Abstract
The third variable region (V3) of HIV-1 envelope glycoprotein gp120 plays a key role in determination of viral coreceptor usage (tropism). However, which combinations of mutations in V3 confer a tropism shift is still unclear. A unique pattern of mutations in antiretroviral therapy-naive HIV-1 patient was observed associated with the HIV-1 tropism shift CCR5 to CXCR4. The insertion of arginine at position 11 and the loss of the N-linked glycosylation site were indispensable for acquiring pure CXCR4-tropism, which were confirmed by cell-cell fusion assay and phenotype analysis of recombinant HIV-1 variants. The same pattern of mutations in V3 and the associated tropism shift were identified in two of 53 other patients (3.8%) with CD4+ cell count <200/mm3. The combination of arginine insertion and loss of N-linked glycosylation site usually confers CXCR4-tropism. Awareness of this rule will help to confirm the tropism prediction from V3 sequences by conventional rules.
Since the introduction of maraviroc, a specific CCR5 antagonist, into clinical practice, scientific and clinical studies have focused on the coreceptor usage of human immunodeficiency virus type 1 (HIV-1)1. Evidence indicates the presence of a homogeneous population of predominantly CCR5-tropic variants2,3 early in HIV-1 infection4,5. CXCR4-tropic variants6,7, against which specific CCR5 antagonists are inefficient, can be distinguished from R5-tropic variants by their tendency for higher replication kinetics and a broader target cell range8. Their presence in vivo has been associated with accelerated fall in CD4+ cell count and rapid disease progression9,10. CXCR4-tropic variants evolve from CCR5-tropic ones in the natural course of HIV-1 infection, though the exact mechanism of viral tropism evolution is not known yet. Long-term observation of natural course, which is indispensable for understanding the mechanism of tropism evolution, is usually difficult, because early use of antiretroviral therapy (ART) is highly recommended11. In this study, untreated natural course of one hemophiliac, who acquired HIV-1 infection through contaminated blood product before 1985 and exhibited slow disease progression, was followed until a rapid fall in CD4+ cell count in 2007. The sequence change in the HIV-1 envelope (Env) glycoprotein gp120 V3 region (V3), the main determinant of HIV-1 tropism12, was analyzed between 2003 and 2007. The results identified a unique change in 2007 associated with change in HIV-1 tropism. The same kind of sequence change in V3 was also identified in two other patients and in some of the registered sequences in the Los Alamos HIV sequence database.
Results
V3 sequence changes in Case 1
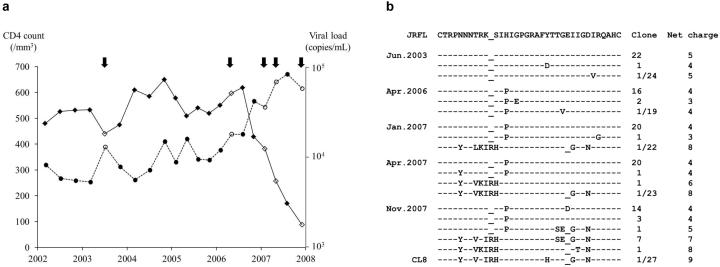
Case 1 was an ART-naive Japanese hemophiliac who acquired HIV-1 subtype B infection through contaminated blood product before 1985 and exhibited a slow disease progression. We reported previously the emergence of an escape mutation in HIV-1 Pol from cytotoxic T-lymphocyte (CTL) response in association with a mild increase in viral load in 1999 in this patient (KI-127)13. The HIV-1 viral load was steady around 104 copies/mL between 2002 and 2006 (Figure 1a). However, at the end of 2006, the viral load began to increase, coupled with a rapid fall in CD4+ cell count. Since the emergence of CXCR4-tropic variants was suspected, changes in the V3 region were analyzed at five time points (June 2003, April 2006, and January, April, and November 2007) by sequencing 19–27 clones. Originally, most of the clones had identical or resembled V3 sequences with CCR5-tropic HIV-1 JRFL (Figure 1b). Interestingly, a unique clone, harboring arginine insertion at position 11 of V3 (Ins11R), one amino acid deletion at position 25 (Del25), and other multiple amino acid substitutions, was identified at a frequency of 1/22 in January 2007, and the frequency of the same kind of the clones subsequently increased to 2/23 in April 2007, and 9/27 in November 2007, which was considered to be associated with the rapid fall in CD4+ cell count.
Figure 1.
(a). Clinical course of Case 1. The CD4+ cell count (diamonds and solid lines) and HIV-1 load (circles and broken lines) from 2002 to 2008 are plotted. Arrows at the top indicate the five time points when V3 sequences were analyzed. Open diamonds and circles indicate the CD4+ cell counts and HIV-1 loads at the same five time points. (b). V3 sequence changes in Case 1.Cloned sequences analyzed at the five time points are shown. The V3 sequence of HIV-1 JRFL is shown at the top column as a reference. Amino acids identical to those of HIV-1 JRFL are indicated as dashes. The numbers of clones harboring the corresponding V3 sequences are shown on the right.
