Summary
The hallmark of a virus is its capsid, which harbors the viral genome and is formed from protein subunits, which assemble following precise geometric rules. dsRNA viruses use an unusual protein multiplicity (120 copies) to form their closed capsids. We have determined the atomic structure of the capsid protein (P1) from the dsRNA cystovirus Φ8. In the crystal P1 forms pentamers, very similar in shape to facets of empty procapsids, suggesting an unexpected assembly pathway that proceeds via a pentameric intermediate. Unlike the elongated proteins used by dsRNA mammalian reoviruses, P1 has a compact trapezoid-like shape and a distinct arrangement in the shell, with two near-identical conformers in nonequivalent structural environments. Nevertheless, structural similarity with the analogous protein from the mammalian viruses suggests a common ancestor. The unusual shape of the molecule may facilitate dramatic capsid expansion during phage maturation, allowing P1 to switch interaction interfaces to provide capsid plasticity.
Graphical Abstract
Highlights
-
•
Crystal structure of the major capsid protein P1 of the Pseudomonas phage Φ8
-
•
Φ8 P1 shares a common ancestor with mammalian reoviruses
-
•
Φ8 P1’s trapezoidal shape may facilitate capsid expansion during maturation
-
•
The pentameric organization of Φ8 P1 suggests a revised assembly pathway
El Omari et al. report a structure of the dsRNA bacteriophage ϕ8 capsid protein P1. P1 crystallizes as a pentamer, suggesting a new pathway for procapsid assembly. P1 displays a novel fold and a trapezoidal shape, distinct from that of other dsRNA virus, which may facilitate capsid expansion during maturation.
Introduction
Mammalian double-stranded RNA (dsRNA) viruses hide their genome from the cellular environment, as dsRNA provokes strong immune responses. This is accomplished by maintaining at all times the segmented dsRNA genome within the confines of the closed capsid. The dsRNA virus capsid is therefore a complex molecular machine capable of specifically encapsidating the plus polarity single-stranded RNA (ssRNA) genomic precursors, synthesizing the minus strands inside the particle (replication), making plus strands from the dsRNA genomes (transcription), and finally extruding the newly made plus strand transcripts to the particle exterior (for review, see Poranen and Bamford, 2012). These transcripts can either enter translation or will be encapsidated into newly assembled capsids closing the viral replication cycle. Bacterial dsRNA viruses follow a similar strategy; however, in common with many other bacterial viruses, the capsid is initially assembled in a “deflated” (procapsid) form, which is “inflated” as the genome is packaged.
The capsid of most dsRNA viruses is a characteristic 120 subunit T = 1 shell, sometimes nicknamed “T = 2.” Depending on the complexity of each virus, additional layers may build on this shell. The first dsRNA virus capsid structure solved to high resolution was that of the mammalian bluetongue virus (BTV), revealing the intriguing way the particle is constructed (Grimes et al., 1998). This structure and subsequently solved structures of other dsRNA virus shells (summarized by Luque et al., 2010) show that the basic building blocks of the T = 2 capsid are capsid protein dimers formed from nonsymmetrically related molecules, A and B. Because of different interactions with their respective neighbors, monomers A and B have identical structural folds but differences in the tertiary structure mainly arising from the movement of domains about hinge points.
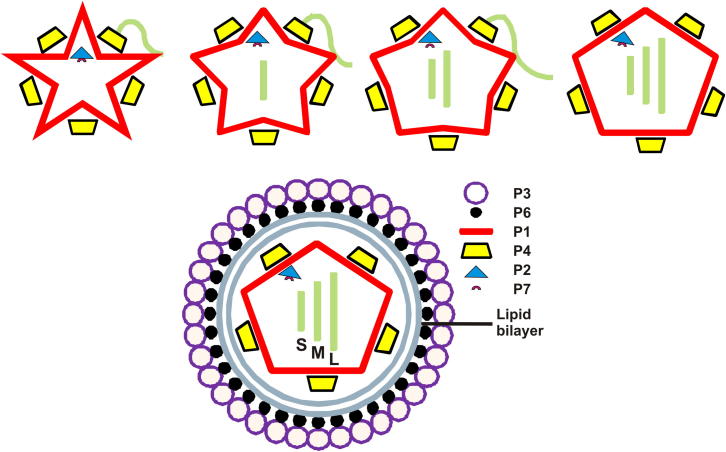
Φ8 is an enveloped bacteriophage with a segmented dsRNA genome, belonging to the Cystoviridae family. Related viruses include those from Φ6 to Φ14 and Φ2954, where Φ6 is the type-virus of the family (Mindich et al., 1999; Qiao et al., 2010). The outer layer of cystoviruses is composed of a membrane envelope (where the lipids are of host origin) containing three to four virus-specific integral membrane proteins, one of them (P6) mediating the fusion between the viral membrane and the host outer membrane (Bamford et al., 1987; Etten et al., 1976; Gottlieb et al., 1988; Laurinavicius et al., 2004) (Figure 1). Under the membrane vesicle, the nucleocapsid shell, composed of protein P8, (Etten et al., 1976) usually forms a middle protein layer (Bamford and Mindich, 1980). In Φ6, P8 trimers form a T = 13 lattice covering the inner capsid, except at 5-fold locations that are occupied by the packaging NTPase P4 (Butcher et al., 1997; Huiskonen et al., 2006). In Φ8, however, the P8 shell seems to be missing and the membrane contacts are carried out mostly by protein P4 (Jäälinoja et al., 2007). The capsid is composed of four proteins P1, P2, P4, and P7, where 120 copies of the capsid protein P1 form a thin icosahedral shell. The RNA-dependent RNA polymerase (P2) is located internally close to the 3-fold symmetry position (Nemecek et al., 2012; Sen et al., 2008), whereas the P4 packaging hexamers reside on the outer surface on the 5-fold axes, creating a mismatch in symmetry (de Haas et al., 1999). The assembly cofactor P7 seems to reside, as a monomer, inside the capsid near the 3-fold symmetry location, possibly competing with the polymerase protein for the binding site (Katz et al., 2012; Nemecek et al., 2010, 2012). The accessory proteins (P2, P4, and P7) only partially occupy their binding sites in the virion (Nemecek et al., 2010, 2012; Sun et al., 2012).
Figure 1.
Schematic Representation of the Organization and Expansion of Bacteriophage Φ8
The three dsRNA genomic segments s, m, and l (small, medium, and large, respectively) are depicted as green rectangles. Proteins discussed in the paper are labeled.
See also Figures S1 and S2.
The cystoviral dsRNA genome is composed of three segments: small (S), medium (M), and large (L) (Figure 1). There is evidence that the segments are packaged in a sequential order (S then M and finally L) by the capsid (Frilander and Bamford, 1995; Qiao et al., 1995). The 5′ end of each ssRNA segment has a phage packaging site (pac) (Gottlieb et al., 1992, 1994; Pirttimaa and Bamford, 2000; Qiao et al., 2005) that initially binds to protein P1 via a specific sequence (Qiao et al., 2003b), followed by translocation into the capsid by the hexameric P4 packaging NTPase (Frilander and Bamford, 1995; Gottlieb et al., 1991; Kainov et al., 2003b; Pirttimaa et al., 2002). Genome packaging and replication induce major conformational changes of the capsid, leading to expansion of the shell (Butcher et al., 1997; Nemecek et al., 2011). The current sequential packaging model proposes that the empty capsid exposes a binding site only for S; when S is packaged, the particle expands somewhat and the S binding site disappears, while that for M appears. The same mechanism applies to allow the packaging of L (Mindich, 2012).
Although the genome organization and virion structure are conserved among cystoviruses, sequence conservation between the members can be very limited. Within the genus, viruses are grouped according to their relationship to the type member Φ6 (Mindich et al., 1999). The most distantly related member of the genus is bacteriophage Φ8, sharing only 15% overall sequence identity, mostly in the polymerase P2 and the core domain of the P4 protein (Hoogstraten et al., 2000), whereas their P1 capsid proteins have an even lower sequence identity of 6%. This is reflected in changes during viral entry: Φ6 enters the periplasmic space with a protein P8 shell, whereas Φ8 does not have this layer (Sun et al., 2003).
Cystoviruses in general and Φ6 in particular are one of the best understood viral assembly systems (Poranen et al., 2001) in which purified viral proteins and RNA constituents can self-assemble to generate infectious viral particles with high efficiency. In addition the RNA-dependent RNA polymerase of Φ6 was the first such polymerase to be isolated (Makeyev and Bamford, 2000). Its structure and mechanistic functions are known in great detail (see, for example, Butcher et al., 2001; Poranen et al., 2008; Salgado et al., 2004; Wright et al., 2012). Also the mechanism of RNA translocation by P4 is known in atomic detail (Kainov et al., 2006; Mancini et al., 2004a, 2004b). There are several electron microscopy (EM)-based studies defining the organization of the P1 shell and its transitions (see Table 1). In bacteriophages Φ6 and Φ8, five A subunits encircle the 5-fold axes, and B subunits interact at the 2-fold and 3-fold axes, with no direct contacts seen between A subunits belonging to different 5-fold vertices. However, a detailed description of the structure of the P1 coat protein is still missing.
Table 1.
