Abstract
Customized TALENs and Cas9/gRNAs have been used for targeted mutagenesis in zebrafish to induce indels into protein-coding genes. However, indels are usually not sufficient to disrupt the function of non-coding genes, gene clusters or regulatory sequences, whereas large genomic deletions or inversions are more desirable for this purpose. By injecting two pairs of TALEN mRNAs or two gRNAs together with Cas9 mRNA targeting distal DNA sites of the same chromosome, we obtained predictable genomic deletions or inversions with sizes ranging from several hundred bases to nearly 1 Mb. We have successfully achieved this type of modifications for 11 chromosomal loci by TALENs and 2 by Cas9/gRNAs with different combinations of gRNA pairs, including clusters of miRNA and protein-coding genes. Seven of eight TALEN-targeted lines transmitted the deletions and one transmitted the inversion through germ line. Our findings indicate that both TALENs and Cas9/gRNAs can be used as an efficient tool to engineer genomes to achieve large deletions or inversions, including fragments covering multiple genes and non-coding sequences. To facilitate the analyses and application of existing ZFN, TALEN and CRISPR/Cas data, we have updated our EENdb database to provide a chromosomal view of all reported engineered endonucleases targeting human and zebrafish genomes.
INTRODUCTION
Engineered endonucleases (EENs), such as zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems, which comprise DNA-recognizing components [ZFP or TALE domains for ZFNs or TALENs; crRNAs or guide-RNAs (gRNAs) for CRISPR/Cas systems] and DNA-cleavage components (FokI domains for ZFNs and TALENs; Cas proteins, e.g. Cas9, for CRISPR/Cas systems), have been shown to be useful for gene targeting by efficient and specific cleavage of genomes in cultured cells, plants and animals, including the zebrafish (Danio rerio) (1–20). TALENs and CRISPR/Cas systems show a predictable one-to-one relationship to the nucleotides in their target sites, i.e. one unit of the tandem repeat in the TALE domain recognizes one nucleotide in the target site, and the crRNA or gRNA of CRISPR/Cas system binds to the complementary sequence in the DNA target strand with the traditional Watson–Crick base pairing, which makes these EENs easy to engineer (21–23). Most current applications use a pair of TALENs or a Cas9 protein with one gRNA to generate DNA double-strand breaks (DSBs) in the target site. These DSBs are then repaired via non-homologous end-joining or homologous recombination (HR). Non-homologous end-joining often results in repair errors that generate small insertions and deletions (indels) that disrupt the target gene’s function. Recently, we and other groups have demonstrated precise modification of the target locus via HR by the addition of a donor template (24,25). Both these strategies focus on manipulating the DNA sequence in a single locus. However, indel mutations are not suitable for all purposes because (i) the existence of multiple transcript variants or alternative start codons in the same gene, whether annotated, make it difficult to determine whether the functional protein product is destroyed by a limited local indel mutation; (ii) for non-coding RNA (ncRNA) genes, untranslated regions or regulatory regions, the single-point–based targeted mutation is powerless to disrupt the functions of these genes or cis-elements; and (iii) it is hard to disrupt more than one gene or a cluster of adjacent genes on the chromosome with only one Cas9/gRNA or one pair of TALENs targeting a single locus.
To some extent, complete deletion of a chromosomal region can help to solve the aforementioned problems. It has been reported that genomic segments up to 15 Mb between two distal target sites in cultured human or other mammalian cells can be deleted by the combination of two pairs of the nucleases or the derived nickases (26–30). Inversions and insertions (duplications of a region) of the segments around the human CCR2 and CCR5 genes were also reported (28,31). In addition, translocations between two chromosomes were also detected by ZFNs and TALENs (32–34). At the organism level, heritable-targeted deletion of an ∼800-bp segment within the BmBLOS2 gene by two pairs of TALENs was reported in silkworm (Bombyx mori) (35). Other studies concerning deletions include the deletion of IgJ locus in rats mediated by two pairs of ZFNs, and deletion of three human genes via HR approach mediated by only one pair of ZFNs together with an ssODN template (36,37). However, in vivo attempts of inducing deletions through EENs in vertebrates have not been reported when this work was carried out, although we have successfully induced deletions via Tol2 jump-out in transgenic zebrafish (38,39). Furthermore, other types of targeted large genomic fragment manipulation, such as chromosomal inversions and insertions/duplications, have not been tested either in vivo or at the inheritable level.
Recently, the bacterial immune system CRISPR/Cas has been used for gene targeting in eukaryotes, including zebrafish (40,41). Cas9/guide-RNA (gRNA), the most frequently used type II CRISPR/Cas system, consists of a non-specific DNA cleavage protein Cas9, and one short RNA oligo for base pairing to the target DNA and recruitment of the Cas9/gRNA complex. The gRNA oligo contains an ∼20-nt sequence at its 5′-end, which is identified to the sequence of one DNA strand of the target sequence (the protospacer sequence) where a special NGGNN motif (the protospacer associated motif; PAM) is located downstream to the 3′-end of the protospacer on the target DNA (22,23,42–46). Comparing with the long and highly repetitive TALENs, the Cas9/gRNA system is much easier for engineering and application, as one only needs to synthesize a specific gRNA oligo of <100 nt for each new target sequence, and the Cas9 protein (or a plasmid or mRNA encoding this protein) is universal for all different target sites. Although this new system is different than ZFNs or TALENs in principle for target site recognition, they all can produce DSBs in their target sequence and induce indel mutations. However, the potentials of producing large genomic deletions by the Cas9/gRNA system have not been determined.
Here, we demonstrated the successful application of two pairs of TALENs to generate a 43.8 kb heritable chromosomal deletion as well as inversion at the same locus (sema3fb) in the zebrafish genome. Segmental deletions of six other loci with lengths ranging from several hundred base pairs to 122 kb of coding and/or non-coding sequences (including miRNA genes and clusters) were also obtained in both injected embryos and F1 fish, and deletions in four more loci of up to ∼1 Mb (981 kb) segment were also detected in somatic cells of the injected embryos. We also achieved targeted genomic deletions of two miRNA gene clusters with Cas9/gRNA systems in injected embryos. Based on these results and given the simplicity of the design of TALENs and CRISPR/Cas systems and particularly the simplicity for the construction of Cas9/gRNAs, targeted large fragment chromosomal manipulations mediated by two EENs can theoretically be achieved throughout the whole genome in vertebrates.
