Abstract
Ribosome assembly is a process fundamental for all cellular activities. The efficiency and accuracy of the subunit assembly are tightly regulated and closely monitored. In the present work, we characterized, both compositionally and structurally, a set of in vivo 50S subunit precursors (45S), isolated from a mutant bacterial strain. Our qualitative mass spectrometry data indicate that L28, L16, L33, L36 and L35 are dramatically underrepresented in the 45S particles. This protein spectrum shows interesting similarity to many qualitatively analyzed 50S precursors from different genetic background, indicating the presence of global rate-limiting steps in the late-stage assembly of 50S subunit. Our structural data reveal two major intermediate states for the 45S particles. Consistently, both states severally lack those proteins, but they also differ in the stability of the functional centers of the 50S subunit, demonstrating that they are translationally inactive. Detailed analysis indicates that the orientation of H38 accounts for the global conformational differences in these intermediate structures, and suggests that the reorientation of H38 to its native position is rate-limiting during the late-stage assembly. Especially, H38 plays an essential role in stabilizing the central protuberance, through the interaction with the 5S rRNA, and the correctly orientated H38 is likely a prerequisite for further maturation of the 50S subunit.
INTRODUCTION
As the largest ribonucleoprotein complex in the cell, the ribosome is responsible for the biosynthesis of proteins in all living organisms. Ribosome biogenesis accounts for a majority of cellular RNA synthesis activities, and is tightly regulated and coupled with growth control pathways (1). Despite the complex composition of ribosomal subunits, earlier pioneering works led by Nomura (2) and Nierhaus (3) have demonstrated that active subunits could be reconstituted in vitro from individual rRNAs and proteins. Through numerous measurements of binding interdependences between different proteins, they have constructed the general in vitro assembly maps for the 30S and 50S subunits (4,5). Their data reveal that ribosomal proteins bind in different orders, displaying both hierarchical and parallel manners.
In the cell, the subunit assembly is highly efficient, facilitated by a variety of cofactors with diverse functions, including ribonucleases, rRNA helicases and chaperones, rRNA and ribosomal protein modification enzymes and RNA-binding GTPases (1,6). Disruption of cofactors by genetic deletion or mutation leads to growth defects and accumulation of subunit precursors. GTPases constitute a large part of assembly cofactors (7,8). Although the precise roles of these GTPases are still not clear, emerging experimental data show that most of them act at relatively late stages of the subunit assembly, and particularly, some of them might function to couple the assembly to other cellular processes, or intervene at different checkpoints to ensure the quality of subunit production (7,9).
Several bacterial GTPases, such as YlqF, YsxC, EngA, ObgE, Era, RsgA and YqeH, have been implicated in the assembly of the 50S or 30S subunit (7). Among them, YlqF (also known as RbgA) was first identified to be indispensable for growth in Bacillus subtilis (10,11). YlqF belongs to an unusual GTPase family, featuring a circularly permuted GTPase domain (12). YlqF homologs are widely present in gram-positive bacteria, archaea and all eukaryotes, and also found in a few gram-negative bacteria (but not Escherichia coli) (13,14). Depletion of YlqF in B. subtilis confers a slow growth phenotype, and induces an abnormality of the in vivo ribosome profile, with an increased level of 45S precursors and a decreased level of 70S ribosomes (15–17). And the premature 45S particles accumulated in YlqF-depleted cells unambiguously lack several proteins, including L16, L27 and L36 (15,16,18), indicating that these 45S particles are a specific category of in vivo assembly intermediates.
To understand the structural change of the 23S rRNA during conformational maturation and to investigate possible quality control mechanism on the 50S subunit assembly, in the present work, we combined quantitative mass spectrometry (QMS) and cryo-electron microscopy (cryo-EM) to characterize the premature 45S particles isolated from an YlqF-deficient B. subtilis strain (16). The QMS data indicate that a few proteins, such as L28, L16, L33, L36 and L35, are dramatically underrepresented in the 45S particles. This protein spectrum shows interesting similarity to a number of qualitatively analyzed 50S precursors from E. coli, indicating the presence of global rate-limiting steps in the late-stage assembly of bacterial 50S subunit. More importantly, the structural analysis reveals several major conformations of the 23S rRNA in the 45S particles, differing in the stability of the functional centers of the 50S subunit and the orientation of a long rRNA helix H38. These structural data indicate that the 45S particles are defective in both subunit association and tRNA binding. Interestingly, further comparison of two intermediate 45S conformations shows that the reorientation of H38 from its intermediate non-native position to the native-like conformation is likely a rate-limiting step, which would lead to a global stabilization of the whole central protuberance (CP) of the 50S subunit. Together with published data, our results suggest that the mutual stabilization between the CP and H38 is crucial for the further maturation of the 23S rRNA in the functional centers, and assembly factors, such as YlqF, come into play to facilitate this transition to accelerate the late-stage 50S subunit maturation.
MATERIALS AND METHODS
Purification of mature and premature 50S ribosomal subunits
Mature 50S subunits were purified from wild-type B. subtilis 168 strain, and premature 45S particles from a previously constructed YM01 strain (16), in which the genomic ylqF gene was placed behind the isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible spac promoter. Both strains were grown in Luria-Bertani medium at 37°C without IPTG. Cells were collected through centrifugation, when OD600 reached 0.4–0.6, to avoid the formation of spores, in which most of the ribosomes are in the form of 100S. Cell pellets were resuspended with opening buffer [20 mM Tris–HCl, pH 7.5, 100 mM NH4Cl, 10.5 mM Mg(OAc)2, 0.5 mM EDTA, 1 mM TCEP (Tris(2-carboxyethyl)phosphine)], and disrupted by sonication. Cell debris was removed by centrifugation at 20 000 rpm (Avanti J-26 XP, Beckman Coulter) for 40 min. The supernatant was applied onto 5 ml sucrose cushion buffer [20 mM Tris–HCl, pH 7.5, 500 mM NH4Cl, 10.5 mM Mg(OAc)2, 0.5 mM EDTA, 1.1 M sucrose, 1 mM TCEP] and centrifuged using a 70Ti rotor (Beckman Coulter) at 28 000 rpm for 19 h. The pellets were washed with 2.0 ml washing buffer [20 mM Tris–HCl, pH 7.5, 500 mM NH4Cl, 10.5 mM Mg(OAc)2, 0.5 mM EDTA, 1 mM TCEP] and subjected to an additional centrifugation using 5 ml sucrose cushion. The final ribosomal pellets were washed and dissolved in buffer I [20 mM Tris–HCl, pH 7.5, 100 mM NH4Cl, 15 mM Mg(OAc)2, 1 mM TCEP].
For mature 50S subunits, the crude ribosomal mixture was subjected to a 10–40% sucrose density gradient [15 mM Mg(OAc)2] centrifugation using a SW32 rotor (Beckman Coulter) at 30 000 rpm for 8 h. Fractions corresponding to the 70S peak were collected and applied onto a 10–40% sucrose gradient [2 mM Mg(OAc)2], to separate the 30S and 50S subunits. Fractions corresponding to the 50S peak were pooled and diluted in buffer II [20 mM Tris–HCl, pH 7.5, 100 mM NH4Cl, 10 mM Mg(OAc)2, 1 mM TCEP].