Summary of Available Cryo-EM Structures of Bacteriophages Φ6 and Φ8
| Virus | Structure | Resolution (Å) | Reference | EMDB Accession Code | |
|---|---|---|---|---|---|
| Φ6 | Nucleocapsid | Detergent-treated virions | 7.5 | Huiskonen et al., 2006 | 1206 |
| Φ6 | Nucleocapsid | Detergent-treated virions | 12.0 | Huiskonen et al., 2006 | 1207 |
| Φ8 | Core | Detergent-treated virions | 15.0 | Huiskonen et al., 2007 | 1256 |
| Φ8 | Virion | 21.0 | Jäälinoja et al., 2007 | 1299 | |
| Φ8 | Core | Detergent-treated virions | 8.7 | Jäälinoja et al., 2007 | 1300 |
| Φ6 | Virion | 18.0 | Jäälinoja et al., 2007 | 1301 | |
| Φ6 | Recombinant procapsid | P1, P2, P4, and P7 | 14.0 | Sen et al., 2008 | 1500 |
| Φ6 | Recombinant procapsid | P1, P2, P4, and P7. Mutation in P1 (E390A) | 19.0 | Sen et al., 2008 | 1501 |
| Φ6 | Recombinant procapsid | P1, P4, and P7 | 11.0 | Sen et al., 2008 | 1502 |
| Φ6 | Recombinant procapsid | P1, P2, and P4 | 16.0 | Sen et al., 2008 | 1503 |
| Φ6 | Recombinant procapsid | P1, P2, P4, and P7 | 8.1 | Nemecek et al., 2012 | 2341 |
| Φ6 | Recombinant procapsid | P1, P2, and P4 | 9.7 | Nemecek et al., 2012 | 2342 |
| Φ6 | Recombinant procapsid | P1, P4, and P7 | 9.6 | Nemecek et al., 2012 | 2344 |
| Φ6 | Recombinant procapsid | P1 and P4 | 12.4 | Nemecek et al., 2012 | 2346 |
| Φ6 | Recombinant expansion intermediate 1 procapsid | P1, P2, P4, and P7 | 19.0 | Nemecek et al., 2011 | 5355 |
| Φ6 | Recombinant expansion intermediate 2 procapsid | P1, P2, P4, and P7 | 20.0 | Nemecek et al., 2011 | 5356 |
| Φ6 | Recombinant expansion intermediate 2 procapsid | P1, P2, P4, and P7 | 20.0 | Nemecek et al., 2011 | 5357 |
| Φ6 | Recombinant capsid | P1, P2, P4, P7, and RNA | 18.0 | Nemecek et al., 2011 | 5358 |
Here, we describe the crystal structure of the major capsid protein P1 of bacteriophage Φ8 at a resolution of 3.7 Å. The oligomeric organization of P1 protein in the crystal and in solution is pentameric, with the pentamers in the crystal structure resembling closely the facet of empty Φ6 procapsids. This suggests an assembly mechanism, proceeding via pentameric intermediates, which is different to that proposed earlier. The atomic details of P1 allow us to make structural comparison with other dsRNA capsid proteins, providing insight into viral evolution. The fit of the crystal structure into previously obtained EM maps revealed conformational changes during virus maturation and shed light on the packaging of the cystoviral genome and the mechanism by which, during genome packaging and replication, the procapsid expands dramatically to form the almost spherical nucleocapsid, with minor internal changes in the plate-like P1 subunits that, however, tilt and twist dramatically.
Results and Discussion
Overall Fold of Φ8 P1
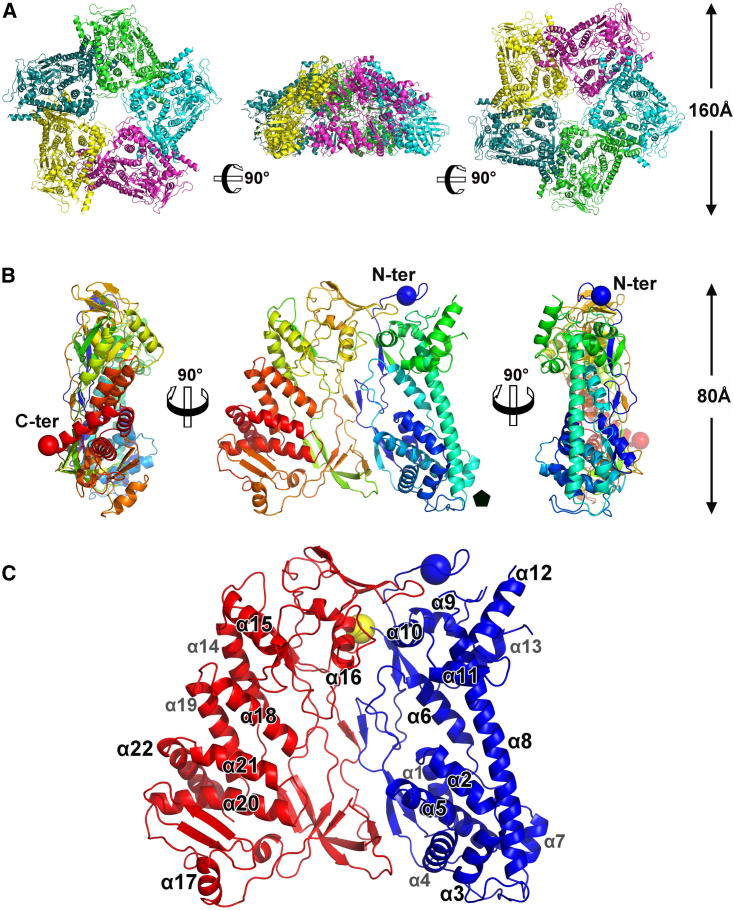
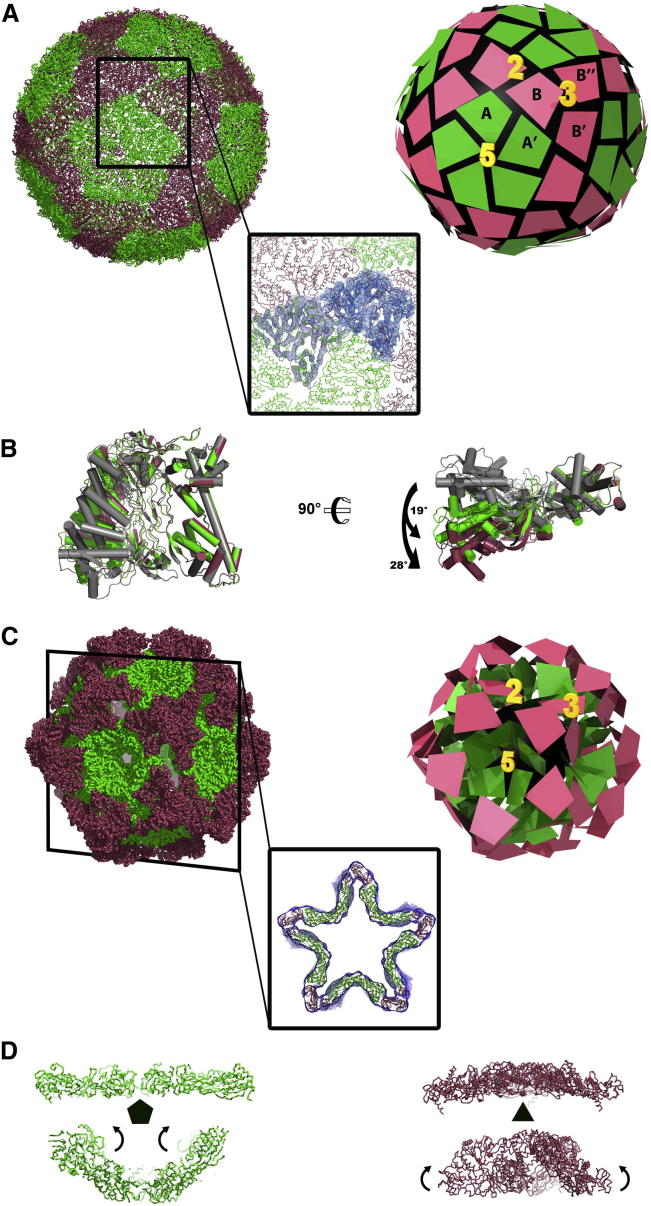
The building block of P1 crystals is a dimer of dome-shaped pentamers, arranged bottom-to-bottom to form a hollow sphere (Figure S1 available online). This leads to an extremely high solvent content in the crystals (80%). Combining solvent flattening and 10-fold noncrystallographic symmetry (NCS) averaging resulted in clearly interpretable electron density maps and a reliable refined atomic model, despite the limited resolution (3.7 Å) (Figure S2). Each dome-like pentamer is composed of virtually identical (root-mean-square deviation [rmsd] ∼0.1 Å) and closely interacting monomers, with an average interface surface area of about 1,800 Å2 (Figure 2A).
Figure 2.
Overall Fold of the Φ8 P1 Structure
(A) Architecture of the pentameric Φ8 P1 structure from the top, side, and bottom views.
(B) Monomeric Φ8 P1 is colored from the N-terminal (blue) to the C-terminal (red). The first and the last residues are represented as spheres. The pentagon shows the region of the monomer that forms the hole at the 5-fold axis.
