Abstract

Worldwide, a number of viable populations of fish are found in environments heavily contaminated with metals, including brown trout (Salmo trutta) inhabiting the River Hayle in South-West of England. This population is chronically exposed to a water-borne mixture of metals, including copper and zinc, at concentrations lethal to naïve fish. We aimed to investigate the molecular mechanisms employed by the River Hayle brown trout to tolerate high metal concentrations. To achieve this, we combined tissue metal analysis with whole-transcriptome profiling using RNA-seq on an Illumina platform. Metal concentrations in the Hayle trout, compared to fish from a relatively unimpacted river, were significantly increased in the gills, liver and kidney (63-, 34- and 19-fold respectively), but not the gut. This confirms that these fish can tolerate considerable metal accumulation, highlighting the importance of these tissues in metal uptake (gill), storage and detoxification (liver, kidney). We sequenced, assembled and annotated the brown trout transcriptome using a de novo approach. Subsequent gene expression analysis identified 998 differentially expressed transcripts and functional analysis revealed that metal- and ion-homeostasis pathways are likely to be the most important mechanisms contributing to the metal tolerance exhibited by this population.
Introduction
Metal contamination of freshwater systems occurs worldwide, in some cases reaching concentrations known to cause acute toxicity, yet a few of these rivers and lakes support viable populations of fish. Yellow perch populations inhabiting lakes in North America contaminated through industrial and mining activity with a number of metals (particularly copper, cadmium and nickel) are an exceptionally well studied example. Gradients in contamination have been used to demonstrate correlations between chronic metal exposure and a number of physiological changes associated with metal toxicity and/or tolerance, including alterations in metabolic processes, the antioxidant system and metal transporting/sequestering pathways [e.g., refs (1−7)]. Additionally, these metal exposures have resulted in impaired growth, reproduction and genetic diversity, with potentially adverse implications for the health of these populations.5,6,8
Brown trout (Salmo trutta) inhabiting the River Hayle in Cornwall (Southwest England) are another population of fish chronically exposed to elevated metal concentrations. Historically, mining in the surrounding area dates back to Neolithic times, peaking during the 1800s and drainage from the disused mines continues to contaminate the river with a mixture of metals.9,10 The middle region of the river Hayle has extremely high metal concentrations, where little fish or invertebrate life is found, however this does not prevent brown trout migration and gene flow between the upper and lower sections.10 Concentrations of metals in the lower region, where brown trout are readily found, have been documented to cause acute toxicity in metal-naive brown trout [e.g., refs (10−13)], including total zinc, copper and iron which averaged 639, 42 and 200 μg/L respectively (data kindly provided from the Environment Agency, Supporting Information Table S1). Despite the persistent and significant levels of metal contamination, the River Hayle appears to support a sustainable population of brown trout that exhibits no evidence of reduced genetic diversity.10 Therefore, it is expected that the brown trout population in the River Hayle may exhibit mechanisms allowing them to tolerate chronic metal exposure. Populations of brown trout inhabiting water systems contaminated with metals are not unique to the River Hayle and other examples include populations found in Norway and the USA.14,15 Despite this, very little is known about the physiological and molecular adaptations that allow this species to survive high concentrations of metals in their environment.
We aimed to address this knowledge gap, using brown trout from the River Hayle as a case study. We adopted an integrative approach combining genomics with analysis of tissue metal accumulation, to understand the molecular mechanisms employed by fish from this population to tolerate the high concentrations of metals in their environment. There is relatively limited gene sequence information for brown trout, therefore our first goal was to sequence, assemble and annotate the transcriptome for this species, using the Illumina sequencing platform to perform RNA-seq. We then used this resource to investigate molecular pathways differentially regulated in fish from the River Hayle, compared to a metal naive brown trout population originating from a relatively un-impacted river within the same geographical region. Our data demonstrated the very significant metal accumulation in tissues of fish originating from the River Hayle and proposes a number of molecular mechanisms employed by this species to cope with the high concentrations of metals in their environment, including the regulation of metal and ion homeostasis pathways and activation of antioxidant systems.
Materials and Methods
Sample Collection
To obtain a comprehensive sequence data set for assembly of the brown trout transcriptome, we collected five embryos at ten different stages of development and a range of tissues from adult fish. In a second phase, to investigate the mechanisms of tolerance to metals in the brown trout population from the River Hayle, five adult fish from this river and ten from a control river (River Teign) were sampled for analysis of tissue metal content and for transcriptomic analysis. A full description of the samples collected is presented in Supporting Information (Table S2).
Metal Analysis
Metal concentrations in the River Hayle and Teign were kindly provided by the Environment Agency and are presented in Supporting Information (Table S1). Portions of gill, gut (stomach and intestine), kidney and liver were dissected from fish obtained from the River Hayle and River Teign for determination of the metal content in these tissues. Samples were dried to a constant weight and digested with 1 mL of concentrated HNO3 (for trace metal analysis, Fisher Chemicals) for 24 h at 60 °C and treated with 60 μL H2O2. The samples were diluted with 9 mL of Milli-Q water and analyzed for Cu, Pb, Zn, As, Cd, Fe and Ni by inductively coupled plasma mass spectrometry (ICP-MS; E:AN 6100DRC, Perkin-Elmer, Cambridge, U.K.). Cluster analysis was performed on the metal concentrations in individual fish tissues from the river Hayle (h1–h5) and river Teign (t1–t10) using Euclidean distance measure and heatmaps were produced using the pheatmap package in R/Bioconductor.16 Associations between individual fish length/weight and tissue metal concentrations were tested using regression analysis.
RNA Extraction, Library Construction and Sequencing
Total RNA was extracted from all individual embryos and adult tissues using TRI reagent (Sigma-Aldrich), according to the manufacturer’s instructions. The concentration and quality of RNA in each sample was determined using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, U.S.A.) and an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., U.S.A.). Equal amounts of RNA from individual samples were pooled to obtain the samples described in Supporting Information Table S2. cDNA libraries were prepared for each pooled sample using the Illumina TruSeq RNA Sample Preparation kit and sequencing was conducted using an Illumina GAIIx Genome Analyzer, generating 100 bp paired end reads for the embryonic library and 76 bp paired end reads for 12× multiplexed adult libraries. A detailed description of these methods is presented in Supporting Information.
