Abstract
The Ras superfamily of proteins consists of five branches: Ras, Rho, Arf, Rab and Ran subfamilies. These proteins are involved in a plethora of biological functions spanning cytoskeletal organization, cell proliferation, transcription and intracellular trafficking. Ras-Binding Domains (RBDs) have classically been identified as autonomous ubiquitin-like folded regions that bind certain activated Ras GTPases of the Ras subfamily. In general, RBDs in many proteins have been tagged with membrane-targeting functions as in the case of the well-characterized c-Raf-RBD/Ras interaction. However, it is becoming apparent that the definition and functions of RBDs need to be revamped in order to reflect the new discoveries associated with this domain. Here, we discuss in more detail the recent advances associated with these RBDs. We highlight research identifying RBDs in formins, ELMOs and the RhoGEF, Syx and discuss the emerging role for RBDs in controlling autoinhibition relief and the newly recognized versatility of RBDs to interact with Rho and Arf family GTPases. In addition, these recent findings raise the exciting hypothesis that functional RBDs remain hidden in the proteome and are ready to be uncovered.
Keywords: ELMO, FHOD1, Ras GTPases, Ras-Binding Domain, Syx
Introduction
One-hundred-fifty or so Ras proteins are subdivided into Ras, Rho, Arf, Rab and Ran subfamilies. Numerous important biological activities have been ascribed to these groups of GTPases: Ras’ regulate cell proliferation, Rhos’ control the actin cytoskeleton and Arfs’ and Rabs’ shape vesicular trafficking.1,2 Yet, this is a gross generalization of their functions and it is important to underscore that many different activities are also present within each subfamily. For example, some Ras and Arf family members can directly impinge on the actin cytoskeleton and play a role in cell migration.3–5 Individual and careful study of each GTPase is therefore warranted to uncover the unique roles of distinct GTPases.
In most cases, members of the Ras superfamily act as molecular switches alternating between active, GTP-bound, and inactive, GDP-bound, conformational states. Because of their high affinity for guanine nucleotides and their low intrinsic GTPase activity, at least two classes of regulatory proteins are indispensable for controlling the biological activity of these small GTPases in cells. The guanine nucleotide exchange factors (GEFs) act as positive regulators by promoting the dissociation of GDP from small GTPases and allowing for their reloading with the GTP nucleotide.6–10 Inversely, the GTPase-activating proteins (GAPs) recognize GTP-bound G-proteins and enhance their intrinsic GTPase activity to favor the inactive GDP-bound state, therefore acting as negative regulators of the cycle.11–13 A third class of regulators, acting only on Rho and Rab G-proteins, are the guanine nucleotide dissociation inhibitors (GDIs). These inhibitory proteins have the double duty of both locking the GTPase in a nucleotide state, most of the time in a GDP conformation, and preventing membrane targeting by masking the membrane targeting signals.14–19 It remains an important area of research to determine exactly how GDIs are uncoupled from Rho/Rab GTPases during GEF-mediated activation.
In their active GTP-loaded state, Ras G-proteins can transmit signals by binding to a specific set of proteins termed effectors. The structural basis for such interactions is in many cases now well established: the most significant structural change between the GDP and GTP bound state of a G-protein is a conformational change in two surface loops known as Switch I and Switch II regions.20,21 Remarkably, this GTP-induced remodeling of the switch regions increases the affinity of a given GTPase for its own set of specific effectors allowing for physical interaction and signal transduction.20,21 A number of domains in effector proteins have now been demonstrated to mediate specific binding to Ras [Ras Association domains (RAs), Ras-Binding Domains (RBDs), Ral-Binding Domains (RalBDs), etc.], Rho [Cdc42/Rac Interactive Binding domains (CRIB), GTPase-Binding Domains (GBDs), Anti-Parallel Coiled-Coil domains (ACCs), split PH, etc.], Arf [GGA and Tom1 domains (GATs), Leucine Zippers (LZs), Arf-Binding Domains (ArfBDs), etc.] and Rab [Rab-Binding Domains (RabBDs), Zinc Fingers (ZFs), RUN-PLAT domains, etc.] GTPases. In Table 1, we summarize some of the interactions between GTPases and their effectors focusing on complexes where structural data are available.
Table 1. List of solved crystal structures for small Ras superfamily GTPase/effector domain complexes.
| GTPase superfamily subdivisions |
Effector binding proteins |
Crystal structures |
GTPase effector protein/effector domain crystal structures |
|---|---|---|---|
| Arf subfamily | |||
|
Arf1 |
ARHGAP21 |
ARHGAP21ArfBD (PH domain and helical region) in complex with Arf1 [17347647] |
Arf1/ARHGAP21 Arf-binding domain (ArfBD) |
| |
GGA |
GGAGAT domain (N-terminus) in complex with Arf1 [12679809] |
Arf1/GGA GAT domain |
|
Arf6 |
Cholera toxin (CT) |
CTA1 subunit in complex with Arf6 [16099990] |
- |
| |
JIP4 |
JIP4LZII in complex with Arf6 [19644450] |
Arf6/JIP4 leucine zipper II (LZII) |
| |
MKLP1 |
MKLP1C in complex with Arf6 [22522702] |
- |
|
Arl1 |
Arfaptin-2 |
Arfaptin-2BAR Domain in complex with Arl1 [22679020] |
Arl1/Arfaptin-2 BAR domain |
| |
Golgin-245 |
Golgin-245GRIP Domain in complex with Arl1 [14718928] |
Arl1/Golgin-245 GRIP domain |
|
Arl2 |
BART |
BART in complex with Arl2 [19368893] |
- |
| |
PDEδ |
PDEδC in complex with Arl2 [11980706] |
- |
| |
UNC119a |
UNC119a in complex with Arl2 [22960633] |
- |
|
Arl3 |
UNC119a |
UNC119a in complex with Arl3 [22960633] |
- |
|
Rho subfamily |
|
|
|
|
Cdc42 |
ACK |
ACKCRIB in complex with Cdc42 [10360579] |
Cdc42/ACK CRIB domain |
| |
PAK |
PAKCRIB in complex with Cdc42 [10802735] |
Cdc42/PAK CRIB domain |
| |
Par6 |
Par6CRIB-PDZ in complex with Cdc42 [12606577] |
Cdc42/Par6 CRIB-PDZ domains |
| |
WASP |
WASPCRIB in complex with Cdc42 [10360578] |
Cdc42/WASP CRIB domain |
|
Rac1 |
Phospholipase C-β2 |