(C) N- and C-terminal domains of Φ8 P1 colored in blue and red, respectively. The single main-chain connection between the two domains is shown as a yellow sphere at residue 370.
The overall shape of the P1 monomer is that of a thin trapezoidal plate (∼30 × 70 × 80 Å) (Figure 2B). The atomic model includes the majority of the 792 residues, lacking only the first 23, the last 2, and residues 325 to 346, due to disorder. The structure can be divided into two roughly equal domains (Figure 2C). The N-terminal domain (residues 1–370) is composed of thirteen α helices, including the remarkably long, 40-residue helix α-8, which spans one side of the trapezoid, and three β sheets, each made of two or three β strands. The C-terminal domain (residues 371–790) contains nine α helices and four β sheets made of either three or four β strands. The N- and C-terminal are linked by a single connection around residue 370. The buried area between the two domains is approximately 2,100 Å2, and the contacts are mainly hydrophobic interactions and hydrogen bonds linking β strands from each domain.
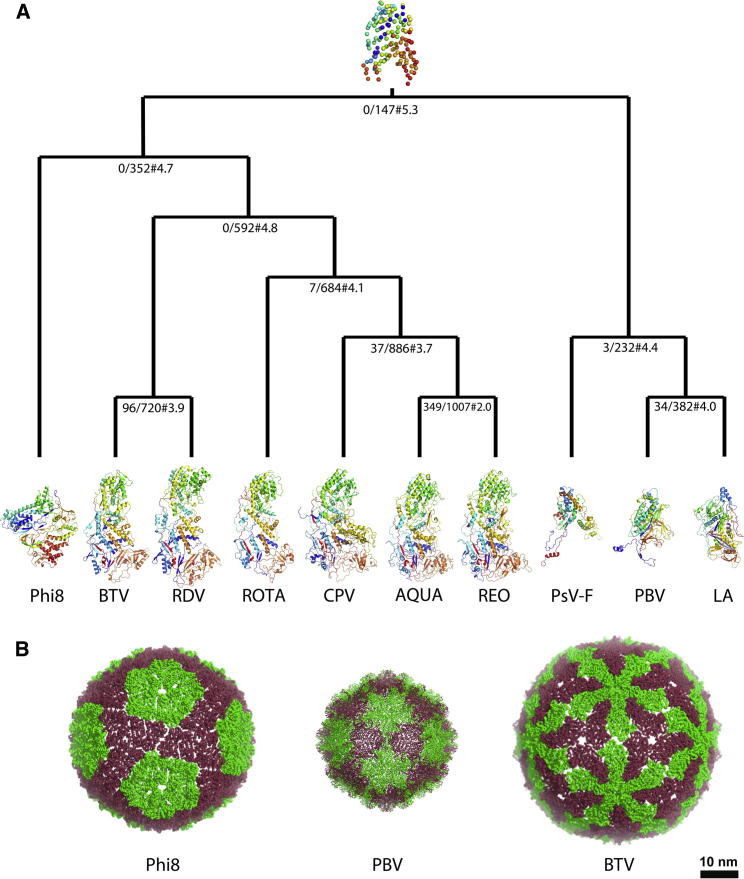
When compared to capsid proteins of other members of the dsRNA virus lineage, Φ8 P1 has a similar molecular weight and high α-helical content but a very different overall shape (Figure 3A). Previous analyses have shown that although the capsid folds differ between members of the lineage, there are discernible similarities in the arrangement of the structural elements within the proteins (Abrescia et al., 2012). Analysis using the program HSF (Ravantti et al., 2013) confirms this and places P1 as intermediate between eukaryotic virus capsid proteins (such as BTV) and the smaller proteins from other dsRNA viruses (such as PBV) (Figure 3B).
Figure 3.
Structural Comparison of Φ8 P1 with Other dsRNA Virus Capsid Proteins
(A) Structure-based phylogenetic tree of the dsRNA virus capsid proteins. Each node in the tree is labeled x/y#z, where x is the number of identical residues within the equivalent residues, y is the number of equivalent residues in the core, and z is the average rmsd of the aligned equivalent residues to the corresponding average residue. Capsid proteins are colored from the N terminus (blue) to the C terminus (red) and are labeled as followed with the protein and Protein Data Bank codes inside parentheses: Phi8, bacteriophage Φ8 (P1, 4BTP), BTV, bluetongue virus (VP3, 2BTV), RDV, Rice dwarf virus (P3, 1UF2), ROTA, rotavirus (VP2, 3KZ4), CPV, cytoplasmic polyhedrosis virus (VP1, 3IZX), AQUA, Aquareovirus (VP3, 3IYL), REO, orthoreovirus (λ1, 1EJ6), PSV-F, partitivirus (3ES5), PBV, picobirnavirus (2VF1), and LA, L-A virus (1M1C). The protein at the top of the tree corresponds to a communal core.
(B) The capsid shells of Φ8, picobirnavirus, and bluetongue virus are shown as ribbons. Monomers A and B are colored in green and red, respectively.
Overall Architecture of the Expanded Φ8 Capsid
The atomic structure of the Φ8 P1 monomer was fitted into the 8.7 Å cryo-electron microscopy (cryo-EM) map of the Φ8 expanded capsid (Jäälinoja et al., 2007) to generate a pseudoatomic model (Figure 4A). Fitting was performed using VEDA (Navaza et al., 2002), a reciprocal-space fitting software package that allows refinement of the fit, while taking icosahedral symmetry into account. Densities for 14 out of the 22 α helices are resolved in the cryo-EM reconstruction, allowing a good initial rigid-body fitting of the P1 monomer in the A and B positions (CC = 0.58). The A and B subunits were subsequently allowed to freely rotate about the hinge residue between the N and C termini, producing an improvement of the fit (CC = 0.61). Compared to the crystal structure, there is a rotation of the C terminus of 19° and 28°, respectively, in the A and B subunits, if the structures are superimposed on their N-terminal domain (Figure 4B).
Figure 4.
Fitting of the Φ8 P1 Monomers A and B Colored in Green and Red, Respectively, in Cryo-EM Reconstructions
(A) Fitting of the Φ8 P1 in the 8.7 Å reconstruction of the Φ8 expanded capsid. On the left, ribbon representation of the Φ8 expanded capsid. The box shows the fit of the structure in the EM map. On the right, model of the Φ8 expanded capsid showing the symmetry axes. P1 monomers are represented by trapezoids.
(B) Rotation movement observed between monomeric Φ8 P1 from the crystal structure (gray) and the monomers A and B of the Φ8 expanded capsid (green and red, respectively). Structures have been superimposed on their N-terminal domain.
(C) Fitting of the Φ8 P1 structure in the 14 Å reconstruction of the Φ6 procapsid. On the left, ribbon representation of the Φ8 procapsid. The box shows the fit of the structure in the Φ6 EM map. On the right, model of the procapsid showing the symmetry axes.
(D) Movement observed during the capsid expansion at the 5-fold (pentagon) and 3-fold (triangle) of a conformer A pentamer (green) and a conformer B trimer (red).
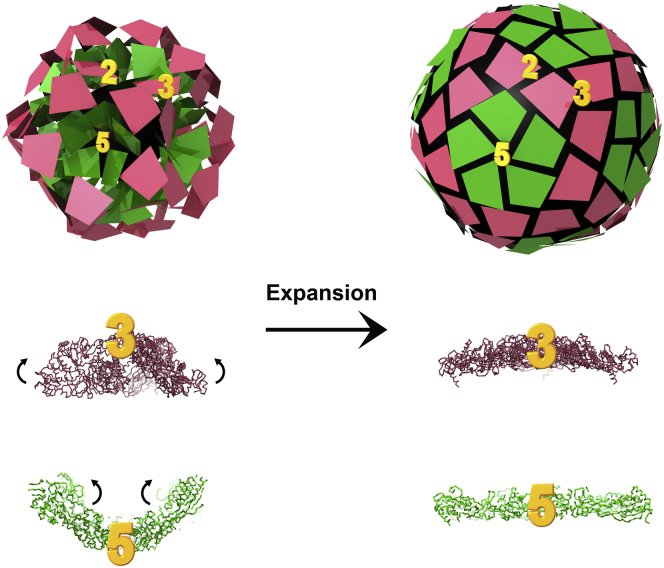
The arrangement of the 120 P1 subunits on the icosahedrally symmetric lattice is shown in Figure 4A. The A and B subunits are located in distinct environments on the capsid surface, making nonequivalent contacts with neighboring subunits. Five closely associated A subunits (average interface area circa 1,000 Å) radiate from each 5-fold axis, giving rise to 12 noninterlocking pentamers, each forming a regular pentagon. An individual B subunit is associated with each side of these pentagons, filling in the spaces between them. The placement of the B subunit relative to A can be described as follows: the whole of each side of a pentamer (formed by equal contributions from two subunits A and A’) is in contact with a B subunit, which is in essentially the same orientation as one of the A subunits that it contacts (Figure 4A, close-up). In total circa 2,000 Å2 of each B subunit is buried in each such interaction. B subunits form trimeric associations on the 3-fold icosahedral axis and the extent of the interface area, circa 2,000 Å2 (B-B’ and B-B’’), is similar to that seen for the A-B and A-B’ interaction, suggesting somewhat similar strength associations. The arrangement of the 120 P1 subunits differs somewhat from the corresponding T = 2 lattice seen in mature cores of reoviruses. The prototype of such proteins, VP3 from BTV, assembles as decamers, with five A subunits clustered around the icosahedral 5-fold axis of symmetry and five B subunits interdigitating between them, a little further from the 5-fold (Figure 3B) (Grimes et al., 1998). In BTV A subunits span icosahedral 2-fold axes, linking adjacent pentamers to form a continuous scaffold, while B subunits form isolated trimeric plugs. In addition, the significant distortion observed between the A and B subunits in BTV, to overcome the conformational restriction preventing the formation of a genuine “T = 2” shell, is present to a much smaller extent in Φ8 P1, where an rmsd of 1.7Å is measured between quasiequivalent subunits. The organization of the P1 lattice is reminiscent of that of the capsid protein of PsV-F, a partivirus, and PBV, a picobirnavirus, two small dsRNA viruses with a simple one-layer structure capsid (Duquerroy et al., 2009; Pan et al., 2009). In both cases, the 60 A subunits surround the icosahedral 5-fold axes and form 12 flower-shaped pentamers that contact only at the icosahedral 2-fold axes, while B subunits pack into 20 trimeric clusters isolated at the 3-fold axes. Dimers of capsid proteins are the proposed building blocks.