Insertion and deletion confer CXCR4-tropism
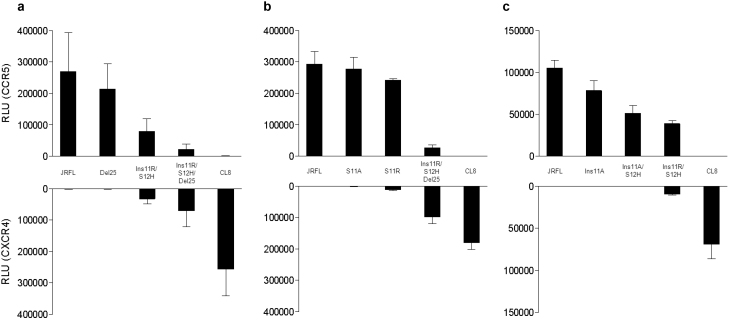
In the next step, cell-cell fusion assay was performed to analyze the effect of the observed V3 changes on viral tropism, using Env-expressing 293 T cells and CD4+ and CCR5+/CXCR4+ COS-7 cells. One V3 clone harboring Ins11R and Del25 identified in November 2007, named CL8-V3 (Figure 1b), was incorporated into JRFL Env-expressing plasmid. The cell-cell fusion assay demonstrated that CL8-V3 was purely CXCR4-tropic whereas JRFL was purely CCR5-tropic (Figure 2a). Ins11R occurred by the insertion of ‘ACA’ between ‘G’ and ’T’ of ‘AGT’ at position 11 at nucleotide sequence level, and therefore, the substitution of serine (S) with histidine (H) at position 12 (S12H) was also associated with Ins11R in Case 1 (‘AGT’ → ‘AGACAT’ at nucleotide level; ‘S’ → ‘RH’ at amino acid level [Ins11R/S12H]). To identify the determinant of observed tropism change, the plasmids harboring Ins11R/S12H, Del25, and Ins11R/S12H/Del25 were also constructed using JRFL backbone. In the cell-cell fusion assay, Ins11R/S12H decreased CCR5-tropism of JRFL-V3 and conferred CXCR4-tropism, resulting in dual-tropic (Ins11R/S12H vs. JRFL). Del25 further decreased the CCR5-tropism of Ins11R/S12H-V3 and increased CXCR4-tropism (Ins11R/S12H/Del25 vs Ins11R/S12H), though Del25 alone did not significantly change the JRFL-V3 tropism (Del25 vs JRFL). Considered together, the results suggest that Ins11R/S12H is indispensable for CXCR4-tropism of CL8-V3 and that Del25 strengthened the CXCR4-tropism in the presence of Ins11R/S12H. However, their combination was not enough to confer JRFL-V3 pure CXCR4-tropism (Ins11R/S12H/Del25-V3-expressing 293 T cells still fused with CD4+/CCR5+ COS-7 cells at low level), and some other mutations were necessary for pure CXCR4-tropism of CL8-V3.
Figure 2.
Effect of Ins11R/S12H and Del25 in cell-cell fusion assay (a). Cell-cell fusion assay was performed using Env-expressing 293 T cells and CD4+ and CCR5+/CXCR4+ COS-7 cells. Data are mean±SD values in relative luminescent unit (RLU) of six experiments (performed in duplicate and repeated three times). Analysis of two elements of Ins11R in cell-cell fusion assay (b and c). Effects of placing R at position 11 (b) and one amino acid elongation of V3 (c) were analyzed. Cell-cell fusion assay was performed using Env-expressing 293 T cells and CD4+ and CCR5+/CXCR4+ COS-7 cells. Data are mean±SD values in relative luminescent unit (RLU) of six experiments (performed in duplicate and repeated three times).
Which is important, substitution or elongation? The above results suggested that Ins11R/S12H was indispensable for CXCR4-tropism of CL8-V3. According to the 11/25 rule, a basic amino acid residue (R or lysine [K]) at either position 11 or 25 of V3 is associated with CXCR4-tropism, whereas acidic or neutral amino acid residues at these positions are associated with CCR5-tropism12,14,15. Ins11R/S12H has two elements: one is to place R at position 11 and the other is one amino acid elongation of V3. To determine whether positioning R at 11 is sufficient for conferring CXCR4-tropism or whether V3 elongation is also necessary for this process, S at position 11 of JRFL-V3 was substituted with R (S11R) and alanine (A) (S11A) as reference. However, both substitutions did not alter the pure CCR5-tropism of JRFL (Figure 2b), indicating that not only R at position 11 but also one amino acid elongation was indispensable for dual tropism caused by Ins11R/S12H.
Is one amino acid elongation sufficient to induce CXCR4-tropism or is positioning R at 11 is also necessary? To answer this question, two V3-expressing plasmids were constructed; one harbored Ins11A only and the other harbored Ins11A and S12H (Ins11A/S12H). The cell-cell fusion assay indicated that Ins11A decreased and Ins11A/S12H further decreased infectivity with CCR5 though neither of them conferred CXCR4-tropism (Figure 2c). These results indicate that not only one amino acid elongation of V3 but also positioning R at 11 is indispensable for dual tropism caused by Ins11R/S12H.
Effect of net charge of V3
Ins11R/S12H conferred CXCR4-tropism and Del25 strengthened it. However, Ins11R/S12H/Del25-V3 was still dual-tropic, though CL8-V3 was purely CXCR-4 tropic. The next question was which mutation is necessary for Ins11R/S12H/Del25-V3 to become purely CXCR4-tropic, like CL8-V3 (to lose CCR5-tropism)? There are six amino acid substitutions in CL8-V3 (substitution of asparagine [N] with tyrosine [Y] at position 5 [N5Y], substitution of threonine [T] with valine [V] at position 8 [T8V], substitution of K with isoleucine [I] at position 10 [K10I], substitution of Y with H at position 22 [Y22H], substitution of V with glycine [G] at position 26 [V26G], and substitution of aspartic acid [D] with N at position 29 [D29N]), compared with Ins11R/S12H/Del25-V3. According to the net charge rule, the higher net charge of V3 is associated with CXCR4-tropism when calculated by subtracting the number of negatively charged amino acid residues (D and glutamic acid [E]) from the number of positively charged ones (K and R)12,14. D29N was the only amino acid substitution that increased the net charge of V3 among the six amino acid substitutions described above. Therefore, we analyzed the effect of D29N by adding D29N to Ins11R/S12H/Del25-V3 (Ins11R/S12H/Del25/D29N) and reverting it in CL8-V3 (CL8/N29D). In the cell-cell fusion assay, D29N reduced CCR5-tropism of Ins11R/S12H/Del25-V3 though it remained dual-tropic (Ins11R/S12H/Del25/D29N vs Ins11R/S12H/Del25), and the reversion of D29N did not change CL8-V3 tropism (CL8/N29D vs CL8) (see Supplementary Figure S1). These results indicate that D29N does not cause tropism difference between Ins11R/S12H/Del25-V3 and CL8-V3, indicating that the net charge rule did not work well.