Phospholipase C-β2ΔC (PH-EF-TIM-C2 domain-containing fragment) in complex with Rac1 [17115053] |
Rac1/Phospholipase C-β2 PH domain |
| |
Plexin-B1 |
Plexin-B1ΔN in complex with Rac1 [ 21912513] |
- |
|
Rac2 |
Phospholipase C-γ2 |
Phospholipase C-γ2spPH in complex with Rac2 [19394299] |
Rac2/Phospholipase C-γ2 split PH domain |
|
RhoA |
PKN |
PKNN (ACC finger domain) in complex with RhoA [10619026] |
RhoA/PKN atypical coiled-coil (ACC) domain |
| |
ROCK1 |
ROCK1RhoBD (coiled-coil) in complex with RhoA [14660612] |
- |
|
RhoC |
Dia1 |
Dia1N (GBD) in complex with RhoC [15864301; 16292343; 16472745] |
RhoC/Dia1 GTPase-binding domain (GBD) |
|
Rnd1 |
Plexin A2 |
Plexin A2RhoBD (ubiquitin-like fold) in complex with Rnd1 [21610070] |
Rnd1/Plexin A2 Rho-binding domain (RhoBD) or RBD-like |
| |
Plexin B1 |
Plexin B1RhoBD in complex with Rnd1 [21610070] |
Rnd1/Plexin B1 Rho-binding domain (RhoBD) or RBD-like |
|
Rnd3 |
ROCK1 |
ROCK1N in complex with Rnd3 [18946488] |
- |
|
Ras subfamily |
|
|
|
|
H-Ras |
PI3K |
PI3KγRBD in complex with H-Ras [11136978] |
H-Ras/PI3Kγ Ras-binding domain (RBD) |
| |
RalGDS |
RalGDSRBD in complex with H-Ras [9628477; 10371160] |
H-Ras/RalGDS Ras-binding domain (RBD) |
|
RalA |
C3bot1 |
C3bot1 in complex with RalA [15809419] |
- |
| |
Exo84 |
Exo84RalBD (PH-FOLD) in complex with RalA [15920473] |
RalA/Exo84 PH domain |
| |
Sec5 |
Sec5RalBD (immunoglobulin-like β-sandwich) in complex with RalA [12839989] |
RalA/Sec5 immunoglobulin-like β-sandwich |
|
Rap1A |
c-Raf1 |
c-Raf1RBD (ubiquitin-like fold) in complex with Rap1A [7791872] |
Rap1A/c-Raf1 Ras-binding domain (RBD) |
| |
Krit1 |
Krit1FERM domain in complex with Rap1 [22577140] |
Rap1/Krit1 FERM F1 and F2 lobes |
|
Rab subfamily |
|
|
|
|
Rab5A |
EEA1 |
EEA1C2H2 ZF in complex with Rab5A [20534488] |
Rab5A/EEA1 C2H2 zinc finger (ZF) |
|
Rab6 |
Rab6IP1 |
Rab6IP1α-helical RUN-PLAT domains in complex with Rab6 [19141279] |
Rab6/Rab6IP1 RUN domain |
|
Rab8A |
OCRL1 |
OCRL1ASH domain in complex with Rab8A [21378754] |
Rab8A/OCRL1 ASH domain |
| Rab22A | Rabenosyn5 | Rabenosyn5C2H2 ZF in complex with Rab22A [20534488] | Rab22A/C2H2 zinc finger (ZF) |
ACC, antiparallel coiled-coil; ArfBD, Arf-binding domain; ASH, ASPM/SPD-2/Hydin; BAR, Bin1/Amphiphysin/Rvs167; C, C-terminus; CRIB, Cdc42/Rac interactive binding; FERM, 4.1 protein/Ezrin/Radixin/Moesin; GAT, GGA and Tom1; GBD, GTPase-binding domain; GRIP, Golgin-97/RanBP2alpha/Imh1p/p230/golgin-245; LF, leucine finger; LZ, leucine zipper; N, N-terminus; PDZ, PSD-95 and ZO-1; PH, pleckstrin homology; RBD, Ras-binding domain; RhoBD, Rho-binding domain; RUN, RPIP8/UNC-14/NESCA; spPH, split PH; ZF, zinc finger. Numbers in brackets denote Pubmed IDs (PMIDs).
Through various mechanisms, including but not limited to release of auto-inhibition, membrane targeting or direct stimulation of catalytic activity, effector proteins transmit signals leading to the appropriate biological function. What are the structural features of effectors responsible for binding small GTPases? A number of motifs have now been shown to mediate such interaction of effectors with activated Ras family proteins: here, we will discuss in more detail recent advances on the first such domain originally identified in c-Raf and involved in Ras-GTP interaction, the self-titled Ras-Binding Domain (RBD).
Defining Features of the Ras-Binding Domain
Historically, experiments using an activated and oncogenic version of Ras were instrumental to establish the notion that effector proteins are essential to transmit G-protein signaling.22,23 Indeed, activated mutants of Ras with point mutations in the Switch I region were unable to transform cells implying that this region in Ras is critical to bind to downstream proteins.24 Following these observations, evidence that c-Raf acts downstream of Ras was accumulating and the domain mediating the direct interaction between c-Raf and Ras was uncovered: residues 51–131 of c-Raf defined the first effector domain, termed the RBD.25–29 Simply put, the role of this interaction is to facilitate the Ras-dependent recruitment of c-Raf to the membrane where it can optimally be positioned for activation by a collection of additional proteins. Despite the minimalism of this signaling event, the Ras pathway, in particular at the level of c-Raf activation, is known to be deregulated in cancer and has been under intense investigation for several decades.30
The RBD is currently found in a number of signaling intermediates including well-studied proteins such as Rafs’, PI 3-kinases and Regulator of G-protein Signaling (RGS). Interestingly, while initially thought to be an additional type of Ras-interacting module, the more abundant RalGDS/AF6 or Ras Association (RA) domain folds comparably to the RBD.31,32 This so-called RA domain is found in many proteins including RIN1/2, EPAC and PDZGEF.33–35 Here, and in agreement with other reports, we will refer globally to both the RBD and RA modules as the RBD. Interestingly, as of today, more than 90 human proteins are cataloged to contain such RBDs in the SMART database. While partners have been identified for a number of them, others remain orphan.32 In fact, it has been postulated that it is unlikely that all RBDs will turn out to be bona fide Ras subfamily effectors.32
Structurally, the RBD presents itself as a ubiquitin superfold.28 This structural unit, with a β1-, β2-, α1-, β3-, β4-, α2- and β5- topology, is frequent in the proteome and can be observed within signaling domains such as in the F1 lobe of the FERM domain.36 Multiple structures of Ras proteins bound to RBDs have been solved and the residues involved in the interaction are identified. In general, most of the contacts on the RBD side involve charged residues of β2 with some residues in β1 and α1 also touching the Ras GTPases.28,37 Conversely, charged residues found in β2/β3 (within the Switch I region) of the Ras family GTPase are implicated in contacting the RBDs.37
Until recently, the RBD was viewed as a Ras subfamily-specific recognition module. Here, we will briefly review a series of recent papers reporting interaction of newly discovered RBDs with affinities for GTPases of the Rho and Arf families.