MATERIALS AND METHODS
Zebrafish husbandry
The wild-type strain of zebrafish used in this study is Tübingen. All the fish were maintained at 28.5°C under standard conditions.
TALEN construction and microinjection
TALEN target sites were designed using the web-tool TALEN-NT (47). TALEN expression vectors were constructed using the ‘Unit Assembly’ method with Sharkey-AS and Sharkey-R forms of FokI cleavage domains as described previously (16,48). TALEN expression vectors were linearized by NotI and used as templates for TALEN mRNA synthesis with SP6 mMESSAGE mMACHINE Kit (Ambion). To evaluate the targeting efficiency of TALEN pairs, 50–200 pg TALEN mRNAs encoding each monomer were injected in pairs into one cell-stage zebrafish embryos. The dosage showing the highest indel-inducing efficiency and acceptable low-level cytotoxicity was then used for the deletion or inversion experiments by co-injecting two pairs of TALEN mRNAs.
Cas9 and gRNA construction and microinjection
Unless otherwise specified, the Cas9 mRNA and gRNAs were synthesized as described by Chang et al. (41). Briefly, the Cas9 mRNA was synthesized by in vitro transcription using T7 mMESSAGE mMACHINE Kit (Ambion). The primers for the generation of DNA templates of gRNAs by polymerase chain reaction (PCR) were designed manually, and a T7 or SP6 promoter sequence was added to the 5′-upstream of the gRNA sequence. The gRNAs were in vitro transcribed and purified using T7 or SP6 Riboprobe Systems (Promega) and mirVana miRNA Isolation Kit (Ambion), respectively. About 300 pg of Cas9 mRNA and 50 pg of gRNA were co-injected into one cell-stage zebrafish embryos. For the deletion experiment, the same dosage of Cas9 mRNA and a pair of gRNAs, ∼50 pg each, were injected together.
Efficiency analyses of EENs and detection of large deletion and inversion events in injected embryos
The efficiency of indel mutations of a single pair of TALENs or individual Cas9/gRNA was evaluated by a restriction-endonuclease–resistant restriction fragment length polymorphism (RFLP) assay, where applicable, as described previously, and the best dosage was chosen for further experiments (16). To analyze the deletions or inversions induced by two pairs of TALENs or two Cas9/gRNAs, injected and morphologically normal embryos at 24 hpf (hours post-fertilization) were collected in groups (three to six embryos per group). The embryo groups were treated with 50 μl of 50 mmol/l NaOH at 95°C for 10 min, and then neutralized by adding 1/10 volume of 1 mol/l pH 8.0 HCl–Tris buffer. The resulting genomic DNA preparation was either used as PCR template immediately or stored at −20°C. For each injection, five to seven groups of embryos were analyzed by PCR. The forward primer of the upstream target site and the reverse primer of the downstream target site were used to detect deletions, and two forward primers or two reverse primers were used to amplify the regions spanning the inversion junctions. All the expected PCR products were gel extracted and confirmed by sequencing. The targeting efficiencies of TALEN pairs or Cas9/gRNAs are summarized in Supplementary Table S1. The sequences of PCR primers are listed in Supplementary Table S2.
Screening for germ line transmission
Genomic DNA was isolated from either F1 embryos obtained from the outcross of founder fish or tail clips from adult F1 fish by using the similar method described in the last section. For each founder, 24 hpf F1 embryos were collected in groups (6–10 embryos per group), and 4–14 groups were screened for germ line transmission by PCR analyses after genomic DNA extraction. The F1 embryos of positive founders were raised to adulthood, and tail clips of these adult F1 were collected and analyzed individually.
Evaluation of deletion rates in Cas9/gRNAs-injected embryos
To evaluate the frequency of deletion alleles in the embryos injected by Cas9 mRNA together with two gRNAs, the genomic DNA of the heterozygous F1 embryos individually selected from the offspring obtained by outcross of the founders injected with two pairs of TALEN mRNAs targeting the same locus (founder #M13 for targeting dre-mir-126a and founder #M25 for targeting miRNA Cluster Chr. 9) were used as the reference genome. The heterozygous genomic DNA (1/2 or 50% of the target alleles are deletion alleles) was diluted by the wild-type genome preparation of the same concentration to obtain a series of proportions of deletion alleles, such as 1/4 (25%), 1/8 (∼13%) and so forth. The genomic DNA of embryos injected with Cas9/gRNAs, together with the series of reference genomes (0.2–50% deletion alleles) and the wild-type genome (0% deletion alleles), were used as PCR templates with the same quantity (90 ng of total DNA) for each sample and amplified in parallel under the same conditions with the same primer pairs.
Chromosomal views of EENs in the EENdb database
The EENdb database (http://eendb.zfgenetics.org) was updated to add known gRNAs of the CRISPR/Cas system and their target sites. Web pages of chromosomal view of human and zebrafish, the two species that are targeted by most of the reported EENs, were developed with PHP script. EENdb is running on a public server and more information for this database was described previously (1).