In silico prediction of the effect of substitutions
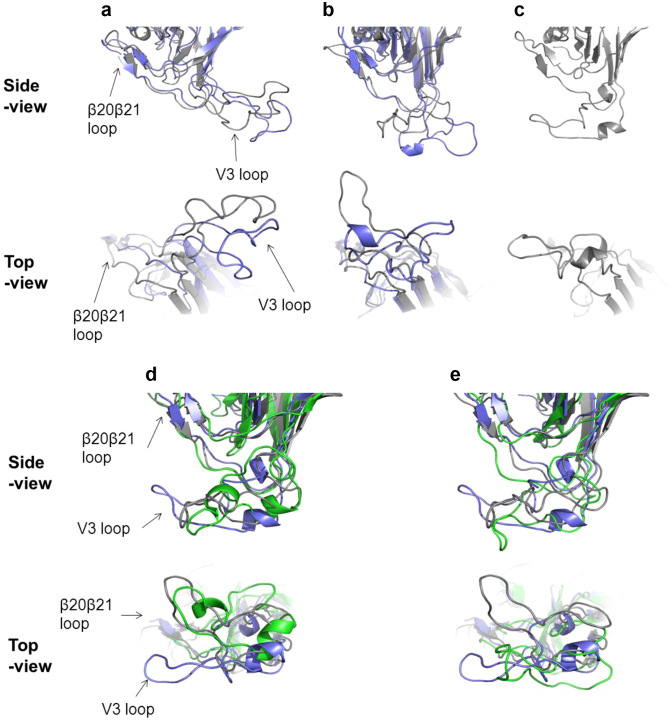
Adding D29N failed to alter the tropism of Ins11R/S12H/Del25-V3 from dual-tropic to purely CXCR4-tropic. There were five other amino acid substitutions between Ins11R/S12H/Del25-V3 and CL8-V3. Because the V3 conformation is important for coreceptor interactions16 and because conformation of V3 loop is sensitive to V3 mutations17,18, we examined how these V3 mutations could influence conformation of V3 in solution using molecular dynamics (MD) simulation19. In our MD simulation study, V3-loops of JRFL and Del25 (both CCR5-tropic) were placed in the opposite direction from the β20-β21 loop (Figure 3a), while CL8-V3 loop (CXCR4-tropic) was closed to and in the same direction with the β20-β21 loop (Figure 3c). The results were consistent with those obtained with gp120LAI recombinant outer domains containing CCR5-tropic and CXCR4-tropic V3 loop, respectively17,18. The loops of dual-tropic Ins11R/S12H-V3 and Ins11R/S12H/Del25 were located between Del25-V3 and CL8-V3 (Figure 3b). In fact, when the structural differences at the tip of the V3 tip region, i.e., the ‘GPGR’ amino acid residues were quantitatively measured with the root mean square deviation (RMSD) of the main chain17, CL8-V3 was found to be located far from the loop of JRFL-V3 and Del25-V3, while those of Ins11R/S12H-V3 and Ins11R/S12H/Del25-V3 were between them (Table 1). These results suggest that our MD simulation could predict the V3 tropism based on the magnitude of the RMSD values of the V3 loop tip. In the next step, each of the six amino acid substitutions of CL8-V3 was incorporated into Ins11R/S12H/Del25-V3, and the location and conformation of the constructed loop was analyzed. When D29N was incorporated, the RMSD from JRFL-V3 decreased and that from CL8-V3 increased (Table 2), and the loop orientation was still similar to that of Ins11R/S12H/Del25 (Figure 3d), suggesting that D29N does not seem to change the tropism, compatible with the results of the cell-cell fusion assay (see Supplementary Figure S1). Among other single amino acid substitutions, only T8V was found to increase the RMSD from JRFL-V3 and decrease that from CL8-V3 (Table 2), and caused a positional shift of the V3 resembling that of the CL8-V3 (Figure 3e). N5Y did not change the orientation of the V3 loop (see Supplementary Figure S2a) though the RMSD from CL8-V3 increased and that from JRFL-V3 decreased (Table 2). K10I, Y22H, and V26G decreased the RMSD from JRFL-V3 and increased that from CL8-V3 (Table 2), and the V3 loop orientation was distinct from both Ins11R/S12H/Del25-V3 and CL8-V3 (see Supplementary Figure S2b, S2c, and S2d). These results suggest that among the six amino acid substitutions, T8V has the greatest impact on the tropism shift toward CXCR4-tropic.
Figure 3.
Structural models of V3 loops on HIV-1 gp120 outer domains (a, b and c). MD simulations were performed for the HIV-1 JRFL gp120 outer domain with various V3 loops for CCR5 (a), dual (b), and CXCR4 (c) tropism. The most frequently appeared structures during 5–10 ns of MD simulations were extracted, and the top and side views of the structures around V3 loops are highlighted. (a) JRFL (gray) and Del25 (navy). (b) Ins11R/S12H/Del25 (gray) and Ins11R/S12H (navy). (c) CL8 (gray). Structural models of V3 loops of Ins11R/S12H/Del25-derived mutants (d and e). MD simulations were performed for the HIV-1 Ins11R/S12H/Del25 gp120 outer domain with D29N (d) or T8V (e) substitution in V3 loop. The most frequently appeared structures during 5–10 ns of MD simulations were extracted and superimposed with those of Ins11R/S12H/Del25 and CL8. (d) Superimposition of D29N (green), Ins11R/S12H/Del25 (gray), and CL8 (navy). (e) Superimposition of Ins11R/S12H/Del25/T8V (green), Ins11R/S12H/Del25 (gray), and CL8 (navy). Top and side views of the structures around V3 loops are shown.
Table 1. Overall structural differences between the two V3 loop tips of various HIV-1 variants.
| RMSD (Å)* | ||||
|---|---|---|---|---|
| ID of V3 | JRFL | Del25 | Ins11R/S12H | Ins11R/S12H/Del25 |
| Del25 | 13.8 | - | - | - |
| Ins11R/S12H | 17.4 | 8.6 | - | - |
| Ins11R/S12H/Del25 | 29.4 | 28.7 | 23.6 | - |
| CL8 | 38.9 | 37.5 | 33.1 | 14.2 |
*RMSD values of the main chain atoms at V3 tips (GPGR) of two gp120 outer domain models from MD simulations. A smaller RMSD value means a closer conformation between two gp120s.