Assembly and Annotation of the Brown Trout Transcriptome
Raw sequences were processed to remove Illumina adapter sequences and filter out sequences that did not meet the quality thresholds. Sequences less than 30 bp in length were removed. All paired reads of the adult tissue and embryo libraries were pooled and assembled de novo using Velvet (version 1.2.08; ref (17)) and Oases (version 0.2.08; ref (18)) using a range of k-mers (see Supporting Information for full details). The resulting transcripts were annotated using Blastn and Blastx and a selection of fish and mammalian nucleotide and protein databases and using an e-value cut off <1 × 10–15. Gene expression was determined in the gill, gut, kidney and liver of fish inhabiting the metal-contaminated river Hayle and the reference river Teign using RSEM.19 Reads were mapped against the brown trout reference transcriptome using the “--no_polyA” parameter and using default settings. Subsequent analyses in RSEM were conducted using a selection of scripts provided as part of the Trinity assembly package (version r2012–10–05;.20 Statistical differences in gene expression levels between tissues of the two rivers were calculated using edgeR.21 Genes were considered differentially expressed when FDR < 0.1 (Benjamini–Hochberg correction). A 4-way Venn diagram showing overlapping differentially expressed genes was produced using VennDiagram22 in R/Bioconductor. All analyses were carried out on a local server running under the NEBC Bio-Linux 7 environment (23) unless stated otherwise. A flow diagram describing the transcriptome assembly and gene expression analysis is presented in Figure 1 and a full description of the methodology is presented in the Supporting Information.
Figure 1.

Flow diagram illustrating the workflow employed for sequencing, assembling and annotating the brown trout transcriptome and for determining changes in gene expression profiling between brown trout populations. Red text indicates results at each stage of the analysis pipeline.
The sequence data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE45637.
Transcriptomic Analysis
Functional analysis was then performed for differentially expressed genes from each tissue using the Database for Annotation, Visualization and Integrated Discovery (DAVID version 6.7; (24)), using the brown trout transcriptome as background list. Gene Ontology (GO) Fat terms for Biological Process, Cellular Component and Molecular Function were considered significantly over-represented when P < 0.05.
In order to validate the quantitative analysis of differential gene expression between Hayle and Teign trout, a selection of four transcripts (mtb, gpx1b, cat, slc40a1) were analyzed via real time quantitative PCR (RT-QPCR) on gill, gut, kidney and liver samples from all individual fish, according to previously described methods.25,26 These transcripts encode proteins involved in metal homeostasis and oxidative stress response and are therefore potentially differentially regulated by metal exposure. They include transcripts that were found to be both differentially expressed and not differentially expressed in the RNA-seq data, to corroborate both of these scenarios. Transcript expression levels were normalized using the control gene Actin-related protein 2/3 complex 3 (arpc3) which was selected from the RNA-seq data set based on its consistent expression between Hayle and Teign fish in all tissues (Supporting Information Table S3a). Full details are presented in the Supporting Information.
Results and Discussion
Tissue Metal Accumulation
In the gills, liver and kidney the concentration of all seven metals measured (Cu, Pb, Zn, As, Cd, Fe, Ni) was significantly higher in the Hayle trout than the Teign trout. Across all metals the fold change was highest in the gill (mean 62.6-fold) followed by the liver (mean 33.7-fold), then the kidney (mean 18.5-fold). In contrast, in the gut there was no significant difference in the concentration of any of the metals measured (Figure 2). The considerable increase in Hayle gill metal concentration contrasts sharply with the lack of difference in the gut and suggests that the gills are the principal route of metal uptake in these fish. This is because of their large surface area in direct contact with water and abundance of metal specific carriers (e.g., for essential metals copper, zinc and iron), as well as other ion/metal transporters that allow uptake of a number of metals through ionic mimicry (e.g., Cu+ via Na+ uptake routes and Zn2+ and Cd2+ via Ca2+ uptake routes).27,28
Figure 2.
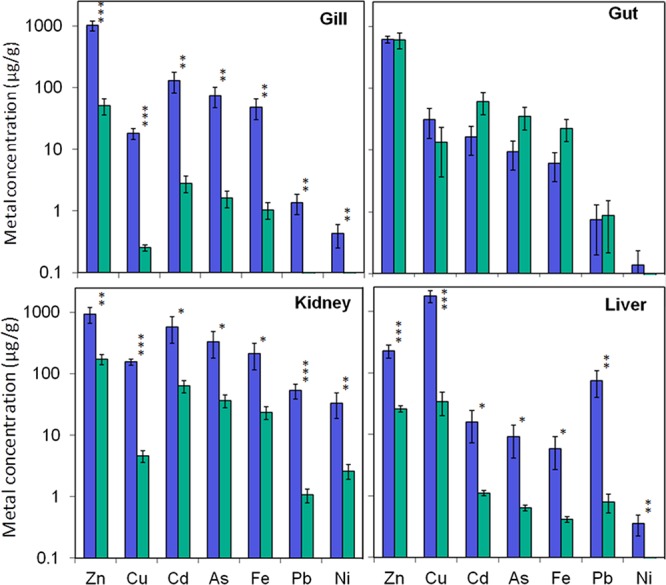
Concentration of six metals measured by ICP-MS, in the gill, gut, kidney and liver of fish from the rivers Hayle and Teign. Values are expressed as mean ± SEM. Blue bars represent data from fish originating from the metal contaminated river Hayle (n = 5) and green bars represent data from fish originating from the relatively unimpacted river Teign (n = 10). Asterisks indicate significant differences in concentration of each metal between fish from each population, * P < 0.05, **P < 0.01, ***P < 0.001.
After uptake, metals are transported in the bloodstream throughout the body. The considerable accumulation and greatest total concentration, of metals in the kidney and liver reflects the essential role of these tissues in metal processing, detoxification, storage and excretion. In both Hayle and Teign fish, zinc was the most abundant metal in the gill, gut and kidney, while copper was found at the highest concentration in the liver. Copper and zinc were also the metals that increased to the greatest extent, in terms of absolute concentration, in the gills, liver and kidney. Corresponding with this, water concentrations of zinc and copper were elevated to the greatest extent of all metals in the River Hayle compared to the Teign, by approximately 60- and 40-fold, respectively (Supporting Information Table S1). The tissue distribution and accumulation patterns of iron, cadmium and arsenic reveal some striking correlations between these three metals and this is supported by cluster analysis on individual fish (Supporting Information Figure S1). This strongly suggests that the uptake, storage and metabolism of these metals, in particular, are linked. No significant correlation was found between fish length or weight and metal concentration in any tissue, suggesting the difference in size/age of the sampled populations is unlikely to have influenced the metal accumulation patterns.
