Abstract
Recruitment of cofactors to specific DNA sites is integral for specificity in gene regulation. As a model system, we examined how targeting and transcriptional control of the sulfur metabolism genes in Saccharomyces cerevisiae is governed by recruitment of the transcriptional co-activator Met4. We developed genome-scale approaches to measure transcription factor (TF) DNA-binding affinities and cofactor recruitment to >1300 genomic binding site sequences. We report that genes responding to the TF Cbf1 and cofactor Met28 contain a novel ‘recruitment motif’ (RYAAT), adjacent to Cbf1 binding sites, which enhances the binding of a Met4–Met28–Cbf1 regulatory complex, and that abrogation of this motif significantly reduces gene induction under low-sulfur conditions. Furthermore, we show that correct recognition of this composite motif requires both non-DNA-binding cofactors Met4 and Met28. Finally, we demonstrate that the presence of an RYAAT motif next to a Cbf1 site, rather than Cbf1 binding affinity, specifies Cbf1-dependent sulfur metabolism genes. Our results highlight the need to examine TF/cofactor complexes, as novel specificity can result from cofactors that lack intrinsic DNA-binding specificity.
Keywords: cofactor, DNA-binding affinities, DNA binding site, sulfur metabolism, transcription factor
Introduction
Individual transcription factors (TFs) typically bind to a relatively broad set of DNA binding site sequences (Badis et al, 2009), yet must coordinate the exquisitely specific gene expression responses fundamental to cellular function. Therefore, a variety of mechanisms exist to differentiate the binding of a TF at different genomic loci, such as TF binding site affinity (Jiang and Levine, 1993; Gaudet and Mango, 2002; Rowan et al, 2010), TF binding site clustering (Berman et al, 2002; Frith et al, 2002; Markstein et al, 2002; Pramila et al, 2002; Giorgetti et al, 2010), cooperative interactions between TFs (Stein and Baldwin, 1993; Joshi et al, 2007; Mann et al, 2009), and synergistic recruitment of cofactors by TFs (Carey, 1998; Merika and Thanos, 2001). However, despite the known functions of many TFs in recruiting non-DNA-binding transcriptional cofactors to target sites in the genome (Dilworth and Chambon, 2001; Struhl, 2005), the sequence dependence of cofactor recruitment has remained largely unexplored.
To address this issue, we examined the roles of TF binding site affinity and differential cofactor recruitment in regulating a set of target genes. As a model system, we selected the Met4-dependent genes that control sulfur metabolism in the yeast S. cerevisiae as both the recruited cofactors (Met4 and Met28) and the sequence-specific DNA-binding TFs (Cbf1, Met31, and Met32) had been characterized. Met4 is the sole transcriptional activator of the sulfur metabolism genes but exhibits no intrinsic DNA-binding activity (Lee et al, 2010). To promote transcription, Met4 is recruited to target gene promoters by the TFs Cbf1, Met31, or Met32 (Kuras et al, 1997; Blaiseau and Thomas, 1998). Cbf1 is a basic helix-loop-helix (bHLH)-containing TF that binds as a homodimer to a palindromic E-box site with a consensus CACGTG core, while Met31 and Met32 are paralogous C2H2 zinc finger-containing TFs that bind to sites with a TGTGGC core (Kuras et al, 1996, 1997; Blaiseau et al, 1997; Blaiseau and Thomas, 1998; Badis et al, 2008; Zhu et al, 2009). An additional transcriptional cofactor, Met28, has been shown to bind with Met4 to these TFs in DNA-bound, multi-protein complexes (Blaiseau et al, 1997; Kuras et al, 1997; Blaiseau and Thomas, 1998). Like Met4, Met28 does not exhibit intrinsic DNA-binding activity, but binding of Met28 has been shown to stabilize DNA-bound Met4–Met28–Cbf1 complexes (Kuras et al, 1997).
In a recent comprehensive analysis of the Met4 transcriptional system, examining gene expression and TF promoter occupancy in multiple yeast strains deficient for key regulators of sulfur metabolism genes, Lee et al (2010) described a set of 45 sulfur metabolism genes that are induced under two different Met4-related conditions: Met4 hyperactivation and sulfur limitation. This gene set, referred to as the Met4 core regulon, comprises a comprehensive set of genes regulated by the cofactor Met4 under both of these conditions. It was further demonstrated that induction of every Met4 core regulon gene is abrogated in both a met4Δ strain and met31Δmet32Δ double knockout strain, while induction is affected for only a subset of genes in cbf1Δ or met28Δ strains. Based on their comprehensive gene expression analysis, the Met4 regulon was subdivided into three classes: genes whose transcription is strictly dependent on Cbf1 and Met28 in both conditions (Class 1); genes with intermediate dependency on Cbf1 and Met28 (Class 2); genes whose expression is independent of Cbf1 and Met28 (Class 3) (Lee et al, 2010).
Here, we have examined the contributions of TF binding site affinity and cofactor recruitment to the cis-regulatory logic governing the expression of the Met4 core regulon genes. We developed genome-scale approaches to measure both protein-DNA binding affinities (Kds) and sequence specificity in Met4 recruitment using the protein-binding microarray (PBM) technology (Bulyk et al, 2001; Mukherjee et al, 2004; Berger et al, 2006b). Our results suggest that two different modes of Met4 recruitment are used to target the Met4 regulon genes: (1) recruitment of Met4 by Met31 or Met32 to high-affinity Met31/Met32 DNA binding sites specifies the Class 2 and 3 subsets of the regulon genes; (2) recruitment of Met4 by Cbf1 and Met28 to variant Met4 ‘recruitment sites’ specifies the Class 1, Cbf1-dependent subset of the Met4 regulon genes.
Examining the site-specific recruitment of Met4 by Cbf1 and Met28, we identified a strict requirement for a composite DNA binding site composed of the Cbf1 E-box sequence (CACGTG) flanked by a newly discovered Met4 ‘recruitment motif’ (RYAAT), separated by a 2-bp spacer. Reporter assays confirmed the importance of this recruitment motif in vivo; mutation of this RYAAT motif significantly reduces induction of Cbf1-dependent (Class 1) regulon genes in low-sulfur conditions. The identification of this motif was unexpected as Cbf1 binding is not affected by the presence of the recruitment motif, and neither Met4 nor Met28 exhibit any specific DNA binding either individually or together. Instead, selective binding to the composite DNA binding site occurs only with the full trimeric complex. Therefore, the non-DNA-binding cofactors Met4 and Met28 operate synergistically to direct their own recruitment to specific DNA sites, and thereby discriminate between Cbf1 bound at different sites. These results reveal an under-appreciated and powerful mechanism for enhancing DNA sequence specificity in transcriptional cofactor recruitment that is distinct from traditional allosteric mechanisms. Our work highlights the need to examine the DNA binding of cofactor/TF complexes, since novel specificity can arise even when cofactors do not bind DNA on their own. Furthermore, we demonstrate how the PBM technology can be used to examine these phenomena at genome scale.
Results
Determining protein-DNA binding affinities (Kds) using PBMs
To perform a comprehensive, genome-scale biophysical characterization of the roles exhibited by Cbf1, Met31, and Met32 in regulation of the Met4 core regulon, we sought an accurate characterization of the binding affinities (Kds) of these TFs to all predicted DNA binding sites in the S. cerevisiae genome (a description of how these sites were identified is provided below in the section ‘Genome-wide characterization of Cbf1 and Met32 DNA-binding affinities’). Furthermore, to account for any potential dependence on the sequences flanking the individual DNA binding sites, we chose to measure these TFs' binding affinities to each DNA binding site within the context of its native genomic flanking sequence. The number of such unique binding sites (thousands) precluded the use of conventional approaches for determining affinities (e.g., electromobility shift assay or surface plasmon resonance (SPR)). Therefore, we utilized the PBM technology (Bulyk et al, 2001; Mukherjee et al, 2004; Berger and Bulyk, 2006a) to determine protein-DNA binding affinities in a high-throughput manner.
PBMs are an in-vitro, double-stranded DNA (dsDNA) microarray technology that allows the simultaneous characterization of a protein's DNA-binding preference to tens of thousands of unique DNA sequences in a single experiment (Bulyk et al, 2001; Mukherjee et al, 2004; Berger et al, 2006b). PBM fluorescence signal intensities and derived scores for individual DNA binding site sequences have been shown to correlate with prior protein-DNA binding affinity measurements (i.e., Kd values; Bulyk et al, 2001; Berger and Bulyk, 2006a; Badis et al, 2009). To account for the protein concentration dependence of binding to DNA, we performed PBM experiments using purified Cbf1 or Met32 at eight different protein concentrations, ranging from ∼10 nM to 30 μM (Supplementary Table S1; Supplementary Figure S1), and we fit saturation binding curves to the eight fluorescence measurements for each probe on the microarray (Figure 1A; Materials and methods). This follows an approach used successfully by Jones et al (2006) to measure the affinities of phosphopeptides binding to protein domains immobilized on a protein microarray. We identified Cbf1 and Met32 binding sites in the S. cerevisiae genome using previously published universal PBM data (Zhu et al, 2009) (see details below), and we incorporated those binding sites into DNA probe sequences on custom arrays that we designed for this study. This customized Cbf1/Met32 PBM design allowed us to better control for the effects of binding site sequence context by putting the Cbf1 and Met32 binding sites at a constant position relative to the surface of the glass slide and within constant flanking sequences.