Table 2. Effect of a single amino acid substitution on overall structure of the gp120 V3 tip.
| RMSD (Å)¶ | |||
|---|---|---|---|
| ID of V3 | Added mutations* | JRFL | CL8 |
| Ins11R/S12H/Del25 | None | 29.4 | 14.2 |
| Ins11R/S12H/Del25/N5Y | N5Y | 32.5 | 14.1 |
| Ins11R/S12H/Del25/T8V | T8V | 33.6 | 12.6 |
| Ins11R/S12H/Del25/K10I | K10I | 26.3 | 39.4 |
| Ins11R/S12H/Del25/Y22H | Y22H | 28.6 | 27.0 |
| Ins11R/S12H/Del25/V26G | V26G | 28.4 | 19.6 |
| Ins11R/S12H/Del25/D29N | D29N | 28.4 | 17.0 |
*Added amino acid substitution in the V3 loop of the Ins11R/S12H/Del25 gp120.
¶RMSD values of the main chain atoms at V3 tips (GPGR) of two gp120 outer domain models from MD simulations.
Impact of T8V
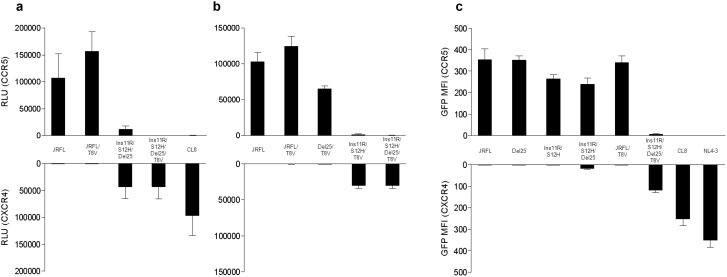
Our in silico modeling predicted that T8V could alter the tropism of Ins11R/S12H/Del25-V3. In the next series of experiments, we incorporated T8V into JRFL-V3 (JRFL/T8V) and Ins11R/S12H/Del25-V3 (Ins11R/S12H/Del25/T8V), and analyzed the effect of such incorporation on their tropism using the cell-cell fusion assay. The incorporation of T8V into JRFL-V3 increased CCR5-tropism though it did not confer CXCR4-tropism (Figure 4a and 4b). However, T8V abrogated CCR5-tropism of Ins11R/S12H/Del25-V3 and converted it to purely CXCR4-tropic, although it did not increase CXCR4-tropism and Ins11R/S12H/Del25/T8V-V3 still had smaller CXCR4-tropism than CL8-V3 (Figure 4a). The combination of Ins11R/S12H/T8V was sufficient to confer CXCR4-tropism, although Del25/T8V did not (Figure 4b). T8V breaks the N-linked glycosylation motif ‘NXT’ at position 6–8 of V3, the loss of which was reported with tropism shift towards CXCR4-tropic20,21. Our results indicated that T8V was indispensable for pure CXCR4-tropism of CL8, which seemed to support the previous findings of the importance of the loss of N-linked glycosylation motif for CXCR4-tropism. The loss of the glycan moiety in V3 stem might lead to change gp120 interaction surface for coreceptor binding and influence coreceptor tropism. However, available structural information was against this possibility, because the glycosylation site was exposed toward an opposite direction from the putative coreceptor binding site on V316,22,23. Accordingly, presence or absence of the glycan moiety in V3 stem did not cause significant differences in V3 configuration in our MD simulation system17,24. Probably, amino acid substitution itself altered V3 configuration and coreceptor tropism.
Figure 4.
Effects of T8V in cell-cell fusion assay (a and b). The effects of T8V were analyzed in combination with Ins11R/S12H/Del25 (a), and Del25 and Ins11R/S12H (b). Cell-cell fusion assay was performed using Env-expressing 293 T cells and CD4+ and CCR5+/CXCR4+ COS-7 cells. Data are mean±SD values in relative luminescent unit (RLU) of six experiments (performed in duplicate and repeated three times). Tropism of recombinant HIV-1 variants harboring mutations identified in Case 1 (c). Tropism of HIV-1 variants was assessed in CCR5+ GHOST Hi5 and CXCR4+ GHOST CXCR4 cells. The mean fluorescent intensity (MFI) of infected cells expressing green fluorescent protein (GFP) was measured. Data are mean±SD values of six experiments (performed in duplicate and repeated three times).
GHOST cell infection assay
Our cell-cell fusion assay indicated that Ins11R/S12H and T8V were indispensable for pure CXCR4-tropism of CL8. The next series of experiments were designed to confirm the findings using HIV-1 infection assay in GHOST cells25,26. HIV-1 JRFL and the recombinant HIV-1 variants harboring Del25-V3 and T8V-V3 had the same level of CCR5-tropism, although none could infect CXCR4+ GHOST cells (Figure 4c). In comparison, Ins11R/S12H-V3- and Ins11R/S12H/Del25-V3-harboring variants had lower levels of CCR5-tropism. The latter variant, but not the former, infected CXCR4+ GHOST cells though at low level. The Ins11R/S12H/Del25/T8V-V3-harboring variant lost the CCR5-tropism and acquired CXCR4-tropism, although the level of CXCR4-tropism was still lower than those of CL8-V3-harboring variant and HIV-1 NL4-3 (a CXCR4-tropic experimental strain). These results were compatible with the abovementioned results of the cell-cell fusion assay, though the CCR5-tropism of Ins11R/S12H/Del25-V3 seemed stronger in the cell infection assay. Dual-tropic Ins11R/S12H/Del25-V3 might have decreased susceptibility to AMD3100 used in the CCR5+ GHOST Hi5 assay compared with pure CXCR4-tropic CL8-V3 and NL4-3-V3.