The levels of metal accumulation in the tissues of brown trout from the River Hayle were considerably higher than that measured in other chronically exposed fish from metal-contaminated regions. For example, Hayle trout had accumulated 156, 1800 and 18 μg/g copper and 929, 229 and 1020 μg/g zinc in the kidney, liver and gills respectively, compared with 4.88, 242 and 3.85 μg/g copper and 186, 47.9 and 73.7 μg/g zinc in the same tissues of brown trout in copper and zinc rich Norwegian rivers.29 Highest recorded values of 256.6 μg/g copper and 157 μg/g zinc in liver of yellow perch from metal contaminated Canadian lakes (30) are also far lower than the concentrations of these metals in the liver of the Hayle brown trout (1800 and 229 μg/g for copper and zinc, respectively). This highlights both the extent of metal contamination in the River Hayle and the high degree of metal tolerance of its resident brown trout population.
Assembly of the Brown Trout Transcriptome
Sequencing generated 68.8 M 100 bp reads from the embryonic library and a total of 78.7 M 76 bp reads from the multiplexed libraries (ranging from 5.1 to 7.7 M reads per library). Following raw sequence read processing and quality filtering a total of 60.1 M (9.7% orphans) embryonic reads and 66.5 M (8.7% orphans) multiplexed reads were retained and input into the transcriptome assemblies. The de novo assembly consisted of 202,994 transcripts (136,848 loci), with an average length of 821 bp and an N50 of 1853 bp, 48% of which were annotated by Blast (e-value <1 × 10–15) (Figure 1). The final transcriptome assembly provides a high quality template for global gene expression profiling in this study and also provides a valuable tool for wider research on brown trout, which is an ecologically and economically important fish species with limited existing genomic resources.
Transcriptome Profiling
Transcript profile analysis revealed that 73 881 transcripts were expressed in at least one of the eight Hayle and Teign libraries tested. The total number of genes present and their expression level distribution were generally consistent between libraries, although some tissue-specific differences were evident. For example, liver and gut tissues expressed a greater proportion of rare genes and fewer genes in total, than gill and kidney (Supporting Information Table S5). The gene expression patterns for the four selected tissues for the Hayle and Teign fish were examined to identify potential mechanisms of toxicity and/or tolerance to the metal exposure in the River Hayle. A total of 998 transcripts were differentially expressed in at least one tissue (Figure 3). The greatest number of differentially expressed genes (792) occurred in the kidney. Perhaps surprisingly, given its role in metal uptake, substantial metal accumulation and known susceptibility to acute metal toxicity, fewest genes (183) were differentially expressed in the gill. In contrast, despite no increase in metal accumulation a considerable number of genes (288) were differentially expressed in the gut, but a large proportion of these genes are linked to digestion and likely to be related to dietary differences between the sites (see below). RT-QPCR analysis was in full agreement with the RNA-seq transcriptional profiling data, confirming the reliability of the quantitative data obtained from sequencing analysis on pooled samples (Supporting Information Table S6). Significantly over-represented GO FAT terms (P < 0.05), among the differentially expressed gene lists are shown in Supporting Information Table S7.
Figure 3.
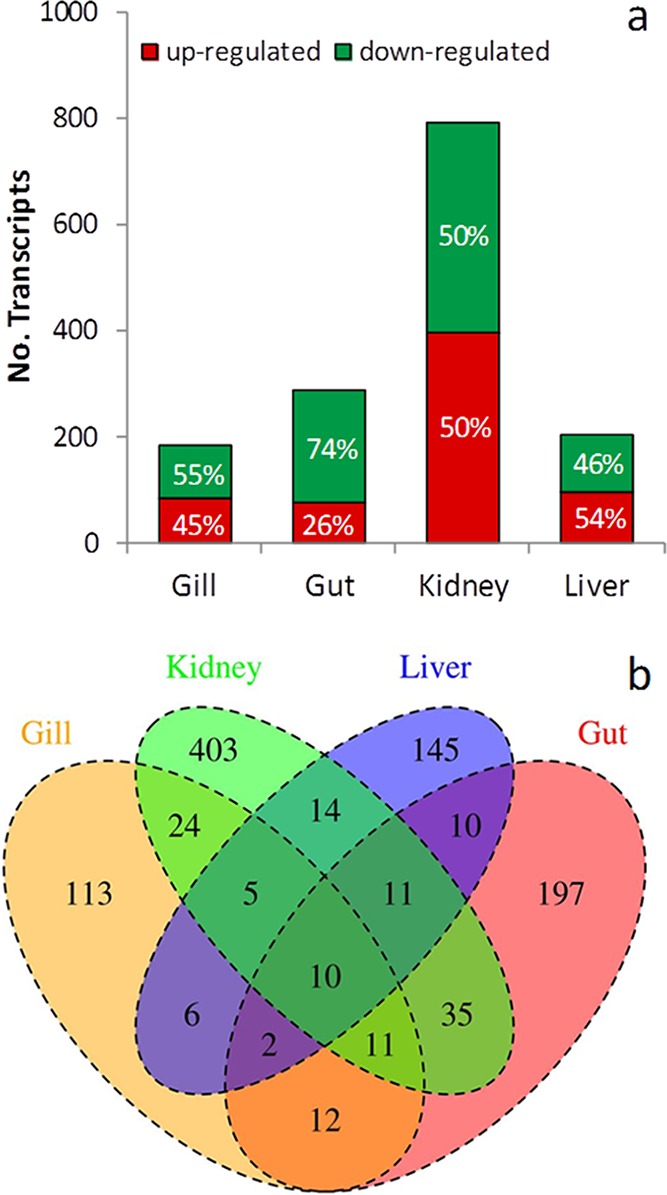
(A) Number of differentially expressed genes between populations obtained using EdgeR with a FDR <0.1 in each tissue. Numbers within bars represent the percentage of the total that were up/down-regulated in brown trout originating from the river Hayle compared to the river Teign. (B) Venn diagram displaying the number of differentially expressed genes in each tissue and the overlay between these gene lists across tissues.
Metal Homeostasis
Fundamentally, metal homeostasis consists of ensuring an adequate supply of essential metals for metabolic processes and controlling the level of essential and nonessential free metal ions to prevent toxicity. This involves regulating uptake from the environment, distribution through the bloodstream and delivery to target organs, supply to metabolic pathways, biotransformation, storage and excretion.28 Exposure to elevated metal concentrations in the River Hayle and the resulting increase in metal tissue accumulation, would therefore be expected to be associated with changes in the activity of components involved in this homeostatic system.