RESULTS
Targeted chromosomal deletions and inversions at the zebrafish sema3fb locus mediated by TALENs
We selected zebrafish sema3fb as the first target gene to test whether TALENs can efficiently induce large genomic modifications, such as deletions or inversions in vivo. This gene has three splice variants as annotated by current Ensembl genome assembly (Zv9) and release (e71, April 2013) (49). Two of the transcripts contain different translation start codons with a long distance from each other, and the third one has no annotation on the translation start site. Two pairs of TALENs were constructed to target two sites in this gene, one at the fourth exon and the other at the last common (18th) exon, separated by ∼43 kb (Figure 1A). Injection of each pair of TALEN mRNAs targeting a single site into single-cell zebrafish embryos showed that both TALEN pairs have a mutagenesis rate of ∼40% leading to small indels in the corresponding target sites as evaluated by using similar restriction enzyme-detection method as described before (16). After co-injection of the mRNA mixture of the two pairs of TALENs, deletion events in the genomic region between the two target sites were detected by PCR with primers across this region in 24 hpf embryos, in which the left half of the first TALEN site and the right half of the last TALEN site were joined together, which were confirmed by sequencing results (Figure 1B). Positive PCR results of the two new potential junctions of the inversion of the same chromosome region between the two sites were also obtained (Figure 1C). Surviving embryos were grown to adulthood. Of 19 tested founder fish, three showed stable germ line transmission of the distal deletion to F1 fish, and another founder showed germ line transmission of the chromosomal inversion (Supplementary Figure S1). The ratios of F1 fish carrying deletions (mosaicism) varied from 0.2 to 7.5%, and ∼14.6% of F1 fish from the positive founder carried inversions (Table 1, row #1).
Figure 1.
Heritable chromosomal deletions and inversions in zebrafish in the region of sema3fb mediated by two pairs of TALENs. (A) The annotated structure of the zebrafish sema3fb gene and the locations and sequences of the TALEN target sites. The binding sites for the four TALEN monomers of TALEN pairs E4 and E18 are highlighted with different colors. (B) Graphic views of the target sites and PCR primers in the wild-type (wt) chromosome or the chromosomes with deletions or inversions in the sema3fb locus. The primers (E4S, E4A, E18S and E18A) are marked with similar colors as the corresponding TALEN-binding sites. (C and D) PCR results of the genomic DNA from embryos injected with two (E4 + E18), one (E4 or E18) or none (WT) of these pairs of TALENs, with the sequencing results of the PCR products from the embryos injected with the two pairs of TALENs (F0) and their progeny (F1) carrying deletions (C) or inversions (D).
Table 1.
Summary of the chromosomal manipulations induced by the two types of EENs
| No. | Targeted loci in chromosomes and genes in the regiona | EENdb IDb | Distance | Manip.c | Ratio of positive foundersd | Ratio of positive F1e | Average Ratio |
|---|---|---|---|---|---|---|---|
| 1 | Chr. 11: 36.3 Mb | TNT01 + TNT02 | 43.8 kb | Del. | 3/19 | #F3: 5/(10 × 14), 10/296* | 3.7% |
| sema3fb | |||||||
| #F4: 1/(10 × 51) | |||||||
| #M4: 6/(10 × 8) | |||||||
| Inv. | 1/19 | #F6: 2/(10 × 4), 6/41* | 10% | ||||
| 2 | Chr. 8: 12.1 Mb | TNT03 + TNT04 | 0.82 kb | Del. | 2/21 | #M12: 3/14 | 21% |
| dre-mir-126a | #M13: 3/14 | ||||||
| TNT03 + TNT05 | 0.82 kb | Del. | ND | ||||
| TNT03 + TNT06 | 0.70 kb | Del. | ND | ||||
| CNT01 + CNT02 | 0.60 kb | Del. | ND | ||||
| CNT01 + CNT04 | 0.61 kb | Del. | ND | ||||
| CNT03 + CNT02 | 0.60 kb | Del. | ND | ||||
| CNT03 + CNT04 | 0.61 kb | Del. | ND | ||||
| 3 | Chr. 11: 38.5 Mb | TNT07 + TNT08 | 0.19 kb | Del. | 1/9 | #M2: 1/(6 × 7) | 2% |
| dre-mir-126b | |||||||
| 4 | Chr. 1: 2.8 Mb | TNT09 + TNT10 | 1.5 kb | Del. | 5/15 | #M6: 6/14 | 23% |
| dre-mir-17a-1–dre-mir-92a-1 | #M7: 5/(6 × 7) | ||||||
| (miRNA Cluster Chr. 1, including 6 miRNA genes) | #M12: 5/14 | ||||||
| #M14: 5/(6 × 7) | |||||||
| #M15: 2/14 | |||||||
| TNT11 + TNT10 | 2.1 kb | Del. | 1/3 | #M2: 7/(6 × 7) | 20% | ||
| TNT09 + TNT12 | 2.6 kb | Del. | 2/9 | #M1: 2/14 | 21% | ||
| #M9: 4/14 | |||||||
| TNT11 + TNT12 | 3.2 kb | Del. | 2/11 | #M1: 5/28 | 13% | ||
| #M10: 2/28 | |||||||
| 5 | Chr.9: 55.4 Mb | TNT13 + TNT14 | 1.5 kb | Del. | 3/36 | #M19: 2/28 | 16% |
| dre-mir-17a-2–dre-mir-92a-2 | #M25: 2/28 | ||||||
| (miRNA Cluster Chr. 9, including 6 miRNA genes) | #M32: 5/14 | ||||||
| CNT05 + CNT06 | 1.5 kb | Del. | ND | ||||
| CNT05 + CNT08 | 1.5 kb | Del. | ND | ||||
| CNT07 + CNT06 | 1.5 kb | Del. | ND | ||||
| CNT07 + CNT08 | 1.5 kb | Del. | ND | ||||
| 6 | Chr. 11: 18.8 Mb | TNT15 + TNT16 | 30.2 kb | Del. | 2/40 | #M9: 3/(6 × 8), 10/64* | 13% |
| fgd5 and ENSDARG00000070653 | |||||||
| #M43: 5/(6 × 8) | |||||||
| 7 | Chr. 6: 51.7–51.8 Mb | TNT17 + TNT18 | 122 kb | Del. | 5/55 | #M26: 1/(8 × 8) | 14% |
| ENSDARG00000076787 to psmf1 | #M42: 4/16 | ||||||
| (3 coding and 1 miRNA genes) | #M46: 1/(8 × 8) | ||||||
| #M49: 3/16 | |||||||
| #M51: 4/16 | |||||||
| 8 | Chr. 23: 31.3–31.8 Mb | TNT19 + TNT20 | 436 kb | Del. | ND | ||
| tbx18 to tcf21 | |||||||
| (12 coding and 2 snoRNA genes) | |||||||
| 9 | Chr. 25: 15.5–16.5 Mb | TNT21 + TNT22 | 981 kb | Del. | ND | ||
| wt1a to mical2b | |||||||
| (27 coding and 1 miRNA genes) | |||||||
| 10 | Chr. 1: 5.1–5.2 Mb | TNT23 + TNT24 | 73.6 kb | Del. | ND | ||
| gtf3c3 and ENSDARG0000 0063253 | |||||||
| 11 | Chr. 22: 18.9 Mb–Tol2 | TNT25 + TNT26 | 57.5 kb | Del. | 0/40 | ||
| (in a transgenic line) | Inv. | ND | |||||
| midn and eGFP (exogenous) | TNT25 + TNT27 | 55.5 kb | Del. | ND |
aGenome assembly and gene annotation are based on Ensembl Zv9, release 71 (April 2013); ZFIN- or miRBase-approved gene symbols or Ensembl IDs are used.