dsRNA virus capsid proteins thus share a similar “T = 2” shell architecture and a high content of helices. They can, however, adopt very different shapes, which might reflect differences in their assembly/maturation pathways. Φ8 P1, which is trapezoidal in shape, is different from the other known dsRNA virus capsid proteins of comparable size, which are rather elongated (Abrescia et al., 2012) (Figure 3A). Nevertheless, structural alignments surprisingly show relationships between these capsid proteins and reveal that Φ8 P1 is more similar to the reoviral capsid proteins than to the simple-capsid dsRNA viruses with smaller capsid proteins.
Overall Architecture of the Procapsid
The pentamer observed in the crystal structure of Φ8 P1 is very reminiscent of the facet of the empty Φ6 procapsid, where P1 pentamers point toward the inside of the procapsid (Figure 2A). Although Φ6 and Φ8 P1 share low sequence similarity (no more than 6%, rendering reliable sequence alignment in the absence of structure problematic), they both possess a similar flat trapezoidal shape and appear similar at the tertiary structure level, when examined at low resolution by cryo-EM (Jäälinoja et al., 2007). To analyze this, the atomic structure of Φ8 P1 was fitted into the 14Å resolution cryo-EM reconstruction of the Φ6 procapsid to generate a pseudoatomic model (Figure 4C). It was possible to fit the complete P1 pentamer from our crystal structure as a rigid body at the 5-fold vertices of the procapsid and individual P1 subunits around the 2- and 3-fold axes. Fitting the entire pentamer led to a CC of 0.89, while allowing the individual subunits to move only increased the CC to 0.93 with a ∼13° rotation of the P1 subunit (Figure S3). These subunits correspond to the A subunits in the expanded capsid; however, they are radically rearranged (∼45° rotation and ∼84 Å translation) (Figure 4D). The surrounding B subunits are also in a very different position and orientation between the two structures (20° rotation and 30 Å translation) (Figure 4D). The changes between these two structures give insight into virus maturation, as discussed below.
Procapsid Assembly
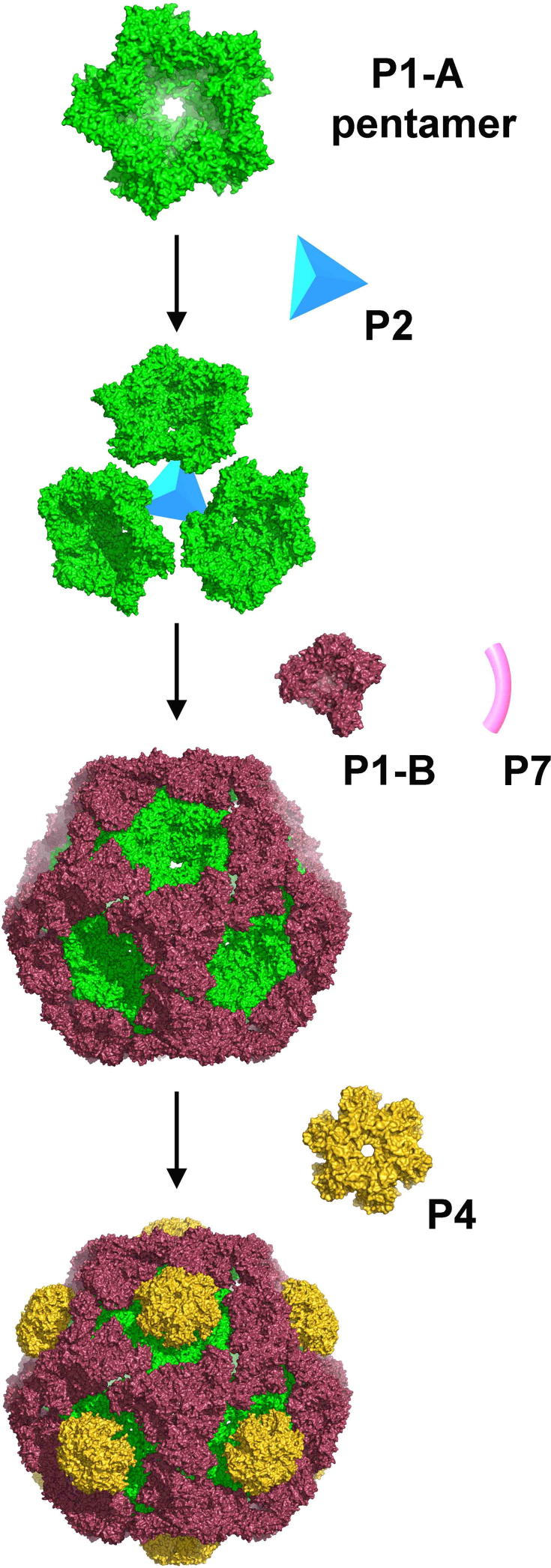
It has been proposed that the cystovirus procapsids are assembled from building blocks of tetramers of P1. This hypothesis was based on gel filtration and light-scattering experiments, showing that in solution Φ8 P1 forms assemblies with a hydrodynamic radius of 5.5 nm (Kainov et al., 2003a). Because this proposal is not in accord with our results, we suggest instead that the basic building block for viral assembly is a pentamer of P1 subunits (Figure 5). The presence of pentamers in the crystal structure, in a shape essentially indistinguishable from that of the 5-fold facet of the procapsid (Figure S3), suggests that the pentamers are present in solution prior to assembly. Indeed, we would expect the crystal structure to select the lowest energy conformation, subject to crystal packing forces (which are negligible for the P1 crystals, where the water content exceeds that in a living cell). Further support for the relevance of this assembly unit comes from negative stain electron micrographs of purified protein, which supports the presence of Φ8 P1 pentameric assemblies in solution, similar in shape to the ones seen in the crystal (Figure S4).
Figure 5.
Proposed Model of the Assembly Pathway of the Bacteriophage Φ8
See also Figure S4.
It has been previously proposed that P1 building blocks nucleate larger assemblies in the presence of P2 protein (ratio 3:1) and that the complex is further stabilized by P4 proteins (Kainov et al., 2003a). If the P1 building block is a pentamer, then a single P2 molecule would be able to stabilize three such pentamers, in line with the proposed location of P2 under the 3-fold axis in the procapsid of Φ6 (Nemecek et al., 2012; Sen et al., 2008). If one of the three attachment sites to P1 is maintained during partial expansion, then P2 would be naturally transported toward the 5-fold axis, its proposed location in the expanded capsid shell (Sen et al., 2008). It was also previously demonstrated that Φ8 P1 alone could, at high enough concentrations, assemble into procapsid-like dodecahedral cages (Kainov et al., 2003a). The high concentration threshold for cage formation (∼5 mg/ml) and the instability of these particles suggested, however, that other proteins are needed to stabilize intermediates and to lower the nucleation threshold.
Taking these pieces of evidence together, we propose a revised model for the assembly of the cystovirus procapsid, where preformed P1 pentamers, in the shape of collapsed 5-fold facets (A subunits), accumulate in solution and form trimeric higher order assemblies, with the assembly being stabilized by B subunits recruited from a pool of P1 monomers (Figure 5). This process is probably accelerated in vivo by the presence of the P2 protein, which sitting under the 3-fold axis facilitates the association of three pentamers. The fact that the copy number of P2 is between 3 and 10 instead of 20 (Sen et al., 2008; Sun et al., 2012) indicates that most of the 3-fold axes are not occupied by P2 and that substoichiometric numbers are needed for assembly. Moreover if, following expansion, P2 migrates toward the 5-fold axes, its copy number should not exceed 12. P4 is attached to the outside of the procapsid, further stabilizing the complex, perhaps acting as a wedge to ensure that the P1 pentamers adopt the right curvature. The role of P4 is discussed further below.