Same V3 pattern in two other cases
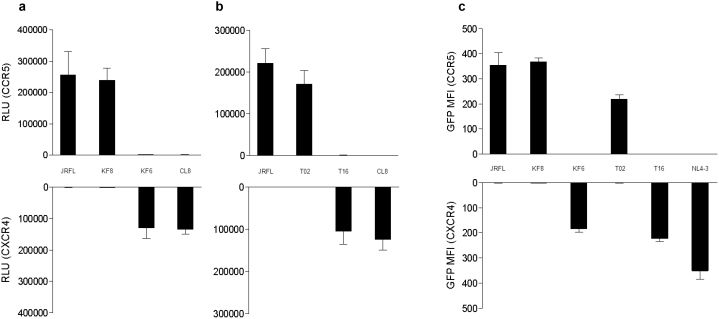
The analysis of V3 sequence changes in Case 1 demonstrated that Ins11R and the loss of N-linked glycosylation site indispensably contribute to a shift toward CXCR4-tropism. To determine whether this finding was true only in Case 1 or could be generalized to other cases, HIV-1 subtype B V3 sequences were analyzed in 53 other treatment-naïve patients with CD4+ cell count < 200/mm3. The same pattern of mutations was identified in two cases (3.8%). In one case (Case 2), four of twenty analyzed sub-clones of V3 sequences harbored Ins11R associated with S12H, Del25, and N6A resulting in the loss of N-linked glycosylation site, compared with JRFL-V3 (Figure 5). In the other case (Case 3), three of twenty-two sub-clones harbored Ins11R associated with S11R, Del25, and T8V, resulting in the loss of N-linked glycosylation site. To delineate the tropism of the V3 abovementioned clones, two V3 clones in each case, one harboring Ins11R and the loss of N-linked glycosylation site (KF6 in Case 2, T16 in Case 3 [see Figure 5]) and the other harboring none of them (KF8 in Case 2, T02 in Case 3 [see Figure 5]), was incorporated into JRFL Env-expressing plasmid. As expected, cell-cell fusion assay indicated that the clones harboring Ins11R and the loss of N-linked glycosylation site (KF6 and T16) were purely CXCR4-tropic, although the clones harboring none of them (KF8 and T02) were purely CCR5-tropic (Figure 6a and 6b). The results of the GHOST cell infection assay using V3-incorporated HIV-1 JRFL (Figure 6c) were similar to those of the cell-cell fusion assay. Accordingly, it was concluded that the findings of the indispensability of Ins11R and the loss of N-linked glycosylation site for CXCR4-tropism were not only true in Case 1 but also in other cases.
Figure 5. Cloned V3 sequences in Cases 2 and 3.
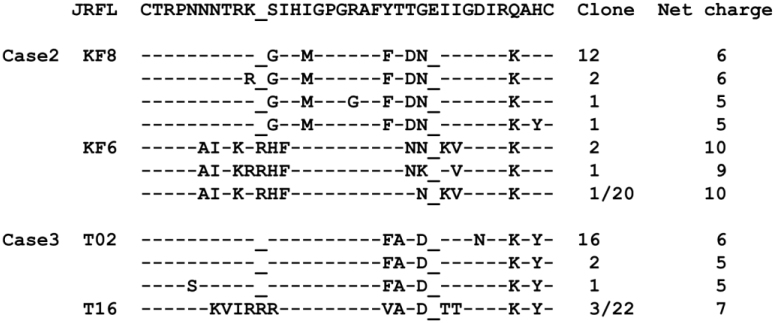
The V3 sequence of HIV-1 JRFL is shown at the top column as a reference. Amino acids identical to those of HIV-1 JRFL are indicated as dashes. The numbers of clones harboring the corresponding V3 sequences are shown on the right.
Figure 6.
Tropism of cloned V3 incorporated into JRFL gp120 backbone (a and b). Two distinct V3 clones from each of Case 2 (a) and Case 3 (b) were analyzed with the reference of JRFL-V3 and CL8-V3. Cell-cell fusion assay was performed using Env-expressing 293T cells and CD4+ and CCR5+/CXCR4+ COS-7 cells. Data are mean±SD values in relative luminescent unit (RLU) of six experiments (performed in duplicate and repeated three times). Tropism of recombinant HIV-1 variants harboring V3 sequences derived from Cases 2 and 3 (c). Tropism of HIV-1 variants was assessed in CCR5+ GHOST Hi5 and CXCR4+ GHOST CXCR4 cells. The mean fluorescent intensity (MFI) of infected cells expressing green fluorescent protein (GFP) was measured. Data are mean±SD values of six experiments (performed in duplicate and repeated three times).
Discussion
The phenotypic assay Trofile™ (Monogram Bioscience, South San Francisco, CA), which is based on recombinant virus technology, has been the most widely used diagnostic test for the detection of CXCR4-tropic HIV-1 variants27. However, this method has logistical and technical limitations that make it far from convenient as a diagnostic test in clinical practice. Genotypic methods based on V3 sequence represent a more feasible alternative28 and are progressively replacing phenotypic assays, though their clinical use requires good genotypic-phenotypic correlations. The 11/25 rule and the net charge rule were proposed for the tropism prediction from V3 sequence12,14,15, although they show only a moderate correlation with the results of phenotypic assays12,15,28. The results of specific genotypic tools, such as geno2pheno (Max-Planck Institute, Munich, Germany)29,30 and position-specific scoring matrix (PSSM)31,32 are comparable to those of phenotypic assays, suggesting that there should be some more genetic determinations for viral tropism. In this study, we successfully demonstrated two rules other than the 11/25 rule and the net charge rule on the association with CXCR4-tropic variants. One was that R insertion at position 11 of V3, not just placing R at position 11 but also one amino acid elongation, strongly shifted the HIV-1 tropism towards CXCR4-tropic. The other was that the loss of N-linked glycosylation site in V3 also shifted viral tropism towards CXCR4-tropic, which was previously described in some reports20,21. In the V3 analysis in our index case, R insertion at position 11 conferred dual-tropism to originally CCR5-tropic V3, and the loss of N-linked glycosylation site altered it totally CXCR4-tropic (see Supplementary Figure S3). We identified these mutation patterns not only in the index case but also in two other cases. When we surveyed V3 sequences with tropism confirmed by phenotypic assay registered at the Los Alamos HIV sequence database (Los Alamos National laboratory, Los Alamos, NM) (http://www.hiv.lanl.gov. as of September 25, 2012), 28 sequences had R insertion at position 11; 7 of 199 (3.5%) CXCR4-tropic, 14 of 513 (2.7%) dual-tropic, and 7 of 3301 (0.2%) CCR5-tropic sequences. Their frequency was significantly higher in CXCR4-tropic and dual-tropic sequences than CXCR5-tropic ones (p < 0.0001; Chi-square test). (All of the 7 CCR5-tropic sequences with R insertion at position 11 were sub-clones derived from one pair of a transmitter mother and her transmitted child33, and the sequences were so unique that it was actually difficult to determine the exact site of one amino acid insertion). Interestingly, all of the 28 V3 sequences with R insertion at position 11, had lost the N-linked glycosylation site and had one amino acid deletion in the C-terminal half of V3 (one amino acid deletion at position 24 or 25 in 18 sequences [64.3%]), similar to our three cases. No other amino acid elongation patterns were found in the N-terminal half of V3 in the Los Alamos database. There were 3,301 CCR5-tropic V3 sequences registered in the database. Among them, 18 sequences had a basic amino acid residue at position 11 and therefore they were misjudged as CXCR4-tropic by the 11/25 rule. Only 7 of them had R insertion and the other 11 were recognized as CCR5-tropic by our rules. Therefore using our rules increased the specificity from 99.5% [(3,301–18)/3,301] to 99.8% [(3,301–7)/3,301] in identifying CXCR4- or dual-tropic V3 sequences in the Los Alamos database.