bEfficiency of individual pair of TALENs and Cas9/gRNA are listed in Supplementary Table S1.
cManip. = manipulation; Del. = deletion; Inv. = inversion.
dND = not determined.
eMosaicism (the ratio of F1 offspring from each founder) was evaluated by different ways and presented in three patterns: (i) a/(b × c): PCR results from c wells of mixed genomic DNA (b F1 embryos per well) were obtained, and a wells were positive (i.e. no less than a F1 embryos were positive); (ii) a/b: PCR results of b single F1 embryos were obtained and a of them were positive; (iii) a/b*: PCR results of single tail clips of b F1 adult fish were obtained, and a of them were positive. In case the data from both embryos and adults are available, the latter are used to calculate the average ratios for the last column.
Targeted chromosomal deletions at non-coding loci of zebrafish genome mediated by TALENs
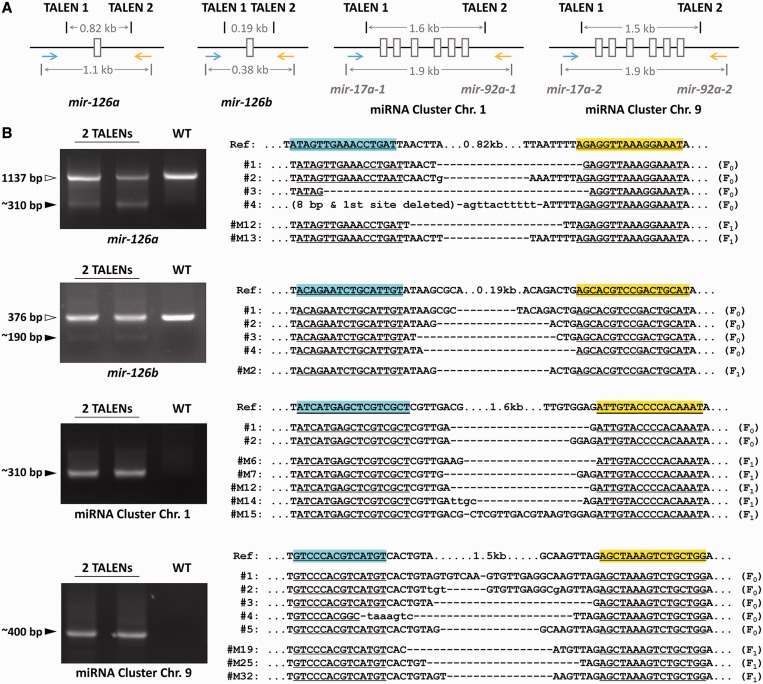
To test the possibility of generating deletions of non-coding sequences, we constructed TALENs targeting the 5′-upstream and 3′-downstream sequences of two microRNA (miRNA) genes, dre-mir-126a and dre-mir-126b, and two miRNA gene clusters, one comprising six miRNAs on chromosome 1 (Cluster Chr. 1, from dre-mir-17a-1 to dre-mir-92a-1) and another of six miRNAs on chromosome 9 (Cluster Chr. 9, from dre-mir-17a-2 to dre-mir-92a-2) (Figure 2A). Complete deletions of the ∼1.5–3.1-kb region, containing all the DNA sequence corresponding to the pre-miRNA, were detected by PCR in the embryos co-injected with the mRNAs encoding the corresponding TALENs. The TALEN target sites around the loci of dre-mir-126a and dre-mir-126b were close enough to get the PCR results of both wild-type sequences and sequences with deletions (Figure 2B and Supplementary Figures S2–S5). We tested three combinations of TALEN pairs for dre-mir-126a, one for dre-mir-126b, four for miRNA Cluster Chr. 1 and one for miRNA Cluster Chr. 9; all of the attempts were successful with deletions being detected in the mosaic-injected embryos. Furthermore, seven of the nine combinations targeting each of the four loci have been efficiently inherited to the F1 offspring as 2/21 founders for dre-mir-126a, 1/9 for dre-mir-126b, 2/11-1/3 for Cluster Chr. 1 and 3/36 for Cluster Chr. 9 (Table 1, rows #2–5).
Figure 2.
Heritable chromosomal deletions in zebrafish in the region of miRNA genes and miRNA gene clusters mediated by two pairs of TALENs. (A) The structure of the four loci of two zebrafish miRNA genes dre-mir-126a and dre-mir-126b and two miRNA gene clusters located on Chr. 1 and Chr. 9, with the approximate locations and distances of TALEN target sites and primers for PCR detection. (B) PCR results of the genomic DNA from injected (two TALENs) and un-injected (WT) embryos. For the loci of dre-mir-126a and dre-mir-126b, the PCR products of both wild-type sequences (white arrowheads) and sequences containing deletions (black arrowheads) can be seen. For the loci of the two miRNA gene clusters (the two lower panels), only the sequences carrying deletions (black arrowheads) can be amplified with the PCR program used in this experiment. Only one combination of TALEN pairs is shown here for each locus. Results of other combinations of TALENs for the same locus can be found in Supplementary Figures S2 and S4.