Symmetry Mismatch at the 5-Fold Vertex
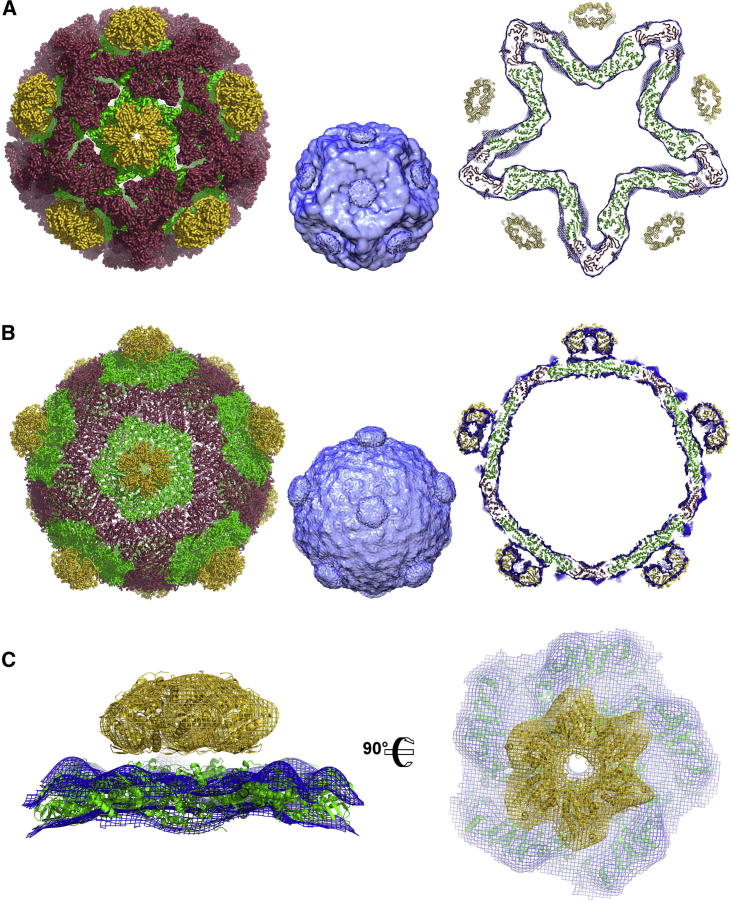
Hexameric P4 protein is responsible for RNA translocation and is located at the 5-fold vertices (Figure 6). It has been suggested that the 6/5-fold symmetry mismatch is critical for the packaging function and might facilitate the P4 ring opening required for RNA loading prior to translocation (Huiskonen et al., 2007). The structure of Φ8 P4 has been determined by X-ray crystallography (K.E., C. Meier, D. Kainov, G.S., J.M.G., M.M. Poranen, D.H.B., R. Tuma, D.I.S., and E.J.M., unpublished data); thus, in addition to the Φ8 P1 structure reported here, we have been able to fit Φ8 P4 into the available EM reconstructions. Although the broad localization of P4 hexamers on the EM reconstructions of the Φ6 procapsid and Φ8 expanded capsid is straightforward (Figures 6A and 6B), a detailed study of P4 is limited by the incorrect averaging at the 5-fold vertices due to the icosahedral three-dimensional reconstruction techniques. In order to study the symmetry mismatch, a different approach has been used to obtain a 15 Å resolution cryo-EM reconstruction of the Φ8 P1:P4 interaction (Huiskonen et al., 2007) (Figure 6C).
Figure 6.
Fitting of the Φ8 P4 Structure in Cryo-EM Reconstructions, Where Φ8 P4 Is Colored in Yellow and Φ8 P1 Monomers A and B Are Colored in Green and Red, Respectively
(A) On the left, ribbon representation of Φ8 P1 and Φ8 P4 forming the procapsid. In the middle, cryo-EM map (EMD 1500) used for the fitting. On the right, Φ8 P4 is fitted at the 5-fold vertices of the Φ6 procapsid reconstruction.
(B) On the left, ribbon representation of Φ8 P1 and Φ8 P4 forming the Φ8 expanded capsid. In the middle, cryo-EM map (EMD 1300) used for the fitting. On the right, fitting of Φ8 P4 at the 5-fold vertices of the Φ8 expanded capsid reconstruction.
(C) Fitting of the Φ8 P4 and Φ8 P1 in the asymmetric 5-fold vertex. The left is a side view, and the right is a top view.
In the latter reconstruction fitted with the Φ8 P1 and P4 crystal structures, the hexameric P4 seems to be floating above the P1 shell. Lower map contour levels reveal that one P4 monomer is closer to P1 than the others and might interact with P1 (Huiskonen et al., 2007). These “floating” hexamers are also seen in Φ6 procapsid reconstructions, suggesting that a similar mechanism of attachment is employed for Φ8 and Φ6. The C-terminal face of P4 is orientated toward the P1 shell, and in the four known P4 structures, the C terminus is disordered (K.E., C. Meier, D. Kainov, G.S., J.M.G., M.M. Poranen, D.H.B., R. Tuma, D.I.S., and E.J.M., unpublished data). A very flexible C-terminal region would not be seen by X-ray crystallography or cryo-EM techniques; moreover, the available P4 structures are mostly complete apart from the C terminus so that the only part of P4 available to contact P1 is the C terminus. It could be argued that similarly P1 might provide a flexible region to contact P4, but in our P1 structure, the missing residues (1–23 and 325–346) are pointing toward the inside of the shell and would not be in contact with P4 hexamers. Thus, we postulate that the flexible P4 C termini are used by P4 hexamers to bind to the P1 shell. Flexibility in binding confers a number of advantages: (1) not all the monomers need to bind at the same time, which would solve the symmetry mismatch, (2) the P4 hexameric ring could open while still bound to the P1 shell, and (3) P4 would stay bound to P1 during the large conformational changes of the expansion step.
For Φ8 P4 the C terminus is seen folding inside the central channel via the top of the hexamer, on the most distant side from the P1 shell (K.E., C. Meier, D. Kainov, G.S., J.M.G., M.M. Poranen, D.H.B., R. Tuma, D.I.S., and E.J.M., unpublished data). In agreement with deuterium exchange experiments (Lísal et al., 2005), we believe that the C terminus is rather flexible and is displaced from the central channel by RNA loading. Here, we propose that at least a single P4 subunit would contact to P1 shell, as proposed from the EM reconstruction of the asymmetric vertex (Huiskonen et al., 2007), and that at least one of the remaining ones would remain folded in the central channel preventing ATP hydrolysis. Upon RNA loading, the remaining C termini would be displaced from the central channel and activate ATP hydrolysis and thus RNA translocation. At the same time, some of the displaced C termini will be able to bind the P1 shell and increase the stability of P4 for P1 during RNA translocation. This is in agreement with the known biochemical properties of the capsid.
Procapsid Expansion
Bacteriophages Φ6 and Φ8 undergo radical conformational changes upon packaging of the genomic RNA segments (Figures 4 and 6; Movie S1) with a volume increase reported to be ∼2.4 fold (Huiskonen et al., 2006). However, based on our modeling and incorporating the substoichiometric number of P2 and P7 proteins, we estimate that the increase in volume accessible to RNA is likely to be circa 5-fold (data not shown). Our crystal structure of Φ8 P1, closely replicates the arrangement of the A subunits in the Φ6 procapsid (Figure S3), and this, together with the fitting of the B subunits to the procapsid and the articulated fitting to the A and B subunits of the EM reconstruction of the expanded Φ8 capsid, reveals that the N- and C-terminal domains of P1 undergo very significant relative rotations during expansion. If the N-terminal domain of the A and B subunits of the expanded Φ8 capsid are superimposed on the N-terminal domain of the A subunit of the Φ6 procapsid, the rotations of the C-terminal domain with respect to the N-terminal domains are 19° and 28°, respectively (Figure 4D). While these rotations are occurring during expansion, the A subunits at the 5-fold axis undergo an iris-like motion. In the procapsid, the P1 subunits form recessed 5-fold facets by overlapping of neighbors within the pentamer (Figure 4C) to give an interface area between adjacent subunits in excess of 2,000 Å2. Upon expansion, each subunit moves away from its neighbors so that eventually it makes only side-to-side contacts (Figure 4A). In the procapsid, interactions at the 2-fold axes are almost nonexistent, whereas at the 3-fold, three subunits form a spike that flattens upon expansion (Figure 4C). The trapezoidal shape of the cystoviral capsid protein might confer higher flexibility to the virus over the elongated molecule, such as those found in the reoviruses that would have to undergo much larger internal conformational changes, because they span from the 5- to the 2-fold axes and rigidify the capsid.
RNA Recognition and Packaging
Along with its structural role, P1 is also required for the specific recognition of the single-stranded genomic segments S, M, and L and their sequential packaging following handover to the NTPase P4 (Qiao et al., 2003a). This process is strictly regulated for Φ6, where only segment S can be packaged alone, whereas M requires prior packaging of S, and L requires prior packaging of M. The pac sites are specific for each segment and have limited sequence identity. To explain how sequential recognition and packaging occurs, a model has been put forward for Φ6, in which the conformational changes occurring after packaging of each segments lead to the consecutive exposure of new RNA binding surfaces (Qiao et al., 1997).