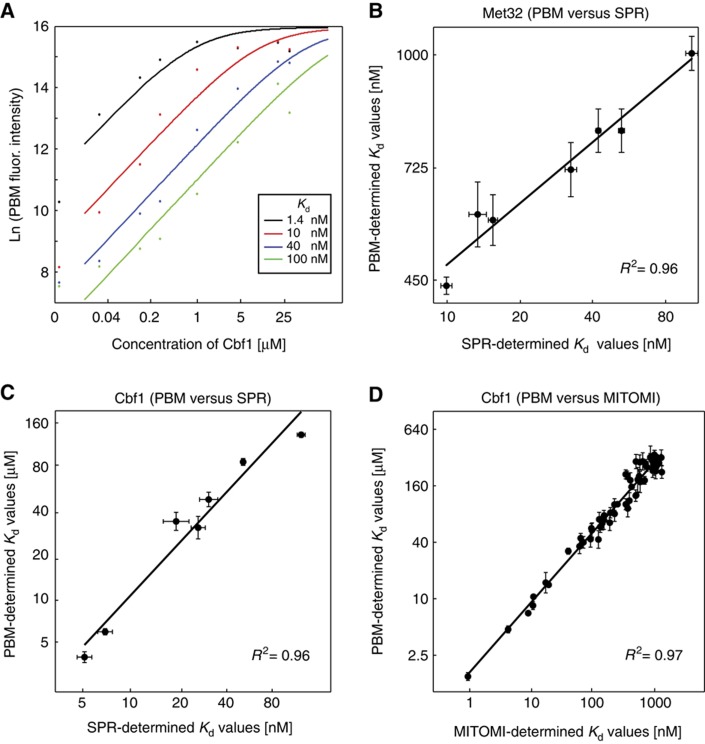
Figure 1.
PBM-determined protein-DNA binding affinities (Kd's) and comparison with SPR- and MITOMI-determined values. (A) Fitted saturation binding curves to PBM fluorescence values at eight concentrations of applied Cbf1 protein are shown for four representative PBM probes (i.e., sequences) with varying SPR-PBM-determined affinities. (B, C) Comparison of PBM- and SPR-determined Kd values for Met32 and Cbf1, respectively. Error bars indicate the standard deviation calculated over replicate measurements (n=4 for PBM, n=2 for SPR). (D) Comparison of PBM- and MITOMI-determined (Maerkl and Quake, 2007) Kd values for Cbf1 to 64 binding site variants of the form GTCACNNN. Plots are shown on a log-log scale. See Supplementary information for linear regression parameters.
To assess the accuracy of the PBM-determined affinity measurements, we measured equilibrium binding affinities for a subset of the PBM probe sequences by SPR (Materials and methods) and compared them with the PBM data. We observed excellent linear agreement between the natural log values of our PBM-determined Kds (i.e., the binding energies) and the SPR-determined Kds (R2=0.96) over an affinity range of 10-fold (Met32; Figure 1B) to 20-fold (Cbf1; Figure 1C). Our PBM-determined values are also in excellent agreement (R2=0.97) with data obtained using a high-throughput microfluidic approach (MITOMI) for Cbf1 binding to 64 variant sites (Maerkl and Quake, 2007) over an ∼300-fold range in Kd (Figure 1D). Despite the strong linear correlation with independent measurements, the absolute Kds derived solely from the PBM data are consistently higher (i.e., weaker affinity) than Kds determined by SPR or MITOMI (Figure 1; see Supplementary information for extended discussion). Therefore, we implemented a hybrid strategy whereby a linear transformation is applied to the PBM-determined energies based on a set of SPR measurements. We assessed the accuracy of this approach using a standard cross-validation analysis where the linear transformation of the PBM data is performed using n−1 of the SPR measurements and the accuracy is assessed on the remaining measurement. Using the ratio of the SPR affinity to the transformed PBM affinity as an indicator of accuracy, we observed mean values of 1.05 (±0.24) for Met32 and 1.08 (±0.34) for Cbf1. Thus, the majority of the transformed PBM affinity measurements (i.e., Kd values) are within ∼30% of the SPR-determined absolute Kd values. Therefore, the hybrid SPR-PBM approach provides a practical approach to accurately measure the absolute binding affinity (Kd) of a protein (or protein complex) to thousands of unique DNA sites simultaneously.
Genome-wide characterization of Cbf1 and Met32 DNA-binding affinities
To characterize the DNA-binding affinity landscape of Cbf1 and Met32 across the yeast genome, we used the hybrid SPR-PBM approach to measure the in-vitro DNA-binding affinities (absolute Kds) of Cbf1 and Met32 to predicted DNA binding sites (673 and 685, respectively), identified in ∼4900 intergenic regions of the S. cerevisiae genome (Materials and methods). This set of intergenic regions contains the upstream and downstream intergenic regions surrounding the 45 Met4 regulon genes described in Lee et al (2010), and all intergenic regions identified as ‘bound’ (P<0.005) by any of 203 S. cerevisiae TFs examined in a chromatin immunoprecipitation (ChIP) survey of in-vivo TF binding by Harbison et al (2004). We reasoned that the ChIP-‘bound’ regions from this large data set represented a reasonable estimate of these TFs' potential gene regulatory regions in the genome. We measured the binding affinities for Cbf1 and Met32, separately, to all 1358 DNA binding sites in the context of their native genomic flanking sequences (Figure 2A and B; Materials and methods).
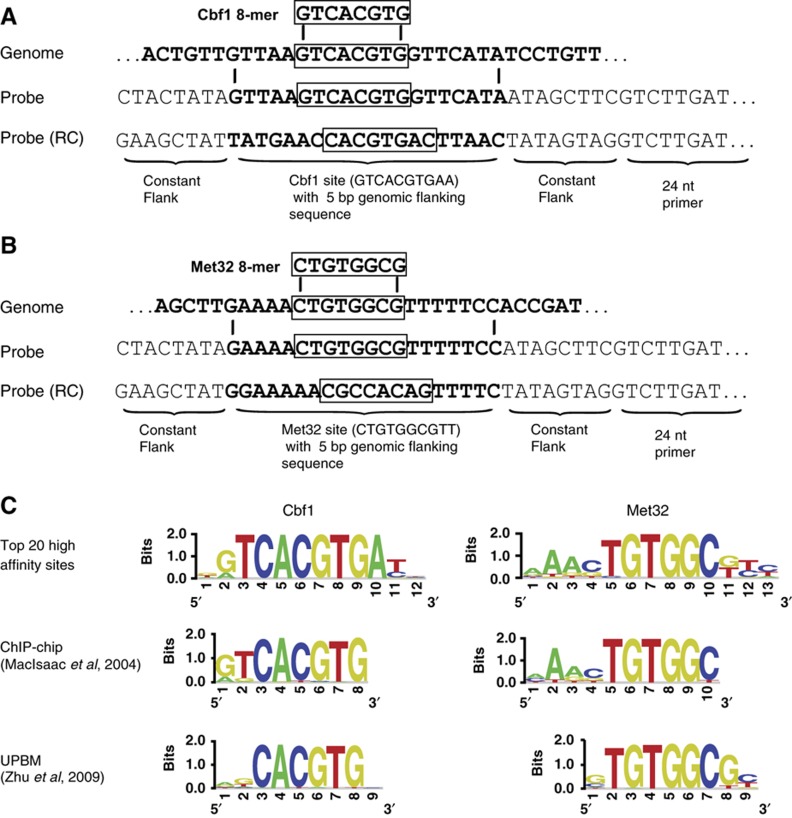
Figure 2.
Genome-wide binding affinity analysis: PBM probe design and sequence specificity of high-affinity sites. (A, B) Schematic illustrating the design of PBM probe sequences from binding site sequences identified in the yeast genome. Boxed are high-scoring 8-mers described for Cbf1 or Met32 (Zhu et al, 2009), 8-mers for each factor are put in a common register based on binding motifs from Zhu et al and the 20-bp genomic sequence is incorporated into the probe within constant flanking sequence (see Materials and methods for details). All 20 bp binding sites are present on the PBM in duplicate and in their reverse complement (RC) orientation (four probes in total). (C) Logos for Cbf1 and Met32 constructed by aligning the top 20 highest affinity sites (top); determined by MacIsaac et al (2006) by analyzing ChIP-chip data for each factor (middle); and determined by Zhu et al (2009) using a universal PBM approach (bottom). Palindromic Cbf1 sites were randomly oriented in constructing the logo, Met32 sites were oriented with respect to the motif from Zhu et al.