Targeted large genomic deletions of zebrafish miRNA genes and miRNA gene clusters mediated by CRISPR/Cas systems
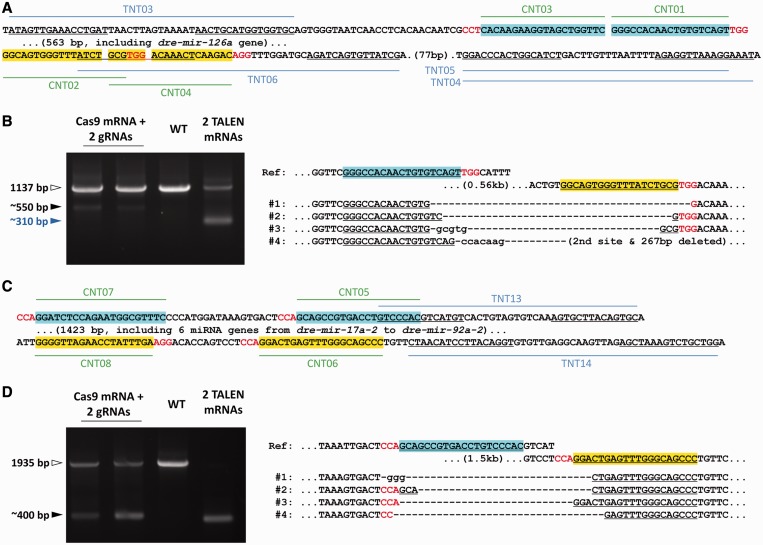
To see whether the newly developed CRISPR/Cas gene-targeting system could also be used for chromosomal large fragmental manipulation similar to TALENs, we constructed pairs of gRNAs with 19–21 nt at the 5′-end corresponding to the target recognition protospacer sequences with a PAM adjacently located at the 5′-upstream and 3′-downstream of the dre-mir-126a or miRNA Cluster Chr. 9, close to the TALEN target sites described earlier in the text (Figure 3A and C). The efficiency of indel mutations induced by individual Cas9/gRNA varies among different sites, ranging from ∼3 to ∼70% (Supplementary Table S1). Co-injection of two gRNAs together with the mRNA encoding Cas9 successfully induced relatively large deletions in this chromosomal region between the two protospacer in the mosaic-injected embryos, comparable with the deletions induced by the two pairs of TALENs. Comparing the PCR results of wild-type sequences and sequences containing deletions, the efficiencies of deletions by Cas9/gRNAs were lower than those of corresponding TALENs (Figure 3B and D and Supplementary Figure S5). The deletion rates in TALEN mRNA-injected embryos were no <25% in both loci, whereas those injected with Cas9/gRNAs were ∼1–3%, as roughly evaluated by PCR amplification and comparison with the serial dilutions of reference genomes from F1 heterozygous embryos targeting the same chromosome region by TALENs (Supplementary Figure S6).
Figure 3.
Chromosomal deletions in zebrafish in the region of an miRNA gene and an miRNA gene cluster mediated by Cas9 and gRNA pairs. (A) Sequences of the TALEN and Cas9/gRNA target sites around the miRNA gene dre-mir-126a. The protospacer target sequences of the Cas9/gRNA are highlighted with different colors; the 3-nt sequence of PAM (-NGG, or shown as its reversed sequence CCN-) is shown in red. The binding sites of TALEN monomers are underlined. (B) Chromosomal deletions induced by Cas9 and two gRNAs targeting this locus. The result from one of the four combinations of gRNA pairs used to generate deletions in this region is shown here, and the results for other gRNA combinations can be found in Supplementary Figure S2E. Deletion induced by two pairs of TALENs is shown in the last lane for comparison (as a positive control). The PCR products of wild-type sequences (white arrowhead), sequences with deletions induced by Cas9/gRNAs (blue arrowhead) or TALENs (black arrowhead) are indicated. (C) Sequences of the TALEN and Cas9/gRNA target sites around the miRNA gene cluster on Chr. 9 (from dre-mir-17a-2 to dre-mir-92a-2). The target sites of Cas9/gRNA and TALENs are indicated as shown in (A). (D) Chromosomal deletions induced by Cas9 and two gRNAs targeting this locus. The result from one of the four combinations of gRNA pairs used to generate deletions in this region is shown here, and the results for other gRNA combinations can be found in Supplementary Figure S5C. Deletions induced by two pairs of TALENs are shown in the last lane for comparison (as a positive control). The PCR products of wild-type sequences (white arrowhead) and sequences with deletions induced by Cas9/gRNAs or TALENs (black arrowhead) are indicated. A relatively longer extension time for PCR was used in this experiment to simultaneously amplify the wild-type allele.
Updating EENdb to help searching for candidate loci of EEN-mediated chromosomal modification
We used our recently published database EENdb to obtain more candidate loci with different distances in zebrafish genome to test the universal applications of large genomic deletions by two pairs of EENs. The database and knowledge base of EENdb (http://eendb.zfgenetics.org) was built to collect information of all reported ZFNs and TALENs tested in different species and the related information, uses and resources for EEN engineering (1). With the development of the new gene-targeting technology based on the CRISPR/Cas system and the predictable rapid increasing of Cas9/gRNA target sites, we have started to collect the information of Cas9/gRNA in EENdb. In addition, we also updated EENdb to provide an intuitional view of all reported target sites of EENs on the chromosomes of human and zebrafish, currently the two species that are targeted by most of the reported EENs. One can easily identify hotspots being targeted, with the help of the chromosomal view of EEN target sites; for example, human CCR5 locus has been targeted by many different ZFNs and TALENs. The chromosome view of EENdb provides a centralized view of all types of EENs targeting the same chromosome with different colors and the distances to any other EEN target sites relative to the one of the user’s choice will be calculated and displayed automatically (Supplementary Figure S13). This view allows users to easily find EEN target sites around chromosomal regions of interest with links to more detailed information of every EEN entry, and help to design a new pair of TALENs and/or a new Cas9/gRNA target site with appropriate distances from an existing target site. The chromosome views can be expanded to other species when necessary.