For premature 45S particles, the crude ribosomal mixture was incubated with excessive mature 30S subunits at 37°C for 10 min, and subjected to a 10–40% sucrose gradient [15 mM Mg(OAc)2] in a SW32 rotor (Beckman Coulter) centrifuged at 30 000 rpm for 8 h. Fractions corresponding to the 45S peak were pooled and diluted in buffer II. Two independent biological replicates (batch 1 and batch 2) of the 45S samples were similarly obtained and subjected to further analysis.
Quantitative mass spectrometry
To quantify the relative ratios of ribosomal proteins in the two samples, the tandem mass tags (TMTs) labeling method was used (19). The detailed procedures were described previously (20). For normalization purpose, samples with same A260 absorption value were resolved by protein gel. Target proteins were recovered and digested using trypsin (Promega). Peptides were labeled with TMT reagents (Thermo, Pierce Biotechnology). TMT 126 and 127 were used for the 45S (batch 1) and 50S samples, respectively. Labeled peptides were subjected to Liquid chromatography-tandem mass spectrometry analysis. Ratios of two or more tryptic peptides from the same protein were used to calculate the means and standard deviations (Supplementary Table S1).
Cryo-EM and image processing
Cryo-grids were prepared as previously described (20). All images were collected using an FEI Titan Krios at 300 kV under low-dose conditions (20 e−/Å2) on an FEI Eagle 4K × 4K CCD camera. Preprocessing of micrographs and particle selection were performed with SPIDER (21). For the mature 50S sample, 4841 micrographs were collected at a magnification of 75 000×. A total of 152 540 particles were obtained and subjected to a standard reference-matching refinement using SPIDER (21). The final resolution is 7.6 Å (FSC 0.5 criterion) for the mature 50S map. For the premature 45S sample, 2019 (batch 1) and 2925 (batch 2) micrographs were collected at a magnification of 59 000×. Particles of the 45S samples were subjected to reference-free 2D and 3D classifications using RELION (22). Different parameters and class numbers have been tested. On the basis of the classification results, 30S, 70S and junk particles were discarded (Supplementary Figure S1). For the batch 1 data, 3D classification of final 91 323 particles rendered three conformational states (Supplementary Table S2 and Supplementary Figure S2). For the batch 2 data, final 201 876 particles were subjected to 3D classification, and four conformational states of the 45S particles were obtained (Supplementary Table S2). These intermediate structures were further grouped into two major states (I and II), according to their map features. All the density maps were sharpened using a B-factor approach (23). The statistics of image processing and summary of the map features are presented in Supplementary Table S2. According to the 3D classification results, ∼19% of the particles from the batch 2 data are mature 50S particles, but virtually no mature 50S particles were found in the batch 1 data (Supplementary Table S2). This is probably due to experimental variations during the sucrose gradient fractionation.
Atomic model of the B. subtilis 50S subunit and flexible fitting
Models of the 23S and 5S rRNAs were built using the crystal structures of the 50S subunits from E. coli (PDB ID: 2AW4) (24) and Thermus thermophilus (PDB ID: 2J01) (25). Sequence alignments of the 23S and 5S rRNAs were constructed with the software S2S (Sequence to Structure) (26). ModeRNA (27) was used for building RNA models. Models of ribosomal proteins, L1, L3, L4, L6, L13, L14, L15, L17, L19, L20, L21, L22, L23, L24, L27, L29, L30, L31, L32, L33, L34, L35 and L36 were downloaded from the SWISS-MODEL Repository (28). The others, including L2, L5, L11, L16, L18 and L28 were modeled using MODELLER (29) with crystal structures of E. coli (24) and T. thermophilus (25) 50S subunits as templates.
The atomic model of the B. subtilis 50S subunit was docked into the 7.6 Å mature 50S density map, optimized by the means of Molecular Dynamics Flexible Fitting (30). Atomic models of the structures of the 45S particles were similarly obtained. The temperature maps of the premature 45S structures were constructed using an extension module of PyMOL (31). The scripts (rmsd_b.py and color_b.py) were downloaded from http://pldserver1.biochem.queensu.ca/∼rlc/work/pymol/. Three-dimensional distance deviations of the 23S rRNA atoms in the 45S models, relative to the mature 50S model, were computed and stored in the B-factor column. The ColoRNA (32) was used to draw different colors to 2D secondary structure diagrams of the 23S rRNA based on values of the B-factor column. PyMOL and Chimera (33) were used for structural visualization.
RESULTS
Compositional analysis of the 45S particles from the YlqF-deficient strain
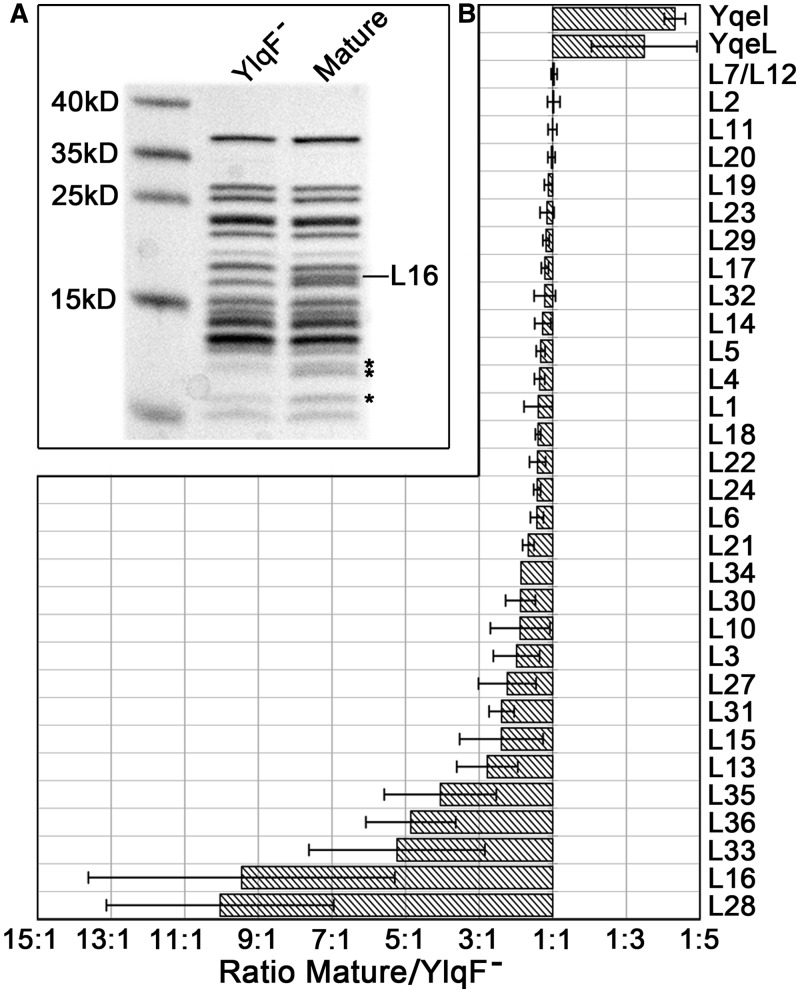
We isolated the premature 45S particles from a previously established B. subtilis YM01 strain (18), in which the genomic ylqF gene was conditionally inactivated by an IPTG-inducible spac promoter. Tricine-sodium dodecyl sulphate-polyacrylamide gel electrophoresis analysis of the 45S particles and mature 50S subunits dissociated from the 70S ribosomes confirmed that at least L16 and a few other proteins are apparently missing from the 45S particles (Figure 1A). To characterize possible underrepresented proteins that are beyond the polyacrylamide gel electrophoresis resolution, an isobaric tag-based mass spectrometry (34) was used to determine the occupancies of ribosomal proteins in the 45S particles relative to the mature 50S subunits.
