Abstract
As a central regulator for cell cycle arrest, apoptosis, and cellular senescence, p53 requires multiple layers of regulatory control to ensure correct temporal and spatial functions. It is well accepted that Mdm2-mediated ubiquitination plays a crucial role in p53 regulation. In addition to proteasome-mediated degradation, ubiquitination of p53 by Mdm2 acts a key signal for its nuclear export. Nuclear export has previously been thought to require the disassociation of the p53 tetramer and exposure of the intrinsic nuclear export signal. To elucidate the molecular mechanism of degradation-independent repression on p53 by Mdm2, we have developed a two-step approach to purify ubiquitinated forms of p53 induced by Mdm2 from human cells. Surprisingly, however, we found that ubiquitination has no effect on the tetramerization/oligomerization of p53, arguing against this seemingly well accepted model. Moreover, nuclear export of p53 alone is not sufficient to completely abolish p53 activity. Ubiquitination-mediated repression of p53 by Mdm2 acts at least, in part, through inhibiting the sequence-specific DNA binding activity. Thus, our results have important implications regarding the mechanisms by which Mdm2 acts on p53.
The p53 tumor suppressor is a critical regulator of many cellular functions including cell growth arrest, apoptosis, and cellular senescence (1). Its importance is underscored by the observation that it is frequently mutated in ~40–50% of all human tumors (2– 4). p53 protein levels within the cell are controlled predominantly through the ubiquitin-proteasome pathway, and several E3 ubiquitin ligases have been described as having specificity for p53 (5). However, the predominant regulator of p53 levels remains Mdm2, a RING E32 ubiquitin ligase that specifically ubiquitinates and degrades p53 to maintain the protein at low levels during normal cellular resting conditions. Upon DNA damage events and other types of stress stimuli, p53 is quickly stabilized and activated. The exact mechanisms leading to p53 stabilization remain poorly understood, although Mdm2 destabilization as well as post-translational modifications on p53 is thought to play a role (6).
The ubiquitin-proteasome pathway consists of El-activating enzymes, E2-conjugating enzymes, and E3 ubiquitin ligases (7). In the case of p53, Mdm2 acts as the specific E3 ubiquitin ligase for p53 by mediating the transfer of an ubiquitin moiety from E2-ubiquitin to the p53 substrate. Mdm2 has the capability of catalyzing both monoubiquitination and polyubiquitination of p53, and the particular choice for one or the other has been shown to be dependent on the levels of Mdm2 (8). When Mdm2 levels are low, Mdm2 preferentially catalyzes monoubiquitination of p53. When the levels are high, p53 polyubiquitination occurs. The fates of these different ubiquitinated forms of p53 have significantly different consequences as well. Monoubiquitination acts as a signal for p53 nuclear export, whereas polyubiquitinated p53 is quickly and efficiently degraded by nuclear 26 S proteasomes.
Recent studies have shown that ubiquitination, in particular monoubiquitination, serves as an important occurrence for a variety of cellular functions including transcriptional activation, protein-protein interactions, and intracellular localization (9). The movement of proteins between various cellular compartments is an important mechanism for functional regulation. Movement of p53 from the nucleus to the cytoplasm not only removes it from its transcriptional targets, but it also allows for further post-translational modifications to occur. In addition, moving p53 to the cytoplasm places it near mitochondria where its transcription-independent pro-apoptotic functions can take place (10–12).
The well accepted model for the nuclear export of p53 requires the dissociation of the tetramer and exposure of the nuclear export sequence for accessibility and recognition by nuclear export machinery such as CRM1. We have shown previously that monoubiquitination is a signal for nuclear export. Here, we have expanded on this notion and shown for the first time that monoubiquitination of p53 has no effect on its ability to tetramerize. These data are significant conclusions in the mechanistic studies of p53 nuclear export, as they show that dissociation of the p53 tetramer as a mechanism for NES exposure is not necessary for efficient nuclear export.
Although it is clear that monoubiquitination is an important event for mediating the nuclear export of p53, it is unclear if monoubiquitination of p53 has any effect on its function as a transcriptional activator. In this study, we have developed a two-step approach for purifying pure ubiquitinated p53 from cells. Interestingly, purified monoubiquitinated p53 retains its ability to tetramerize but loses transcriptional activity. The loss of transcriptional activity stems from the inability of p53 to bind DNA and activate target promoters.
EXPERIMENTAL PROCEDURES
Plasmid Construction
To construct the p53, p53-UB, Mdm2, p21-Luc, and TK-Luc expression vectors, the DNA sequences corresponding to the full-length proteins were amplified by PCR from Marathon-Ready HeLa cDNA (Clontech) or other templates, and subcloned either into pCIN4 or pcDNA3.1/V5-His-TOPO vector (Invitrogen) for expression in mammalian cells. The FLAG and His6 sequences were introduced at the N terminus of p53 and UB, respectively, by PCR. To make the p53-UB fusion construct, p53 was amplified from a pCIN4 template and subcloned into a pCIN4-UB construct lacking the ubiquitin start codon. The p53Δ325–354 construct was a generous gift from Z. Ronai.