Unexpected Binding Partners for RBDs: Beyond Ras Proteins
FHOD1 binds Rac-family GTPases via a RBD
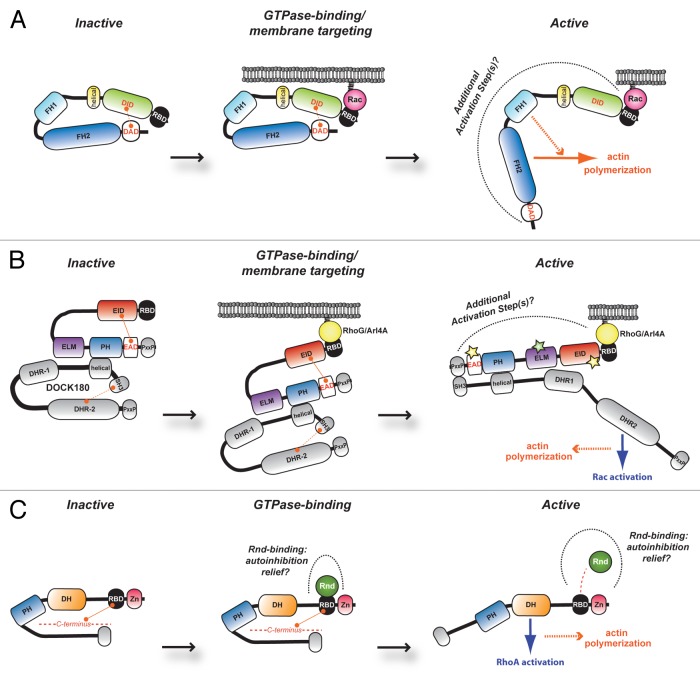
FHOD1 is a formin involved in promoting actin polymerization via its formin homology (FH)-1 and FH-2 module.38,39 Like several members of this family, FHOD1 exists in a repressed state due to intramolecular interactions between its DID (diaphanous inhibitory domain) and DAD (diaphanous autoregulatory domain) domains39,40 (Fig. 1A). Rac GTPase-binding activity was known to be present in the N-terminus of this formin. Likewise, close family members, Dia-related formins, were well established to bind to RhoA/B/C GTPases via their N-terminus in a manner that facilitated the release of the DID-DAD inhibitory contacts to favor activation of proteins.41–45 Schulte et al. therefore opted to use a structural approach to gain insight into how FHOD1 may become activated in cells.39 Not surprisingly, the DID domain of FHOD1 folded in a manner similar to the related domains in Dia formins and the authors could define a single point mutation capable of disrupting DID-DAD binding therein producing an activated form of FHOD1. Surprising, however, was the discovery that a ubiquitin-like fold forms the region involved in Rac GTPase recognition. As discussed above, this ubiquitin-like domain belongs to the RBD family of motifs and was previously found guilty of mediating interaction with Ras subfamily GTPases. These findings therefore established for the first time that RBD domains are not limited to Ras subfamily GTPases in terms of specificity. From a functional point a view, the RBD of FHOD1 is critical for targeting the formin to the membrane upon Rac activation. However, this essential recruitment step is not sufficient to fully activate the formin activity; this is in contrast to Rho-mediated recruitment of Dia1 to the membrane where this appears sufficient to also release DID-DAD interactions41,46 (Fig. 1A). In the case of FHOD1, it is consequently believed that additional steps, such as its phosphorylation by ROCK,47 are also required to maximally activate this formin. Searches for structural homologs revealed striking similarity of the RBD of FHOD1 to RBDs of RalGDS and PI 3-Kinase. Homology to the F1 lobe of the FERM domain of Moesin was also noted. It remains unclear as to how the RBD of FHOD1 binds to Rac. No mutagenesis was performed to determine the critical residues responsible for Rac-binding. A prediction would be that it binds to GTPases in a manner similar to Ras/Raf via charged amino acids. Co-crystallization of activated Rac and the FHOD1 RBD domain would be one way to reveal the residues critical to the contact of these molecules. Such an approach may also uncover if there is crosstalk between the RBD and DID (as is the case for Dia1), as such to propose a new hypothesis on the role of Rac-binding with respect to changing the inactive conformation of FHOD1 to an active one.