A number of cellular metal binding proteins serve to detoxify and store metal ions through binding and removal of their redox potential. Glutathione and metallothioneins (MTs) act as buffers for metal ions entering cells; both have very high affinity for most metals and glutathione is generally present at high concentrations. MTs are cysteine-rich, thiol-containing proteins and are widely acknowledged to account for a major portion of the cellular storage of zinc, copper, cadmium and to a lesser extent iron, lead and nickel in fish.28 MT-bound metals contribute to the metabolically detoxified cellular fraction and can also be stored in metal rich granules for even more stable, long-term storage.31 Increased MT synthesis has been extensively shown to occur in response to many metals, in both short-term laboratory exposures and in chronically exposed wild fish and it is the most consistent and sometimes only, mechanism of metal tolerance in fish [e.g., refs (14, 15 and 32−34)]. A single MT isoform is predominantly induced by metals in fish; free metal ions bind mtf1 transcription factors, which then bind metal response elements (MREs) in its promoter region, stimulating transcription.35 Corresponding with this, we found that one MT (metallothionein b) was among the most strongly up-regulated genes in the Hayle trout (significantly up-regulated by 8.2-, 7.7- and 5.6-fold in the gill, gut and liver, respectively, as well as 2.2 fold in the kidney), indicating sequestration of metals by MT represents a very important mechanism of metal tolerance in this population. Another MT isoform was also present in the brown trout transcriptome assembly but only expressed at very low levels.
Information available in the existing literature shows that acute metal exposures alter the expression of genes encoding metal-specific transporting proteins in the gill, gut, kidney and liver. These include the main cellular transporters of copper (copper transporter 1 (slc31a1), divalent metal transporter 1 (slc11a2), copper-transporting ATPases (atp7a, atp7b)); zinc (various members of the ZnT (or slc30) and ZIP (or slc39) families); and iron (slc11a2, ferroportin (slc40a1)).35−38 This suggests limiting uptake of metals from the environment, to slow onset of toxicity and increasing delivery to organs involved in metal metabolism, storage and excretion may be a mechanism of metal tolerance in short-term exposures. Less information is available on response to chronic exposure, but Xie and Klerks39 found a reduced rate of cadmium uptake in a tolerant laboratory population of killifish and Gale et al.40 showed a reduction in copper uptake rates in the gill contributed to copper tolerance of rainbow fish. In contrast, other studies have found no evidence of altered metal uptake and distribution kinetics during acclimation [e.g., ref (34)]. For Hayle trout, we hypothesized that gene pathways related to metabolism of copper and zinc were the most likely to be altered, given that concentrations of these metals were the most elevated in both river water and tissues. Of these genes, only the zinc transporter slc39a2 was differentially expressed (down-regulated in the kidney). There were no apparent trends in altered regulation of the other copper and zinc specific transporting proteins listed above. However, potential changes in copper and zinc transporters at the protein level, for example through post-transcriptional modification or changes in tissue/cellular localization, should not be ruled out. In contrast, there were increasing trends in expression levels of iron transporters in Hayle trout; slc11a2 was up-regulated 2.2 and 3.5 fold in the liver and kidney respectively, while slc40a1 was up-regulated 4.3 fold in the kidney, although these differences were not statistically significant. A number of genes encoding proteins involved in wider iron transport and storage were differentially expressed, particularly in the kidney, liver and gut. These include transferrin, a precursor of serotransferrin, as well as transferrin receptor 1b, which is responsible for cellular uptake of metals from transferrin and a form of ferritin, the main cellular iron-binding protein. Additionally, hemopexin, heme-binding protein 2 and heme transporter, which are involved in wider iron homeostasis though hemoglobin regulation, were differentially expressed. Moreover, the main regulator of iron homeostasis, the hormone hepcidin, was down-regulated in the liver. These changes in iron-metabolism related genes are particularly marked in contrast to the lack of change in those specific to copper and zinc. Furthermore, these changes are occurring in the absence of long-term significant elevation in the concentrations of Fe in the Hayle, compared to the Teign, river water (Supporting Information Table S1), but in the presence of significant accumulation of Fe in the liver, kidney and gill of Hayle fish (Figure 2). We hypothesize that these iron-homeostasis genes may be regulated by other metals present in the water, or that iron-homeostasis is a target of metal toxicity. An alternate hypothesis may be that a peak in Fe in the river water occurred close to the sample collection and was not recorded in the water sampling conducted by the Environment Agency. It is impossible to ascertain if this was the case and, therefore, we cannot conclusively interpret the reasons for the alterations of iron related pathways in the Hayle fish. Despite this, the striking association between the concentrations of Fe and several other metals (particularly As and Cd), together with the alteration in the expression of genes involved in iron-homeostasis, suggest that these iron-handling pathways play an important role in the response of the Hayle fish to metal exposure. This is supported by previous reports suggesting an association of other metals, including Cd, Cu and Pb, with binding and regulation of transcription of various components of the iron homeostatic system.41−44 Less is known about arsenic distribution pathways in fish, although arsenic is capable of being transported by transferrin in human plasma.45
Ion homeostasis
One major mechanism of toxicity common to a number of metals is disruption of ion homeostasis, particularly in the gills. Zn2+, Cd2+ and Pb2+ inhibit Ca2+ uptake, through competition for Ca2+ uptake pathways and direct inhibition of Ca2+ ATPase, leading to hypocalcaemia. Cu+ reduces Na+ uptake, both competitively and by interference with Na+/K+ ATPase. Copper, zinc, cadmium and lead also inhibit carbonic anhydrase, reducing supply of H+ and HCO3– ions for Na+ and Cl– uptake exchange. Lower plasma NaCl levels lead to increased blood viscosity and can cause circulatory collapse.46−49 Several laboratory studies have demonstrated acclimation of fish to metals, including copper, cadmium and lead, following chronic exposure, characterized by a restoration of plasma ionic balance and physiological condition. Increased synthesis of Na+/K+ ATPase to restore total cellular Na+/K+ ATPase activity, as well as morphological changes in the gill contribute to acclimation to copper.50−52
Although not significant, in the kidney our results show a trend of up-regulation for both of these ATPases known to be inhibited by metals, particularly for the most highly expressed (and therefore probably functionally most important) isoforms in this tissue. Na+/K+ ATPases atp1a and atp1b1a were both up-regulated by 2 fold, while Ca2+ ATPase atp2b1a was up-regulated by 3.3 fold. Additionally there was a significant up-regulation of carbonic anhydrase in the liver. This suggests that the up-regulation of these enzymes in the Hayle brown trout may be employed to counter their inhibition by metals. A number of other genes encoding proteins important in maintaining ion balance were differentially expressed. In the kidney Na+-Cl– cotransporter (slc12a3), which reabsorbs NaCl from urine, was significantly up-regulated by 4.8 fold. There was also a general trend of up-regulation in the kidney of a number of other transporters including those in the slc12 family, although a low-expressed isoform slc12a9 had significantly reduced expression, as well as those in the slc4 (sodium-bicarbonate transporter) and slc9 (Na+H+ exchanger) families. However, slc24a6, a Na+Ca2+K+ exchanger was significantly down-regulated in both the kidney and liver. Several other genes responsible for ion transport were down-regulated including chloride intracellular channel related proteins in the kidney and serum/glucocorticoid regulated kinase, which has a role in activating ion channels, in the liver. Three aquaporins, which contribute to maintenance of osmotic balance, were also differentially expressed in the kidney and gut. Additionally, a number of genes with a role in maintaining calcium homeostasis were differentially expressed, predominantly being down-regulated in the kidney, gut and liver. These include two calcium binding proteins and two s100-calcium binding proteins, calmodulin and calmodulin binding transcription activator, which are involved in calcium signaling and three isoforms of stanniocalcin, which regulates calcium flux. Additionally, two chemokine receptors which are related to calcium flux and signaling, together with calcium binding proteins with specific roles in muscle contraction (calponin, calsequestrin and caldesmon) were also down-regulated. Overall, these results indicate an integrated response of the ion-homeostatic system, which may contribute to the metal tolerance of this population through compensation of metal-induced ion balance disturbance. The most pronounced response appears to be in the kidney, reflecting the important role of this organ in regulating plasma ion and water balance. However, it is surprising that so few genes related to ion homeostasis were differentially expressed in the gill given that it is the main target of metal-disrupted ion balance.