Figure 1.
Compositional analysis of the 45S particles by QMS. (A) Protein gel analysis of the mature 50S (Mature) and premature 45S samples (YlqF−). The ribosomal protein bands that are missing or weak in the 45S sample are labeled with asterisks. (B) QMS analysis of the occupancies of the 50S proteins in the 45S sample. Standard deviations are shown by error bars.
As a result, QMS data show a specific pattern, with a dozen of the 50S proteins underrepresented at varying levels in the 45S sample (Supplementary Table S1). In contrast, the rest 20 proteins are comparable in both samples. This distinctive protein spectrum indicates that the 45S particles from the YlqF-deficient strain are a specific set of assembly intermediates, and secondary effect from the impaired ribosomal protein supply is minimal. Among underrepresented proteins, L28, L16, L33, L36 and L35 were detected to be dramatically reduced in the 45S particles (Figure 1B). Especially, L28 and L16 are virtually absent (∼10%) from the 45S particles, considering the inevitable contamination of free 50S subunits during the 45S sample preparation (Supplementary Figure S1). In addition, proteins L13, L15, L31 and L27 show moderately reduced levels, ranging from 36% to 45%. Structural mapping of these proteins indicates that most of them are in the neck region of the CP (Supplementary Figure S3). In fact, all of the CP proteins are underrepresented, except two 5S rRNA-binding proteins L5 and L18. These results agree well with early in vitro data (5,35) showing that these underrepresented proteins are at the branch ends of the in vitro 50S assembly map. Also, taking into consideration of the previous data that the 5S rRNA, L5 and L18 incorporate as a preformed ribonucleoprotein complex at the beginning stage of the CP assembly (5,36,37), it demonstrates that these 45S intermediates represent a late stage of the 50S assembly after the incorporation of the 5S rRNA.
In addition, we also found that two proteins, YqeI and YqeL, are enriched in the 45S sample (Figure 1B). The E. coli homologs of them (YhbY and YbeB, respectively) were also reported to cofractionate with the large subunit fractions (34,38). This indicates that these two proteins are likely involved in the ribosome-related function as well.
Image analysis of the 45S particles from the YlqF-deficient strain
Next, we used cryo-EM to probe the structural viability of the 45S particles. At the 2D level, unsupervised classification of particles results in nicely resolved class averages (Supplementary Figure S1). A prominent feature of these average images is that all of them display a stable body domain with discernible fine details, but most of them also exhibit large flexibility at the CP, L1 and L7/L12 stalks. In some cases, densities at these regions are nearly smeared out.
It is known that the two peripheral stalks of the 50S subunit are intrinsically dynamic. However, the proper function of the ribosome requires the rigidity of the CP, as it not only participates in the intersubunit association, but also directly interacts with the D-loops of the A-site and P-site tRNAs to form the essential tRNA corridor on the 70S ribosome (24). Therefore, from the 2D analysis alone, it is likely that the 45S particles are functionally defective and unable to associate with the 30S subunit.
Cryo-EM structures of the 45S assembly intermediates
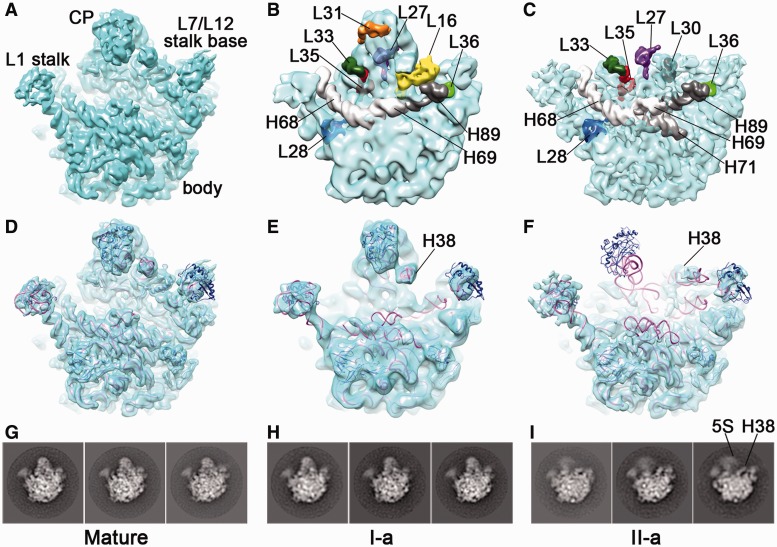
At the 3D level, with extensive tuning of classification parameters, we were able to sort the heterogeneous 45S particles into several well-defined groups, and obtain a series of structures at near 1 nm resolution (Supplementary Table S2). Classification of two independent biological replicates of the 45S samples produced similar results (Supplementary Table S2 and Supplementary Figure S2). On these maps, many secondary structural elements of the body domain can be easily identified, demonstrating the validity of the classification (Figure 2). Based on the map features, these intermediate maps were grouped into two major conformational states (I and II) (Figure 2, Supplementary Figure S2 and Supplementary Table S2). The first state (I-a), accounting for nearly one-third of particles, is in a conformation generally similar to the mature 50S subunit (Figure 2B and E). In contrast, the second state (II-a and II-b), derived from ∼70% of particles, displays a floppy CP and a well-resolved body domain (Figure 2C and F). When the II-a map is displayed at normal threshold, the whole CP domain is invisible (Figure 2F), indicating that the CP in the 45S particles is highly dynamic. At relatively low threshold, bulk densities of the CP begin to show up and appear to have flipped toward the L1 stalk direction (Figure 2I, Supplementary Figure S2).
Figure 2.
Structural analysis of the 45S particles. (A) Surface representation of the cryo-EM map of the mature 50S subunit. (B–C) Surface representation of the 45S maps in the I-a (B) and II-a (C) states, superimposed with their missing components. (D–F) Surface representation of the cryo-EM maps of the mature 50S subunit (D), I-a (E) and II-a (F) states of the 45S particles, with their respective flexibly fitted atomic models superimposed. For simplification, only I-a and II-a states are shown. (G–I) Representative class averages of the cryo-EM raw images for the three conformational states (Mature, I-a and II-a), produced by a reference-free classification.
Highly consistent with the QMS data, analysis of the two states of the 45S particles reveals that a number of proteins are evidently missing from the density maps (Figure 2B and C and Supplementary Movie S1–S3). Most of these missing proteins bind at multiple helical junctions, underlining their contribution to the stabilization of the 23S rRNA conformation. Therefore, our structural and compositional analysis suggests that the incorporation of the CP-binding proteins is rate-limiting in the late-stage maturation of the 50S subunit, and the tertiary interactions of these proteins with the 23S rRNA are likely crucial for the maturation of the 23S rRNA at the CP.
Functional centers of the 45S particles are highly flexible
In addition to the floppy CP domain, these assembly intermediates also differ at varying extents in other regions. Especially, functional centers of the 50S subunit, as well as rRNA helices involved in tRNA binding, deviate largely from the structure of the mature 50S subunit.