Figure 1. Representative models of FHOD1, ELMO family and Syx protein regulation. Schematic depicting proposed model of RBD-mediated regulation of (A) FHOD1, (B) ELMO family and (C) Syx. (A) At basal levels, the formin FHOD1 is repressed via intramolecular contacts between the DID and the DAD obscuring its actin nucleation function. RBD engagement by active Rac tethers the molecule at the cell membrane and relief of autoinhibition is suggested to occur through additional activation steps. A similar model of autoinhibition relief is proposed for (B) ELMO family proteins. These latter molecules are found constitutively in complex with DOCK proteins. It is suggested that binding of the ELMO RBD via active RhoG or Arl4A results in membrane targeting of the complex, release of DOCK180 autoinhibition (resulting in Rac activation through the DOCK GEF activity) and concomitant cytoskeletal reorganization. (C) In unstimulated conditions, it is hypothesized that Syx exists in an autoinhibited conformation through as yet undetermined portions in its N- and C-terminus. Relief of this closed conformation may come in the form of Rnd1/3-binding to the Syx RBD, thereby exposing its DH domain to catalyze nucleotide exchange specifically on RhoA and induce cytoskeleton rearrangement. RBD, Ras-binding domain; DID, diaphanous inhibitory domain; DD, dimerization domain; CC, coiled-coil region; FH, the actin nucleation module of formin-homology-1 and FH-2 regions; DAD, diaphanous autoregulatory domain; EID, ELMO inhibitory domain; ELM, ELMO homology; PH, pleckstrin homology; EAD, ELMO-autoregulatory domain; PxxP, proline-rich motif; DHR, DOCK homology region-1 and DHR-2; Zn, zinc-finger domain; DH, Dbl homology
A versatile RBD in ELMO proteins interacts with RhoG and Arl4A GTPases
ELMO proteins interact with a subset of the family of DOCK180 atypical GEFs and contribute to coordinate Rac signaling.7 These proteins are evolutionarily conserved and recent data in mammalian models highlight that activation of Rac by DOCK180 is required for myoblast fusion and cardiovascular development.48,49 ELMO1, in vivo, appears to be a key player in engulfment of apoptotic germ cells in the testes and in the clearance of apoptotic neurons during neurogenesis.50,51 At the molecular level, ELMOs couple DOCK180 for Rac activation within various cellular compartments whether at the plasma membrane via small GTPase-binding or integrins/Integrin-Linked Kinase recruitment, or directly to receptors, such as through their interaction with the G-protein coupled receptor, BAI1.52,53
When screening for novel partners of activated RhoG, Katoh and Negishi identified ELMO1 and how this RhoG/ELMO complex co-recruited DOCK180 for Rac activation.54 Mapping studies revealed that a unique extreme N-terminal portion of ELMO is involved in the nucleotide-state specific interaction with RhoG.54 We recently found that ELMO proteins exist in a closed conformation similar to what is described above for formins such as Dia1 and FHOD1 (Fig. 1B). These investigations led to the identification of three novel domains involved in controlling the ELMO conformation state. ELMO’s newly identified domains consist of the ELMO Inhibitory Domain (EID) and ELMO Autoregulation Domain (EAD), mimicking the formins’ DID and DAD function, while the RhoG-binding site on ELMO is defined as a RBD-like domain55 (Fig. 1B). Our bioinformatics analyses and structural superimposition model found that the ELMO RBD more closely resembles the ubiquitin-like fold of the FHOD1 RBD and also shares weak sequence homology to the RBDs of c-Raf and PI3-K.55 We found a similar superfold at the extreme N-terminus of mammalian ELMO and its orthologs in Drosophila and C. elegans, showing its evolutionarily conserved nature in these proteins. Additionally, biochemical analyses of a critical conserved residue (L43 in ELMO1) supported our structural model and further cemented the RBD nature of the ELMO N-terminus.55
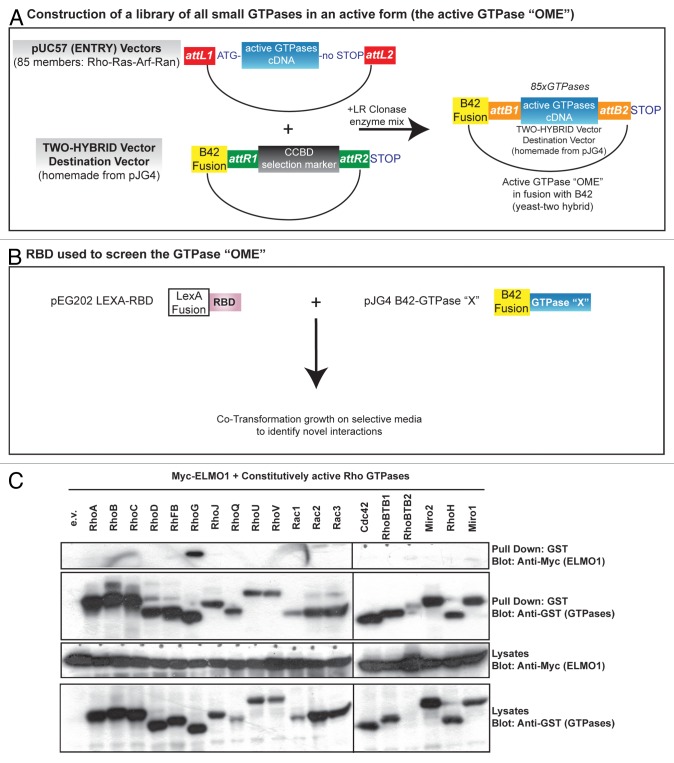
Since we suspected that the RBD domain of ELMO might specifically interact with GTPase(s) other than RhoG to broaden the spectrum of DOCK180-mediated Rac activation, we developed a screening strategy to systematically interrogate the GTPase-RBD coupling. More specifically, we generated a library of cDNA coding for 85 GTPases (covering completely the Ras, Rho, Ran and Arf families) in their active conformation in the GATEWAY system (Fig. 2). With these tools in hand, we constructed a library of B42 prey constructs compatible with the yeast two-hybrid system to perform interaction screens with RBDs of interest expressed as LexA fusion proteins as shown in Figure 256 (these reagents are available to the GTPase community by contacting us directly). Intriguingly, in our attempt to uncover novel ELMO RBD-binding partners, we identified active Arl4A, an Arf family GTPase, which relied specifically on the conserved ELMO RBD for complex formation.56 The fact that the Arl4A binding site coincided with that of RhoG led to the discovery of a polyvalent RBD in ELMO that demonstrates cross-selectivity for Rho and Arf family GTPases.56 Arl4A, a constitutively active GTPase, was found to facilitate the recruitment of ELMO/DOCK180 to the membrane to promote cytoskeletal changes such as actin stress fiber depolymerization and membrane ruffles.56 The exact biological processes regulated by Arl4A remain poorly understood. So far, using the ELMO RBD as bait, we found that initial screening by the yeast-two hybrid system only gave 2 positive interactions (RhoG, Arl4A) out of 85 tested for which the interaction could be reproduced in cells and were functionally important. When screening additional RBDs, it will be important to take into account that the yeast-two hybrid screens could uncover weak and non-physiological interactions. In these cases, it will be important to perform interaction assays and functional experiments to fully establish the importance of GTPase/RBD coupling. Accordingly, the library of GTPases we generated in the GATEWAY system might prove very useful to rapidly shuttle the GTPases’ cDNAs in various expression vectors designed to carry out specific experiments (GST, GFP, TAP-TAG, FLAG, etc.). For example, to assay the specificity of the ELMO/RhoG interaction, we created a library of 85 GST-tagged GTPases in a mammalian expression vector (pDEST27) and rapidly tested the ability of ELMO to bind to all Ras, Rho and Arf family members. In Figure 2C, we show such an example where we screened all the Rho proteins for ELMO-binding. Likewise, we also created a library of 85 eGFP-Rho, eGFP-Ras and Arf-eGFP family members to test for GTPases that could recruit ELMO to the membrane in an RBD-dependent manner. These co-localization experiments led us to confirm a specific recruitment of Myc-ELMO1 to the membrane when co-expressed with RhoG and Arl4A (data not shown). A similar library was generated by Tobias Meyer’s lab and was successfully used to identify GTPases mediating cytoskeletal changes and activation of PI 3-Kinase signaling.57,58 In contrast to our library, traditional cloning would need to be done to swap the Meyer lab’s entire library into new vectors.