Targeted chromosomal deletions and inversions at six other loci of zebrafish genome mediated by TALENs
We selected five chromosomal segments sandwiched by two known TALEN target sites, with the distance ranging from 30.2 kb to ∼1 Mb (981 kb) by searching EENdb (Table 1, rows #6–10). Using these pre-constructed TALENs, we tested the range of efficiency and distance of TALEN-mediated deletions. Distal deletions were detected in the embryos injected with the TALEN mRNAs targeting two sites separated by long distances, including the deletion of a 436-kb genomic region between the TALEN sites located in the tbx18 and tcf21 genes on chromosome 23, spanning 11 other annotated genes, and deletion of a nearly 1 Mb region between wt1a and mical2b genes on chromosome 25, which contained 26 additional genes (Supplementary Figures S9 and S10). Three other combinations of TALEN pairs used to induce large chromosomal deletions were also tested in injected embryos, including regions containing multiple coding and non-coding genes. Two of them, with a distance of 30.2 kb containing only two genes and a distance of 122 kb spanning three coding and one miRNA genes, produced heritable germ line transmission in 2/40 and 1/30 founders, with mosaicism of 13% in average and ∼2%, respectively (Supplementary Figures S7, S8 and S11).
An additional chromosomal region between an endogenous locus of midn gene and an exogenous locus in the 5′-arm of Tol2 transposon in a transgenic zebrafish line with a distance of ∼57.5 kb including an eGFP reporter gene was tested, in which both deletions and inversions were detected in injected embryos (Table 1, row #11; Supplementary Figure S12).
DISCUSSION
In this study, we were able to create heritable distal deletions (up to 122 kb) and inversions (43.8 kb) mediated by TALENs in the genome of an in vivo vertebrate model organism, and by using the CRISPR/Cas system, large fragment deletions were also induced in the zebrafish genome for the first time. Through this approach, we have obtained heritable inversion mutations in addition to deletions, and also fully disrupted several single miRNA genes and gene clusters in the F1 offspring. Attempts to delete regions >122 kb and up to ∼1 Mb were also successful in injected embryos. Taken together, our results support the concept that using two pairs of EENs can manipulate the genome at the chromosomal level.
In the progress of this work, another group reported the success of heritable-targeted deletions of coding genes in zebrafish and somatic deletions of non-coding sequences mediated by two pairs of TALENs in injected embryos, as well as inversions in mosaic-injected embryos, which is consistent with our findings (50). In addition, we provide the first evidence that CRISPR/Cas system can also be used to create comparable segmental deletions in zebrafish as TALENs.
In the current Ensembl release of zebrafish genome annotation (e71, April 2013), 10 665 of 26 241 protein-coding genes have more than one transcript, in which 6914 have at least two different positions of translation start sites (the annotated start codon, or the available first codon if the start codon has not been annotated) of all their transcripts (Supplementary Methods). Unlike the well-annotated genomes, such as human or mouse, cis-elements in zebrafish chromosomes are rarely annotated with evidences. To fully analyze and understand the function of these complex genes and potential regulatory elements, single-site indel mutations are certainly not enough. Furthermore, there are currently 333 miRBase annotated and 407 predicted miRNA genes in the zebrafish genome, as well as many other ncRNA genes. Two publications have predicted 1132 and 691 long (intergenic) non-coding RNA (lincRNA or lncRNA) genes in the zebrafish genome, respectively, in which few have been studied concerning their functions, partially because of the lack of an efficient mutagenesis strategy (51,52). The method of chromosomal deletions mediated by EENs reported in this work provide a robust and efficient tool to complete knockout of either coding or non-coding genes, or any other desirable genomic sequences, and will be helpful for targeting the genome for functional and mechanism studies in more flexible ways.
It should be cautious when screening for large deletions of multiple genes or regions spanning long distance, including long intergenic sequences. In our effort to screen for a 57.5-kb deletion between the midn gene and a Tol2 transposon insertion, no positive germ line transmission event was obtained from 40 founders, although there seems to have no other genes annotated in this region. This may be due to low efficiency of deletions, or haploinsufficiency of certain genes or elements in the region between the two target sites, either annotated or still unknown. We, therefore, suggest designing the TALEN or Cas9/gRNA target sites as closer as possible to the target genes, gene clusters or any other sequences to be removed, to avoid disrupting other potentially functional regions when using the large genomic deletion strategy.
The Cas9/gRNA system has been reported to have comparable mutagenesis efficiency with TALENs in zebrafish. Although the efficiencies varied dramatically based on our data and more characteristics of this system still need to be investigated, it has shown some advantages over ZFNs and TALENs. Cas9/gRNA system is simple to design and quick to construct. In vitro transcription and purification of a gRNA as short as ∼100 nt can be completed within a few hours, and this is the only specific reagent needed for a specific target site, whereas the Cas9 mRNA or protein is common for all loci, whereas construction of a pair of TALEN expression plasmids encoding >700 amino acids proteins (for TALENs containing at least 15 tandem repeats) is required for each new target site. We believe that the application of Cas9/gRNA system, which is complementary to the TALEN technology, as well as the strategies of large fragment chromosomal manipulations mediated by two EENs, hold new promise for functional genomics studies.
PCR has been used to approximately determine the relative efficiencies of genomic deletions induced by pairs of EENs targeting similar loci (i.e. similar sequence and length of deletions), e.g. the relative intensities of PCR products from embryos injected with three different combinations of TALEN pairs targeting the dre-mir-126a locus, correlated well to the efficiencies of individual TALEN pair (Figure 2B, upper panel, Supplementary Figure S2C and D and Supplementary Table S1). However, in most cases, PCR is not sufficient to estimate the actual ratio of alleles bearing deletions, inversions or other positive-selection–based genome modifications (such as HR induced by EENs). In the case of detecting deletions, even if two amplicons/bands with different length were obtained in a single PCR reaction with a single pair of primers, the ratio of the intensities between the two bands (one corresponding to the deletion allele and the other corresponding to the wild-type allele) usually can not reflect the actual ratio of the two templates before amplification, as the efficiency of PCR amplification heavily relies on the length of template DNA, where longer templates are usually poorly amplified and, therefore, underrepresented, especially in the cases of two amplicons with different sizes. For example, the genome of F1 fish heterozygous for certain deletions should contain equal amount of deletion allele and the wild-type allele as templates for PCR amplification. However, when detecting the F1 fish heterozygous for the deletion of dre-mir-126a or miRNA Cluster Chr. 9, a much stronger band corresponding to the deletion allele (∼310 bp for dre-mir-126a and ∼400 bp for miRNA Cluster Chr. 9) was dominant in the PCR product, whereas the band corresponding to the wild-type allele (1137 bp for dre-mir-126a and 1935 bp for miRNA Cluster Chr. 9) was faint and barely visible, which was notably deviated from the actual 1:1 ratio of initial templates for amplification (Supplementary Figures S2B and S5B).