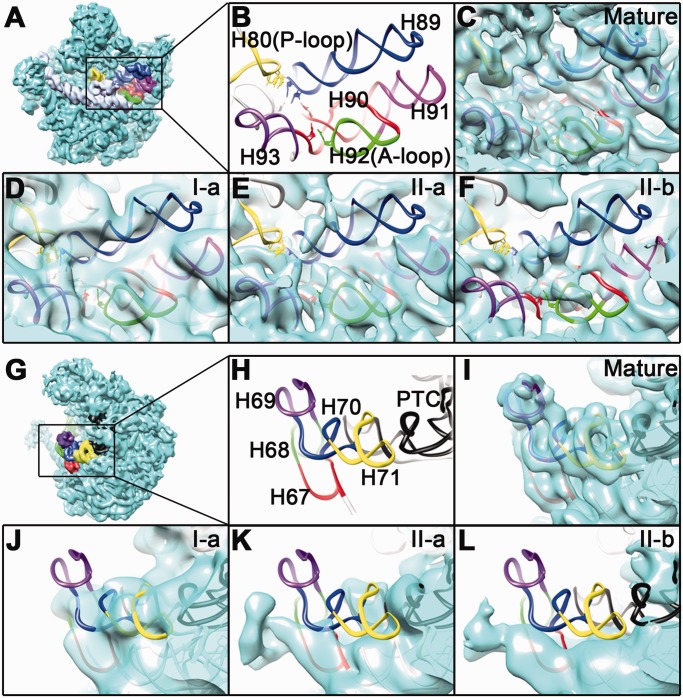
Firstly, the peptidyl-transferase center (PTC), which spans RNA helices 89–93, shows significant conformational differences among these structures. This functional center is the most important enzymatic center of the ribosome, which interacts with the CCA-ends of the A-site and P-site tRNAs, and directly participates in the peptide elongation and release reactions (39). None of the 45S maps exhibits completely resolved PTC, and especially, H89 appears to be extremely dynamic (Figure 3A).
Figure 3.
Conformational variability of the PTC and H69 regions in the 45S structures. (A) Thumbnail of the mature 50S subunit viewed from the intersubunit side. (B) Close-up view of the PTC. H80, H89, H90, H91, H92 and H93 are colored yellow, blue, red, magenta, green and purple, respectively. All the maps in (C–F) are superimposed with their respective atomic models. As shown, none of the intermediate maps (I-a, II-a and II-b) display a completely resolved PTC, indicating a large flexibility at these helices. (G) Thumbnail of the mature 50S subunit viewed from the L7/L12 side. (H) Close-up view of the H69 region. H67, H68, H69, H70, H71 and the PTC are colored red, green, purple, blue, yellow and black, respectively. All the maps in (I–L) are superimposed with the atomic model of the mature 50S structure.
Secondly, the helices 67–71 of the 23S rRNA are also highly unstable. The long rRNA stem H68 is invisible in all the structures of the 45S particles (Figure 2 and Supplementary Figure S2). H69 is highly flexible in both conformational states (Figures 2 and 3), and appears to be more bent toward the 30S direction in the maps of the state II (Figure 3B). Notably, H69 is an essential component of the intersubunit bridge (B2a) and also directly involved in coordinating tRNA movement during translation (40).
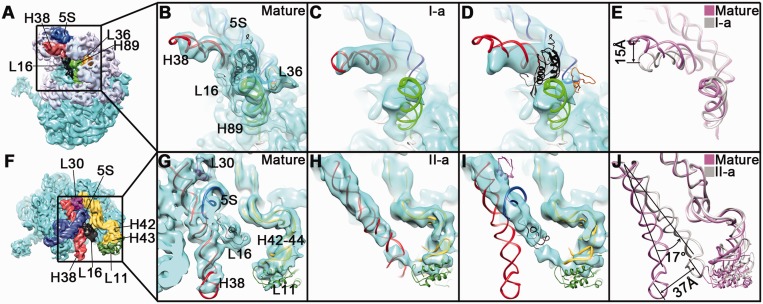
Thirdly, a significant difference lies in the position of H38 of the 23S rRNA. H38 is also called the A-site finger, which interacts with the A-site tRNA during translation, and has been demonstrated to have a number of functional implications (41–43). The terminal loop of H38 also interacts with the 30S subunit to form an intersubunit bridge B1a (24). In the I-a map of the state I, H38 is in a native-like conformation, with its tip bent toward the body domain by ∼15 Å (Figure 4A–E). In contrast, in the structures of the state II, H38 is seen to have a massive displacement, rotated ∼17° toward the L7/L12 stalk direction, forming a non-native interaction with L11 (Figure 4F–J). Notably, this ‘trapped’ conformation of H38 in the state II totally blocks the binding site of L16 (Figure 4I), which readily explains why the L16 level is extremely low in the 45S particles.
Figure 4.
Conformational variability of H38 of the 23S rRNA in the 45S intermediates. Structural comparison of H38 in the I-a (B–E) or II-a (G–J) structures, with that of the mature 50S structure. Two different thumbnails of the mature 50S subunit (A and F), with a few components highlighted in different colors, are shown in left panels. (B, C, G and H) Close-up view of the mature 50S (B and G), the I-a (C) and II-a (H) structures in the same area as in their respective thumbnails, with their atomic models superimposed. (D and I) The I-a (D) and II-a (I) structures are superimposed with the atomic model of the mature 50S structure. (E and J) Conformational change of H38 between the mature and the I-a (E) or II-a (J) state. The 5S rRNA, H38, H89 and H42–44 are colored blue, red, green and yellow, respectively. Ribosomal proteins, L16, L36, L30 and L11 are colored black, golden, purple and dark green, respectively. For clarification, the 23S rRNA helices colored gray in the upper left thumbnail (A) are not shown in (B–E).
Taking together, our structural analyses show that the functional centers of the 45S particles are not correctly assembled, indicating that these 45S particles are defective not only in intersubunit association but also in tRNA binding.
Domains IV and V of the 23S rRNA are not correctly assembled in the 45S particles
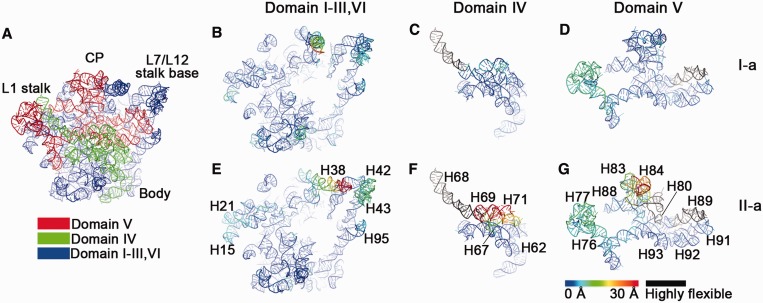
To understand the 23S rRNA folding in a global scale, we computed temperature factors for the 23S rRNAs in these 45S structures according to their deviations from the structure of the mature 50S subunit (Figure 5). The temperature factors were mapped to both the 3D structures (Figure 5 and Supplementary Figure S4) and 2D secondary maps (Supplementary Figure S5).
Figure 5.
Temperature maps of the 23S rRNA in the 45S intermediates. (A) Cartoon representation of the 23S rRNA, with its domains colored separately. (B–G) Temperature maps of the domains of the 23S rRNA, colored according to their deviations from the conformation of the mature 50S subunit. For simplification, only I-a (B–D) and II-a (E-G) states are shown. Individual rRNA helices are labeled. The rRNA helices that are not well resolved in cryo-EM maps are colored black (highly flexible). CP, central protuberance; L7/L12 SB, L7/L12 stalk base.