Markers of Oxidative Stress and Cellular Damage
Metals induce cellular oxidative stress by several different mechanisms. Redox-active metal ions, including copper and iron, generate reactive oxygen species (ROS) via Fenton chemistry. Several metals, including copper, cadmium, arsenic, nickel and lead, can deplete and inhibit the cellular antioxidants. Copper and cadmium can also generate ROS via disruption of the electron transport chain.28,53 Oxidative stress can lead to lipid peroxidation, DNA and protein damage with associated adverse health effects at the cellular level. To counteract this, increased cellular ROS stimulate an up-regulation of the cellular antioxidant defense system, comprised of reduced glutathione (GSH) and a suite of enzymes, to limit oxidative damage. There is evidence from gene expression studies that various metals stimulate a response of the antioxidant system during short-term exposures [e.g., refs (11, 36 and 43)]. In chronically exposed wild fish populations there is some evidence of damage caused by oxidative stress, including lipid peroxidation in brown trout,15 but gene expression profiling has found inconsistent changes of the antioxidant system.1,14
Although gene expression profiling revealed that one isoform of the antioxidant superoxide dismutase was significantly down-regulated in all tissues, other, more highly expressed isoforms showed no change in expression level. No significant difference in expression was found for any other antioxidants, although in the gill, there was >2-fold apparent up-regulation of GSH-related antioxidants glutathione peroxidase (gpx1b) and glutathione-S-transferases (gsta5, gsto1, gstz1). There were no differences in expression level of glutathione synthetase or reductase, which are essential for GSH synthesis and restoration from its oxidized form, although a number of oxidoreductase enzymes, involved in supplying NADPH for restoring GSH, were significantly up-regulated. We found significant up-regulation of cytochrome C oxidases (cox4i2, cox7a) in the liver and kidney, which have been linked with oxidative stress generation and response, but are also associated with metal-induced metabolic changes (see below). 70 kD heat shock protein (hsp70), which temporarily binds and stabilizes damaged proteins was also significantly up-regulated in the gill and gut, while other heat shock proteins (hsp40, hsp27) were differentially expressed in the liver and gut, respectively. Overall, this suggests a modest response of the antioxidant system of Hayle trout, particularly in the gill. This is perhaps because after uptake from the environment and before being bound and/or transported elsewhere, the concentration of toxic free ions is likely to be higher in the gills compared to other tissues, leaving it more susceptible to oxidative stress. However, the response of the antioxidant system as a whole is much less pronounced than expected to occur following acute metal exposures, suggesting that other mechanisms of tolerance are likely to play a key role in reducing oxidative stress caused by metal ions in the Hayle brown trout population. These mechanisms may include increased synthesis of MTs, which have a high affinity for ROS and are therefore powerful cellular antioxidants and may offer significant protection against oxidative stress.46 This mechanism is likely to be a consistent and significant contributor to the metal tolerance of this brown trout population. Corresponding with this, we found no evidence of changes in pathways related to cellular repair mechanisms or evidence of cellular toxicity, such as changes in expression of apoptotic gene markers.
Metabolic Processes
A large proportion of the genes differentially expressed in the gut are related to digestion. This is particularly obvious from the GO term analysis; various peptidases dominate molecular function over-representation and proteolysis is the only over-represented biological process in this organ. Protein metabolism, including proteolysis specifically, has been shown to be altered in response to both short-term and chronic metal exposures. Furthermore, it has been suggested that this facilitates enhanced protein turnover which is important in the replacement of damaged proteins and proteins involved in metal detoxification, repair and storage.1−3,54 However, in the present study differentially expressed genes related to proteolysis were predominantly proteases, carboxypeptidases, chymotrypsins and elastases, which are more specifically associated with digestion and almost exclusively occurred in the gut. This strongly suggests that these differences are most likely related to diet. Dietary differences between the Hayle and Teign fish may be influenced by many environmental variables, although may not be entirely unrelated to metal contamination because metal exposure is well-known to alter food webs through changes in river species assemblage [e.g., ref (55)].
The mechanisms of metal tolerance employed by the Hayle trout are likely to result in increased energetic demand. Metal exposure has induced metabolic changes that have been suggested to be compensatory, in order to facilitate metal tolerance and detoxification. For example, chronically exposed wild yellow perch show some evidence of enhanced aerobic respiration, potentially increasing ATP production.1,56 Conversely, metal exposure has also been associated with down-regulation of various metabolic processes, due to the energetic demands of metal detoxification and/or through impairment of metabolic enzymes. In chronically exposed wild yellow perch there is evidence of lower aerobic capacity (3) and down-regulation of various components of lipid synthesis and transport, which have been associated with depletion of lipid reserves leading to adverse impacts on fish health and condition.2,5
In the Hayle trout, there was some evidence of changes in aerobic respiration, in particular down-regulation of genes with a role in oxidative phosphorylation including down-regulation in all tissues of several NADH dehydrogenases (mt-nd1, mt-nd1b, mt-nd4, mt-nd5) and up-regulation of cytochrome c oxidases (cox4i2, cox7a) in the liver and kidney respectively. With regard to other metabolic processes, in particular there were changes in expression of a number of genes involved in lipid, fatty acid and steroid synthesis and transport, mainly in the liver and kidney. These include peroxisome proliferator-activated receptor alpha, low-density lipoprotein receptor, lysophosphatidylglycerol acyltransferase, Cyp46, stearoyl-CoA desaturase and five apolipoproteins. In contrast to previous studies, these genes were almost exclusively up-regulated. A likely explanation for the up-regulation of lipid metabolism in the Hayle trout is that the sample population of five fish consisted of three reproductively mature females and two immature fish, while the Teign population sample was dominated by reproductively immature fish with no maturing females. Lipid metabolic pathways are essential for the synthesis of the egg yolk precursor protein vitellogenin and other egg shell proteins, in the liver of females as they undergo gonadal development and maturation. The energetic costs of reproductive maturation are also likely to influence other metabolic processes (i.e., energy generation through aerobic respiration and diversion from proteolysis), therefore the differences in metabolic processes observed can by no means be exclusively attributed to metal exposure. Consistent with this, genes associated with reproductive development in females were strongly up-regulated, including of genes encoding vitellogenin and egg outer membrane zona pellucida proteins in the liver of Hayle fish.