Considered together, amino acid elongation may be a rare event, but R insertion at position 11 sometimes occurs. The occurrence of such insertion seems to be always accompanied by loss of the N-linked glycosylation site and deletion of one amino acid in the C-terminal half of V3. The combination of these mutations usually confers CXCR4-tropism. Awareness of this rule will help to confirm the tropism prediction from V3 sequences by conventional rules.
Methods
Patients
Case 1 was an ART-naive Japanese hemophiliac who acquired HIV-1 subtype B infection through contaminated blood product before 1985 and exhibited slow disease progression, as described previously (KI-127)13. The study also included 53 other treatment-naïve HIV-1 subtype B-infected patients with CD4+ cell count < 200/mm3, who were newly diagnosed in 2008. The ethics committee of The National Center for Global Health and Medicine approved the study and all participants provided written informed consent.
Cells
The 293 T and COS-7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco, Grand Island, NY) with 10% fetal bovine serum (FBS; Equitech-Bio, Kerrville, TX). Parental GHOST cells34 were cultured in DMEM supplemented with 10% FBS, 500 μg/ml G418 and 100 μg/ml hygromycin B. CCR5+ GHOST Hi5 and CXCR4+ GHOST CXCR4 cells34 were cultured in DMEM supplemented with 10% FBS, 500 μg/ml G418, 100 μg/ml hygromycin B and 1 μg/ml puromycin.
Amplification, cloning and sequencing of Env V3 region
Total RNA was extracted from 200 μl of plasma using High Pure Viral RNA Kit (Roche, Indianapolis, IN) according to the instructions supplied by the manufacturer. HIV-1 cDNA was obtained by reverse transcriptase-polymerase chain reaction (RT-PCR) using the One Step RNA PCR kit (TaKaRa Bio, Kyoto, Japan). The DNA fragments were amplified by using the Ex Taq Hot Start Version (TaKaRa Bio) with the primer sets as follows. The Env fragment containing V3 region was amplified by RT-PCR with primers of C2 (5′ - AATGTCAGCACAGTACAATGTACAC - 3′) and C3 (5′ - ACAATTTCTGGGTCCCCTCCTGAGGA - 3′). S1 (5′ - ATGGAATTAGGCCAGTAGT - 3′) and A1 (5′ - CTCTTAATTTTATAACTATC - 3′) primer sets were used for nested PCR. The amplified PCR products were purified using QIAquick PCR purification kit (Qiagen, Valencia, CA) and cloned by using the TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA) according to the protocol provided by the manufacturer. At least 19 colonies were selected, inoculated into 4 ml of L broth, and incubated at 37°C overnight under vigorous agitation. In the next step, plasmids were isolated by using the QIAprep Spin Miniprep Kit (Qiagen). The purified plasmids were sequenced by using the ABI BigDye Terminator v3.1 Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA) and processed with an automated ABI 3730 DNA Analyzer (Applied Biosystems).
Plasmid construction
The pcDNA6.2-CCR5 and pcDNA6.2-HIV-tat plasmids were constructed as described previously35. Briefly, the entire human CCR5 gene including a stop codon was amplified using pZeoSV-CCR536 as a template. The PCR product was ligated into pcDNA6.2/cLumio-DEST vector (Invitrogen), cloned using the method recommended by the manufacture, and termed as pcDNA6.2-CCR5. The CD4 expression vector (pcDNA6.2-CD4) and CXCR4 expression vector (pcDNA6.2-CXCR4) were generated using the same method. The CCR5-tropic HIV-1 JRFL37 Env expression vector (pCXN-JRenv) and pLTR-LucE were used as described previously35. The full length Env and part of the Nef encoding regions of the HIV-1 genome was amplified using pHIV-1 JRFL. The PCR product was ligated into pGEM-T Easy Vector System (Promega, Madison, WI), cloned using the protocol supplied by the manufacturer, and termed as pGEM-T Easy-Env. Amino acid substitutions, insertion and deletion were introduced into the V3 region of pGEM-T Easy-Env using the Quikchange Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA) and applying the protocols supplied by the manufacturer. The V3 regions of pGEM-T Easy-Env containing mutations were digested with StuI and XhoI, and the obtained fragments were introduced into pCXN-JRenv or pHIV-1 JRFL.
Cell-cell fusion assay
The assay was conducted as described in detail previously35. Briefly, the JRFL Env expression vector (WT or mutant) and Tat expression vector (0.5 μg each) were cotransfected into 293 T cells (2 × 105) using Lipofectamine 2000 (Invitrogen), while the CD4, CCR5 or CXCR4 expression vector and a reporter (luciferase) gene containing plasmid, pLTR-LucE (0.5 μg each) were cotransfected into COS-7 cells (2 × 105). On the next day, both cotransfected cells were harvested and mixed in a well of 96-well plates (2 × 104 cells each). The cotransfected cells were incubated further for 6 hrs and the luciferase activity in each well was detected using Bright-Glo Luciferase Assay System (Promega) and its luminescence level was measured using Wallac ARVO Sx 1420 multilabel counter (Perkin-Elmer, Waltham, MA).