Our data are in excellent agreement with previously published data for both Cbf1 and Met32. DNA binding site motifs constructed from the top 20 highest affinity sites agree well with both ChIP-chip-derived (Harbison et al, 2004; MacIsaac et al, 2006) and other PBM-determined (Berger et al, 2006b; Badis et al, 2009; Zhu et al, 2009) motifs (Figure 2C). For Cbf1, consistent with prior MITOMI data (Maerkl and Quake, 2007), we also identified many high-affinity sequences that deviated from the consensus sequence (G/A)TCACGTG. For example, many sequences with variant E-box sequences (CACATG, not consensus G), or variant flanking bases (GCACGTG, not consensus T) had Kd values within five-fold of the highest affinity site.
For Met32, the in-vitro binding data suggested a longer binding site than the TGTGGCG core previously defined by universal PBM experiments (Badis et al, 2008; Zhu et al, 2009; Figure 2C). The A-rich sequence preference observed 5′ to the core agrees well with the ChIP-chip-derived motif (Harbison et al, 2004; MacIsaac et al, 2006; Figure 2C), demonstrating that the ChIP-identified sequence preferences are in fact consistent with affinity differences in Met32 monomer binding. These results also demonstrate that the previously described AAACTGTGGC consensus (Lee et al, 2010), which had been motivated by identification of AAACTGTGG sequences upstream of many Met genes (Blaiseau et al, 1997), is consistent with high-affinity Met32 binding. However, our motif analysis identified additional sequence preferences 3′ to the consensus site (positions 11–13, Figure 2C); in fact, the affinity distribution of the 17 genomic sequences containing the consensus AAACTGTGGC (e.g., NNAAACTGTGGCNNNNNNNNN) ranges from 9.0 to 64.4 nM (>6-fold range), demonstrating that flanking bases beyond this high-affinity consensus sequence can have a considerable effect on Met32 binding affinity.
High-affinity Met31/Met32 sites specify the Met4 regulon genes with Cbf1-independent expression
To explore the relative contributions of Cbf1 and Met31/Met32 to the transcriptional regulation of the Met4 regulon genes, we constructed a simple biophysical model of gene regulation based on the binding of each factor to gene promoter regions. Cbf1, Met31, and Met32 have all been shown to recruit Met4 to DNA (Kuras et al, 1997; Blaiseau and Thomas, 1998); therefore, we used the predicted probability of finding a factor bound to at least one site in the gene promoter as a direct measure for the strength of Met4 recruitment to each promoter, and consequently for the level of gene regulation. The binding of proteins to sites was treated using an equilibrium thermodynamic model parameterized with our genome-scale binding affinity data (see Supplementary information). Here, and for the rest of this analysis, we have used Met32 binding data to model binding of both Met31 and Met32. Universal PBM experiments for these factors identified no detectable differences in their DNA-binding specificities (Badis et al, 2008).
We generated models using either Met31/Met32 or Cbf1 binding (i.e., single-TF models). We scored the promoter regions of the Met4 regulon genes as well as 4824 additional intergenic regions from the Harbison et al (2004) ChIP-chip data set as described above. For analysis, we divided the Met4 regulon genes into the three classes described by Lee et al (2010) based on the Cbf1 dependence of their expression: Cbf1 dependent (Class 1), partially Cbf1 dependent (Class 2), and Cbf1 independent (Class 3). Scores for Met4 regulon genes were compared with the 500 top-scoring background genes to provide a stricter assessment of specificity and to better resolve differences among the regulon gene classes (Figure 3A and B). Receiver-operating characteristic (ROC) curve analyses were used to assess the sensitivity and specificity of the model predictions (Figure 3C and D).
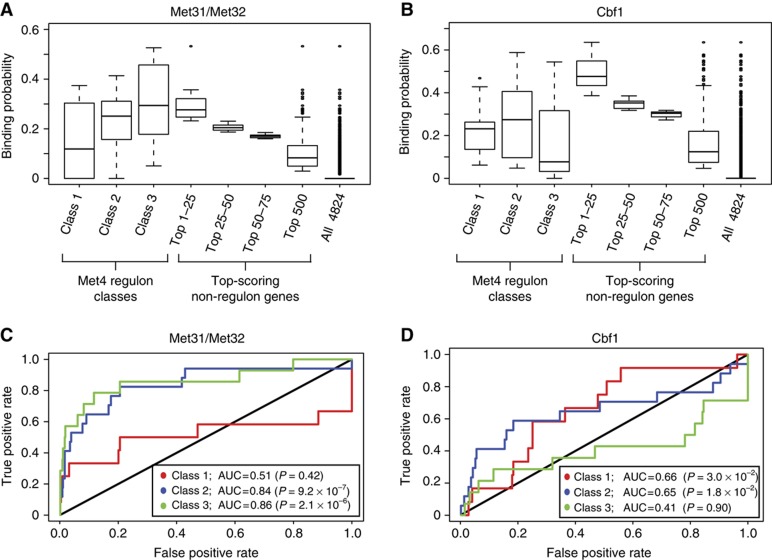
Figure 3.
Specificity of Met32- and Cbf1-specific binding models for Met4 regulon genes. (A, B) Probability distribution for Met32 and Cbf1 binding to gene promoters from different gene sets. Met4 regulon genes are grouped according to Met4 regulon class designation of Lee et al (2010) and non-regulon genes are grouped into the top-scoring gene sets of 25 genes (3 sets) and top-scoring 500 genes (1 set). (C, D) ROC curve analysis for the prediction of each regulon gene class using the 500 top-scoring background (i.e., non-regulon) genes as false positives. Genes are scored and ranked according to the Met32-specific and Cbf1-specific binding probabilities calculated for each gene promoter (Materials and methods). Wilcoxon–Mann–Whitney U-test was applied to each regulon gene set to calculate significance of the AUC values.
We found that the Met31/Met32-specific model of binding was strongly predictive of Class 3 (area under ROC curve (AUC)=0.86) and Class 2 (AUC=0.84) regulon genes, but a poor predictor for Class 1 genes (AUC=0.51) (Figure 3A and C). These results were robust to the concentration of Met31/Met32 (the single free parameter) used in our modeling (Supplementary Table S9). Therefore, the Met31/Met32 binding affinity provides a highly predictive measure for two gene classes of the Met4 regulon.
In contrast to the results from the Met31/Met32-specific model, the Cbf1-specific model yielded moderate predictions for Class 1 (AUC=0.66) and Class 2 (AUC=0.65), but poor predictions for the Cbf1-independent Class 3 genes (AUC=0.41). These results were robust for nuclear Cbf1 concentrations modeled from 0.5 to 5 nM; however, at much higher concentrations, we found that predictions for Class 2 genes improved (AUC=0.79, [Cbf1]=250 nM, see Supplementary Table S9), suggesting the existence of lower affinity Cbf1 sites in Class 2 gene promoters that become important in regulating Class 2 genes at higher Cbf1 concentrations. Paradoxically, however, the Cbf1-specific model is only moderately predictive for the most Cbf1-dependent class of regulon genes (Class 1). Therefore, we hypothesized that some additional cis-regulatory feature must specify this class of genes and explain their observed Cbf1 dependence.
Met4 is recruited equally to all Met32-bound sites
An assumption in our affinity-dependent binding models was that the Met4 cofactor was recruited equally well to any DNA-bound Met31/Met32 or Cbf1 protein (Supplementary information). However, it has been demonstrated, using purified recombinant proteins, that the multi-protein Met4–Met28–Cbf1 complex can assemble on the MET16 UAS element, but not on the MET28 UAS element, despite both of these elements having a Cbf1 binding site (Kuras et al, 1997). Therefore, we examined the possibility of DNA sequence requirements for the assembly of Met4-containing protein complexes. We performed a genome-scale analysis of sequence specificity in Met4 recruitment by Met32, Cbf1, and Met28. To do this, we adapted the standard PBM experimental approach to examine the recruitment of Met4 to the ∼1300 Cbf1 or Met32 sites on our custom, genomic microarray; specifically, we examined the DNA binding of Met4 by PBM experiments performed in the presence or absence of Met32, Cbf1, and Met28 (Materials and methods).
We observed that in the absence of Met32 (Figure 4B), Met4 binds weakly and non-specifically to all 685 Met32 sites in the PBM experiments, consistent with the reported absence of an intrinsic DNA-binding ability (Lee et al, 2010). However, in the presence of Met32, binding by Met4 scales with the binding affinity (Kd) of Met32 to each site (Figure 4A). Therefore, it is the concentration of Met32 bound to each PBM spot that determines the concentration of bound (recruited) Met4. Addition of Met28 had no effect on Met4 recruitment by Met32 (Supplementary Figure S2A and B). These results demonstrate that DNA-bound Met32 recruits Met4 equally to all sites in a Met28-independent manner.