Recently, a more reliable approach for the evaluation of deletion efficiencies was reported, in which the genome of verified targeted cell clones was serially diluted with the wild-type genome in different proportions and used as references, and then the genome of treated cells was examined together with these benchmarks by using the same quantity of templates for PCR amplification (26). By using similar methods, the relative efficiency of deletions or inversions in injected zebrafish embryos should be able to be evaluated with the benchmarks of genomes from heterozygous or homozygous embryos or adult fish. As germ line transmission of the deletions induced by Cas9/gRNAs was temporarily not available in this work, the F1 embryos heterozygous for deletions induced by TALENs targeting the same chromosome region were used as the reference genome to roughly compare and evaluate the deletion efficiencies in the embryos injected with Cas9/gRNAs in this work (Supplementary Figure S6).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–13 and Supplementary Methods.
FUNDING
The 973 programs [2012CB945101, 2011CBA01000]; National Natural Science Foundation of China [31110103904, 30730056]; NIH [DK54508 to S.L.]. Funding for open access charge: [31110103904].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are thankful to Dr I. Bruce and Dr M. Veldman for proofreading and editing the manuscript; J. Xiong of Institute of Molecular Medicine at Peking University for providing the CRISPR/Cas system; Y. Feng, D. Liu and S. He for construction of some of the TALENs and help in fish screening; Y. Shen, Y. Gao and J. Zhang for technical support and laboratory management; Y. Jia, J. Chen, X. Yang and H. Cui for zebrafish maintenance.
REFERENCES
- 1.Xiao A, Wu Y, Yang Z, Hu Y, Wang W, Zhang Y, Kong L, Gao G, Zhu Z, Lin S, et al. EENdb: a database and knowledge base of ZFNs and TALENs for endonuclease engineering. Nucleic Acids Res. 2013;41:D415–D422. doi: 10.1093/nar/gks1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foley JE, Yeh JR, Maeder ML, Reyon D, Sander JD, Peterson RT, Joung JK. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by Oligomerized Pool ENgineering (OPEN) PLoS One. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siekmann AF, Standley C, Fogarty KE, Wolfe SA, Lawson ND. Chemokine signaling guides regional patterning of the first embryonic artery. Genes Dev. 2009;23:2272–2277. doi: 10.1101/gad.1813509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta A, Meng X, Zhu LJ, Lawson ND, Wolfe SA. Zinc finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc finger nucleases. Nucleic Acids Res. 2011;39:381–392. doi: 10.1093/nar/gkq787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu C, Smith T, McNulty J, Rayla AL, Lakshmanan A, Siekmann AF, Buffardi M, Meng X, Shin J, Padmanabhan A, et al. Evaluation and application of modularly assembled zinc-finger nucleases in zebrafish. Development. 2011;138:4555–4564. doi: 10.1242/dev.066779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore FE, Reyon D, Sander JD, Martinez SA, Blackburn JS, Khayter C, Ramirez CL, Joung JK, Langenau DM. Improved somatic mutagenesis in zebrafish using transcription activator-like effector nucleases (TALENs) PLoS One. 2012;7:e37877. doi: 10.1371/journal.pone.0037877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xing L, Hoshijima K, Grunwald DJ, Fujimoto E, Quist TS, Sneddon J, Chien CB, Stevenson TJ, Bonkowsky JL. Zebrafish foxP2 zinc finger nuclease mutant has normal axon pathfinding. PLoS One. 2012;7:e43968. doi: 10.1371/journal.pone.0043968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kok FO, Taibi A, Wanner SJ, Xie X, Moravec CE, Love CE, Prince VE, Mumm JS, Sirotkin HI. Zebrafish rest regulates developmental gene expression but not neurogenesis. Development. 2012;139:3838–3848. doi: 10.1242/dev.080994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S, Oikonomou G, Chiu CN, Niles BJ, Liu J, Lee DA, Antoshechkin I, Prober DA. A large-scale in vivo analysis reveals that TALENs are significantly more mutagenic than ZFNs generated using context-dependent assembly. Nucleic Acids Res. 2013;41:2769–2778. doi: 10.1093/nar/gks1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Bebber F, Hruscha A, Willem M, Schmid B, Haass C. Loss of Bace2 in zebrafish affects melanocyte migration and is distinct from Bace1 knock out phenotypes. J. Neurochem. 2013 doi: 10.1111/jnc.12198. March 11 (doi:10.1111/jnc.12198; Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 14.Taibi A, Mandavawala KP, Noel J, Okoye EV, Milano CR, Martin BL, Sirotkin HI. Zebrafish churchill regulates developmental gene expression and cell migration. Dev. Dyn. 2013;242:614–621. doi: 10.1002/dvdy.23958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sood R, Carrington B, Bishop K, Jones M, Rissone A, Candotti F, Chandrasekharappa SC, Liu P. Efficient methods for targeted mutagenesis in zebrafish using zinc-finger nucleases: data from targeting of nine genes using CompoZr or CoDA ZFNs. PLoS One. 2013;8:e57239. doi: 10.1371/journal.pone.0057239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 17.Cade L, Reyon D, Hwang WY, Tsai SQ, Patel S, Khayter C, Joung JK, Sander JD, Peterson RT, Yeh JR. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 2012;40:8001–8010. doi: 10.1093/nar/gks518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, Weis AM, Voytas DF, Grunwald DJ. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 2012;8:e1002861. doi: 10.1371/journal.pgen.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navis A, Marjoram L, Bagnat M. Cftr controls lumen expansion and function of Kupffer's vesicle in zebrafish. Development. 2013;140:1703–1712. doi: 10.1242/dev.091819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben J, Elworthy S, Ng AS, van Eeden F, Ingham PW. Targeted mutation of the talpid3 gene in zebrafish reveals its conserved requirement for ciliogenesis and Hedgehog signalling across the vertebrates. Development. 2011;138:4969–4978. doi: 10.1242/dev.070862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei C, Liu J, Yu Z, Zhang B, Gao G, Jiao R. TALEN or Cas9 - rapid, efficient and specific choices for genomic modifications. J. Genet. Genomics. 2013;40:281–289. doi: 10.1016/j.jgg.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrangou R. RNA-mediated programmable DNA cleavage. Nat. Biotechnol. 2012;30:836–838. doi: 10.1038/nbt.2357. [DOI] [PubMed] [Google Scholar]