MD simulation
MD simulation of gp120 outer domain containing V3 loop was performed as described previously17,18 with some modifications. Initially, the gp120 outer domain structures with various V3 elements were constructed by homology modeling38,39 using the Molecular Operating Environment (MOE) ver. 2008.10 (Chemical Computing Group Inc., Montreal, Quebec, Canada), as described previously17,18. As a modeling template, we used the crystal structure of HIV-1 gp120 containing the entire V3 element (PDB code: 2B4C)19. Subsequently, MD simulations were performed for individual models using the SANDER module in the AMBER 9 program package40,41. Heating calculations were achieved for 100 picoseconds until 310 K and MD simulations were subsequently executed at 310 K for 10 nanoseconds. The time step was set to 2.0 femtoseconds. The AMBER ff03ua force field42 and the GB implicit solvent function by Hawkins, Cramer, and Truhlar43,44 were applied. The "no cutoff" calculation was applied for the non-bonded energy calculation. In this study, we analyzed most frequently observed conformation among 5,000 snapshots obtained from 5.0–10.0 ns of MD simulation, which was selected by the Bayesian clustering algorithm45.
Calculation of the RMSD
We compared the orientation of V3 loop between two gp120 outer domain models by the following procedure. We first superimposed two models by coordinating main chain atoms (N, Cα, and C) in amino acid residues other than those in the V3 loop using PyMOL ver. 0.99 rc6 (Schrödinger LLC, Portland, OR, http://www.pymol.org/). Subsequently, the RMSD values for the V3 loop tip (GPGR) between the two models were calculated using the coordinates of the main chain atoms using the in-house program.
Viral tropism assay
The wild type CCR5-tropic HIV-1 strain, pHIV-1 JRFL, CXCR4-tropic HIV-1 strain, pHIV-1 NL4-346, and each pHIV-1 JRFL Env derived from mutations containing the V3 region of pGEM-T Easy-Env were transfected into 293 T cells with Lipofectamine 2000 (Invitrogen), and the obtained infectious clonal viruses were harvested 48 hrs after transfection and stored at −80°C until use. The GHOST cell infection assay25,47 was performed by incubating 1 ml containing 20 ng of p24 antigen of each virus with GHOST cells (2 × 104). Parental GHOST, CCR5+ GHOST Hi5, and CXCR4+ GHOST CXCR4 cells were infected for 72 hrs and then harvested. The mean fluorescent intensity (MFI) of infected cells expressing green fluorescent protein (GFP) was measured on a flow cytometer (FACSCalibur; BD Bioscience, San Jose, CA). GHOST cells express low levels of CXCR4 and therefore infection of GHOST Hi5 alone was performed in presence of the CXCR4 antagonist AMD3100 (Sigma-Aldrich, St. Louis, MO) at dose of 1 μM.
Author Contributions
K.T. designed and performed the research, analyzed the data and wrote the manuscript. H.O. and H.S. performed MD simulation and calculation of the RMSD, wrote the manuscript. T.H. and J.K. performed the cloning and sequencing. S.O. designed and supervised the study. H.G. designed the study, analyzed the data, wrote and edited the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Supplementary Figure S1, Supplementary Figure S2, Supplementary Figure S3
Acknowledgments
We thank Dr. Kenji Maeda, Experimental Retrovirology Section, HIV and AIDS Malignancy Branch, National Cancer Institute, National Institutes of Health, for providing plasmids and helpful suggestion, and Drs. Hiroaki Mitsuya and Hirotomo Nakata, Department of Infectious Diseases, Kumamoto University School of Medicine, for providing GHOST cell lines. We also thank the clinical and laboratory staffs of the AIDS Clinical Center, National Center for Global Health and Medicine, for their helpful support. This work was supported by a Grant-in-Aid for AIDS research from the Japanese Ministry of Health, Labour and Welfare (H23-AIDS-001), and the Global Center of Excellence Program (Global Education and Research Center Aiming at the Control of AIDS) from the Japanese Ministry of Education, Science, Sports, and Culture.
Footnotes
S.O. has received honorariums and research grants from MSD, Janssen Pharmaceutical, Abbott, Roche Diagnostics, and Pfizer; has received honorariums from ViiV Healthcare, Torii Pharmaceutical, Bristol-Myers, Astellas Pharmaceutical, GlaxoSmithKline, Taisho Toyama Pharmaceutical, Dainippon Sumitomo Pharma, and Daiichisankyo. The remaining authors declare to have no conflict of interest.
References
- Parra J. et al. Clinical utility of maraviroc. Clin. Drug Invest. 31, 527–542 (2011). [DOI] [PubMed] [Google Scholar]
- Alkhatib G. et al. CC CKR5: a RNATES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272, 1955–1958 (1996). [DOI] [PubMed] [Google Scholar]
- Dragic T. et al. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381, 667–673 (1996). [DOI] [PubMed] [Google Scholar]
- van't Wout A. B. et al. Macrophage-tropic variant initiate human immunodeficiency virus type 1 infection after sexual, parenteral and vertical transmission. J. Clin. Invest. 94, 2060–2067 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T. et al. Genotypic and phenotypic characterization of HIV-1 in patients with primary infection. Science 261, 1179–1181 (1993). [DOI] [PubMed] [Google Scholar]
- Björndal A. et al. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 71, 7478–7487 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlatti G. et al. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine mediated suppression. Nat. Med. 3, 1259–1265 (1997). [DOI] [PubMed] [Google Scholar]
- Blaak H. et al. In vivo HIV-1 infection of CD45RA+ CD4+ T cells is established primarily by syncytium-inducing variants and correlates with the rate of CD4+ T cell decline. Proc. Natl. Acad. Sci. U. S. A. 97, 1269–1274 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor R. I., Sheridan K. E., Ceradini D., Choe S. & Landau N. R. Change in copeceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185, 621–628 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koot M. et al. Prognostic value of human immunodeficiency virus type 1 biological phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann. Intern. Med. 118, 681–688 (1993). [DOI] [PubMed] [Google Scholar]
- The HHS Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the Use of Antiretroviral Agents in HIV-1-Infected Adults and Adolescents. U.S. Department of Health and Human Services (2011). [Google Scholar]
- Poveda E. et al. Genotype determination of HIV tropism – clinical and methodological recommendations to guide the therapeutic use of CCR5 antagonists. AIDS Rev. 12, 135–148 (2010). [PubMed] [Google Scholar]