Protein Purification of Monoubiquitinated p53
To isolate monoubiquitinated p53 from cells, FLAG-p53 and His-UB were transfected into three 15-cm plates containing 293 cells at 70% confluence. 24 h post-transfection, the cells were split 1:4 and incubated for an additional 48 h. The cells were then harvested on ice and lysed in 6 m guanidine buffer (6 m guanidine HCl, 0.1 m Na2PO4/NaH2PO4, 0.01 m Tris/HCl, pH 8.0, 10 mm β-mercaptoethanol, 5 mm imidazole, 0.2% Triton X-100, and fresh protease inhibitors) with mild sonication. Ni2+-nitrilotriacetic acid-agarose beads (Qiagen) were then added and lysates were rotated at room temperature overnight. The beads were then washed for 5 min with each of the following buffers as previously described (13) with some modifications: 6 m guanidinium HCl, 0.1 m Na2PO4/NaH2PO4, 0.01 m Tris/HCl, pH 8.0, 10 mm β-mercaptoethanol, 5 mm imidazole, 0.2% Triton X-100; 8 m urea, 0.1 m Na2PO4/NaH2PO4 0.01 m Tris/HCl, pH 8.0, 10 mm β-mercaptoethanol, 5 mm imidazole; 8 m urea, 0.1 m Na2PO4/NaH2PO4 0.01 Tris/HCl, pH 6.3, 10 mm β-mercaptoethanol, 5 mm imidazole; (BC100) 20 mm Tris/HCl, pH 7.3, 10% glycerol, 0.1 mm EDTA, and 100 mm NaCl. Ubiquitinated proteins were eluted using 500 mm imidazole at room temperature. The elutions were pooled and dialyzed overnight against BC100 with several changes of the dialysis buffer, and then subjected to a second round of purification with anti-FLAG monoclonal antibody-conjugated agarose beads (M2, Sigma). These beads were then eluted twice using FLAG peptide and the eluates pooled for purified monoubiquitinated p53.
Gel-filtration Chromatography
Purification and size fractionation was performed on a Superose-12 column using the SMART/FPLC system (GE Healthcare). For fractionation of the fusion constructs, H1299 cells were transiently transfected with p53wt, p53-UB, P531–290, or p53Δ325–254 and harvested on ice 24 h post-transfection. The cells were then lysed with FLAG lysis buffer for 1 h, cleared by centrifugation, and dialyzed against hypotonic buffer (10 mm Tris/HCl, pH 7.3, 10 mm KCl, and 1.5 mm MgCl2) overnight. The dialyzed protein was then passed on the column and 40-µl fractions collected. The samples were resolved on SDS-PAGE and analyzed by Western blots using the α-p53 monoclonal antibody (DO-1) (Santa Cruz).
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was performed essentially as described (14) with some modifications. The long probe (220 bp) was generated by PCR with 5′-tgctgcctgcttcccaggaaca-3′ (sense) and 5′-ccatccccttcctcac-ctgaaa-3′ (antisense) primers. The probe was 32P end-labeled by using T4 polynucleotide kinase and then purified by placing it on a Bio-Spin P-30 column (Bio-Rad). The binding reactions (20 µl) contained 20 mm Tris/HCl, pH 7.3, 50 mm KCl, 5 mm MgCl2, 1 mm dithiothreitol, 0.5 mm EDTA, 10% glycerol, 0.5 mg/ml bovine serum albumin (BSA), 15 ng of salmon sperm DNA, and proteins as indicated. The reaction mixtures were incubated for 20 min at room temperature before adding the 32P-labeled DNA probe and further incubated for an additional 20 min. Samples were then resolved on a native 4% polyacrylamide gel at 4 °C followed by autoradiography.
ImmunofluorescentStaining
For immunofluorescent staining of transiently transfected fusion proteins, p53wt or p53-Ub were transfected into the p53-null H1299 cell line grown on coverslips in 6-well plates. 24 h post-transfection, the cells were fixed with 4% paraformaldehyde for 15 min at 37 °C and then rehydrated for 5 min at room temperature with serum-free Dulbecco’s modified Eagle’s medium (Cellgro). The cells were then permeablized using 0.1% Triton X-100 (Fisher) for 10 min at room temperature. Cells were then incubated with 1% BSA (Sigma)/Dulbecco’s phosphate-buffered salt solution (DPBS) (Cellgro) for 1 h. Primary antibody DO-1 (Santa Cruz) in 1% BSA/DPBS was incubated with the cells for 1 h at room temperature and then washed with 1% BSA/DPBS for 20 min. Alexa 488-conjugated anti-mouse antibody (Molecular Probes) was added and incubated for 30 min at room temperature. Finally, after a 15-min 1% BSA/DPBS wash, the cells were counterstained with DAPI in the last wash to visualize the nuclei.
