [Google Scholar]
- [63].Cooks RG, Jo SC, Green J. Collisions of organic ions at surfaces. Appl Surf Sci. 2004;231:13–21. [Google Scholar]
- [64].Takats Z, Wiseman JM, Cooks RG. Ambient mass spectrometry using desorption electrospray ionization (DESI): instrumentation, mechanisms and applications in forensics, chemistry, and biology. J Mass Spectrom. 2005;40:1261–1275. doi: 10.1002/jms.922. [DOI] [PubMed] [Google Scholar]
- [65].Wiseman JM, Ifa DR, Zhu YX, Kissinger CB, Manicke NE, Kissinger PT, Cooks RG. Desorption electrospray ionization mass spectrometry: Imaging drugs and metabolites in tissues. P Natl Acad Sci USA. 2008;105:18120–18125. doi: 10.1073/pnas.0801066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Cotte-Rodriguez I, Takats Z, Talaty N, Chen HW, Cooks RG. Desorption electrospray ionization of explosives on surfaces: Sensitivity and selectivity enhancement by reactive desorption electrospray ionization. Analytical chemistry. 2005;77:6755–6764. doi: 10.1021/ac050995+. [DOI] [PubMed] [Google Scholar]
- [67].Wu C, Ifa DR, Manicke NE, Cooks RG. Rapid, direct analysis of cholesterol by charge labeling in reactive desorption electrospray ionization. Anal Chem. 2009;81:7618–7624. doi: 10.1021/ac901003u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Touboul D, Brunelle A, Laprevote O. Structural analysis of secondary ions by post-source decay in time-of-flight secondary ion mass spectrometry. Rapid communications in mass spectrometry : RCM. 2006;20:703–709. doi: 10.1002/rcm.2362. [DOI] [PubMed] [Google Scholar]
- [69].Campbell DI, Ferreira CR, Eberlin LS, Cooks RG. Improved spatial resolution in the imaging of biological tissue using desorption electrospray ionization. Anal Bioanal Chem. 2012;404:389–398. doi: 10.1007/s00216-012-6173-6. [DOI] [PubMed] [Google Scholar]
- [70].Stoeckli M, Staab D, Schweitzer A, Gardiner J, Seebach D. Imaging of a beta-peptide distribution in whole-body mice sections by MALDI mass spectrometry. J Am Soc Mass Spectr. 2007;18:1921–1924. doi: 10.1016/j.jasms.2007.08.005. [DOI] [PubMed] [Google Scholar]
- [71].Enthaler B, Pruns JK, Wessel S, Rapp C, Fischer M, Wittern KP. Improved sample preparation for MALDI-MSI of endogenous compounds in skin tissue sections and mapping of exogenous active compounds subsequent to ex-vivo skin penetration. Anal Bioanal Chem. 2012;402:1159–1167. doi: 10.1007/s00216-011-5562-6. [DOI] [PubMed] [Google Scholar]
- [72].Kreye F, Hamm G, Karrout Y, Legouffe R, Bonnel D, Siepmann F, Siepmann J. MALDI-TOF MS imaging of controlled release implants. J Control Release. 2012;161:98–108. doi: 10.1016/j.jconrel.2012.04.017. [DOI] [PubMed] [Google Scholar]
- [73].Drexler DM, Tannehill-Gregg SH, Wang LF, Brock BJ. Utility of quantitative whole-body autoradiography (QWBA) and imaging mass spectrometry (IMS) by matrix-assisted laser desorption/ionization (MALDI) in the assessment of ocular distribution of drugs. Journal of Pharmacological and Toxicological Methods. 2011;63:205–208. doi: 10.1016/j.vascn.2010.10.003. [DOI] [PubMed] [Google Scholar]
- [74].Heeren RM, Smith DF, Stauber J, Kukrer-Kaletas B, MacAleese L. Imaging mass spectrometry: hype or hope? J Am Soc Mass Spectrom. 2009;20:1006–1014. doi: 10.1016/j.jasms.2009.01.011. [DOI] [PubMed] [Google Scholar]