- Kawashima Y. et al. Long-term control of HIV-1 in hemophiliacs carrying slow-progressing allele HLA-B*5101. J. Virol. 84, 7151–7160 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delobel P. et al. Population-based sequencing of the V3 region of env for predicting the coreceptor usage of human immunodeficiency virus type 1 quasispecies. J. Clin. Mcirob. 45, 1572–1580 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandekerckhove L. P. et al. European guidelines on the clinical management of HIV-1 tropism testing. Lancet Infect. Dis. 11, 394–407 (2011). [DOI] [PubMed] [Google Scholar]
- Huang C. C. et al. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 317, 1930–1934 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama M., Naganawa S., Yoshimura K., Matsushita S. & Sato H. Structural dynamics of HIV-1 envelope gp120 outer domain with V3 loop. PLoS One 7, e37530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganawa S. et al. Net positive charge of HIV-1 CRF01_AE V3 sequence regulates viral sensitivity to humoral immunity. PLoS One 3, e3206 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. C. et al. Structure of a V3-containing HIV-1 gp120 core. Science 310, 1025–1028 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevestig P., Pramanik L., Leitner T. & Ehrnst A. CCR5 use by human immunodeficiency virus type 1 is associated closely with the gp120 V3 loop N-linked glycosylation site. J. Gen. Virol. 87, 607–612 (2006). [DOI] [PubMed] [Google Scholar]
- Van Baelen K. et al. HIV-1 coreceptor usage determination in clinical isolates using cloncal and population-based genotypic and phenotypic assays. J. Virol. Methods 146, 61–73 (2007). [DOI] [PubMed] [Google Scholar]
- Schnur E. et al. The conformation and orientation of a 27-residue CCR5 peptide in a ternary complex with HIV-1 gp120 and a CD4-mimic peptide. J. Mol. Biol. 410, 778–797 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R. et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334, 1097–1103 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwata T. et al. Conformational epitope consisting of the V3 and V4 loops as a target for potent and broad neutralization of simian immunodeficiency viruses. J. Virol. 87, 5424–5436 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecilia D. et al. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J. Virol. 72, 6988–6996 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B. K. et al. Biologic and genetic characterization of a panel of 60 human immunodeficiency virus type 1 isolates, representing clades A, B, C, D, CRF01_AE, and CRF02_AG, for the development and assessment of candidate vaccines. J. Virol. 79, 6089–6101 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcomb J. M. et al. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob. Agents Chemother. 51, 566–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M. A. & van't Wout A. B. Predicting HIV-1 coreceptor usage with sequence analysis. AIDS Rev. 5, 104–112 (2003). [PubMed] [Google Scholar]
- Sing T. et al. Predicting HIV coreceptor usage on the basis of genetic and clinical covariates. Antivir. Ther. 12, 1097–1106 (2007). [PubMed] [Google Scholar]
- Jensen M. A. et al. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J. Virol. 77, 13376–13388 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low A. J. et al. Current V3 genotyping algorithms are inadequate for predicting X4 co-receptor usage in clinical isolates. AIDS 21, F17–24 (2007). [DOI] [PubMed] [Google Scholar]
- Poveda E. et al. Design and validation of new genotypic tools for easy and reliable estimation of HIV tropism before using CCR5 antagonists. J. Antimicrob. Chemother. 63, 1006–1010 (2009). [DOI] [PubMed] [Google Scholar]
- Arroyo M. A. et al. Virologic risk factors for vertical transmission of HIV type 1 in Puerto Rico. AIDS Res. Hum. Retroviruses 18, 447–460 (2002). [DOI] [PubMed] [Google Scholar]
- Mörner A. et al. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 73, 2343–2349 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K. et al. Involvement of the second extracellular loop and transmembrane residues of CCR5 in inhibitor binding and HIV-1 fusion: insights into the mechanism of allosteric inhibition. J. Mol. Biol. 381, 956–974 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y., Foda M., Matsushita S. & Harada S. Involvement of both the V2 and V3 regions of the CCR5-tropic human immunodeficiency virus type 1 envelope in reduced sensitivity to macrophage inflammatory protein 1alpha. J. Virol. 74, 1787–1793 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi Y. et al. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science 236, 819–822 (1987). [DOI] [PubMed] [Google Scholar]
- Martí-Renom M. A. et al. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29, 291–325 (2000). [DOI] [PubMed] [Google Scholar]
- Baker D. & Sali A. Protein structure prediction and structural genomics. Science 294, 93–96 (2001). [DOI] [PubMed] [Google Scholar]
- Kollman P. A. et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 33, 889–897 (2000). [DOI] [PubMed] [Google Scholar]
- Pearlman D. A. et al. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comp. Phys. Commun. 91, 1–41 (1995). [Google Scholar]
- Hsieh M. J. & Luo R. Balancing Simulation Accuracy and Efficiency with the Amber United Atom Force Field. J. Phys. Chem. B 114, 2886–2893 (2010). [DOI] [PubMed] [Google Scholar]
- Hawkins G. D., Cramer C. J. & Truhlar D. G. Parametrized models of aqueous free energies of solvation based on pairwise descreening of solute atomic charges from a dielectric medium. J. Phys. Chem. 100, 19824–19839 (1996). [Google Scholar]
- Hawkins G. D., Cramer C. J. & Truhlar D. G. Pairwise solute descreening of solute charges from a dielectric medium. Chem. Phys. Lett. 246, 122–129 (1995). [Google Scholar]
- Shao J., Tanner S. W., Thompson N. & Cheatham T. E. 3rd Clustering molecular dynamics trajectories: 1. Characterizing the performance of different clustering algorithms. J. Chem. Theory Comput. 3, 2312–2334 (2007). [DOI] [PubMed] [Google Scholar]
- Westervelt P., Gendelman H. E. & Ratner L. Identification of a determinant within the human immunodeficiency virus 1 surface envelope glycoprotein critical for productive infection of primary monocytes. Proc. Natl. Acad. Sci. U. S. A. 88, 3097–3101 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jekle A. et al. Coreceptor phenotype of natural human immunodeficiency virus with nef deleted evolves in vivo, leading to increased virulence. J. Virol. 76, 6966–6973 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1, Supplementary Figure S2, Supplementary Figure S3