Figure 2. A high throughput system to probe GTPases/RBD interactions. (A) Schematic representing the construction of a new library of all small GTPases in an active form (the active GTPase “OME” of the Ras, Rho, Ran and Arf subfamily of Ras GTPase) using the GATEWAY system. In principle, this library can be shuttled into any type of expression vector needed for experimental testing using the appropriate destination vector. (B) We shuttled the library of cDNA in a yeast two-hybrid compatible vector and isolated a novel interaction between ELMO-RBD and Arl4A (see text). This system can be using to identify partners of RBDs or other types of GTPases binding domain. Yeast two-hybrid screening for novel interactions is based on co-transformation of tagged constructs (LexA-RBD and B42-GTPase) in yeast and then grown on selective media. (C) Example of shuttling the library to a different expression system; in this case, the cDNAs of the GTPases were recombined in the pDEST27 vector for mammalian cell expression of GST-tagged GTPases. We probed all Rho family GTPases for binding to Myc-ELMO1 and we confirm that activated RhoG is the only Rho GTPase specifically binding ELMO1.
We have demonstrated that “opening” of ELMO is integral for the biological function of the DOCK180/ELMO tandem during cell migration.55 However, thus far, the signals that unhinge the closed ELMO molecule evade us. Whether active RhoG or Arl4A can induce direct conformational changes that evoke opening of the ELMO protein remains to be formally determined. In our attempt to study such direct effects of RhoG on ELMO conformation, we were confronted with technical limitations in our ELMO biosensor, namely that the tagging of the N-terminus of ELMO2 with either GFP10 or Renilla Luciferase II (RLucII) created steric constraints that prevented efficient RhoG/RBD interactions.55 Structural analysis of a RhoG-GTP/ELMO or Arl4A/ELMO complex would clarify if this interaction can perturb the closed conformation of ELMO proteins. What we know at this point is that engagement of either Arl4A or RhoG to ELMO targets and tethers the complex to the cell membrane55,56 (Fig. 1B). It would also be of interest to test known ELMO-binding partners (i.e., IpgB1, BAI1 and ERM proteins)52,59,60 in the event that these partners are responsible for endorsing the opening of ELMO through the RBD and/or adjacent regions. As it is suggested for the formins FHOD1, DAAM1 and partially for Dia, we propose that yet to be identified factors in the form of novel interactors and/or post-translational regulation, contribute to unlock ELMO in a spatio-temporal manner.
A previously unidentified RBD in Syx specifically binds to the Rho family member Rnd3
Syx, also referred to as GEF720/PLEKHG5/TECH, is a member of the Dbl RhoGEF family.61–63 In vivo studies demonstrate that Syx plays an important role during neuronal cell differentiation and zebrafish development.62,64 Structurally, Syx is a multidomain protein where a zinc-finger domain is present at the N-terminus and a Dbl Homology-Pleckstrin Homology (DH-PH) module is found centrally64 (Fig. 1C). The enzymatic activity of Syx relies on the DH domain and has been shown to catalyze nucleotide exchange specifically on RhoA.64 Interestingly, the Manser lab discovered, through co-purification, that Syx is also a Rnd3-associated protein.64
Rnd3 is labeled as an atypical Rho GTPase, diverging from most conventional GTPases in that it does not require catalysis via GEFs for activation. Rather, the Rnd branch of Rho GTPases (Rnd1–3) has low intrinsic GTPase activity and is thus found primarily in a GTP-bound active conformation.65,66 It would seem plausible that Syx would act as a GEF for Rnd3 in this case. However, Goh and Manser found that Syx is rather an effector for Rnd3 signaling and acts as a stabilizing agent for Rnd proteins.67 Such a GTPase cascade, Rnd3→Syx→RhoA-GTP is reminiscent of the signaling by RhoG and Arl4 to activate Rac via recruitment and activation of the ELMO/DOCK complex. Intriguingly, visual inspection and manual sequence alignments led the authors to conclude that Syx contains an ubiquitin-like RBD involved in Rnd3-binding (Fig. 1C). Importantly, mutation of key residues in the Syx RBD, correlating with the Raf1 side chains responsible for Ras-binding, abolished Syx-Rnd3 complex formation. Much like for the RBDs of ELMO and FHOD1, the RBD of Syx provides additional evidence for the ability of RBDs to interact broadly with activated GTPases.
Furthermore, Goh and Manser’s biochemical results indicated that a N-terminal truncation mutant rather than full-length Syx bound more intensely to Rnd1/3, suggesting that structurally Syx may exist in an autoinhibited conformation that is relieved when the C-terminal portion of Syx is deleted64 (Fig. 1C). It is quite possible then that at basal level, Syx exists in an autoinhibited conformation and Rnd1/3 binding to the Syx RBD may act as a release signal for this molecule (Fig. 1C). It will be exciting to test such models in the future. Interestingly, Syx is not the first identified RhoGEF to contain a RBD. For example, the RBDs of TIAM1/2 act as recognition sequences for activated Ras and such interactions have important consequences for tumorigenesis.68
What is unique about the Rho- and Arf-Binding RBDs?
The work discussed here suggests that a collection of RBDs may currently rest uncovered in the proteome. The crystal structure of a fragment of FHOD1 was instrumental in the discovery of a RBD involved in Rac activation.39 The detection of the ELMO RBD was possible due to sequence homology to FHOD1.55 Finally, Manser et al. had to visually inspect the primary sequence of Syx to uncover its RBD.64 Improving bioinformatics methods to detect RBDs would be of great interest to better understand the signaling networks controlled by Ras-family GTPases.