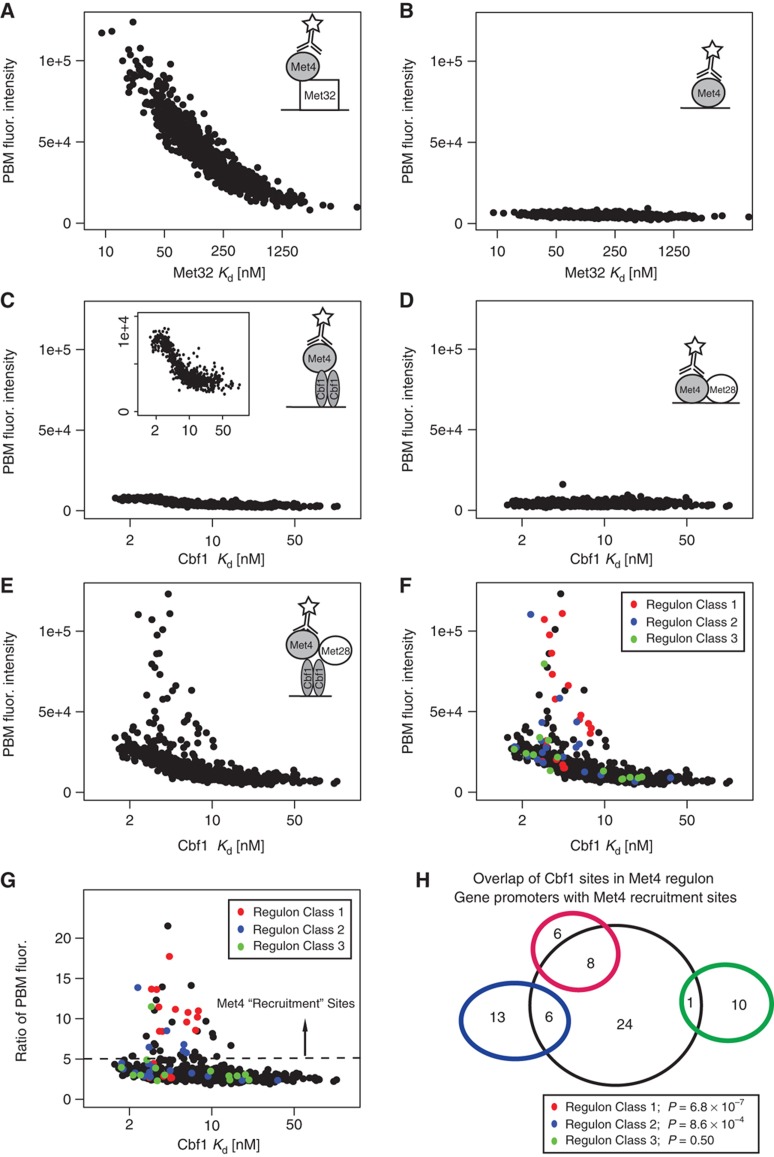
Figure 4.
Sequence dependence of Met4 recruitment. (A–E) The median PBM probe fluorescence intensities for GST-tagged Met4 binding to 685 Met32 sites (A, B) and 673 Cbf1 sites (C–E) in the presence of different 6xHis-tagged proteins are shown: (A) Met4 binding to Met32 sites assayed in the presence of Met32; (B) Met4 binding to Met32 sites by itself; (C) Met4 binding to Cbf1 sites assayed in the presence of Cbf1; (D) Met4 binding to Cbf1 sites in the presence of Met28; (E) Met4 binding to Cbf1 sites in the presence of Met28 and Cbf1. X-axis coordinates are the PBM/SPR-determined Kd values for Met32 and Cbf1 binding to the respective sites. Cartoons in each panel represent the hypothesis being tested. (F) The plot from (E) with Cbf1 sites identified in the promoters of Met4 regulon genes highlighted according to Met4 regulon Class designations of Lee et al (2010) is shown. (G) Ratio of PBM fluorescence values for the Met4/Met28/Cbf1 experiment (E) over the Met4/Cbf1 experiment (C). Individual sites are colored as in (F). Met4 ‘recruitment sites’ are indicated as sites having a ratio >5.0. (H) Overlap of Cbf1 sites identified in upstream promoter region of Met4 regulon genes and Met4 recruitment sites in (G). Promoter regions are defined as 1500 bp upstream of TSS or until next coding region. Significance of observed overlap is calculated using Fisher's one-tail exact test (hypergeometric distribution).
Met4–Met28–Cbf1 complex enhances Met4 recruitment to specific DNA sites
In striking contrast to the results for Met32 recruitment of Met4, we found that the Cbf1–Met28–Met4 complex assembles preferentially in a sequence-dependent manner (Figure 4E). Cbf1 recruits Met4 weakly to the 673 Cbf1 binding sites (Figure 4C), and we observed that the weak Met4 recruitment correlates with Cbf1 binding affinity (Kd). Met28 does not recruit Met4 to DNA (Figure 4D), nor does Met4 bind specifically to Cbf1 sites on its own (Supplementary Figure S2E), consistent with the reported absence of intrinsic DNA-binding activity for Met4 or Met28. However, when Met4 recruitment was examined in combination with both Cbf1 and Met28, we observed both (1) a stabilization of Met4 at all Cbf1 sites (bottom ‘cloud’ in Figure 4E that correlates with Cbf1 Kd values) and (2) an even stronger stabilization at a distinct set of Cbf1 sites with Kd values ranging from high (2 nM) to moderate (10 nM) affinity. Normalizing the PBM fluorescence values from the full Met4/Met28/Cbf1 experiment (Figure 4E) by the non-specific signal from the Met4/Cbf1 experiment (Figure 4C) makes it apparent that addition of the Met28 cofactor enhances binding of Met4 to all 673 Cbf1 sites by ∼2- to 3-fold, but to a small subset of ∼35 sites by 5- to 22-fold (Figure 4G), hereafter referred to as Met4 ‘recruitment’ sites.
Sequence specificity of Met4–Met28–Cbf1 complex binding requires all three factors
Selective binding of the Met4–Met28–Cbf1 complex to a small subset of Cbf1 sites (Met4 recruitment sites) does not correlate with binding affinity of Cbf1. In fact, many of the sites had Kd values 5- to 10-fold lower than the highest affinity Cbf1 sites (Figure 4C). Preferred binding to the Met4 recruitment sites was similarly not observed in the Met4/Cbf1 (Figure 4C) or Met4/Met28 (Figure 4D; Supplementary Figure S2D) experiments. It was previously shown that in-vitro Met28 could stabilize Cbf1 binding to DNA (Kuras et al, 1997). Therefore, we examined whether specification of Met4 recruitment sites could be due to a Met28–Cbf1 complex. PBM experiments with Met28 and Cbf1, however, demonstrated no enhanced specificity for these sites (Supplementary Figure S2C). These results demonstrate that selectivity for Met4 recruitment sites requires the full Met4–Met28–Cbf1 complex.
Promoters of Cbf1-dependent regulon genes are enriched for binding sites with enhanced Met4–Met28–Cbf1 complex binding
We examined whether the Met4 recruitment sites that enhance the binding of the Met4–Met28–Cbf1 complex are found in the promoters of the Met4 regulon genes, and therefore might have a role in their regulation. We found that many Cbf1 sites found in Class 1 and Class 2 genes' upstream regions are Met4 recruitment sites (Figure 4F and G; Supplementary Table S2). We assessed the statistical significance of the overlap between promoter Cbf1 sites and Met4 recruitment sites using Fisher's one-tailed exact test (i.e., using a hypergeometric distribution) (Figure 4H) and found that Cbf1 sites in Class 1 and Class 2 genes are highly enriched for Met4 recruitment sites; 8/14 (P=6.8 × 10−7) and 6/19 (P=8.6 × 10−4), respectively. These recruitment sites occur in the promoters of 8/12 Class 1 genes (67%) and 5/19 Class 2 genes (26%). We note that while both Class 1 and Class 2 gene promoters are enriched for Met4 recruitment sites, the enhanced binding of the Met4–Met28–Cbf1 complex is stronger to the sites in Class 1 gene promoters (Figure 4F and G), which correlates with the increased Cbf1 dependency of the expression for this gene class. Our analysis reveals that the promoters of Met4 regulon genes that exhibit Cbf1-dependent expression are highly enriched for specialized Met4 recruitment sites that enhance the binding of the Met4–Met28–Cbf1 complex.