Our work illustrates in detail the rearrangements of P1 following packaging and therefore suggests a mechanism for the RNA packaging specificity. During expansion we see dramatic tectonic movements of the P1 plates, especially at the 5-fold and at the 3-fold axes, where the interfaces between plates go from angled to almost parallel. We propose that the RNA binding sites are situated at the interfaces between P1 plates, in a way that they will be sequentially exposed or buried in accordance with the packaging state of the virus. Specifically, we think that the S segment RNA binding site is situated at the 3-fold axis because in the “deflated” state (1) the 5-fold plates are collapsed, pointing inward, and almost totally covered by P4, while the 3-fold plates form bulging ridges on the surface, and (2) the 3-fold plates are in close spatial proximity to P4 and remain close throughout the expansion process. In this model, the S segment binds around the 3-fold axis of P1 and is then handed over to the P4 hexamer, which undergoes ring opening. Closing of the ring and binding of the RNA trigger ATP hydrolysis and translocation of the RNA inside the capsid, causing its expansion and exposure of the M segment binding site. This process would be repeated with packaging of M and L. Given the size of the pac elements, it is likely that the RNA packaging sites are formed by the quaternary structure arrangement of the three P1 monomers on the 3-fold and not simply by the tertiary structure of the individual monomers. This configuration would also curb the number of binding sites on the capsid and mitigate the unproductive binding of RNA prior to assembly.
In previous studies, a Φ8 mutant virus was isolated, capable of bypassing the strict sequential recognition of segments and package chimeric M segments containing “L” pac sites. Sequencing showed that a single point mutation, valine 242 to alanine, had occurred in protein P1. Interestingly, valine 242 is in the middle of the long α8 helix, which essentially constitutes one of the sides of the P1 monomer and is therefore found at the interface between P1 plates.
Conclusions
In summary the structure of a cystovirus P1 shell protein reveals a tertiary structure, which can be convincingly aligned with that of the apparently quite different proteins that form the corresponding shell in dsRNA viruses. Indeed the cystovirus P1 structure is more similar to the reovirus proteins than it is to proteins of the other simpler members of the dsRNA virus lineage (such as L-A virus). This suggests that phages such as the cystoviruses either represent an ancestral form of the coat protein that has then formed specialist adaptations for the reoviruses (where particle expansion is no longer required) and the simplified dsRNA viruses or else the cystoviruses are a serial homolog in the evolution that led from the simpler viruses to the more complex reoviruses. The remarkable reconfiguration of the P1 subunits during particle expansion is possible only due to the more compact form of the trapezoidal subunit, compared to the elongated homologs of the reoviruses, and the crystal structure and EM observations of the subunits in solution, taken with earlier experimental results, allows us to propose an augmented pentamer-based assembly mechanism for these complex particles.
Experimental Procedures
Cloning, Expression, and Purification
Cloning of Φ8 P1 has been reported elsewhere (Kainov et al., 2003a). Briefly, the gene encoding Φ8 P1 was PCR-amplified from the plasmid pLM2424 (Hoogstraten et al., 2000) and cloned into pT7-7, resulting in the plasmid pDK5. The expression plasmid was transformed into BL21 (DE3) cells for protein expression and grown at 37°C in Luria broth starter culture supplemented with 150 μg/ml ampicillin until the A540 reached 0.5. The culture was then diluted 1/50 into fresh medium and grown at 37°C to an A540 of 0.6. The cultures were induced with 1 mM isopropyl-beta-D-thiogalactopyranoside (IPTG) and further incubated at 18°C for 12 hr. The cells were harvested by centrifugation, and the pellets were resuspended in buffer A (20 mM Tris [pH 8], 50 mM NaCl, 7.5 mM MgCl2) supplemented with protease inhibitors, prior to cell disruption using sonication. The lysed cells were clarified by centrifugation at 27,000 g for 1 hr at 15°C. Ammonium sulfate was added to the supernatant to 20% saturation, and after 15 min on ice, precipitated P1 was collected by centrifugation at 20,000 g for 15 min at 4°C. Precipitated P1 was resuspended in buffer A and then centrifuged again to discharge residual material at 20,000 g for 15 min at 4°C. The P1 protein-containing solution was applied to a Heparin column (GE Healthcare) pre-equilibrated with buffer A. P1 was eluted by a linear gradient of NaCl, from 0.05 to 1 M, buffered with 20 mM Tris [pH 8] containing 7.5 mM MgCl2. The fractions containing P1 were pooled and diluted 5-fold prior to repeating the same experiment by anion-exchange chromatography (Q-sepharose column, GE Healthcare). Fractions of interest were further purified by size exclusion chromatography (Superdex S200, GE Healthcare), pooled, and concentrated to 5 mg/ml for crystallization experiments.
Crystallization and Data Collection
Crystallization trials were performed by hand in 24-well Linbro plates using the sitting-drop vapor-diffusion method at 15°C, mixing 1 μl of protein solution with 1 μl reservoir solution and equilibrating the drop against 1,000 μl reservoir solution. Native and selenomethionine-labeled Φ8 P1 crystals appeared within 1 week of setup in 100 mM Tris [pH 8.5], 30 mM sodium citrate, and between 34% and 39% PEG 200. Mercury incorporation was obtained by soaking the crystals in a drop containing Baker’s dimercurial (1, 4-diacetoxymercuri-2, 3 -dimethoxybutane) at a final concentration of 3 mM for 60 min; crystals were then back-soaked against the reservoir solution containing 20% glycerol. X-ray diffraction data were collected at the European Synchrotron Radiation Facility (ESRF) and the Swiss Light Source (SLS). Diffraction data were indexed, integrated, and scaled with HKL2000 (Otwinowski and Minor, 1997). The crystals belonged to space group P41212 with unit cell parameters a = b = 313.1, c = 524.8Å, α = β = γ = 90° (Table 2).
Table 2.
Data Collection and Refinement Statistics
| Data Collection Details | Native | Se | Hg |
|---|---|---|---|
| Space group | P41212 | ||
| Unit cell dimensions (Å) | 315.1, 315.1, 529.9 | 314.1, 314.1, 521.8 | 313.1, 313.1, 524.7 |
| Wavelength (Å) | 0.9326 | 0.9787 | 1.005 |
| Resolution range (Å) | 30.0–3.7 (3.83–3.70) | 80.0–4.8 (4.97–4.80) | 35.0–6.0 (6.21–6.00) |
| Number of unique reflections | 271,956 (19,751) | 128,534 (11,157) | 65,583 (6,426) |
| Redundancy | 11.1 (4.8) | 7.6 (7.7) | 17.4 (17.0) |
| Completeness (%) | 97.1 (71.3) | 100.0 (100.0) | 100.0 (100.0) |
| Average I/σ(I) | 15 (2.1) | 8.3 (4.4) | 32.3 (6.0) |
| Rmerge | 0.17 (1.1) | 0.26 (0.56) | 0.10 (0.56) |
| CC1/2 | 0.99 (0.66) | ||
| Refinement Statistics | |||
| Resolution range (Å) | 30.0–3.7 | ||
| R-factor (Rwork/Rfree) | 0.245/0.258 | ||
| Rmsd bond (Å) | 0.007 | ||
| Rmsd angle (°) | 1.2 | ||
| Mean B-factor/Wilson plot (Å2) | 140/141 | ||
| Ramachandran plot (%) favored/allowed/outliers | 95.6/100/0 | ||
Values in parentheses are for the highest resolution shell.
The P1 structure was solved using a single anomalous dispersion (SAD) experiment at the peak wavelength of the mercury X-ray absorption edge. HKL2MAP (Sheldrick, 2010) was used to detect heavy atom sites, which were then input in PHENIX AUTOSOL (Adams et al., 2002), resulting, after density modification, in a low-resolution (6 Å) interpretable electron density map. Inspection of the map revealed two pentamers of P1 in the crystallographic asymmetric unit, clearly visible due to the unusually high solvent content (80%). The portion of the map corresponding to the asymmetric unit was cut out, placed in an oversized P1 cell, and used for molecular replacement using PHASER (McCoy et al., 2007) with both the selenomethionine-labeled Φ8 P1 data set (at 4.8 Å resolution), in order to find the selenium site positions, and the higher resolution (3.7 Å) native data set, for phase extension. Initial manual building with polyalanine chains was carried out with the program COOT (Emsley and Cowtan, 2004), and the sequence assignment was facilitated by the location of selenium sites. Because of the excellent observation-to-parameter ratio of 4.5, arising from the 10-fold NCS and extremely high solvent content, it was possible to use phase and NCS-restrained individual atom refinement with REFMAC5 (Murshudov et al., 2011), which resulted in final Rwork and Rfree of, respectively, 24.5% and 25.8% and good stereochemistry. The final structure was validated with MOLPROBITY (Davis et al., 2007), and detailed data collection and refinement statistics are presented in Table 2.