We began exploring if unique characteristics could be attributed to RBDs binding Rho and Arf proteins in comparison to classical Ras-binding RBDs. Our manual sequence analyses and secondary structure predictions highlight similarities of the FHOD/ELMO/Syx RBDs to the c-Raf RBD, and further accentuates conserved charged and hydrophobic residues in the predicted secondary structure of the ubiquitin fold (Fig. 3). One difference we could observe in the case of the RBD of ELMO and FHOD1 is the presence of an acidic residue (Asp) at the end of β2 whereas a basic residue is found in c-Raf at this position (Arg), and several other Raf-like RBDs (RGS12_RBD1/2 and RGS14_RBD1/2) contain a Lys.31,32 We also noted that α1 tends to contain more hydrophobic residues in comparison to Raf1 (Fig. 3) and a panel of other RBDs (not shown). The identification of novel RBDs binding Rhos and Arfs would provide a better framework to clearly establish what is unique about these interactions. Crystallizing complexes of novel RBDs bound to GTPases may also uncover whether the binding mode is identical to canonical RBDs’ binding to Ras or if special requirements have evolved.

Figure 3. Sequence homology in the RBDs of ELMO, c-Raf1, FHOD1 and Syx. Secondary structure prediction and sequence comparison between c-Raf1, FHOD1, ELMO-family proteins and Syx indicates an evolutionarily conserved Ras-Binding Domain (RBD) characterized by the presence of a ubiquitin-like subdomain. ELMO secondary structure was predicted with Jpred3. FHOD1 (Protein Data Bank ID code 3DAD) and Raf1 (Protein Data Bank ID code 1GUA) structures were used for the manual alignment with the ELMO RBDs. Conserved hydrophobic residues are highlighted in yellow, and conserved positively and negatively charged residues are indicated in blue and purple, respectively. Green residues depict the conserved Leucine residue in the ELMO proteins shown to be critical for RhoG and Arl4A GTPase binding. Charged residues in c-Raf involved in contacting Ras are shown in red. Residues highlighted in bold lettering indicate sequences that fold as helical. Asterisks indicate residues in Syx suggested to be involved in GTPase binding. E indicates β strand, H indicates α-helical.
Interestingly, the RBD domain of Grb14, a member of the Grb7-family of adaptor proteins, was recently shown to bind Rab5 in a nucleotide-dependent manner.69 This raises the exciting possibility that selected RBDs could also be specifically interacting with Rab family members therefore extending the functionality of the RBD domain. Likewise, it was also recently observed that the Lobe 1 (L1) of the FERM domain of Krit1, a region known to fold as a ubiquitin-like domain, is involved in binding to active Rap1. Structural studies also uncovered that the F2 lobe of the Krit1 FERM domain also contributes to this interaction.70 Globally, this finding suggests that a subset of FERM domains could act as RBD-like domains and further broaden mechanisms of GTPases’ signaling.
Conclusions
Studying the expanding realm of RBDs highlights the truly divergent nature of these domains. This is true not only in terms of structure, but also in effector binding and biological function. It remains to be determined whether a clear signature in RBDs exists that will establish if a RBD will bind GTPases, and if so, which ones. For this, we herein describe a library of cDNAs coding for all GTPases of the Ras, Rho, Ran and Arf families in their active form that can be used to systematically interrogate RBD/GTPase interaction (Fig. 2). Collectively, with each new discovery of RBD-containing proteins, we are getting closer to a clearer understanding of their regulation and biological functions.
Acknowledgments
We thank Dr Yoran Margaron and Viviane Tran for generating some of the eGFP-tagged and B42-tagged Ras superfamily GTPases discussed in this review. M.P is a recipient of a FRQ-S post-doctoral fellowship and is currently training in Dr Courtneidge’s laboratory (Sandford-Burnham Institute, La Jolla, CA). J.-F.C. holds a FRQ-S “Junior 2” career award. Work in the Côté laboratory is funded by operating grants from the Canadian Cancer Society (#019104), Canadian Institute of Health Research (MOP 77591) and Quebec Breast Cancer Foundation to J.-F.C.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/24298
References
- 1.Colicelli J. Human RAS superfamily proteins and related GTPases. Sci STKE. 2004;2004:RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 3.Myers KR, Casanova JE. Regulation of actin cytoskeleton dynamics by Arf-family GTPases. Trends Cell Biol. 2008;18:184–92. doi: 10.1016/j.tcb.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall A, Lalli G. Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harb Perspect Biol. 2010;2:a001818. doi: 10.1101/cshperspect.a001818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsumoto K, Asano T, Endo T. Novel small GTPase M-Ras participates in reorganization of actin cytoskeleton. Oncogene. 1997;15:2409–17. doi: 10.1038/sj.onc.1201416. [DOI] [PubMed] [Google Scholar]
- 6.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–80. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 7.Côté JF, Vuori K. GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol. 2007;17:383–93. doi: 10.1016/j.tcb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erickson JW, Cerione RA. Structural elements, mechanism, and evolutionary convergence of Rho protein-guanine nucleotide exchange factor complexes. Biochemistry. 2004;43:837–42. doi: 10.1021/bi036026v. [DOI] [PubMed] [Google Scholar]
- 9.Yang J, Zhang Z, Roe SM, Marshall CJ, Barford D. Activation of Rho GTPases by DOCK exchange factors is mediated by a nucleotide sensor. Science. 2009;325:1398–402. doi: 10.1126/science.1174468. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt A, Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16:1587–609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- 11.Sprang SR. G proteins, effectors and GAPs: structure and mechanism. Curr Opin Struct Biol. 1997;7:849–56. doi: 10.1016/S0959-440X(97)80157-1. [DOI] [PubMed] [Google Scholar]
- 12.Moon SY, Zheng Y. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 2003;13:13–22. doi: 10.1016/S0962-8924(02)00004-1. [DOI] [PubMed] [Google Scholar]