RYAAT sequence motif located 5′ to the Cbf1 E-box enhances Met4–Met28–Cbf1 complex binding
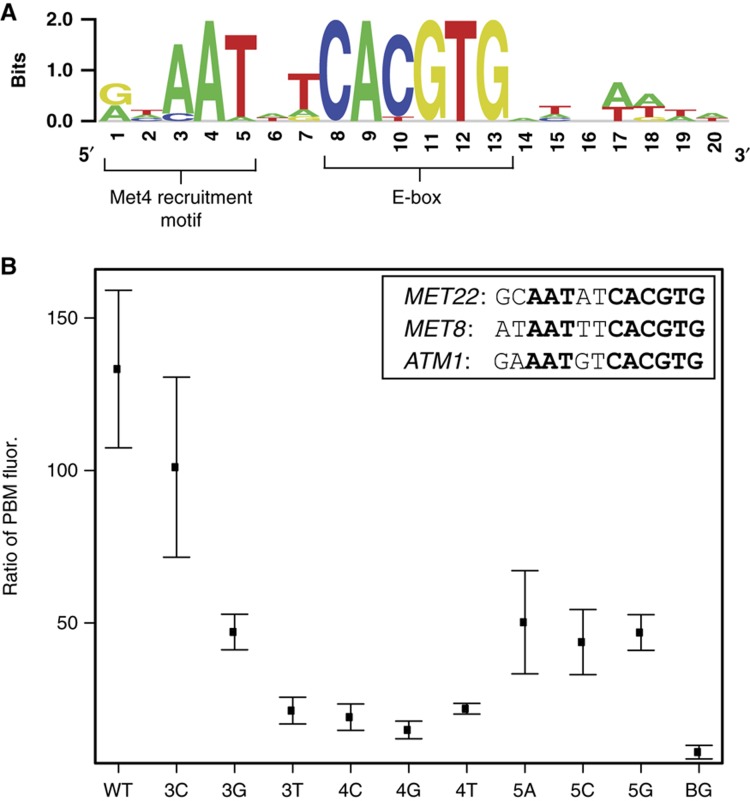
To determine whether specific sequence features of the Met4 recruitment sites account for the enhanced Met4–Met28–Cbf1 binding, we inspected the top-scoring Met4 recruitment sites for any shared sequence features. We found a prominent RYAAT sequence motif located 2 bp 5′ to the canonical CACGTG E-box site and also a weaker sequence motif located more distally on either side of the E-box core (Figure 5A; Supplementary Table S2). We note that the top 20 Met4 recruitment sites, which include Cbf1 sites from 8 of 12 Class 1 regulon gene promoters, all have the RYAAT sequence motif (or RYCAT variant, two sequences) (Supplementary Table S2). To investigate the role of the RYAAT sequence motif, we designed new PBM arrays and examined Met4–Met28–Cbf1 binding to all variants of the AAT submotif (positions 3, 4, and 5 in Figure 5A) for three Met4 recruitment sites (Figure 5B). Deviation from Ade at position 4 reduced binding to near background levels. Deviation from Thy at position 5 also reduced binding, although to a lesser extent. Mutations at position 3 exhibited varied effects, with the Ade to Cyt substitution being tolerated best. To account for any potential artifact that might arise due to the orientation of the RYAAT motif in our PBM probes (i.e., proximal or distal to the glass slide; Supplementary Figure S4), we analyzed the enhanced binding of the Met4–Met28–Cbf1 to recruitment sites for probes in both orientations and found that the effect was preserved.
Figure 5.
Sequence specificity of the Met4 recruitment motif. (A) Logo determined from top 20 Met4 recruitment sites (Supplementary Table S2). These sequences were manually oriented to align the common AAT motif. (B) Ratio of PBM probe fluorescence values (Met4/Met28/Cbf1 PBM experiment over the Met4/Cbf1 PBM experiment) are shown for wild-type and mutant versions of three Met4 recruitment sites (shown in box). Mean and standard deviation are shown for measurements to wild-type or mutant versions of the three sequences. X-axis indicates identity of mutated base (numbering as in (A)). BG indicates measurements over 200 Cbf1 sites with the lowest ratio scores (i.e., background).
To determine the full width of the composite Met4 recruitment/Cbf1 binding site, we designed new custom PBM arrays to make systematic mutations of both 5′ and 3′ distal nucleotide positions. For the Met4 recruitment sites identified in the ADE3 and MET16 gene promoters, we exhaustively tested Met4–Met28–Cbf1 binding to 256 variants that differed at nucleotide positions −2 through 2 (Supplementary Table S3). Met4–Met28–Cbf1 binding to these mutant sequences varied considerably; examination identified a strong sequence preference for a purine (Ade or Gua) at position 1 followed by a pyrimidine (Cyt or Thy) at position 2 (Supplementary Figure S3A). This sequence preference was consistent with the preferences observed for strong Met4–Met28–Cbf1 binding sites identified in the genome (Figure 5A; Supplementary Figure S3A). Mutations at positions 3′ to the E-box (i.e., positions 15–22 in Figure 5A) had no effect on Met4–Met28–Cbf1 binding (data not shown). To rule out a sequence preference at more distal positions, we re-examined Met4–Met28–Cbf1 binding to the 673 Cbf1 sites in the presence of an additional 5 bp of the genomic flanking sequence on either site (positions −5 to 25 in Figure 5A). We observed no significant difference in Met4–Met28–Cbf1 binding (data not shown) and a binding motif constructed from the top 20 ‘extended flank’ recruitment sites showed no additional sequence preference beyond the RYAAT motif (Supplementary Figure S3A). These results demonstrate that enhanced Met4 recruitment in vitro by the Met4–Met28–Cbf1 complex is dependent on the 5-bp Met4 recruitment motif RYAAT (positions 1–5 in Figure 5A) located 5′ to the E-box motif.
The RYAAT sequence motif must occur at a fixed distance from the Cbf1 E-box to enhance Met4–Met28–Cbf1 complex binding
Given the conserved spacing of the Met4 recruitment motif relative to the E-box in the genomic sequences, we tested the importance of the spacing between these two motifs for enhanced Met4–Met28–Cbf1 binding. For the Met4 recruitment sites in the ADE3 and MET16 promoters, we systematically varied the spacing of the Met4 recruitment motif relative to the E-box from 0 bp (i.e., ACAATCACGTG) to 2 bp (i.e., ACAATNNCACGTG, 16 variants) and examined the effect on Met4–Met28–Cbf1 binding (Supplementary Table S3; Supplementary Figure S3B). Binding was reduced to near background levels for all spacing variants except for the native 2-bp spacing, suggesting a strict requirement for exact 2 bp spacing between the AAT of the Met4 recruitment motif and the Cbf1 E-box motif for enhanced Met4–Met28–Cbf1 binding. Therefore, the Met4 recruitment motif is a highly specific composite binding motif with strong spacing and sequence requirements for functionality.
A second RYAAT sequence motif can further enhance Met4–Met28–Cbf1 binding
Motivated by the observation that Cbf1 binds the E-box as a homodimer (Kuras et al, 1996), we asked whether adding a second Met4 recruitment motif on the opposite (3′) side of the E-box would result in a binding site with even stronger Met4–Met28–Cbf1 binding. We observed that adding a second, symmetrically positioned Met4 recruitment motif significantly improves Met4–Met28–Cbf1 binding (Supplementary Figure S3B). Furthermore, as Met28 concentration increases, the PBM signal is enhanced more greatly for sites with a second recruitment motif than to sites with a single recruitment motif. These results demonstrate that the increased Met4 binding (i.e., PBM signal) is due to additional Met28 binding (or recruitment) to the second recruitment site and suggests a direct role for Met28 in the recognition of the Met4 recruitment motif.
Mutations to the RYAAT motif compromise induction of genes in low-sulfur conditions
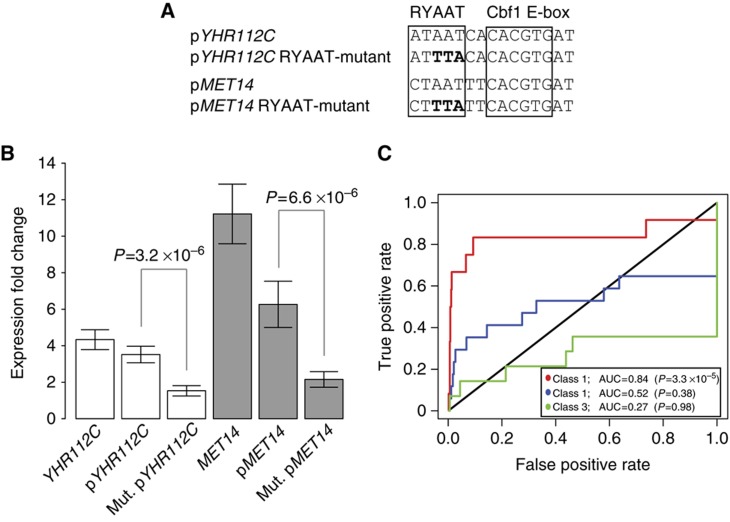
We examined the contribution of the RYAAT recruitment motif to gene induction under conditions of low-sulfur growth. Yeast strains were constructed in which wild-type or RYAAT-mutant versions of the promoter regions from two Class 1 genes, YHR112C and MET14, were inserted upstream of LYS2, which we employed here as a reporter gene (Materials and methods; Figure 6A, Supplementary Figure S5). Both YHR112C and MET14 contained high-scoring Met4 recruitment sites (Supplementary Table S2). The ability of the wild-type and mutant promoters to drive LYS2 gene expression was examined under low-sulfur growth conditions. Mutations to the promoter regions were limited to the RYAAT motif (i.e., RYAAT to RYTTA; see Figure 6A) so as not to perturb Cbf1 binding itself. We observed significant reduction in the promoter activity for RYAAT-mutant versions of the promoters: YHR112C (∼2-fold reduced; P=3.2 × 10−6) and MET14 (∼3-fold reduced; P=6.6 × 10−6) (Figure 6B). Many Class 1 gene promoters contain a moderate affinity Met31/Met32 binding site in addition to a composite Met4 recruitment site. To examine the potential dependence on the proximity of Met31/Met32 sites, we chose MET14 and YHR112C as examples of promoters in which these sites are proximal to each other (MET14, 30 bp; Supplementary Figure S5) or distal (YHR112C, 186 bp; Supplementary Figure S5). While both mutant promoters exhibited considerably reduced activity, some activity remained, which might have resulted from Met4 recruitment to these moderate affinity Met31/Met32 sites. Our results demonstrate that the RYAAT motif is a bone fide cis-regulatory element necessary for the full induction of Class 1 target genes of the Met4–Met28–Cbf1 complex under conditions of low-sulfur growth.