- 24.Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, 2nd, Tan W, Penheiter SG, Ma AC, Leung AY, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zu Y, Tong X, Wang Z, Liu D, Pan R, Li Z, Hu Y, Luo Z, Huang P, Wu Q, et al. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat. Methods. 2013;10:329–331. doi: 10.1038/nmeth.2374. [DOI] [PubMed] [Google Scholar]
- 26.Lee HJ, Kim E, Kim JS. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 2010;20:81–89. doi: 10.1101/gr.099747.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu PQ, Chan EM, Cost GJ, Zhang L, Wang J, Miller JC, Guschin DY, Reik A, Holmes MC, Mott JE, et al. Generation of a triple-gene knockout mammalian cell line using engineered zinc-finger nucleases. Biotechnol. Bioeng. 2010;106:97–105. doi: 10.1002/bit.22654. [DOI] [PubMed] [Google Scholar]
- 28.Kim E, Kim S, Kim DH, Choi BS, Choi IY, Kim JS. Precision genome engineering with programmable DNA-nicking enzymes. Genome Res. 2012;22:1327–1333. doi: 10.1101/gr.138792.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CB, Fahrenkrug SC. Efficient TALEN-mediated gene knockout in livestock. Proc. Natl Acad. Sci. USA. 2012;109:17382–17387. doi: 10.1073/pnas.1211446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim Y, Kweon J, Kim A, Chon JK, Yoo JY, Kim HJ, Kim S, Lee C, Jeong E, Chung E, et al. A library of TAL effector nucleases spanning the human genome. Nat. Biotechnol. 2013;31:251–258. doi: 10.1038/nbt.2517. [DOI] [PubMed] [Google Scholar]
- 31.Lee HJ, Kweon J, Kim E, Kim S, Kim JS. Targeted chromosomal duplications and inversions in the human genome using zinc finger nucleases. Genome Res. 2012;22:539–548. doi: 10.1101/gr.129635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunet E, Simsek D, Tomishima M, DeKelver R, Choi VM, Gregory P, Urnov F, Weinstock DM, Jasin M. Chromosomal translocations induced at specified loci in human stem cells. Proc. Natl Acad. Sci. USA. 2009;106:10620–10625. doi: 10.1073/pnas.0902076106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simsek D, Brunet E, Wong SY, Katyal S, Gao Y, McKinnon PJ, Lou J, Zhang L, Li J, Rebar EJ, et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7:e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piganeau M, Ghezraoui H, De Cian A, Guittat L, Tomishima M, Perrouault L, Rene O, Katibah G, Zhang L, Holmes M, et al. Cancer translocations in human cells induced by zinc finger and TALE nucleases. Genome Res. 2013;23:1182–1193. doi: 10.1101/gr.147314.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma S, Zhang S, Wang F, Liu Y, Liu Y, Xu H, Liu C, Lin Y, Zhao P, Xia Q. Highly efficient and specific genome editing in silkworm using custom TALENs. PLoS One. 2012;7:e45035. doi: 10.1371/journal.pone.0045035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menoret S, Iscache AL, Tesson L, Remy S, Usal C, Osborn MJ, Cost GJ, Bruggemann M, Buelow R, Anegon I. Characterization of immunoglobulin heavy chain knockout rats. Eur. J. Immunol. 2010;40:2932–2941. doi: 10.1002/eji.201040939. [DOI] [PubMed] [Google Scholar]
- 37.Chen F, Pruett-Miller SM, Huang Y, Gjoka M, Duda K, Taunton J, Collingwood TN, Frodin M, Davis GD. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang P, Xu L, Liang W, Tam CI, Zhang Y, Qi F, Zhu Z, Lin S, Zhang B. Genomic deletion induced by Tol2 transposon excision in zebrafish. Nucleic Acids Res. 2013;41:e36. doi: 10.1093/nar/gks1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang P, Zhu Z, Lin S, Zhang B. Reverse genetic approaches in zebrafish. J. Genet. Genomics. 2012;39:421–433. doi: 10.1016/j.jgg.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong JW, Xi JJ. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 44.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramalingam S, Annaluru N, Chandrasegaran S. A CRISPR way to engineer the human genome. Genome Biol. 2013;14:107. doi: 10.1186/gb-2013-14-2-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, Zhang X, Zhang P, Huang X. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013;23:720–723. doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doyle EL, Booher NJ, Standage DS, Voytas DF, Brendel VP, Vandyk JK, Bogdanove AJ. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40:W117–W122. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, Li C, Yu Z, Huang P, Wu H, Wei C, Zhu N, Shen Y, Chen Y, Zhang B, et al. Efficient and specific modifications of the Drosophila genome by means of an easy TALEN strategy. J. Genet. Genomics. 2012;39:209–215. doi: 10.1016/j.jgg.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 49.Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, et al. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gupta A, Hall VL, Kok FO, Shin M, McNulty JC, Lawson ND, Wolfe SA. Targeted chromosomal deletions and inversions in zebrafish. Genome Res. 2013;23:1008–1017. doi: 10.1101/gr.154070.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22:577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.