- [75].Hattori K, Kajimura M, Hishiki T, Nakanishi T, Kubo A, Nagahata Y, Ohmura M, Yachie-Kinoshita A, Matsuura T, Morikawa T, Nakamura T, Setou M, Suematsu M. Paradoxical ATP Elevation in Ischemic Penumbra Revealed by Quantitative Imaging Mass Spectrometry. Antioxid Redox Sign. 2010;13:1157–1167. doi: 10.1089/ars.2010.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Sokoloff L, Reivich M, Kennedy C, Des Rosiers MH, Patlak CS, Pettigrew KD, Sakurada O, Shinohara M. The [14C]deoxyglucose method for the measurement of local cerebral glucose utilization: theory, procedure, and normal values in the conscious and anesthetized albino rat. J. Neurochem. 1977;28:897–916. doi: 10.1111/j.1471-4159.1977.tb10649.x. [DOI] [PubMed] [Google Scholar]
- [77].Koeniger SL, Talaty N, Luo YP, Ready D, Voorbach M, Seifert T, Cepa S, Fagerland JA, Bouska J, Buck W, Johnson RW, Spanton S. A quantitation method for mass spectrometry imaging. Rapid Commun Mass Sp. 2011;25:503–510. doi: 10.1002/rcm.4891. [DOI] [PubMed] [Google Scholar]
- [78].Nilsson A, Fehniger TE, Gustavsson L, Andersson M, Kenne K, Marko-Varga G, Andren PE. Fine Mapping the Spatial Distribution and Concentration of Unlabeled Drugs within Tissue Micro-Compartments Using Imaging Mass Spectrometry. Plos One. 2010;5 doi: 10.1371/journal.pone.0011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Pirman DA, Yost RA. Quantitative Tandem Mass Spectrometric Imaging of Endogenous Acetyl-L-carnitine from Piglet Brain Tissue Using an Internal Standard. Analytical chemistry. 2011;83:8575–8581. doi: 10.1021/ac201949b. [DOI] [PubMed] [Google Scholar]
- [80].Vismeh R, Waldon DJ, Teffera Y, Zhao ZY. Localization and Quantification of Drugs in Animal Tissues by Use of Desorption Electrospray Ionization Mass Spectrometry Imaging. Analytical chemistry. 2012;84:5439–5445. doi: 10.1021/ac3011654. [DOI] [PubMed] [Google Scholar]
- [81].Lechene C, Hillion F, McMahon G, Benson D, Kleinfeld AM, Kampf JP, Distel D, Luyten Y, Bonventre J, Hentschel D, Park KM, Ito S, Schwartz M, Benichou G, Slodzian G. High-resolution quantitative imaging of mammalian and bacterial cells using stable isotope mass spectrometry. Journal of biology. 2006;5:20. doi: 10.1186/jbiol42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Setou M, Shrivas K, Sroyraya M, Yang H, Sugiura Y, Moribe J, Kondo A, Tsutsumi K, Kimura Y, Kurabe N, Hayasaka T, Goto-Inoue N, Zaima N, Ikegami K, Sobhon P, Konishi Y. Developments and applications of mass microscopy. Med Mol Morphol. 2010;43:1–5. doi: 10.1007/s00795-009-0489-0. [DOI] [PubMed] [Google Scholar]
- [83].Chandra S, Lorey DR, Smith DR. Quantitative subcellular secondary ion mass spectrometry (SIMS) imaging of boron-10 and boron-11 isotopes in the same cell delivered by two combined BNCT drugs: In vitro studies on human glioblastoma T98G cells. Radiat Res. 2002;157:700–710. doi: 10.1667/0033-7587(2002)157[0700:qssims]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- [84].Altelaar AFM, Luxembourg SL, McDonnell LA, Piersma SR, Heeren RMA. Imaging mass spectrometry at cellular length scales. Nat Protoc. 2007;2:1185–1196. doi: 10.1038/nprot.2007.117. [DOI] [PubMed] [Google Scholar]
- [85].Spengler B, Hubert M. Scanning microprobe matrix-assisted laser desorption ionization (SMALDI) mass spectrometry: Instrumentation for sub-micrometer resolved LDI and MALDI surface analysis. J Am Soc Mass Spectr. 2002;13:735–748. doi: 10.1016/S1044-0305(02)00376-8. [DOI] [PubMed] [Google Scholar]