Figure 6.
RYAAT motif is critical to promoter activity and specification of Class 1 regulon genes. (A) Schematic of wild-type and mutant versions of the composite Met4 recruitment sites from YHR112C and MET14 gene promoters (pYHR112C and pMET14) cloned upstream of LYS2 reporter gene. (B) Expression fold change, under switch to low-sulfur growth conditions, for endogenous genes (YHR112C and MET14) and the LYS2 reporter gene driven by wild-type (pYHR112C and pMET14) and RYAAT-mutant (Mut. pYHR112C and Mut. pMET14) gene promoters. (C) ROC analysis for the prediction (identification) of the Met4 regulon genes using the Met4 recruitment ‘strength’ of the 673 Cbf1 sites used in our genome-wide affinity analysis. Met4 recruitment strength is the ratio of PBM fluorescence intensities shown in Figure 3G (i.e., ratio of PBM fluorescence intensities for the Met4/Met28/Cbf1 and Met4/Cbf1 PBM experiments). ROC analysis was performed using 500 top-scoring non-regulon genes as false positives. Wilcoxon–Mann–Whitney U-test was applied to each regulon gene set to calculate significance of the AUC value. Source data is available for this figure in the Supplementary information.
Met4 recruitment sites specify Cbf1-dependent Met4 regulon genes
The presence of the RYAAT motif next to the Cbf1 binding site, in addition to enhancing Met4–Met28–Cbf1 complex binding, provides a means to functionally distinguish Cbf1 sites within the genome. This suggested that Met4 recruitment ability of a Cbf1 site (i.e., the presence of an adjacent RYAAT motif) rather than Cbf1 binding site affinity may specify the Class 1 genes within the genome. To investigate this, we scored genes by the Met4 recruitment strength of Cbf1 sites present in their promoters, and compared the regulon genes with the top 500 scoring non-regulon genes as was done previously (Figure 3C and D). Met4 recruitment strength of Cbf1 sites was scored as in Figure 4G. We found that the Class 1 regulon genes are predicted strongly by Met4 recruitment strength alone (AUC=0.84) (Figure 6C). While Class 2 genes do contain Met4 recruitment sites (Figure 4G and H), the class as a whole is not predicted well (AUC=0.52). Scoring genes based on the presence of an RYAAT motif adjacent to Cbf1 sites, as a proxy for Met4 recruitment strength, performed identically (data not shown). These results demonstrate that Met4 recruitment strength of Cbf1 sites, rather than Cbf1 binding site affinity, is what distinguishes Class 1 regulon genes within the genome.
Discussion
Achieving specificity in transcriptional regulation requires that TFs are able to identify specific genomic loci. However, in eukaryotes the degenerate binding of TFs and large genome sizes means that single binding sites occur too often to explain the specificity observed for gene transcription (Wunderlich and Mirny, 2009). As a model system, we have examined the role of TF binding site affinity and sequence-specific cofactor recruitment in specifying the previously described Met4 regulon genes (Lee et al, 2010). Our results suggest that at least two distinct mechanisms are used to achieve specific recruitment of the Met4 transcriptional activator to Met4 regulon gene promoters. For Class 2 and Class 3 Met4 regulon genes (those with expression only weakly dependent or independent of Cbf1, respectively), the presence of high-affinity Met31/Met32 binding sites (which represent binding by either Met31 or Met32) provides specificity and distinguishes these Met4 regulon genes from other genes in the genome. Consistent with this, we found that Met32 can recruit Met4 equally well to any binding site; therefore, it is the binding of Met31 or Met32 itself that provides the specificity. In contrast, for the strongly Cbf1-dependent (Class 1) regulon genes, the presence of novel Met4 recruitment sites that enhance binding by the Met4–Met28–Cbf1 complex provides specificity. We find that the ability of Cbf1 sites to be bound by the Met4–Met28–Cbf1 complex is considerably more predictive of this gene class than is Cbf1 binding affinity alone (AUC=0.84 versus 0.65, Figures 3 and 6, respectively). Furthermore, our demonstration that the recognition of the Met4 recruitment sites requires the full trimeric complex provides an explanation for the observed Cbf1 and Met28 dependence of the Class 1 subset of the Met4 regulon genes: deletions of either Cbf1 or Met28 will abrogate the trimeric complex required to recognize the Met4 recruitment sites present in Class 1 gene promoters. These results demonstrate that TF targeting specificity (Met4 targeting in this system) can be achieved by different mechanisms even within a tightly co-expressed set of genes.
Previous work has described still additional mechanisms for achieving specificity, such as stabilized binding of Met4–Met28–Met32 by proximally bound Cbf1 (Blaiseau and Thomas, 1998) and differential reporter gene expression based on altered spacing of Met31/Met32 and Cbf1 binding sites (Chiang et al, 2006). Future work examining these additional mechanisms of specificity should lead to an even more complete model of transcriptional regulatory control for the Met4 regulon genes.
To investigate the role of DNA-binding affinity, we developed a hybrid SPR-PBM methodology that readily allows the measurement of absolute binding affinities (Kds) of a TF or TF complex to thousands of individual DNA binding sequences. With currently available array densities (e.g., Agilent 1 × 1 M array format), this approach could be extended readily to hundreds of thousands of sites. In this study, we applied this approach to measure the binding affinities of Met32 and Cbf1 to >1300 unique DNA binding sites from the S. cerevisiae genome. We demonstrate that this approach can provide accurate affinity measurements, which are in excellent agreement with other published methods (Figure 1D).
The cooperative assembly of the Met4–Met28–Cbf1 complex on DNA that we report is consistent with results of Kuras et al (1997). Moreover, our results provide an explanation for the differential binding that they observed in vitro for the Met4–Met28–Cbf1 complex on E-box sites from the MET16 (ATCATTTCACGTG) and the MET28 (TAAGTCACGTGCACTCAG) gene promoters: the E-box (shown in bold) from the MET16 gene promoter has a Met4 recruitment motif adjacent to it (underlined), while the site from the MET28 promoter does not. However, in contrast to their observation that Met4–Met28–Cbf1 would not assemble on the MET28 E-box sequence, we find that there is weak non-specific stabilization of the Met4–Met28–Cbf1 complexes to all E-box sequences, and that this stabilization correlates with the DNA-binding affinity of the Cbf1 site (compare Figure 4C and D with Figure 4E). This inconsistency may be due to the different protein concentrations or experimental approaches that were employed in our study versus theirs, or may be due to the different Cbf1 protein constructs that were used; we used GST-tagged, full-length Cbf1, whereas Kuras et al used a 6xHis-tagged, N-terminally truncated version of Cbf1.
The ability of the Met4 recruitment motif RYAAT (Figure 5A) to enhance the assembly of the Met4–Met28–Cbf1 complexes on E-box sites was unexpected. Cbf1 does not preferentially bind to sites adjacent to the Met4 recruitment motif, nor do the pairwise complexes of Met4–Cbf1, Met28–Cbf1, or Met28–Met4 (Figure 4B and D; Supplementary Figure S2). Therefore, specific recognition of the Met4 recruitment motif requires all three proteins to be present in the bound complex. While it remains unclear what part of the Met4–Met28–Cbf1 complex recognizes the Met4 recruitment motif, we find it unlikely that some unknown portion of Cbf1 protein confers the specific recognition of the RYAAT motif. First, it was previously shown that the region of Cbf1 N-terminal to the bHLH DNA-binding domain (amino acids 1–209) was unnecessary for differential recognition of the MET28 and MET16 UAS elements by a Met4–Met28–Cbf1ΔN complex (Kuras et al, 1997). Second, the Cbf1 bHLH DNA-binding domain is itself unlikely to make strong DNA contacts 7 bp from the E-box core, and the ∼80 amino-acid long region C-terminal to the bHLH domain does not contain any known DNA-binding domains.