Structural Comparisons
Structural comparisons were performed in SHP (Stuart et al., 1979) and HSF (Ravantti et al., 2013). In the HSF method, a large number of different properties (e.g., simple residue-residue sequence similarities and Cα-Cα distances) are considered. For a multiple structure comparison, a full pairwise set of comparisons is first assembled on the basis of this extensive set of properties and then a recursive process is used, successively grouping structures. At each grouping stage, the common features of the pair are identified, defining a common “core.” In the next recursion the common core is used instead of the two contributing structures, thus gradually reducing the number of items to be compared and implicitly defining a tree of similarities. Finally a single core remains, which defines the structure common to all the proteins being compared. In this study, the common core for all classified structures (containing 147 equivalent residues with average rmsd of 5.3 Å) is shown in the root of the tree (Figure 3).
Fitting of Atomic Coordinates into EM Reconstructions
Monomers of Φ8 P1 were fitted into the Φ8 capsid reconstruction (EMD-1300) (Jäälinoja et al., 2007) and the Φ6 procapsid reconstruction (EMD-1500) (Sen et al., 2008) using VEDA, the graphical version of URO (Navaza et al., 2002).
In the 8.7 Å resolution EM reconstruction of the Φ8 capsid, it was possible to establish the orientation of the A and B subunits on the icosahedral surface by fitting the α helices to their corresponding density. Fitting to either A or B gave similar correlation coefficients (CCs) of about 0.92 (obtained from CHIMERA), and allowing the subunits to move as two rigid body domains separated at residue 370 resulted in a slightly better fit and a CC of 0.93.
Icosahedral symmetry was then applied with VEDA to generate the complete complement of 120 subunits with a CC of 0.61.
The fitting of P1 subunits in the 14 Å resolution Φ6 procapsid reconstruction was performed as described above, except that P1 was not separated into two domains. A CC of 0.64 was obtained after fitting the A and B subunits with VEDA.
The crystal structure of the hexameric Φ8 P4 (K.E., C. Meier, D. Kainov, G.S., J.M.G., M.M. Poranen, D.H.B., R. Tuma, D.I.S., and E.J.M., unpublished data) and of the monomeric Φ8 P1 were fitted with CHIMERA (Pettersen et al., 2004) into a 15 Å asymmetric reconstruction of the 5-fold vertex of the Φ8 capsid (EMD-1256) (Huiskonen et al., 2007), with respective CCs of 0.74 and 0.78. This asymmetric “in situ” reconstruction was obtained by using icosahedral orientations and origins for both vertex selection and orientation, subtracting away the icosahedral information from the images before classifying the hexamers in order to calculate asymmetric reconstructions of individual vertices. This allows the study of the symmetry mismatch between hexameric P4 and pentameric P1 proteins of the Φ8 capsid.
Acknowledgments
This work was supported by the UK Medical Research Council (MRC) and by Academy Professor (Academy of Finland) funding grants (256197 and 256518 to D.H.B.). The Wellcome Trust provided administrative support (075491/Z/04). E.J.M. is a Royal Society University Research Fellow. K.E.O. is a Junior Research Fellow at Oriel College. We thank the staff of the European Synchrotron Radiation Facility (ESRF) and the Swiss Light Source (SLS) for technical support.
Published: July 25, 2013
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Information includes four figures and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.str.2013.06.017.
Accession Numbers
Coordinates and structure factors of Φ8 P1 have been deposited in the Protein Data Bank under the ID code 4BTP. Coordinates of Φ8 P1 fitted into the Φ8 expanded capsid reconstruction have been deposited under the ID code 4BX4.
Supplemental Information
This movie shows the expansion process. Subunits A and B are coloured in green and red respectively. Movie was made with Maya 2012.
References
- Abrescia N.G., Bamford D.H., Grimes J.M., Stuart D.I. Structure unifies the viral universe. Annu. Rev. Biochem. 2012;81:795–822. doi: 10.1146/annurev-biochem-060910-095130. [DOI] [PubMed] [Google Scholar]
- Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Bamford D.H., Mindich L. Electron microscopy of cells infected with nonsense mutants of bacteriophage phi 6. Virology. 1980;107:222–228. doi: 10.1016/0042-6822(80)90287-1. [DOI] [PubMed] [Google Scholar]
- Bamford D.H., Romantschuk M., Somerharju P.J. Membrane fusion in prokaryotes: bacteriophage phi 6 membrane fuses with the Pseudomonas syringae outer membrane. EMBO J. 1987;6:1467–1473. doi: 10.1002/j.1460-2075.1987.tb02388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher S.J., Dokland T., Ojala P.M., Bamford D.H., Fuller S.D. Intermediates in the assembly pathway of the double-stranded RNA virus phi6. EMBO J. 1997;16:4477–4487. doi: 10.1093/emboj/16.14.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher S.J., Grimes J.M., Makeyev E.V., Bamford D.H., Stuart D.I. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B., 3rd, Snoeyink J., Richardson J.S., Richardson D.C. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35(Web Server issue):W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haas F., Paatero A.O., Mindich L., Bamford D.H., Fuller S.D. A symmetry mismatch at the site of RNA packaging in the polymerase complex of dsRNA bacteriophage phi6. J. Mol. Biol. 1999;294:357–372. doi: 10.1006/jmbi.1999.3260. [DOI] [PubMed] [Google Scholar]
- Duquerroy S., Da Costa B., Henry C., Vigouroux A., Libersou S., Lepault J., Navaza J., Delmas B., Rey F.A. The picobirnavirus crystal structure provides functional insights into virion assembly and cell entry. EMBO J. 2009;28:1655–1665. doi: 10.1038/emboj.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Etten J.V., Lane L., Gonzalez C., Partridge J., Vidaver A. Comparative properties of bacteriophage phi6 and phi6 nucleocapsid. J. Virol. 1976;18:652–658. doi: 10.1128/jvi.18.2.652-658.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frilander M., Bamford D.H. In vitro packaging of the single-stranded RNA genomic precursors of the segmented double-stranded RNA bacteriophage phi 6: the three segments modulate each other’s packaging efficiency. J. Mol. Biol. 1995;246:418–428. doi: 10.1006/jmbi.1994.0096. [DOI] [PubMed] [Google Scholar]
- Gottlieb P., Metzger S., Romantschuk M., Carton J., Strassman J., Bamford D.H., Kalkkinen N., Mindich L. Nucleotide sequence of the middle dsRNA segment of bacteriophage phi 6: placement of the genes of membrane-associated proteins. Virology. 1988;163:183–190. doi: 10.1016/0042-6822(88)90245-0. [DOI] [PubMed] [Google Scholar]
- Gottlieb P., Strassman J., Frucht A., Qiao X.Y., Mindich L. In vitro packaging of the bacteriophage phi 6 ssRNA genomic precursors. Virology. 1991;181:589–594. doi: 10.1016/0042-6822(91)90892-f. [DOI] [PubMed] [Google Scholar]
- Gottlieb P., Strassman J., Qiao X., Frilander M., Frucht A., Mindich L. In vitro packaging and replication of individual genomic segments of bacteriophage phi 6 RNA. J. Virol. 1992;66:2611–2616. doi: 10.1128/jvi.66.5.2611-2616.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb P., Qiao X., Strassman J., Frilander M., Mindich L. Identification of the packaging regions within the genomic RNA segments of bacteriophage phi 6. Virology. 1994;200:42–47. doi: 10.1006/viro.1994.1160. [DOI] [PubMed] [Google Scholar]
- Grimes J.M., Burroughs J.N., Gouet P., Diprose J.M., Malby R., Ziéntara S., Mertens P.P., Stuart D.I. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- Hoogstraten D., Qiao X., Sun Y., Hu A., Onodera S., Mindich L. Characterization of phi8, a bacteriophage containing three double-stranded RNA genomic segments and distantly related to Phi6. Virology. 2000;272:218–224. doi: 10.1006/viro.2000.0374. [DOI] [PubMed] [Google Scholar]
- Huiskonen J.T., de Haas F., Bubeck D., Bamford D.H., Fuller S.D., Butcher S.J. Structure of the bacteriophage phi6 nucleocapsid suggests a mechanism for sequential RNA packaging. Structure. 2006;14:1039–1048. doi: 10.1016/j.str.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Huiskonen J.T., Jäälinoja H.T., Briggs J.A., Fuller S.D., Butcher S.J. Structure of a hexameric RNA packaging motor in a viral polymerase complex. J. Struct. Biol. 2007;158:156–164. doi: 10.1016/j.jsb.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Jäälinoja H.T., Huiskonen J.T., Butcher S.J. Electron cryomicroscopy comparison of the architectures of the enveloped bacteriophages phi6 and phi8. Structure. 2007;15:157–167. doi: 10.1016/j.str.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Kainov D.E., Butcher S.J., Bamford D.H., Tuma R. Conserved intermediates on the assembly pathway of double-stranded RNA bacteriophages. J. Mol. Biol. 2003;328:791–804. doi: 10.1016/s0022-2836(03)00322-x. [DOI] [PubMed] [Google Scholar]
- Kainov D.E., Pirttimaa M., Tuma R., Butcher S.J., Thomas G.J., Jr., Bamford D.H., Makeyev E.V. RNA packaging device of double-stranded RNA bacteriophages, possibly as simple as hexamer of P4 protein. J. Biol. Chem. 2003;278:48084–48091. doi: 10.1074/jbc.M306928200. [DOI] [PubMed] [Google Scholar]