- 13.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–77. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 14.Dovas A, Couchman JR. RhoGDI: multiple functions in the regulation of Rho family GTPase activities. Biochem J. 2005;390:1–9. doi: 10.1042/BJ20050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DerMardirossian C, Bokoch GM. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15:356–63. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 16.Hoffman GR, Nassar N, Cerione RA. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell. 2000;100:345–56. doi: 10.1016/S0092-8674(00)80670-4. [DOI] [PubMed] [Google Scholar]
- 17.Yamashita T, Tohyama M. The p75 receptor acts as a displacement factor that releases Rho from Rho-GDI. Nat Neurosci. 2003;6:461–7. doi: 10.1038/nn1045. [DOI] [PubMed] [Google Scholar]
- 18.Olofsson B. Rho guanine dissociation inhibitors: pivotal molecules in cellular signalling. Cell Signal. 1999;11:545–54. doi: 10.1016/S0898-6568(98)00063-1. [DOI] [PubMed] [Google Scholar]
- 19.Michaelson D, Silletti J, Murphy G, D’Eustachio P, Rush M, Philips MR. Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J Cell Biol. 2001;152:111–26. doi: 10.1083/jcb.152.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 21.Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–6. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 22.Calés C, Hancock JF, Marshall CJ, Hall A. The cytoplasmic protein GAP is implicated as the target for regulation by the ras gene product. Nature. 1988;332:548–51. doi: 10.1038/332548a0. [DOI] [PubMed] [Google Scholar]
- 23.Adari H, Lowy DR, Willumsen BM, Der CJ, McCormick F. Guanosine triphosphatase activating protein (GAP) interacts with the p21 ras effector binding domain. Science. 1988;240:518–21. doi: 10.1126/science.2833817. [DOI] [PubMed] [Google Scholar]
- 24.Milburn MV, Tong L, deVos AM, Brünger A, Yamaizumi Z, Nishimura S, et al. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–45. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 25.Herrmann C, Martin GA, Wittinghofer A. Quantitative analysis of the complex between p21ras and the Ras-binding domain of the human Raf-1 protein kinase. J Biol Chem. 1995;270:2901–5. doi: 10.1074/jbc.270.7.2901. [DOI] [PubMed] [Google Scholar]
- 26.Scheffler JE, Waugh DS, Bekesi E, Kiefer SE, LoSardo JE, Neri A, et al. Characterization of a 78-residue fragment of c-Raf-1 that comprises a minimal binding domain for the interaction with Ras-GTP. J Biol Chem. 1994;269:22340–6. [PubMed] [Google Scholar]
- 27.Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–14. doi: 10.1016/0092-8674(93)90307-C. [DOI] [PubMed] [Google Scholar]
- 28.Nassar N, Horn G, Herrmann C, Scherer A, McCormick F, Wittinghofer A. The 2.2 A crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with Rap1A and a GTP analogue. Nature. 1995;375:554–60. doi: 10.1038/375554a0. [DOI] [PubMed] [Google Scholar]
- 29.Clark GJ, Drugan JK, Terrell RS, Bradham C, Der CJ, Bell RM, et al. Peptides containing a consensus Ras binding sequence from Raf-1 and theGTPase activating protein NF1 inhibit Ras function. Proc Natl Acad Sci USA. 1996;93:1577–81. doi: 10.1073/pnas.93.4.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Udell CM, Rajakulendran T, Sicheri F, Therrien M. Mechanistic principles of RAF kinase signaling. Cell Mol Life Sci. 2011;68:553–65. doi: 10.1007/s00018-010-0520-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiel C, Wohlgemuth S, Rousseau F, Schymkowitz J, Ferkinghoff-Borg J, Wittinghofer F, et al. Recognizing and defining true Ras binding domains II: in silico prediction based on homology modelling and energy calculations. J Mol Biol. 2005;348:759–75. doi: 10.1016/j.jmb.2005.02.046. [DOI] [PubMed] [Google Scholar]
- 32.Wohlgemuth S, Kiel C, Krämer A, Serrano L, Wittinghofer F, Herrmann C. Recognizing and defining true Ras binding domains I: biochemical analysis. J Mol Biol. 2005;348:741–58. doi: 10.1016/j.jmb.2005.02.048. [DOI] [PubMed] [Google Scholar]
- 33.Hunker CM, Giambini H, Galvis A, Hall J, Kruk I, Veisaga ML, et al. Rin1 regulates insulin receptor signal transduction pathways. Exp Cell Res. 2006;312:1106–18. doi: 10.1016/j.yexcr.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 34.Li Y, Asuri S, Rebhun JF, Castro AF, Paranavitana NC, Quilliam LA. The RAP1 guanine nucleotide exchange factor Epac2 couples cyclic AMP and Ras signals at the plasma membrane. J Biol Chem. 2006;281:2506–14. doi: 10.1074/jbc.M508165200. [DOI] [PubMed] [Google Scholar]
- 35.Rebhun JF, Castro AF, Quilliam LA. Identification of guanine nucleotide exchange factors (GEFs) for the Rap1 GTPase. Regulation of MR-GEF by M-Ras-GTP interaction. J Biol Chem. 2000;275:34901–8. doi: 10.1074/jbc.M005327200. [DOI] [PubMed] [Google Scholar]
- 36.Pearson MA, Reczek D, Bretscher A, Karplus PA. Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell. 2000;101:259–70. doi: 10.1016/S0092-8674(00)80836-3. [DOI] [PubMed] [Google Scholar]
- 37.Nassar N, Horn G, Herrmann C, Block C, Janknecht R, Wittinghofer A. Ras/Rap effector specificity determined by charge reversal. Nat Struct Biol. 1996;3:723–9. doi: 10.1038/nsb0896-723. [DOI] [PubMed] [Google Scholar]
- 38.Aspenström P. Formin-binding proteins: modulators of formin-dependent actin polymerization. Biochim Biophys Acta. 2010;1803:174–82. doi: 10.1016/j.bbamcr.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 39.Schulte A, Stolp B, Schönichen A, Pylypenko O, Rak A, Fackler OT, et al. The human formin FHOD1 contains a bipartite structure of FH3 and GTPase-binding domains required for activation. Structure. 2008;16:1313–23. doi: 10.1016/j.str.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 40.Schönichen A, Alexander M, Gasteier JE, Cuesta FE, Fackler OT, Geyer M. Biochemical characterization of the diaphanous autoregulatory interaction in the formin homology protein FHOD1. J Biol Chem. 2006;281:5084–93. doi: 10.1074/jbc.M509226200. [DOI] [PubMed] [Google Scholar]
- 41.Lammers M, Rose R, Scrima A, Wittinghofer A. The regulation of mDia1 by autoinhibition and its release by Rho*GTP. EMBO J. 2005;24:4176–87. doi: 10.1038/sj.emboj.7600879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat Cell Biol. 1999;1:136–43. doi: 10.1038/11056. [DOI] [PubMed] [Google Scholar]