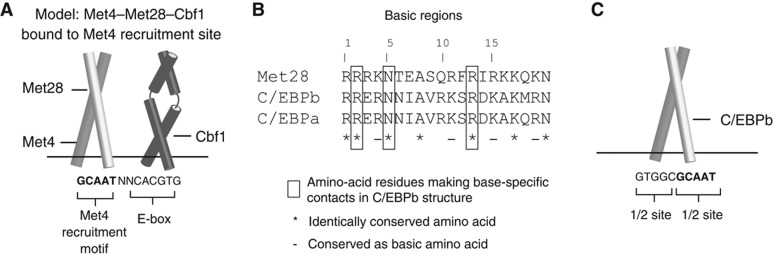
In contrast, despite exhibiting no intrinsic DNA-binding ability, both Met28 and Met4 contain a bZIP DNA-binding motif (Blaiseau and Thomas, 1998). Based on the considerations of protein sequence and structure, we propose a model in which the Met28 subunit of the Met4–Met28–Cbf1 complex makes base-specific contacts to select for the Met4 recruitment motif. Sequence analysis identified a weak homology between the bZIP regions of Met28 and C/EBPa from mouse (BLASTP E-value=0.15, see Materials and methods), and a striking similarity between amino-acid residues of Met28 and those of the C/EBPa paralog C/EBPb (Figure 7B) that make base-specific contacts with a GCAAT binding sequence in an X-ray co-crystal structure (Tahirov et al, 2002). Furthermore, the GCAAT half-site from the C/EBPb crystal structure itself is a perfect match to the RYAAT Met4 recruitment motif (Figure 7C). We favor a model where the Met28 subunit of the Met4–Met28–Cbf1 complex makes base-specific contacts to select for the Met4 recruitment motif. We propose that a plausible configuration for the trimeric complex is one in which a Met4/Met28 bZIP heterodimer, dimerizing via leucine zippers, is positioned adjacent to the Cbf1 homodimer (Figure 7A); this configuration would allow for Met28 to adopt a binding orientation analogous to the C/EBPb subunit that similarly recognizes a GCAAT half-site.
Figure 7.
Proposed model of Met4 recruitment complexes. (A) Diagram of proposed orientation for DNA-binding domains of Met4 (bZIP domain), Met28 (bZIP domain), and Cbf1 (bHLH domain) in the Met4–Met28–Cbf1 complex bound to a Met4 recruitment site. Met4 recruitment motif is in bold. Direct physical contacts that exist between the Met4 and/or Met28 subunits and the Cbf1 homodimer are not explicitly indicated. (B) Sequence alignment of basic region from Met28, human C/EBPb, and human C/EBPa. (C) Diagram of C/EBPb bZIP region homodimer bound to DNA. Binding site sequence for C/EBPb complex is from X-ray crystal structure of C/EBPb complex (PDB code 1H8A) (Tahirov et al, 2002). GCAAT half-site motif is in bold.
Selective binding of the Met4–Met28–Cbf1 complex to the composite (RYAATNNCACGTG) Met4 recruitment site is strikingly similar to the situation described for the Oct-1–HCF-1–VP16 complex that recognizes the consensus site TAATGARAT (Babb et al, 2001). In both situations, non-DNA-binding transcriptional activators (Met4 and VP16) are recruited to DNA by sequence-specific binding TFs (Cbf1 and Oct-1, respectively), and this recruitment is facilitated by non-DNA-binding cofactors (Met28 and HCF-1, respectively). Furthermore, in both situations the multi-protein complex selects for binding sites where a ‘recruitment motif’ (RYAAT and GARAT, respectively) occurs adjacent to the TF binding site motif (CACGTG for Cbf1 and TAAT for Oct-1). The extent to which this shared mechanism exists beyond these two systems remains to be discovered; however, they highlight the need to examine the DNA-binding specificity of multi-protein complexes even when the recruited cofactors are not known to interact with DNA. A direct role for non-DNA-binding cofactors in refining the gene targeting of regulatory complexes might represent a widespread mechanism to achieve greater complexity in eukaryotic gene regulation.
Materials and methods
TF cloning and preparation of protein samples
Full-length CBF1, MET32, MET28, and MET4 open reading frames were cloned into Gateway pDEST15 (N-terminal GST tag) and pDEST17 (N-terminal 6xHis tag) expression vectors. GST-Met32, GST-Cbf1, and GST-Met4 were overexpressed in E. coli BL21 (DE3) cells (New England BioLabs) and purified by FPLC (AKTAprime plus) using 1 ml GSTrap™ FF affinity columns (GE Healthcare). Samples were then concentrated by centrifugation using Amicon Ultra (10 K) filter devices (Millipore) and stored in 10% glycerol at −80°C. Protein concentrations were quantified by standard Bradford assay using Coomassie Plus Protein Assay reagent (Thermo Scientific); stock concentrations of the purified proteins were as follows: GST-Met32 (45 μM), GST-Cbf1 (43 μM), GST-Met4 (5 μM). All 6xHis-tagged proteins produced by in-vitro transcription and translation (IVT) were made using the PURExpress kit (New England BioLabs) from purified plasmids. Western blots were performed for each protein to assess quality and to approximate protein concentration relative to a dilution series of recombinant GST standard (Sigma). See Supplementary information for further details.
Genomic binding site identification and PBM probe construction
We identified potential DNA binding sites in yeast intergenic regions by scanning their sequence with universal PBM data for Cbf1 and Met32 (Zhu et al, 2009). We identified all high-scoring ungapped 8-mers (PBM enrichment score >0.48) in the genome and aligned them to 10 bp position weight matrices (PWMs) defining the core binding site motifs for Cbf1 (GTCACGTGAC) or Met31/Met32 (CTGTGGCGCT) to determine a common sequence register. The identified genomic sequences constituting the core 10 bp motif plus 5 bp of flanking sequence on each side were incorporated into 60 bp probe sequences on a new, custom-designed DNA microarray (Figure 2A and B; Supplementary information).
PBM experiments and analysis
PBM experiments were performed using custom-designed oligonucleotide arrays (Agilent Technologies, Inc., 8 × 15 K array platform; see Supplementary information). Two different custom PBMs were designed and used for this work: design #1 (Agilent Technologies Inc., AMADID #024623) had genomic Cbf1 and Met32 binding sites (Figures 1, 2 and 4; Supplementary Figures S1, S2 and S4); design #2 (Agilent Technologies Inc., AMADID #028293) had mutant versions of Met4 recruitment sites (Figure 5; Supplementary Figure S3; Supplementary Table S3). For PBM experiments used in the hybrid SPR-PBM approach to determine binding affinities, GST-tagged protein (Met32 or Cbf1) was applied at eight different concentrations on a single design #1 array (Supplementary Table S1). For PBM experiments assessing Met4 recruitment (Figure 4), protein samples were applied at the concentrations indicated in Supplementary Table S4. PBM DNA probe sequences are provided in Supplementary File 1. Full PBM data and hybrid SPR-PBM determined Kd values are provided (Supplementary Tables S6 and S7).
SPR experiments
SPR was performed on a Biacore 3000 instrument. Biotinylated oligonucleotides were immobilized onto a Sensor Chip SA (Biacore). Serial concentrations of protein sample were diluted into a running buffer (10 mM Tris–HCl, pH 7.4; 3 mM dithiothreitol (DTT); 0.2 mM EDTA, 0.02% Triton X-100; 120 mM NaCl; 10% glycerol; 0.2 μm filtered and de-gassed) and applied to the Sensor Chip at 25 μl/min (KINJECT option: 250 μl samples/150 s dissociation phase). Binding constants (Kd values) were determined using Scrubber2 software (BioLogic Software). Probes sequences and Kd values are provided (Supplementary Table S5).
Generating yeast strains
Wild-type and RYAAT-mutant promoter constructs were inserted upstream of the native LYS2 gene in the S. cerevisiae genome (yMT-2450 strain; Lee et al, 2010) (Supplementary Figure S5A; Supplementary information). The inserted promoter constructs displace the native LYS2 promoter (i.e., in the 5′ direction relative to the gene) and do not remove it. Wild-type and mutant promoter regions for YHR112C and MET14 (Figure 6A; Supplementary Figure S5B) were constructed by gene synthesis (GenScript). The high-efficiency transformation protocol of Gietz and Woods (2002) was used for all transformations.
Gene expression experiments
Gene expression was examined under conditions of low-sulfur growth as described in Lee et al (2010) (see Supplementary information). Expression was measured in log-phase growth in minimal B-media with 0.5 mM methionine as sole sulfur source (t=0) and 2 h after switching to minimal B-media lacking a sulfur source (t=2 h). Expression was monitored by quantitative PCR (qPCR) for both wild-type and RYAAT-mutant promoter strains. All measurements were performed in biological triplicate (i.e., three independent induction experiments) and technical triplicate (i.e., three independent PCRs).