- [86].Bouschen W, Schulz O, Eikel D, Spengler B. Matrix vapor deposition/recrystallization and dedicated spray preparation for high-resolution scanning microprobe matrix-assisted laser desorption/ionization imaging mass spectrometry (SMALDI-MS) of tissue and single cells. Rapid Commun Mass Sp. 2010;24:355–364. doi: 10.1002/rcm.4401. [DOI] [PubMed] [Google Scholar]
- [87].Richards AL, Lietz CB, Wager-Miller JB, Mackie K, Trimpin S. Imaging mass spectrometry in transmission geometry. Rapid Commun Mass Sp. 2011;25:815–820. doi: 10.1002/rcm.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Richards AL, Lietz CB, Wager-Miller J, Mackie K, Trimpin S. Localization and imaging of gangliosides in mouse brain tissue sections by laserspray ionization inlet. J Lipid Res. 2012;53:1390–1398. doi: 10.1194/jlr.D019711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Zavalin A, Todd EM, Rawhouser PD, Yang J, Norris JL, Caprioli RM. Direct imaging of single cells and tissue at sub-cellular spatial resolution using transmission geometry MALDI MS. J Mass Spectrom. 2012;47:1473–1481. doi: 10.1002/jms.3108. [DOI] [PubMed] [Google Scholar]
- [90].Li LJ, Sweedler JV. Peptides in the Brain: Mass Spectrometry-Based Measurement Approaches and Challenges. Annu Rev Anal Chem. 2008;1:451–483. doi: 10.1146/annurev.anchem.1.031207.113053. [DOI] [PubMed] [Google Scholar]
- [91].Ye H, Greer T, Li L. Probing neuropeptide signaling at the organ and cellular domains via imaging mass spectrometry. J Proteomics. 2012 doi: 10.1016/j.jprot.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Rubakhin SS, Greenough WT, Sweedler JV. Spatial profiling with MALDI MS: Distribution of neuropeptides within single neurons. Analytical chemistry. 2003;75:5374–5380. doi: 10.1021/ac034498+. [DOI] [PubMed] [Google Scholar]
- [93].Guenther S, Rompp A, Kummer W, Spengler B. AP-MALDI imaging of neuropeptides in mouse pituitary gland with 5 mu m spatial resolution and high mass accuracy. Int J Mass Spectrom. 2011;305:228–237. [Google Scholar]
- [94].Hui LM, Zhang YZ, Wang JH, Cook A, Ye H, Nusbaum MP, Li LJ. Discovery and Functional Study of a Novel Crustacean Tachykinin Neuropeptide. Acs Chem Neurosci. 2011;2:711–722. doi: 10.1021/cn200042p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Skold K, Svensson M, Kaplan A, Bjorkesten L, Astrom J, Andren PE. A neuroproteomic approach to targeting neuropeptides in the brain. Proteomics. 2002;2:447–454. doi: 10.1002/1615-9861(200204)2:4<447::AID-PROT447>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- [96].Galli C, Racagni G. Use of microwave techniques to inactivate brain enzymes rapidly. Methods in enzymology. 1982;86:635–642. doi: 10.1016/0076-6879(82)86234-4. [DOI] [PubMed] [Google Scholar]
- [97].Che FY, Lim J, Pan H, Biswas R, Fricker LD. Quantitative neuropeptidomics of microwave-irradiated mouse brain and pituitary. Molecular & cellular proteomics : MCP. 2005;4:1391–1405. doi: 10.1074/mcp.T500010-MCP200. [DOI] [PubMed] [Google Scholar]
- [98].Colgrave ML, Xi L, Lehnert SA, Flatscher-Bader T, Wadensten H, Nilsson A, Andren PE, Wijffels G. Neuropeptide profiling of the bovine hypothalamus: thermal stabilization is an effective tool in inhibiting post-mortem degradation. Proteomics. 2011;11:1264–1276. doi: 10.1002/pmic.201000423. [DOI] [PubMed] [Google Scholar]
- [99].Goodwin RJ, Lang AM, Allingham H, Boren M, Pitt AR. Stopping the clock on proteomic degradation by heat treatment at the point of tissue excision. Proteomics. 2010;10:1751–1761. doi: 10.1002/pmic.200900641. [DOI] [PubMed] [Google Scholar]