- Kainov D.E., Tuma R., Mancini E.J. Hexameric molecular motors: P4 packaging ATPase unravels the mechanism. Cell. Mol. Life Sci. 2006;63:1095–1105. doi: 10.1007/s00018-005-5450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz G., Wei H., Alimova A., Katz A., Morgan D.G., Gottlieb P. Protein P7 of the cystovirus φ6 is located at the three-fold axis of the unexpanded procapsid. PLoS ONE. 2012;7:e47489. doi: 10.1371/journal.pone.0047489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurinavicius S., Käkelä R., Bamford D.H., Somerharju P. The origin of phospholipids of the enveloped bacteriophage phi6. Virology. 2004;326:182–190. doi: 10.1016/j.virol.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Lísal J., Lam T.T., Kainov D.E., Emmett M.R., Marshall A.G., Tuma R. Functional visualization of viral molecular motor by hydrogen-deuterium exchange reveals transient states. Nat. Struct. Mol. Biol. 2005;12:460–466. doi: 10.1038/nsmb927. [DOI] [PubMed] [Google Scholar]
- Luque D., González J.M., Garriga D., Ghabrial S.A., Havens W.M., Trus B., Verdaguer N., Carrascosa J.L., Castón J.R. The T=1 capsid protein of Penicillium chrysogenum virus is formed by a repeated helix-rich core indicative of gene duplication. J. Virol. 2010;84:7256–7266. doi: 10.1128/JVI.00432-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev E.V., Bamford D.H. Replicase activity of purified recombinant protein P2 of double-stranded RNA bacteriophage phi6. EMBO J. 2000;19:124–133. doi: 10.1093/emboj/19.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini E.J., Kainov D.E., Grimes J.M., Tuma R., Bamford D.H., Stuart D.I. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell. 2004;118:743–755. doi: 10.1016/j.cell.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Mancini E.J., Kainov D.E., Wei H., Gottlieb P., Tuma R., Bamford D.H., Stuart D.I., Grimes J.M. Production, crystallization and preliminary X-ray crystallographic studies of the bacteriophage phi 12 packaging motor. Acta Crystallogr. D Biol. Crystallogr. 2004;60:588–590. doi: 10.1107/S0907444904001052. [DOI] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindich L. Packaging in dsRNA viruses. Adv. Exp. Med. Biol. 2012;726:601–608. doi: 10.1007/978-1-4614-0980-9_26. [DOI] [PubMed] [Google Scholar]
- Mindich L., Qiao X., Qiao J., Onodera S., Romantschuk M., Hoogstraten D. Isolation of additional bacteriophages with genomes of segmented double-stranded RNA. J. Bacteriol. 1999;181:4505–4508. doi: 10.1128/jb.181.15.4505-4508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza J., Lepault J., Rey F.A., Alvarez-Rúa C., Borge J. On the fitting of model electron densities into EM reconstructions: a reciprocal-space formulation. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1820–1825. doi: 10.1107/s0907444902013707. [DOI] [PubMed] [Google Scholar]
- Nemecek D., Heymann J.B., Qiao J., Mindich L., Steven A.C. Cryo-electron tomography of bacteriophage phi6 procapsids shows random occupancy of the binding sites for RNA polymerase and packaging NTPase. J. Struct. Biol. 2010;171:389–396. doi: 10.1016/j.jsb.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemecek D., Cheng N., Qiao J., Mindich L., Steven A.C., Heymann J.B. Stepwise expansion of the bacteriophage ϕ6 procapsid: possible packaging intermediates. J. Mol. Biol. 2011;414:260–271. doi: 10.1016/j.jmb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemecek D., Qiao J., Mindich L., Steven A.C., Heymann J.B. Packaging accessory protein P7 and polymerase P2 have mutually occluding binding sites inside the bacteriophage 6 procapsid. J. Virol. 2012;86:11616–11624. doi: 10.1128/JVI.01347-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter C.W., Sweets R.M., editors. Methods in Enzymology. University of Virginia; Charlottesville: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Pan J., Dong L., Lin L., Ochoa W.F., Sinkovits R.S., Havens W.M., Nibert M.L., Baker T.S., Ghabrial S.A., Tao Y.J. Atomic structure reveals the unique capsid organization of a dsRNA virus. Proc. Natl. Acad. Sci. USA. 2009;106:4225–4230. doi: 10.1073/pnas.0812071106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pirttimaa M.J., Bamford D.H. RNA secondary structures of the bacteriophage phi6 packaging regions. RNA. 2000;6:880–889. doi: 10.1017/s1355838200992598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirttimaa M.J., Paatero A.O., Frilander M.J., Bamford D.H. Nonspecific nucleoside triphosphatase P4 of double-stranded RNA bacteriophage phi6 is required for single-stranded RNA packaging and transcription. J. Virol. 2002;76:10122–10127. doi: 10.1128/JVI.76.20.10122-10127.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poranen M.M., Bamford D.H. Assembly of large icosahedral double-stranded RNA viruses. Adv. Exp. Med. Biol. 2012;726:379–402. doi: 10.1007/978-1-4614-0980-9_17. [DOI] [PubMed] [Google Scholar]
- Poranen M.M., Paatero A.O., Tuma R., Bamford D.H. Self-assembly of a viral molecular machine from purified protein and RNA constituents. Mol. Cell. 2001;7:845–854. doi: 10.1016/s1097-2765(01)00228-3. [DOI] [PubMed] [Google Scholar]
- Poranen M.M., Salgado P.S., Koivunen M.R., Wright S., Bamford D.H., Stuart D.I., Grimes J.M. Structural explanation for the role of Mn2+ in the activity of phi6 RNA-dependent RNA polymerase. Nucleic Acids Res. 2008;36:6633–6644. doi: 10.1093/nar/gkn632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X., Casini G., Qiao J., Mindich L. In vitro packaging of individual genomic segments of bacteriophage phi 6 RNA: serial dependence relationships. J. Virol. 1995;69:2926–2931. doi: 10.1128/jvi.69.5.2926-2931.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X., Qiao J., Mindich L. Stoichiometric packaging of the three genomic segments of double-stranded RNA bacteriophage phi6. Proc. Natl. Acad. Sci. USA. 1997;94:4074–4079. doi: 10.1073/pnas.94.8.4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X., Qiao J., Mindich L. Analysis of specific binding involved in genomic packaging of the double-stranded-RNA bacteriophage phi6. J. Bacteriol. 2003;185:6409–6414. doi: 10.1128/JB.185.21.6409-6414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao J., Qiao X., Sun Y., Mindich L. Isolation and analysis of mutants of double-stranded-RNA bacteriophage phi6 with altered packaging specificity. J. Bacteriol. 2003;185:4572–4577. doi: 10.1128/JB.185.15.4572-4577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao J., Qiao X., Mindich L. In vivo studies of genomic packaging in the dsRNA bacteriophage Phi8. BMC Microbiol. 2005;5:10. doi: 10.1186/1471-2180-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X., Sun Y., Qiao J., Di Sanzo F., Mindich L. Characterization of Phi2954, a newly isolated bacteriophage containing three dsRNA genomic segments. BMC Microbiol. 2010;10:55. doi: 10.1186/1471-2180-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravantti J., Bamford D., Stuart D.I. Automatic comparison and classification of protein structures. J. Struct. Biol. 2013 doi: 10.1016/j.jsb.2013.05.007. pii: S1047-8477(13)00121-4. [DOI] [PubMed] [Google Scholar]
- Salgado P.S., Makeyev E.V., Butcher S.J., Bamford D.H., Stuart D.I., Grimes J.M. The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase. Structure. 2004;12:307–316. doi: 10.1016/j.str.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Sen A., Heymann J.B., Cheng N., Qiao J., Mindich L., Steven A.C. Initial location of the RNA-dependent RNA polymerase in the bacteriophage Phi6 procapsid determined by cryo-electron microscopy. J. Biol. Chem. 2008;283:12227–12231. doi: 10.1074/jbc.M710508200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick G.M. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D Biol. Crystallogr. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D.I., Levine M., Muirhead H., Stammers D.K. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 A. J. Mol. Biol. 1979;134:109–142. doi: 10.1016/0022-2836(79)90416-9. [DOI] [PubMed] [Google Scholar]
- Sun Y., Qiao X., Qiao J., Onodera S., Mindich L. Unique properties of the inner core of bacteriophage phi8, a virus with a segmented dsRNA genome. Virology. 2003;308:354–361. doi: 10.1016/s0042-6822(03)00013-8. [DOI] [PubMed] [Google Scholar]
- Sun X., Bamford D.H., Poranen M.M. Probing, by self-assembly, the number of potential binding sites for minor protein subunits in the procapsid of double-stranded RNA bacteriophage Φ6. J. Virol. 2012;86:12208–12216. doi: 10.1128/JVI.01505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., Poranen M.M., Bamford D.H., Stuart D.I., Grimes J.M. Noncatalytic ions direct the RNA-dependent RNA polymerase of bacterial double-stranded RNA virus ϕ6 from de novo initiation to elongation. J. Virol. 2012;86:2837–2849. doi: 10.1128/JVI.05168-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This movie shows the expansion process. Subunits A and B are coloured in green and red respectively. Movie was made with Maya 2012.