As a result, a clear pattern starts to appear. Domains IV and V of the 23S rRNA are the most affected regions. The 3′ half of Domain IV consists of three helices, H68, H69 and H71, and all of them are highly dynamic (Figure 5C and 5F, Supplementary Figure S4). In contrast, the 5′ half of Domain IV, which forms part of the body domain, remains to be relatively stable (Figure 5C and F). Domain V accounts for three major functional components of the 50S subunit, the L1 stalk (H76–78), the PTC (H89–93) and the CP (H80–88). All these three parts display large conformational differences (Figure 5D and 5G, Supplementary Figure S4), especially in the conformational state II. The L1 stalk is visible in all the structures, but in different positions (Figure 5 and Supplementary Figure S4). The other domains of the 23S rRNA (I, II, III and VI) appear to be relatively stable, except that H38 deviates from its mature conformation, and the L7/L12 stalk base (H42–44) also shows some structural viability (Figure 5B and E). These temperature maps show that all the helices involved in the formation of tRNA corridor on the 50S subunit are highly flexible.
On the 2D secondary maps (Supplementary Figure S5), it is clear that the 23S rRNA does not mature in a linear manner, and both the 5′ and 3′ domains of the 23S rRNA in the 45S structures are already correctly assembled. The maturation of the 23S rRNA at Domain IV and V appears to be the slowest step. Together with our QMS data, this indicates that the assembly of the CP is likely rate-limiting, and requires the assistance from assembly factors.
DISCUSSION
Reorientation of H38 is a major rate-limiting step of the late-stage 50S subunit assembly
In the present work, we characterized a set of in vivo 45S assembly intermediates, which reveals two major conformational populations. These two states mainly differ in the stability of the CP and the orientation of H38. Comparison of the two states indicates that the reorientation of H38 to its native-like position leads to a global stabilization of the rRNA structure at the CP (Figure 2E and F). Apparently, this stabilization of the CP in the state I is a direct consequence of the interaction of the 5S rRNA with H38, considering the topology of them in the 50S structure (Supplementary Figure S3). This suggests that the 5S rRNA serves as an important bridge in the mutual stabilization between the CP rRNA components and H38. Consistently, both the omission of the 5S rRNA (44) from the in vitro reconstitution and the depletion of the 5S rRNA binding protein L5 in vivo (36) result in significantly reduced incorporation of the CP proteins into the 50S assembly intermediates. Protein compositions of the two states are similar, both severally lacking the CP-binding proteins (Figure 2), except that L30 is present in the I-a state (Supplementary Figure S6, Supplementary Table S2 and Supplementary Movie S1). L30 binds exactly at the interface between the 5S rRNA and the stem base of H38 (Supplementary Figures S3 and S6), suggesting that the binding of L30 might partially contribute to the reorientation of H38.
The ratios of the 45S particles in the state I to II are 1:2.6 and 1:4.1, for the two biological replicates of the 45S samples, respectively (Supplementary Table S2), indicating that most of the 45S intermediates are trapped in the state II. This suggests that the reorientation of H38 (from states II to I) is likely rate-limiting. Our structural data show that L16 is incompatible with H38 in the state II, implying that the binding of L16 would require the breaking of existing non-native interaction between H38 and the L7/L12 stalk base and the reestablishing the native interaction between H38 and the 5S rRNA (Figure 4). Consistently, L16 is exactly a common protein detected to be missing from various forms of premature particles (45–49) (Supplementary Table S3). Along this line, early in vitro reconstitution experiments showed that the incorporation of L16 into the late-stage particles requires a large activation energy and is temperature dependent (50). Therefore, altogether, it indicates that the native conformation of H38 is probably not established in these different assembly intermediates, suggesting that the reorientation of H38 is a global rate-limiting step in the late-stage assembly pathways. In support of this view, it was already found that mutations on H38 lead to a defect in the 50S subunit assembly in E. coli (43).
Furthermore, we noticed that H38 in the state I is in a near-native conformation, still partially overlapping with the L16 binding sites (Figure 4D). There are 28% and 20% of 45S particles trapped in the state I, for the batch 1 and batch 2 data, respectively (Supplementary Table S2), indicating the presence of unidentified rate-limiting steps after the 45S particles acquire the state I conformation. This suggests that H38 might undergo a series of structural remodeling during the late-stage assembly, and each step could be coupled to certain protein binding events.
It is known that H38 is also dynamic on the 70S ribosome, as required for the reorganization of intersubunit bridge B1a during the ratchet-like motion (51,52). This dynamic behavior is enabled by a unique kink-turn motif (53) at the base of H38 (54). However, its conformational change on the 70S ribosome is in a much smaller scale, compared with its orientations in our 45S structures. Taking together with published data on the function of H38 in translation (41–43,55), it suggests that H38, as an intrinsic flexible element, plays roles in both translation and ribosome assembly.
The late-stage assembly of the 50S subunit in vivo
Protein composition of the 45S particles shows that L28, L16, L33, L36 and L35 are in significantly reduced levels. This pattern of underrepresented proteins shows interesting similarities to the protein spectra of the in vivo 40S or 45S particles isolated from various E. coli or B. subtilis strains with genes for different assembly cofactors disrupted, including csdA, srmB, rrmJ, dnaK, dbpA, engA, yphC, ysxC and obgE (17,45–49,56,57). Among these factors, CsdA, DbpA and SrmB are DEAD-box RNA helicases. DnaK and RrmJ are heat-shock proteins with specialized functions, as general protein chaperone and 23S rRNA methyltransferase, respectively. And ObgE, EngA, YphC and YsxC are all GTPases involved in ribosome biogenesis. These different sets of in vivo assembly intermediates, obtained by perturbation of presumably distinctive assembly events, however, share a similar deficiency in L16, L27, L33, L34, L35 and L36 (Supplementary Table S3). In addition to the perturbation of assembly factors, deletion of certain ribosomal protein genes, such as L5, L27 and L28, also induces the accumulation of in vivo precursors lacking these proteins (36,58–60). Therefore, it indicates that the assembly of these late-stage proteins is likely bottlenecks in vivo and susceptible to various forms of disruption.
However, with a wealth of the above semiquantitative data, it remains impractical to determine the exact binding order of these late-stage proteins because these data only reflect the compositions of various kinetically trapped in vivo intermediates in equilibrium state. The Nierhaus map, on the other hand, did not explicitly test all possible binding dependences among these late-binding proteins. Recently, with the time-resolved pulse-labeling coupled QMS technique, Williamson lab quantified the ribosomal protein levels throughout the whole sucrose gradient of the cell lysate from fast-growing wild-type E. coli cells, and successfully assigned the 50S proteins in six temporal groups (61). All the proteins with highly underrepresented levels in our 45S sample (L28, L16, L33, L36 and L35) are in the latest group, except that L28 and L33 are in the fifth group. Through the integration of all available data, we could isolate a major pathway for the late-stage assembly of the 50S subunit in vivo, with a focus on the CP-binding proteins only (L16, L27, L30, L31, L33, L35 and L36). As shown, there are two major assembly intermediates (state I and II) in this assembly branch (Figure 6). The first transition from the state II to I is facilitated by L30, which places L30 at a relatively earlier stage. This assignment is consistent with the Nierhaus map and the recent time-resolved data (61). It is difficult to unambiguously further time stamp the following CP proteins in the second transition (from state I to mature) owing to the lack of kinetic data with sufficient temporal resolution. Both transitions are rate-limiting because our structural data show that the 45S particles are trapped in both the state I and II. It must be noted that the second transition should consist of multiple steps, of which the crucial rate-limiting ones remain to be investigated.