- [100].Ye H, Greer T, Li LJ. From pixel to voxel: a deeper view of biological tissue by 3D mass spectral imaging. Bioanalysis. 2011;3:313–332. doi: 10.4155/bio.10.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Chen RB, Hui LM, Sturm RM, Li LJ. Three Dimensional Mapping of Neuropeptides and Lipids in Crustacean Brain by Mass Spectral Imaging. J Am Soc Mass Spectr. 2009;20:1068–1077. doi: 10.1016/j.jasms.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Blatherwick EQ, Van Berkel GJ, Pickup K, Johansson MK, Beaudoin ME, Cole RO, Day JM, Iverson S, Wilson ID, Scrivens JH, Weston DJ. Utility of spatially-resolved atmospheric pressure surface sampling and ionization techniques as alternatives to mass spectrometric imaging (MSI) in drug metabolism. Xenobiotica. 2011;41:720–734. doi: 10.3109/00498254.2011.587550. [DOI] [PubMed] [Google Scholar]
- [103].Wang BX, Inutan ED, Trimpin S. A New Approach to High Sensitivity Liquid Chromatography-Mass Spectrometry of Peptides using Nanoflow Solvent Assisted Inlet Ionization. J Am Soc Mass Spectr. 2012;23:442–445. doi: 10.1007/s13361-011-0320-8. [DOI] [PubMed] [Google Scholar]
- [104].Laskin J, Heath BS, Roach PJ, Cazares L, Semmes OJ. Tissue Imaging Using Nanospray Desorption Electrospray Ionization Mass Spectrometry. Analytical chemistry. 2012;84:141–148. doi: 10.1021/ac2021322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Kiss A, Heeren RMA. Size, weight and position: ion mobility spectrometry and imaging MS combined. Anal Bioanal Chem. 2011;399:2623–2634. doi: 10.1007/s00216-010-4644-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Trim PJ, Henson CM, Avery JL, McEwen A, Snel MF, Claude E, Marshall PS, West A, Princivalle AP, Clench MR. Matrix-Assisted Laser Desorption/Ionization-Ion Mobility Separation-Mass Spectrometry Imaging of Vinblastine in Whole Body Tissue Sections. Analytical chemistry. 2008;80:8628–8634. doi: 10.1021/ac8015467. [DOI] [PubMed] [Google Scholar]
- [107].McLean JA, Ridenour WB, Caprioli RM. Profiling and imaging of tissues by imaging ion mobility-mass spectrometry. J Mass Spectrom. 2007;42:1099–1105. doi: 10.1002/jms.1254. [DOI] [PubMed] [Google Scholar]
- [108].Manier ML, Reyzer ML, Goh A, Dartois V, Via LE, Barry CE, 3rd, Caprioli RM. Reagent precoated targets for rapid in-tissue derivatization of the anti-tuberculosis drug isoniazid followed by MALDI imaging mass spectrometry. J Am Soc Mass Spectrom. 2011;22:1409–1419. doi: 10.1007/s13361-011-0150-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Jones EA, Deininger SO, Hogendoorn PC, Deelder AM, McDonnell LA. Imaging mass spectrometry statistical analysis. J Proteomics. 2012;75:4962–4989. doi: 10.1016/j.jprot.2012.06.014. [DOI] [PubMed] [Google Scholar]
- [110].Alexandrov T. MALDI imaging mass spectrometry: statistical data analysis and current computational challenges. BMC bioinformatics. 2012;13(Suppl 16):S11. doi: 10.1186/1471-2105-13-S16-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Thiele H, Heldmann S, Trede D, Strehlow J, Wirtz S, Dreher W, Berger J, Oetjen J, Kobarg JH, Fischer B, Maass P. 2D and 3D MALDI-imaging: Conceptual strategies for visualization and data mining. Biochimica et biophysica acta. 2013 doi: 10.1016/j.bbapap.2013.01.040. doi: 10.1016/j.bbapap.2013.01.040. [DOI] [PubMed] [Google Scholar]
- [112].McDonnell LA, Heeren RM, Andren PE, Stoeckli M, Corthals GL. Going forward: Increasing the accessibility of imaging mass spectrometry. J Proteomics. 2012 doi: 10.1016/j.jprot.2012.05.016. [DOI] [PubMed] [Google Scholar]