Immune system
Impairment of immune function has been extensively demonstrated in previous studies to be associated with exposure to metals. This effect has been attributed, in part, to disruption of energy budget as well as to a direct inhibition of immune function and commonly results in increased susceptibility to bacterial and viral infection.57−59 Gene expression analysis revealed a predominant down-regulation of a number of components of the complement system in the liver, gut and kidney of Hayle trout. These include complement components 1qa, 1qb, 1q2l, 3, 4 and 8; complement factors H, H1 and D and Fanconi anemia, complementation group C. This suggests an impairment of the immune system and is consistent with the findings of Pierron et al.2 and Reynders et al.60 who found inhibited expression of genes in the immune system in fish exposed to metals, especially those involved in the complement system.
We have sequenced, assembled and annotated a transcriptome for the brown trout, providing a useful resource for further research in this species. Using this information, we investigated the molecular mechanisms of tolerance to metals in a brown trout population exposed to significantly elevated concentrations of multiple metals in the River Hayle, U.K. and anchored our data to metal accumulation in tissues and river water concentrations. Tissue metal accumulation patterns indicate the gill represents the major route of metal uptake in Hayle fish, while the accumulation in the kidney and liver reflects their important role in metal storage and detoxification. The considerable tissue metal accumulation observed in the absence of overt toxicity confirms the metal tolerance of Hayle brown trout. Global gene expression profiling revealed that the two broad strategies likely to contribute to the metal tolerance of this population are the regulation of metal homeostasis pathways and ion homeostasis pathways, with up-regulation of the antioxidant system playing a relatively minor role. Within these, several mechanisms appear of particular importance. These include increased synthesis of metallothionein, the prominence of the kidney in regulating ion balance and the putative role of iron-handling pathways in wider metal homeostasis. Although our data set highlighted some potential mechanisms of metal toxicity, particularly inhibition of the immune system, there is little to suggest that the brown trout inhabiting the River Hayle are incurring adverse health effects as a result of the presence of toxic concentrations of metals in their environment. This contrasts with more extensive evidence of metal toxicity from studies on yellow perch, potentially causing adverse impact at both the individual and population levels. A possible explanation for this is that metal contamination in the River Hayle has been present for a greater length of time, perhaps leading to a greater degree of metal tolerance in this population. Whether this is a result of an inherent genetic plasticity, allowing individual acclimation, or local population adaptation, leading to inherited metal-tolerance, is unclear and would require further research to be elucidated.
Acknowledgments
We thank the Environment Agency for facilitating the fish sampling and for provision of historical chemistry data for the rivers studied in this project. We also thank J. Shears and P. Shears for support with the fish sampling, P. Kille for encouraging us to conduct this project, A. Moorhouse, C. Sambles and K. Paszkiewicz for facilitating the sequencing experiments and K. Uren for providing the image of the River Hayle. We acknowledge funding from the Salmon & Trout Association and from the Natural Environment Research Council, grant number NE/1528326/1.
Supporting Information Available
Supplemental experimental section, metal concentrations in the river Hayle and river Teign, description of cDNA libraries sequenced, fold changes between the Hayle and Tein fish of potential control genes for RT-QPCR analysis, target genes, primers and assay details for RT-QPCR analysis, custer diagrams of tissue metal concentration, summary statistics of sequence read processing and QC filtering, transcript abundance level in each tissue, results of RT-QPCR analysis, heatmaps of expression values of differentially expressed genes and gene ontology terms over-represented in the lists of differentially expressed genes between Hayle and Teign fish for each tissue. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Supplementary Material
References
- Pierron F.; Bourret V.; St-Cyr J.; Campbell P. G. C.; Bernatchez L.; Couture P. Transcriptional responses to environmental metal exposure in wild yellow perch (Perca flavescens) collected in lakes with differing environmental metal concentrations (Cd, Cu, Ni). Ecotoxicology 2009, 185620–631. [DOI] [PubMed] [Google Scholar]
- Pierron F.; Normandeau E.; Defo M.; Campbell P.; Bernatchez L.; Couture P. Effects of chronic metal exposure on wild fish populations revealed by high-throughput cDNA sequencing. Ecotoxicology 2011, 2061388–1399. [DOI] [PubMed] [Google Scholar]
- Couture P.; Rajender Kumar P. Impairment of metabolic capacities in copper and cadmium contaminated wild yellow perch (Perca flavescens). Aquat. Toxicol. 2003, 641107–120. [DOI] [PubMed] [Google Scholar]
- Rajotte J. W.; Couture P. Effects of environmental metal contamination on the condition, swimming performance and tissue metabolic capacities of wild yellow perch (Perca flavescens). Can. J. Fish. Aquat. Sci. 2002, 5981296–1304. [Google Scholar]
- Levesque H. M.; Moon T. W.; Campbell P. G. C.; Hontela A. Seasonal variation in carbohydrate and lipid metabolism of yellow perch (Perca flavescens) chronically exposed to metals in the field. Aquat. Toxicol. 2002, 60, 257–267. [DOI] [PubMed] [Google Scholar]
- Levesque H. M.; Dorval J.; Hontela A.; Van Der Kraak G. J.; Campbell P. G. C. Hormonal, morphological and physiological responses of yellow perch (Perca flavescens) to chronic environmental metal exposures. J. Toxicol. Environ. Health, Part A 2003, 667657–676. [DOI] [PubMed] [Google Scholar]