Figure 6.
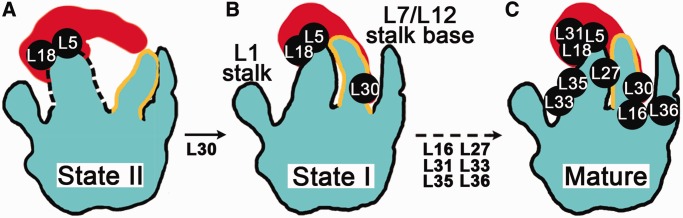
A major branch of the late-stage assembly of the bacterial 50S subunit in vivo. (A) Cartoon representation of the 45S intermediate in the state II, featuring a loosely attached 5S rRNA, a flexible CP domain (dash lines) and a ‘trapped’ conformation of H38 (yellow lines). (B) Cartoon representation of the 45S intermediate in the state I, where the rigidity of the CP is achieved and the interaction between the 5S rRNA and H38 is established. H38 is in a native-like conformation in state I. (C) Cartoon representation of the mature 50S structure. There are two major rate-limiting transitions in this assembly branch, the state II to I, and the state I to the mature conformation. Both transitions involve the structural remodeling of H38.
In terms of the 23S rRNA maturation, our results show that it does not proceed in a 5′–3′ transcriptional order, with the central domains IV and V being the last. Notably, this maturation order was also predicted by the above-mentioned pulse labeling QMS study (61). According to their results, the CP proteins in fact constitute the latest assembly group (61). Interestingly, this non-linear maturation of the 23S rRNA is in sharp contrast to the assembly of the 16S rRNA. Previous in vitro studies indicate that the 16S rRNA matures roughly in a 5′–3′ order (62,63), with the 3′ head domain at last, manifesting a co-transcriptional nature for the 30S subunit assembly in vivo (64). Despite this difference, the co-occurrence of the hypolevels of ribosomal proteins and structural instability of the rRNAs, observed at the CP of the 45S particles, was also found at the 3′ domain of the 16S rRNA during the late-stage assembly of the 30S subunit. Therefore, similar to what is known for the assembly of the 3′ domain of the 30S subunit (62,65–67), there must also be a high degree of cooperativity between the protein binding and RNA remodeling at the CP of the 50S subunit.
Molecular role of YlqF as an rRNA chaperone
Ribosomal subunits are gigantic nucleoprotein complexes, and their structures are rich in RNA–RNA and RNA–protein tertiary interactions. The energy landscape of the assembly of the ribosomal subunits is therefore inevitably decorated with various kinetic traps, corresponding to certain mis-assembled rRNA or slow protein-binding events. Assembly factors, as well as some ribosomal proteins, are believed to act as RNA chaperones to prevent or rescue certain non-native rRNA intermediates (62,68).
Previously, based primarily on the protein composition in the 45S particles from YlqF-deficient cells (15,18,69), the role of YlqF was emphasized in the assembly of L16, L27 and L36. However, these proteins were commonly detected to be missing from various in vivo assembly intermediates (Supplementary Table S3). This means that the most direct role of YlqF is probably not in protein assembly. In the present work, we found that H38 is in an abnormal trapped conformation in the structures of conformational state II, and the reorientation of H38 appears to be a prerequisite for downstream assembly events. This suggests that YlqF likely facilitates the structural remodeling that contributes to the interaction of H38 with the 5S rRNA. Consistent with this view, the putative binding sites of YlqF, mapped by dimethyl sulfate footprinting, are located to the CP (H81 and H85) and H38 (16). Therefore, YlqF might act as a chaperone to lower the energy barrier required for the reorientation of H38. In further support of this proposed role, the low intrinsic GTPase activity of YlqF could be simulated by both the mature 50S and premature 45S subunits (16,18,69). However, the mature 50S subunit has a more marked effect on the GTPase activation of YlqF (69). Thus, combining all these data, sequential actions of YlqF could be reasoned as follows. YlqF in GTP-bound state first binds to the 45S particle and facilitates the reorientation of H38. Once H38 achieves its native-like conformation, the mutual stabilization between the CP and H38 in turn accelerates the further assembly of the CP, allowing an efficient incorporation of the CP proteins. The fully or nearly matured 50S subunit simulates the GTPase activity of YlqF, and YlqF finally releases in GDP-bound state upon GTP hydrolysis.
Intrinsic quality control of the ribosomal subunit production
Translation is such a fundamental process for all cellular activities that the quality of the assembly intermediates along the assembly pathways has to be closely monitored. Some assembly GTPases are believed to act as checkpoint proteins at expense of GTP molecules, e.g. to ensure a certain intermediate conformation or assembly event (9). Although our data is more consistent with an rRNA chaperone role for YlqF, it is possible that an unidentified assembly event in the final stages of the 50S assembly, which is responsible for triggering the GTP hydrolysis on YlqF, could be used as a structural checkpoint. In this alternative mechanism, YlqF might associate with the 45S particle and act as a physical blocker to downstream assembly events until a certain crucial step is finished. This interpretation provides a different view on the role of YlqF, with an emphasis in the quality control of the 50S subunit assembly.
Another interesting finding is that the 45S particles, whether in the state I or II, display large flexibility in intersubunit bridging and tRNA contacting sites. This observation shows intriguing similarity with previous structural information on the premature 30S subunit precursors. Early chemical or hydroxyl radical probing of the 16S rRNA conformation (63,65–67,70), as well as direct visualization of the 30S precursors by cryo-EM (20,71) all indicate that the 3′ domain of the 16S rRNA remains largely immature until a very late stage. Especially, the 3′ minor domain of the 16S rRNA, which includes an essential long helix (h44), is not correctly assembled in the assembly intermediates. Notably, h44 is a functional component of the decoding center on the 30S subunit, and is also indispensable for both subunit association and tRNA interaction. In combination, these data reveal that both the 50S and 30S subunits in premature forms are functionally defective. This could have a physiological relevance, as it might demonstrate an additional level of quality control of protein translation, by preventing the escape of premature subunits into the translation pool.
ACCESSION NUMBERS
Cryo-EM maps of the I-a and II-a structures are deposited in the EMDataBank with accession codes EMD-5642 and EMD-5643, respectively. Atomic models are deposited in the Protein Data Bank with accession codes 3J3V and 3J3W, respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–6, Supplementary Movies 1–3 and Supplementary References [17,21,22,36,45–49,56–60,72–73].
FUNDING
Ministry of Science and Technology of China [2010CB912402, 2013CB910404 and 2010CB912401]; National Natural Science Foundation of China [31170677]. Funding for open access charge: the National Natural Science Foundation of China.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Ogasawara for providing the YM01 strain, and the Tsinghua National Laboratory for Information Science and Technology for providing computing resource.