Biophysical modeling
Met4 recruitment was modeled using an equilibrium thermodynamic model (Bintu et al, 2005; Rowan et al, 2010). Gene activation is modeled as the probability of Met4 being bound at a promoter region. The model was parameterized using our PBM-determined protein-DNA binding affinities (Cbf1 and Met32) and site-specific Met4 recruitment data. The model was implemented in Perl. See Supplementary information for full details.
Sequence analysis
Protein similarity searches for Met28 and Met4 bZIP regions (Met28 a.a. 91–160; Met4 a.a. 581–640) were performed by blastp search from the NCBI BLAST website (http://blast.ncbi.nlm.nih.gov/Blast.cgi) against the non-redundant protein database.
Supplementary Material
Supplementary Figures S1–5, Supplementary Tables S1–9
Acknowledgments
We thank Mike Berger for technical assistance and advice with PBMs and data analysis; Traci Lee for technical assistance and generously providing the yMT-2450 S. cerevisiae strain; Dan Spatt for technical assistance; Fred Winston for the generous gift of pRS plasmids; Steve Gisselbrecht, Leila Shokri, Raluca Gordân, and Jaime Chapoy for helpful discussions; Kevin Struhl for helpful comments on the manuscript. This work was funded by grant # R01 HG003985 from the National Institutes of Health/National Human Genome Research Institute to MLB, the i2b2/HST Summer Institute in Bioinformatics and Integrative Genomics NIH grant #U54 LM008748, and in part by an NSF Postdoctoral Fellowship in Biological Informatics (#630639) to TS.
Author contributions: TS designed all experiments and performed all analyses, generated expression plasmids for Met4, expressed and purified proteins, performed the PBM and SPR experiments, generated reporter yeast strains and performed gene expression experiments. MHD and SK assisted with protein expression and purification. JR generated expression plasmids for Met28, Cbf1, and Met32. MLB supervised the research. TS and MLB wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Babb R, Huang CC, Aufiero DJ, Herr W (2001) DNA recognition by the herpes simplex virus transactivator VP16: a novel DNA-binding structure. Mol Cell Biol 21: 4700–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, Kuznetsov H, Wang CF, Coburn D, Newburger DE, Morris Q, Hughes TR, Bulyk ML (2009) Diversity and complexity in DNA recognition by transcription factors. Science 324: 1720–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, Gebbia M, Talukder S, Yang A, Mnaimneh S, Terterov D, Coburn D, Li Yeo A, Yeo ZX, Clarke ND, Lieb JD et al. (2008) A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell 32: 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Bulyk ML (2006a) Protein binding microarrays (PBMs) for rapid, high-throughput characterization of the sequence specificities of DNA binding proteins. Methods Mol Biol 338: 245–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW III, Bulyk ML (2006b) Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol 24: 1429–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman BP, Nibu Y, Pfeiffer BD, Tomancak P, Celniker SE, Levine M, Rubin GM, Eisen MB (2002) Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc Natl Acad Sci USA 99: 757–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Phillips R (2005) Transcriptional regulation by the numbers: models. Curr Opin Genet Dev 15: 116–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaiseau PL, Isnard AD, Surdin-Kerjan Y, Thomas D (1997) Met31p and Met32p, two related zinc finger proteins, are involved in transcriptional regulation of yeast sulfur amino acid metabolism. Mol Cell Biol 17: 3640–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaiseau PL, Thomas D (1998) Multiple transcriptional activation complexes tether the yeast activator Met4 to DNA. EMBO J 17: 6327–6336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulyk ML, Huang X, Choo Y, Church GM (2001) Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc Natl Acad Sci USA 98: 7158–7163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M (1998) The enhanceosome and transcriptional synergy. Cell 92: 5–8 [DOI] [PubMed] [Google Scholar]
- Chiang DY, Nix DA, Shultzaberger RK, Gasch AP, Eisen MB (2006) Flexible promoter architecture requirements for coactivator recruitment. BMC Mol Biol 7: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth FJ, Chambon P (2001) Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene 20: 3047–3054 [DOI] [PubMed] [Google Scholar]
- Frith MC, Spouge JL, Hansen U, Weng Z (2002) Statistical significance of clusters of motifs represented by position specific scoring matrices in nucleotide sequences. Nucleic Acids Res 30: 3214–3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet J, Mango SE (2002) Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science 295: 821–825 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Woods RA (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350: 87–96 [DOI] [PubMed] [Google Scholar]
- Giorgetti L, Siggers T, Tiana G, Caprara G, Notarbartolo S, Corona T, Pasparakis M, Milani P, Bulyk ML, Natoli G (2010) Noncooperative interactions between transcription factors and clustered DNA binding sites enable graded transcriptional responses to environmental inputs. Mol Cell 37: 418–428 [DOI] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, MacIsaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA (2004) Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Levine M (1993) Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell 72: 741–752 [DOI] [PubMed] [Google Scholar]
- Jones RB, Gordus A, Krall JA, MacBeath G (2006) A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature 439: 168–174 [DOI] [PubMed] [Google Scholar]
- Joshi R, Passner JM, Rohs R, Jain R, Sosinsky A, Crickmore MA, Jacob V, Aggarwal AK, Honig B, Mann RS (2007) Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell 131: 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L, Barbey R, Thomas D (1997) Assembly of a bZIP-bHLH transcription activation complex: formation of the yeast Cbf1-Met4-Met28 complex is regulated through Met28 stimulation of Cbf1 DNA binding. EMBO J 16: 2441–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L, Cherest H, Surdin-Kerjan Y, Thomas D (1996) A heteromeric complex containing the centromere binding factor 1 and two basic leucine zipper factors, Met4 and Met28, mediates the transcription activation of yeast sulfur metabolism. EMBO J 15: 2519–2529 [PMC free article] [PubMed] [Google Scholar]
- Lee TA, Jorgensen P, Bognar AL, Peyraud C, Thomas D, Tyers M (2010) Dissection of combinatorial control by the Met4 transcriptional complex. Mol Biol Cell 21: 456–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIsaac KD, Gordon DB, Nekludova L, Odom DT, Schreiber J, Gifford DK, Young RA, Fraenkel E (2006) A hypothesis-based approach for identifying the binding specificity of regulatory proteins from chromatin immunoprecipitation data. Bioinformatics 22: 423–429 [DOI] [PubMed] [Google Scholar]
- Maerkl SJ, Quake SR (2007) A systems approach to measuring the binding energy landscapes of transcription factors. Science 315: 233–237 [DOI] [PubMed] [Google Scholar]
- Mann RS, Lelli KM, Joshi R (2009) Hox specificity unique roles for cofactors and collaborators. Curr Top Dev Biol 88: 63–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Markstein P, Markstein V, Levine MS (2002) Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci USA 99: 763–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merika M, Thanos D (2001) Enhanceosomes. Curr Opin Genet Dev 11: 205–208 [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Berger MF, Jona G, Wang XS, Muzzey D, Snyder M, Young RA, Bulyk ML (2004) Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat Genet 36: 1331–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramila T, Miles S, GuhaThakurta D, Jemiolo D, Breeden LL (2002) Conserved homeodomain proteins interact with MADS box protein Mcm1 to restrict ECB-dependent transcription to the M/G1 phase of the cell cycle. Genes Dev 16: 3034–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S, Siggers T, Lachke SA, Yue Y, Bulyk ML, Maas RL (2010) Precise temporal control of the eye regulatory gene Pax6 via enhancer-binding site affinity. Genes Dev 24: 980–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein B, Baldwin AS Jr (1993) Distinct mechanisms for regulation of the interleukin-8 gene involve synergism and cooperativity between C/EBP and NF-kappa B. Mol Cell Biol 13: 7191–7198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K (2005) Transcriptional activation: mediator can act after preinitiation complex formation. Mol Cell 17: 752–754 [DOI] [PubMed] [Google Scholar]
- Tahirov TH, Sato K, Ichikawa-Iwata E, Sasaki M, Inoue-Bungo T, Shiina M, Kimura K, Takata S, Fujikawa A, Morii H, Kumasaka T, Yamamoto M, Ishii S, Ogata K (2002) Mechanism of c-Myb-C/EBP beta cooperation from separated sites on a promoter. Cell 108: 57–70 [DOI] [PubMed] [Google Scholar]
- Wunderlich Z, Mirny LA (2009) Different gene regulation strategies revealed by analysis of binding motifs. Trends Genet 25: 434–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, Saulrieta K, Smith Z, Shah MV, Radhakrishnan M, Philippakis AA, Hu Y, De Masi F, Pacek M, Rolfs A, Murthy T, Labaer J, Bulyk ML (2009) High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res 19: 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–5, Supplementary Tables S1–9