- Giguare A.; Campbell P. G. C.; Hare L.; Couture P. Sub-cellular partitioning of cadmium, copper, nickel and zinc in indigenous yellow perch (Perca flavescens) sampled along a polymetallic gradient. Aquat. Toxicol. 2006, 772178–189. [DOI] [PubMed] [Google Scholar]
- Bourret V.; Couture P.; Campbell P. G. C.; Bernatchez L. Evolutionary ecotoxicology of wild yellow perch (Perca flavescens) populations chronically exposed to a polymetallic gradient. Aquat. Toxicol. 2008, 86176–90. [DOI] [PubMed] [Google Scholar]
- Brown B. E. Effects of mine drainage on the river hayle, cornwall a) factors affecting concentrations of copper, zinc and iron in water, sediments and dominant invertebrate fauna. Hydrobiologia 1977, 522221–233. [Google Scholar]
- Durrant C. J.; Stevens J. R.; Hogstrand C.; Bury N. R. The effect of metal pollution on the population genetic structure of brown trout (Salmo trutta L.) residing in the River Hayle, Cornwall, UK. Environ. Pollut. 2011, 159123595–3603. [DOI] [PubMed] [Google Scholar]
- Hansen B. H.; Romma S.; Softeland L. I. R.; Olsvik P. A.; Andersen R. A. Induction and activity of oxidative stress-related proteins during waterborne Cu-exposure in brown trout (Salmo trutta). Chemosphere 2006, 65101707–1714. [DOI] [PubMed] [Google Scholar]
- Beaumont M.; Butler P.; Taylor E. Exposure of brown trout Salmo trutta to a sublethal concentration of copper in soft acidic water: effects upon gas exchange and ammonia accumulation. J. Exp. Biol. 2003, 2061153–162. [DOI] [PubMed] [Google Scholar]
- Everall N.; Macfarlane N.; Sedgwick R. The interactions of water hardness and pH with the acute toxicity of zinc to the brown trout, Salmo trutta L. J. Fish Biol. 2006, 35127–36. [Google Scholar]
- Hansen B. H.; Romma S.; Garmo Ã. A.; Olsvik P. A.; Andersen R. A. Antioxidative stress proteins and their gene expression in brown trout (Salmo trutta) from three rivers with different heavy metal levels. Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol. 2006, 1433263–274. [DOI] [PubMed] [Google Scholar]
- Farag A. d. M.; Stansbury M. A.; Bergman H. L.; Hogstrand C.; MacConnell E. The physiological impairment of free-ranging brown trout exposed to metals in the Clark Fork River, Montana. Can. J. Fish. Aquat. Sci. 1995, 5292038–2050. [Google Scholar]
- Kolde R.Pretty Heatmaps, R package version 0.7.4, 2012.
- Zerbino D. R.; Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 185821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz M. H.; Zerbino D. R.; Vingron M.; Birney E. Oases: Robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 2012, 2881086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.; Dewey C. N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011, 28, 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M. G.; Haas B. J.; Yassour M.; Levin J. Z.; Thompson D. A.; Amit I.; Adiconis X.; Fan L.; Raychowdhury R.; Zeng Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 297644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D.; McCarthy D. J.; Smyth G. K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 261139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.; Boutros P. C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinf. 2011, 24, 801–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field D.; Tiwari B.; Booth T.; Houten S.; Swan D.; Bertrand N.; Thurston M. Open software for biologists: from famine to feast. Nat. Biotechnol. 2006, 247801–804. [DOI] [PubMed] [Google Scholar]
- Huang D. W.; Sherman B. T.; Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008, 4144–57. [DOI] [PubMed] [Google Scholar]
- Uren-Webster T. M.; Lewis C.; Filby A. L.; Paull G. C.; Santos E. M. Mechanisms of toxicity of di(2-ethylhexyl) phthalate on the reproductive health of male zebrafish. Aquat. Toxicol. 2010, 99, 360–369. [DOI] [PubMed] [Google Scholar]
- Filby A. L.; Tyler C. R. Molecular characterization of estrogen receptors 1, 2a and 2b and their tissue and ontogenic expression profiles in fathead minnow (Pimephales promelas). Biol. Reprod. 2005, 734648. [DOI] [PubMed] [Google Scholar]
- Bury N. R.; Walker P. A.; Glover C. N. Nutritive metal uptake in teleost fish. J. Exp. Biol. 2003, 206111–23. [DOI] [PubMed] [Google Scholar]
- Wood C. M., An introduction to metals in fish physiology and toxicology: Basic principles. In Homeostasis and Toxicology of Essential Metals; Wood C. M., Farrel A. P., Brauner C. J., Eds.; Academic Press: London, 2012; pp 1–53. [Google Scholar]
- Olsvik P. l. A.; Gundersen P. l.; Andersen R. A.; Zachariassen K. E. Metal accumulation and metallothionein in brown trout, Salmo trutta, from two Norwegian rivers differently contaminated with Cd, Cu and Zn. Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol. 2001, 1282189–201. [DOI] [PubMed] [Google Scholar]
- Eastwood S.; Couture P. Seasonal variations in condition and liver metal concentrations of yellow perch (Perca flavescens) from a metal-contaminated environment. Aquat. Toxicol. 2002, 58, 43–56. [DOI] [PubMed] [Google Scholar]
- Goto D.; Wallace W. G. Metal intracellular partitioning as a detoxification mechanism for mummichogs (Fundulus heteroclitus) living in metal-polluted salt marshes. Mar. Environ. Res. 2010, 693163–171. [DOI] [PubMed] [Google Scholar]
- Chowdhury M. J.; Baldisserotto B.; Wood C. M. Tissue-specific cadmium and metallothionein levels in rainbow trout chronically acclimated to waterborne or dietary cadmium. Arch. Environ. Contam. Toxicol. 2005, 483381–390. [DOI] [PubMed] [Google Scholar]
- Hogstrand C.; Haux C. Binding and detoxification of heavy metals in lower vertebrates with reference to metallothionein. Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol. 1991, 10012137–141. [DOI] [PubMed] [Google Scholar]
- Dang F.; Zhong H.; Wang W.-X. Copper uptake kinetics and regulation in a marine fish after waterborne copper acclimation. Aquat. Toxicol. 2009, 943238–244. [DOI] [PubMed] [Google Scholar]