REFERENCES
- 1.Kaczanowska M, Ryden-Aulin M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 2007;71:477–494. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Traub P, Nomura M. Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc. Natl Acad. Sci. USA. 1968;59:777–784. doi: 10.1073/pnas.59.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nierhaus KH, Dohme F. Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc. Natl Acad. Sci. USA. 1974;71:4713–4717. doi: 10.1073/pnas.71.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizushima S, Nomura M. Assembly mapping of 30S ribosomal proteins from E. coli. Nature. 1970;226:1214. doi: 10.1038/2261214a0. [DOI] [PubMed] [Google Scholar]
- 5.Rohl R, Nierhaus KH. Assembly map of the large subunit (50S) of Escherichia coli ribosomes. Proc. Natl Acad. Sci. USA. 1982;79:729–733. doi: 10.1073/pnas.79.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shajani Z, Sykes MT, Williamson JR. Assembly of bacterial ribosomes. Annu. Rev. Biochem. 2011;80:501–526. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- 7.Britton RA. Role of GTPases in bacterial ribosome assembly. Annu. Rev. Microbiol. 2009;63:155–176. doi: 10.1146/annurev.micro.091208.073225. [DOI] [PubMed] [Google Scholar]
- 8.Karbstein K. Role of GTPases in ribosome assembly. Biopolymers. 2007;87:1–11. doi: 10.1002/bip.20762. [DOI] [PubMed] [Google Scholar]
- 9.Strunk BS, Karbstein K. Powering through ribosome assembly. RNA. 2009;15:2083–2104. doi: 10.1261/rna.1792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, et al. Essential Bacillus subtilis genes. Proc. Natl Acad. Sci. USA. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morimoto T, Loh PC, Hirai T, Asai K, Kobayashi K, Moriya S, Ogasawara N. Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis. Microbiology. 2002;148:3539–3552. doi: 10.1099/00221287-148-11-3539. [DOI] [PubMed] [Google Scholar]
- 12.Anand B, Verma SK, Prakash B. Structural stabilization of GTP-binding domains in circularly permuted GTPases: implications for RNA binding. Nucleic Acids Res. 2006;34:2196–2205. doi: 10.1093/nar/gkl178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 14.Reynaud EG, Andrade MA, Bonneau F, Ly TB, Knop M, Scheffzek K, Pepperkok R. Human Lsg1 defines a family of essential GTPases that correlates with the evolution of compartmentalization. BMC Biol. 2005;3:21. doi: 10.1186/1741-7007-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uicker WC, Schaefer L, Britton RA. The essential GTPase RbgA (YlqF) is required for 50S ribosome assembly in Bacillus subtilis. Mol. Microbiol. 2006;59:528–540. doi: 10.1111/j.1365-2958.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 16.Matsuo Y, Morimoto T, Kuwano M, Loh PC, Oshima T, Ogasawara N. The GTP-binding protein YlqF participates in the late step of 50 S ribosomal subunit assembly in Bacillus subtilis. J. Biol. Chem. 2006;281:8110–8117. doi: 10.1074/jbc.M512556200. [DOI] [PubMed] [Google Scholar]
- 17.Schaefer L, Uicker WC, Wicker-Planquart C, Foucher AE, Jault JM, Britton RA. Multiple GTPases participate in the assembly of the large ribosomal subunit in Bacillus subtilis. J. Bacteriol. 2006;188:8252–8258. doi: 10.1128/JB.01213-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuo Y, Oshima T, Loh PC, Morimoto T, Ogasawara N. Isolation and characterization of a dominant negative mutant of Bacillus subtilis GTP-binding protein, YlqF, essential for biogenesis and maintenance of the 50 S ribosomal subunit. J. Biol. Chem. 2007;282:25270–25277. doi: 10.1074/jbc.M703894200. [DOI] [PubMed] [Google Scholar]
- 19.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 20.Guo Q, Goto S, Chen Y, Feng B, Xu Y, Muto A, Himeno H, Deng H, Lei J, Gao N. Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process. Nucleic Acids Res. 2013;41:2609–2620. doi: 10.1093/nar/gks1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, Frank J. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protoc. 2008;3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheres SH. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 2012;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandez JJ, Luque D, Caston JR, Carrascosa JL. Sharpening high resolution information in single particle electron cryomicroscopy. J. Struct. Biol. 2008;164:170–175. doi: 10.1016/j.jsb.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 24.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 25.Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 26.Jossinet F, Westhof E. Sequence to Structure (S2S): display, manipulate and interconnect RNA data from sequence to structure. Bioinformatics. 2005;21:3320–3321. doi: 10.1093/bioinformatics/bti504. [DOI] [PubMed] [Google Scholar]
- 27.Rother M, Rother K, Puton T, Bujnicki JM. ModeRNA: a tool for comparative modeling of RNA 3D structure. Nucleic Acids Res. 2011;39:4007–4022. doi: 10.1093/nar/gkq1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinform. 2006 doi: 10.1002/0471250953.bi0506s15. Chapter 5, Unit 5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delano WL. 2008. The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA, USA. [Google Scholar]
- 32.LeBarron J, Mitra K, Frank J. Displaying 3D data on RNA secondary structures: coloRNA. J. Struct. Biol. 2007;157:262–270. doi: 10.1016/j.jsb.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 33.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Jiang M, Sullivan SM, Walker AK, Strahler JR, Andrews PC, Maddock JR. Identification of novel Escherichia coli ribosome-associated proteins using isobaric tags and multidimensional protein identification techniques. J. Bacteriol. 2007;189:3434–3444. doi: 10.1128/JB.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herold M, Nierhaus KH. Incorporation of six additional proteins to complete the assembly map of the 50 S subunit from Escherichia coli ribosomes. J. Biol. Chem. 1987;262:8826–8833. [PubMed] [Google Scholar]
- 36.Korepanov AP, Korobeinikova AV, Shestakov SA, Garber MB, Gongadze GM. Protein L5 is crucial for in vivo assembly of the bacterial 50S ribosomal subunit central protuberance. Nucleic Acids Res. 2012;40:9153–9159. doi: 10.1093/nar/gks676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horne JR, Erdmann VA. Isolation and characterization of 5S RNA-protein complexes from Bacillus stearothermophilus and Escherichia coli ribosomes. Mol. Gen. Genet. 1972;119:337–344. doi: 10.1007/BF00272091. [DOI] [PubMed] [Google Scholar]
- 38.Barkan A, Klipcan L, Ostersetzer O, Kawamura T, Asakura Y, Watkins KP. The CRM domain: an RNA binding module derived from an ancient ribosome-associated protein. RNA. 2007;13:55–64. doi: 10.1261/rna.139607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beringer M, Rodnina MV. The ribosomal peptidyl transferase. Mol. Cell. 2007;26:311–321. doi: 10.1016/j.molcel.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 40.Ali IK, Lancaster L, Feinberg J, Joseph S, Noller HF. Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol. Cell. 2006;23:865–874. doi: 10.1016/j.molcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 41.Komoda T, Sato NS, Phelps SS, Namba N, Joseph S, Suzuki T. The A-site finger in 23 S rRNA acts as a functional attenuator for translocation. J. Biol. Chem. 2006;281:32303–32309. doi: 10.1074/jbc.M607058200. [DOI] [PubMed] [Google Scholar]