- 43.Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, et al. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 1997;16:3044–56. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alberts AS. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J Biol Chem. 2001;276:2824–30. doi: 10.1074/jbc.M006205200. [DOI] [PubMed] [Google Scholar]
- 45.Alberts AS. Diaphanous-related Formin homology proteins. Curr Biol. 2002;12:R796. doi: 10.1016/S0960-9822(02)01309-X. [DOI] [PubMed] [Google Scholar]
- 46.Otomo T, Otomo C, Tomchick DR, Machius M, Rosen MK. Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol Cell. 2005;18:273–81. doi: 10.1016/j.molcel.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 47.Takeya R, Taniguchi K, Narumiya S, Sumimoto H. The mammalian formin FHOD1 is activated through phosphorylation by ROCK and mediates thrombin-induced stress fibre formation in endothelial cells. EMBO J. 2008;27:618–28. doi: 10.1038/emboj.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laurin M, Fradet N, Blangy A, Hall A, Vuori K, Côté JF. The atypical Rac activator Dock180 (Dock1) regulates myoblast fusion in vivo. Proc Natl Acad Sci USA. 2008;105:15446–51. doi: 10.1073/pnas.0805546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanematsu F, Hirashima M, Laurin M, Takii R, Nishikimi A, Kitajima K, et al. DOCK180 is a Rac activator that regulates cardiovascular development by acting downstream of CXCR4. Circ Res. 2010;107:1102–5. doi: 10.1161/CIRCRESAHA.110.223388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elliott MR, Zheng S, Park D, Woodson RI, Reardon MA, Juncadella IJ, et al. Unexpected requirement for ELMO1 in clearance of apoptotic germ cells in vivo. Nature. 2010;467:333–7. doi: 10.1038/nature09356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu Z, Elliott MR, Chen Y, Walsh JT, Klibanov AL, Ravichandran KS, et al. Phagocytic activity of neuronal progenitors regulates adult neurogenesis. Nat Cell Biol. 2011;13:1076–83. doi: 10.1038/ncb2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park D, Tosello-Trampont AC, Elliott MR, Lu M, Haney LB, Ma Z, et al. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature. 2007;450:430–4. doi: 10.1038/nature06329. [DOI] [PubMed] [Google Scholar]
- 53.Ho E, Irvine T, Vilk GJ, Lajoie G, Ravichandran KS, D’Souza SJ, et al. Integrin-linked kinase interactions with ELMO2 modulate cell polarity. Mol Biol Cell. 2009;20:3033–43. doi: 10.1091/mbc.E09-01-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katoh H, Negishi M. RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo. Nature. 2003;424:461–4. doi: 10.1038/nature01817. [DOI] [PubMed] [Google Scholar]
- 55.Patel M, Margaron Y, Fradet N, Yang Q, Wilkes B, Bouvier M, et al. An evolutionarily conserved autoinhibitory molecular switch in ELMO proteins regulates Rac signaling. Curr Biol. 2010;20:2021–7. doi: 10.1016/j.cub.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patel M, Chiang TC, Tran V, Lee FJ, Côté JF. The Arf family GTPase Arl4A complexes with ELMO proteins to promote actin cytoskeleton remodeling and reveals a versatile Ras-binding domain in the ELMO proteins family. J Biol Chem. 2011;286:38969–79. doi: 10.1074/jbc.M111.274191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heo WD, Meyer T. Switch-of-function mutants based on morphology classification of Ras superfamily small GTPases. Cell. 2003;113:315–28. doi: 10.1016/S0092-8674(03)00315-5. [DOI] [PubMed] [Google Scholar]
- 58.Yang HW, Shin MG, Lee S, Kim JR, Park WS, Cho KH, et al. Cooperative activation of PI3K by Ras and Rho family small GTPases. Mol Cell. 2012;47:281–90. doi: 10.1016/j.molcel.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grimsley CM, Lu M, Haney LB, Kinchen JM, Ravichandran KS. Characterization of a novel interaction between ELMO1 and ERM proteins. J Biol Chem. 2006;281:5928–37. doi: 10.1074/jbc.M510647200. [DOI] [PubMed] [Google Scholar]
- 60.Handa Y, Suzuki M, Ohya K, Iwai H, Ishijima N, Koleske AJ, et al. Shigella IpgB1 promotes bacterial entry through the ELMO-Dock180 machinery. Nat Cell Biol. 2007;9:121–8. doi: 10.1038/ncb1526. [DOI] [PubMed] [Google Scholar]
- 61.Liu M, Horowitz A. A PDZ-binding motif as a critical determinant of Rho guanine exchange factor function and cell phenotype. Mol Biol Cell. 2006;17:1880–7. doi: 10.1091/mbc.E06-01-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marx R, Henderson J, Wang J, Baraban JM. Tech: a RhoA GEF selectively expressed in hippocampal and cortical neurons. J Neurochem. 2005;92:850–8. doi: 10.1111/j.1471-4159.2004.02930.x. [DOI] [PubMed] [Google Scholar]
- 63.De Toledo M, Coulon V, Schmidt S, Fort P, Blangy A. The gene for a new brain specific RhoA exchange factor maps to the highly unstable chromosomal region 1p36.2-1p36.3. Oncogene. 2001;20:7307–17. doi: 10.1038/sj.onc.1204921. [DOI] [PubMed] [Google Scholar]
- 64.Goh LL, Manser E. The RhoA GEF Syx is a target of Rnd3 and regulated via a Raf1-like ubiquitin-related domain. PLoS ONE. 2010;5:e12409. doi: 10.1371/journal.pone.0012409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chardin P. Function and regulation of Rnd proteins. Nat Rev Mol Cell Biol. 2006;7:54–62. doi: 10.1038/nrm1788. [DOI] [PubMed] [Google Scholar]
- 66.Foster R, Hu KQ, Lu Y, Nolan KM, Thissen J, Settleman J. Identification of a novel human Rho protein with unusual properties: GTPase deficiency and in vivo farnesylation. Mol Cell Biol. 1996;16:2689–99. doi: 10.1128/mcb.16.6.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goh LL, Manser E. The GTPase-deficient Rnd proteins are stabilized by their effectors. J Biol Chem. 2012;287:31311–20. doi: 10.1074/jbc.M111.327056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Malliri A, van der Kammen RA, Clark K, van der Valk M, Michiels F, Collard JG. Mice deficient in the Rac activator Tiam1 are resistant to Ras-induced skin tumours. Nature. 2002;417:867–71. doi: 10.1038/nature00848. [DOI] [PubMed] [Google Scholar]
- 69.Rajala RV, Rajala A, Gupta VK. Conservation and divergence of Grb7 family of Ras-binding domains. Protein Cell. 2012;3:60–70. doi: 10.1007/s13238-012-2001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li X, Zhang R, Draheim KM, Liu W, Calderwood DA, Boggon TJ. Structural basis for small G protein effector interaction of Ras-related protein 1 (Rap1) and adaptor protein Krev interaction trapped 1 (KRIT1) J Biol Chem. 2012;287:22317–27. doi: 10.1074/jbc.M112.361295. [DOI] [PMC free article] [PubMed] [Google Scholar]