- Craig P. M.; Galus M.; Wood C. M.; McClelland G. B. Dietary iron alters waterborne copper-induced gene expression in soft water acclimated zebrafish (Danio rerio). Am. J. Physiol.: Regul., Integr. Comp. Physiol. 2009, 2962362–373. [DOI] [PubMed] [Google Scholar]
- Minghetti M.; Leaver M. J.; Carpena E.; George S. G. Copper transporter 1, metallothionein and glutathione reductase genes are differentially expressed in tissues of sea bream (Sparus aurata) after exposure to dietary or waterborne copper. Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol. 2008, 1474450–459. [DOI] [PubMed] [Google Scholar]
- Minghetti M.; Leaver M. J.; George S. G. Multiple Cu-ATPase genes are differentially expressed and transcriptionally regulated by Cu exposure in sea bream, Sparus aurata. Aquat. Toxicol. 2011, 97123–33. [DOI] [PubMed] [Google Scholar]
- Zheng D.; Feeney G. P.; Kille P.; Hogstrand C. Regulation of ZIP and ZnT zinc transporters in zebrafish gill: zinc repression of ZIP10 transcription by an intronic MRE cluster. Physiol. Genom. 2008, 342205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L.; Klerks P. L. Changes in cadmium accumulation as a mechanism for cadmium resistance in the least killifish Heterandria formosa. Aquat. Toxicol. 2004, 66173–81. [DOI] [PubMed] [Google Scholar]
- Gale S. A.; Smith S. V.; Lim R. P.; Jeffree R. A.; Petocz P. Insights into the mechanisms of copper tolerance of a population of black-banded rainbowfish (Melanotaenia nigrans) (Richardson) exposed to mine leachate, using 64/67Cu. Aquat. Toxicol. 2003, 622135–153. [DOI] [PubMed] [Google Scholar]
- Craig P. M.; Hogstrand C.; Wood C. M.; McClelland G. B. Gene expression endpoints following chronic waterborne copper exposure in a genomic model organism, the zebrafish, Danio rerio. Physiol. Genom. 2009, 40123–33. [DOI] [PubMed] [Google Scholar]
- Kwong R. W. M.; Andres J. A.; Niyogi S. Molecular evidence and physiological characterization of iron absorption in isolated enterocytes of rainbow trout (Oncorhynchus mykiss): Implications for dietary cadmium and lead absorption. Aquat. Toxicol. 2010, 993343–350. [DOI] [PubMed] [Google Scholar]
- Kwong R. W. M.; Andres J. A.; Niyogi S. Effects of dietary cadmium exposure on tissue-specific cadmium accumulation, iron status and expression of iron-handling and stress-inducible genes in rainbow trout: Influence of elevated dietary iron. Aquat. Toxicol. 2011, 102121–9. [DOI] [PubMed] [Google Scholar]
- Hu Y.-h.; Zheng W.-j.; Sun L. Identification and molecular analysis of a ferritin subunit from red drum (Sciaenops ocellatus). Fish Shellfish Immunol. 2010, 284678–686. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Cornelis R.; De Kimpe J.; Mees L.; Lameire N. Study of arsenicprotein binding in serum of patients on continuous ambulatory peritoneal dialysis. Clin. Chem. 1998, 441141–147. [PubMed] [Google Scholar]
- Grosell M., Copper. In Homeostasis and Toxicology of Essential Metals; Wood C. M., Farrel A. P., Brauner C. J., Eds.; Academic Press: London, 2012; pp 54–135. [Google Scholar]
- Hogstrand C., Zinc. In Homeostasis and Toxicology of Essential Metals; Wood C. M., Farrel A. P., Brauner C. J., Eds.; Academic Press: London, 2012; pp 136–201. [Google Scholar]
- Mager A. M., Lead. In Homeostasis and Toxicology of Non-Essential Metals; Wood C. M., Farrel A. P., Brauner C. J., Eds.; Academic Press: London, 2012; pp 186–237. [Google Scholar]
- McGeer J. C.; Niyogi S.; Smith D. S., Cadmium. In Homeostasis and Toxicology of Non-Essential Metals; Wood C. M., Farrel A. P., Brauner C. J., Eds.; Academic Press: London, 2012; pp 126–185. [Google Scholar]
- Lauren D. J.; McDonald D. G. Acclimation to Copper by Rainbow Trout, Salmo gairdneri: Biochemistry. Can. J. Fish. Aquat. Sci. 1987, 441105–111. [Google Scholar]
- Lauren D. J.; McDonald D. G. Acclimation to Copper by Rainbow Trout, Salmo gairdneri: Physiology. Can. J. Fish. Aquat. Sci. 1987, 44199–104. [Google Scholar]
- Pelgrom S. M. G. J.; Lock R. A. C.; Balm P. H. M.; Bonga S. E. W. Integrated physiological response of tilapia, Oreochromis mossambicus, to sublethal copper exposure. Aquat. Toxicol. 1995, 324303–320. [Google Scholar]
- Valko M.; Morris H.; Cronin M. T. D. Metals, toxicity and oxidative stress. Curr. Med. Chem. 2005, 12101161–1208. [DOI] [PubMed] [Google Scholar]
- De Smet H.; Blust R. Stress responses and changes in protein metabolism in carp Cyprinus carpio during cadmium exposure. Ecotoxicol. Environ. Saf. 2001, 483255–262. [DOI] [PubMed] [Google Scholar]
- Cain D. J.; Luoma S. N.; Wallace W. G. Linking metal bioaccumulation of aquatic insects to their distribution patterns in a mining-impacted river. Environ. Toxicol. Chem. 2004, 2361463–1473. [DOI] [PubMed] [Google Scholar]
- Audet D.; Couture P. Seasonal variations in tissue metabolic capacities of yellow perch (Perca flavescens) from clean and metal-contaminated environments. Can. J. Fish. Aquat. Sci. 2003, 603269–278. [Google Scholar]
- Zelikoff J. T.; Bowser D.; Squibb K. S.; Frenkel K. Immunotoxicity of low level cadmium exposure in fish: An alternative animal model for immunotoxicological studies. J. Toxicol. Environ. Health 1995, 453235–248. [DOI] [PubMed] [Google Scholar]
- Nayak A. S.; Lage C. R.; Kim C. H. Effects of low concentrations of arsenic on the innate immune system of the zebrafish (Danio rerio). Toxicol. Sci. 2007, 981118–124. [DOI] [PubMed] [Google Scholar]
- Esteve C.; Alcaide E.; Urena R. The effect of metals on condition and pathologies of European eel (Anguilla anguilla): In situ and laboratory experiments. Aquat. Toxicol. 2012, 1090176–184. [DOI] [PubMed] [Google Scholar]
- Reynders H.; van der Ven K.; Moens L. N.; van Remortel P.; De Coen W. M.; Blust R. Patterns of gene expression in carp liver after exposure to a mixture of waterborne and dietary cadmium using a custom-made microarray. Aquat. Toxicol. 2006, 802180–193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