- 42.Sergiev PV, Kiparisov SV, Burakovsky DE, Lesnyak DV, Leonov AA, Bogdanov AA, Dontsova OA. The conserved A-site finger of the 23S rRNA: just one of the intersubunit bridges or a part of the allosteric communication pathway? J. Mol. Biol. 2005;353:116–123. doi: 10.1016/j.jmb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 43.Yassin A, Mankin AS. Potential new antibiotic sites in the ribosome revealed by deleterious mutations in RNA of the large ribosomal subunit. J. Biol. Chem. 2007;282:24329–24342. doi: 10.1074/jbc.M703106200. [DOI] [PubMed] [Google Scholar]
- 44.Dohme F, Nierhaus KH. Role of 5S RNA in assembly and function of the 50S subunit from Escherichia coli. Proc. Natl Acad. Sci. USA. 1976;73:2221–2225. doi: 10.1073/pnas.73.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Charollais J, Dreyfus M, Iost I. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 2004;32:2751–2759. doi: 10.1093/nar/gkh603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El Hage A, Alix JH. Authentic precursors to ribosomal subunits accumulate in Escherichia coli in the absence of functional DnaK chaperone. Mol. Microbiol. 2004;51:189–201. doi: 10.1046/j.1365-2958.2003.03813.x. [DOI] [PubMed] [Google Scholar]
- 47.Charollais J, Pflieger D, Vinh J, Dreyfus M, Iost I. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 2003;48:1253–1265. doi: 10.1046/j.1365-2958.2003.03513.x. [DOI] [PubMed] [Google Scholar]
- 48.Hager J, Staker BL, Bugl H, Jakob U. Active site in RrmJ, a heat shock-induced methyltransferase. J. Biol. Chem. 2002;277:41978–41986. doi: 10.1074/jbc.M205423200. [DOI] [PubMed] [Google Scholar]
- 49.Jiang M, Datta K, Walker A, Strahler J, Bagamasbad P, Andrews PC, Maddock JR. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J. Bacteriol. 2006;188:6757–6770. doi: 10.1128/JB.00444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teraoka H, Nierhaus KH. Protein L16 induces a conformational change when incorporated into a L16-deficient core derived from Escherichia coli ribosomes. FEBS Lett. 1978;88:223–226. doi: 10.1016/0014-5793(78)80179-3. [DOI] [PubMed] [Google Scholar]
- 51.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 52.Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 53.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reblova K, Razga F, Li W, Gao H, Frank J, Sponer J. Dynamics of the base of ribosomal A-site finger revealed by molecular dynamics simulations and Cryo-EM. Nucleic Acids Res. 2010;38:1325–1340. doi: 10.1093/nar/gkp1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Piekna-Przybylska D, Przybylski P, Baudin-Baillieu A, Rousset JP, Fournier MJ. Ribosome performance is enhanced by a rich cluster of pseudouridines in the A-site finger region of the large subunit. J. Biol. Chem. 2008;283:26026–26036. doi: 10.1074/jbc.M803049200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharpe Elles LM, Sykes MT, Williamson JR, Uhlenbeck OC. A dominant negative mutant of the E. coli RNA helicase DbpA blocks assembly of the 50S ribosomal subunit. Nucleic Acids Res. 2009;37:6503–6514. doi: 10.1093/nar/gkp711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hwang J, Inouye M. The tandem GTPase, Der, is essential for the biogenesis of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 2006;61:1660–1672. doi: 10.1111/j.1365-2958.2006.05348.x. [DOI] [PubMed] [Google Scholar]
- 58.Wower IK, Wower J, Zimmermann RA. Ribosomal protein L27 participates in both 50 S subunit assembly and the peptidyl transferase reaction. J. Biol. Chem. 1998;273:19847–19852. doi: 10.1074/jbc.273.31.19847. [DOI] [PubMed] [Google Scholar]
- 59.Maguire BA, Wild DG. The roles of proteins L28 and L33 in the assembly and function of Escherichia coli ribosomes in vivo. Mol. Microbiol. 1997;23:237–245. doi: 10.1046/j.1365-2958.1997.2131578.x. [DOI] [PubMed] [Google Scholar]
- 60.Maguire BA, Wild DG. The effects of mutations in the rpmB,G operon of Escherichia coli on ribosome assembly and ribosomal protein synthesis. Biochim. Biophys. Acta. 1997;1353:137–147. doi: 10.1016/s0167-4781(97)00064-x. [DOI] [PubMed] [Google Scholar]
- 61.Chen SS, Williamson JR. Characterization of the ribosome biogenesis landscape in E. coli using quantitative mass spectrometry. J. Mol. Biol. 2013;425:767–779. doi: 10.1016/j.jmb.2012.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mulder AM, Yoshioka C, Beck AH, Bunner AE, Milligan RA, Potter CS, Carragher B, Williamson JR. Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit. Science. 2010;330:673–677. doi: 10.1126/science.1193220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Powers T, Daubresse G, Noller HF. Dynamics of in vitro assembly of 16 S rRNA into 30 S ribosomal subunits. J. Mol. Biol. 1993;232:362–374. doi: 10.1006/jmbi.1993.1396. [DOI] [PubMed] [Google Scholar]
- 64.de Narvaez CC, Schaup HW. In vivo transcriptionally coupled assembly of Escherichia coli ribosomal subunits. J. Mol. Biol. 1979;134:1–22. doi: 10.1016/0022-2836(79)90411-x. [DOI] [PubMed] [Google Scholar]
- 65.Adilakshmi T, Bellur DL, Woodson SA. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature. 2008;455:1268–1272. doi: 10.1038/nature07298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holmes KL, Culver GM. Mapping structural differences between 30S ribosomal subunit assembly intermediates. Nat. Struct. Mol. Biol. 2004;11:179–186. doi: 10.1038/nsmb719. [DOI] [PubMed] [Google Scholar]
- 67.Holmes KL, Culver GM. Analysis of conformational changes in 16 S rRNA during the course of 30 S subunit assembly. J. Mol. Biol. 2005;354:340–357. doi: 10.1016/j.jmb.2005.09.056. [DOI] [PubMed] [Google Scholar]
- 68.Woodson SA. RNA folding pathways and the self-assembly of ribosomes. Acc. Chem. Res. 2011;44:1312–1319. doi: 10.1021/ar2000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Achila D, Gulati M, Jain N, Britton RA. Biochemical characterization of ribosome assembly GTPase RbgA in Bacillus subtilis. J. Biol. Chem. 2012;287:8417–8423. doi: 10.1074/jbc.M111.331322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dutca LM, Culver GM. Assembly of the 5′ and 3′ minor domains of 16S ribosomal RNA as monitored by tethered probing from ribosomal protein S20. J. Mol. Biol. 2008;376:92–108. doi: 10.1016/j.jmb.2007.10.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jomaa A, Stewart G, Martin-Benito J, Zielke R, Campbell TL, Maddock JR, Brown ED, Ortega J. Understanding ribosome assembly: the structure of in vivo assembled immature 30S subunits revealed by cryo-electron microscopy. RNA. 2011;17:697–709. doi: 10.1261/rna.2509811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nanamiya H, Akanuma G, Natori Y, Murayama R, Kosono S, Kudo T, Kobayashi K, Ogasawara N, Park SM, Ochi K, et al. Zinc is a key factor in controlling alternation of two types of L31 protein in the Bacillus subtilis ribosome. Mol. Microbiol. 2004;52:273–283. doi: 10.1111/j.1365-2958.2003.03972.x. [DOI] [PubMed] [Google Scholar]
- 73.Schmalisch M, Langbein I, Stulke J. The general stress protein Ctc of Bacillus subtilis is a ribosomal protein. J. Mol. Microbiol. Biotechnol. 2002;4:495–501. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.