In Vitro Protein Cross-linking Assay
H1299 cells were transiently transfected with p53wt, p53-UB, p53-GFP, or p53Δ325–354 and harvested on ice 24 h post-transfection. After lysis with FLAG lysis buffer for 1 h, equal protein amounts were incubated with 0, 0.01, or 0.1% glutaraldehyde (Fisher) on ice for 30 min. The samples were then boiled with SDS sample buffer and subsequently resolved on 7% SDS-PAGE for Western blot analysis with the anti-p53 monoclonal (DO-1) antibody.
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assay was performed essentially as described with some modifications (15). 1.0 × 107 H1299 cells were transfected with p53wt, p53-UB, or p53-GFP, incubated for 24 h, and cross-linked with 1% formaldehyde for 10 min at room temperature. The cross-linking reaction was stopped by the addition of 0.125 m glycine and the cells were harvested in SDS lysis buffer (0.5% SDS, 100 mm NaCl, 50 mm Tris/HCl, pH 8.0, 5 mm EDTA, and protease inhibitors) and incubated at 4 °C for 10 min. After centrifugation, the cell pellet was resuspended in IP buffer (0.3% SDS, 100 mm NaCl, 50 mm Tris/HCl, pH 8.0, 5 mm EDTA, 2% Triton X-100, and protease inhibitors), followed by sonication to an average DNA length of 500–1000 bp. The samples were cleared by centrifugation, and pre-cleared with protein A-agarose beads pre-coated with salmon sperm DNA (Upstate) for 1 h at 4 °C. The samples were centrifuged and divided into 3 aliquots of 2 ml containing equal protein concentrations, with 30 µl being saved as input control. Antibodies were added to each sample and rotated at 4 °C overnight. 30 µl of protein A-agarose beads pre-coated with salmon sperm DNA were incubated with each sample for an additional 2 h at 4 °C. The beads were then washed three times with 1 ml of mixed Micelle wash buffer (150 mm NaCl, 20 mm Tris/HCl, pH 8.0, 5 mm EDTA, 5% sucrose, 1% Triton X-100, and 0.2% SDS), two times with 1 ml of buffer 500 (0.1% deoxycholic acid, 1 mm EDTA, 50 methanol HEPES, pH 7.5, 500 mm NaCl, and 1% Triton X-100), two times with 1 ml of LiCl/detergent buffer (0.5% deoxycholic acid, 1 mm EDTA, 250 mm LiCl, 0.5% Nonidet P-40, and 10 mm Tris/HCl), and 2 washes with TE buffer (10 mm Tris/Cl, pH 7.5, and 1 mm EDTA). The beads and input were incubated 2 times with 250 µl of elution buffer (1% SDS and 0.1% NaHCO3) for 30 min and incubated at 65 °C overnight to reverse the cross-links. The eluates were then incubated with Proteinase K and RNase for 1 h and extracted with phenol/chloroform followed by ethanol precipitation containing Pellet Paint Co-precipitant (Novagen) to visualize the pellet. After drying, the DNA was dissolved in TE buffer and analyzed by PCR. The full-length p53 polyclonal antibody (FL) (Santa Cruz) was used for all p53 samples. The primers for the PCR corresponded to the 5′ short p21 promoter and had the following sequences: 5′ -ctcacatcctccttct-tcag-3′ (sense) and 5′-cacacacagaatctgactccc-3′ (antisense). The input DNA was diluted 1:10 for the PCR. The PCR products were amplified for 32 cycles.
Western Blot Analysis of Endogenous p21 and Luciferase Reporter Assays
For Western blot analysis of p21 induction, p53-null H1299 cells were transfected with p53wt or p53-UB and harvested on ice 24 h post-transfection. After cell lysis using FLAG lysis buffer, 40 µg of total protein was boiled with SDS sample buffer and loaded on 15% SDS-PAGE gels, followed by Western blot analysis using the anti-p53 monoclonal (DO-l)(Santa Cruz), α-GFP monoclonal (Clontech), or α-p21 (C-19) polyclonal (Santa Cruz) antibodies. For the dual luciferase reporter assays, H1299 cells were transfected with the constructs indicated in the figure legends and included the Renilla luciferase reporter construct in each sample as an internal control. 24 h post-transfection, the cells were lysed in passive lysis buffer and luciferase activities were determined using a dual specific luciferase reporter assay kit (Promega).
RESULTS
Isolation of Pure Ubiquitinated p53 from Cells
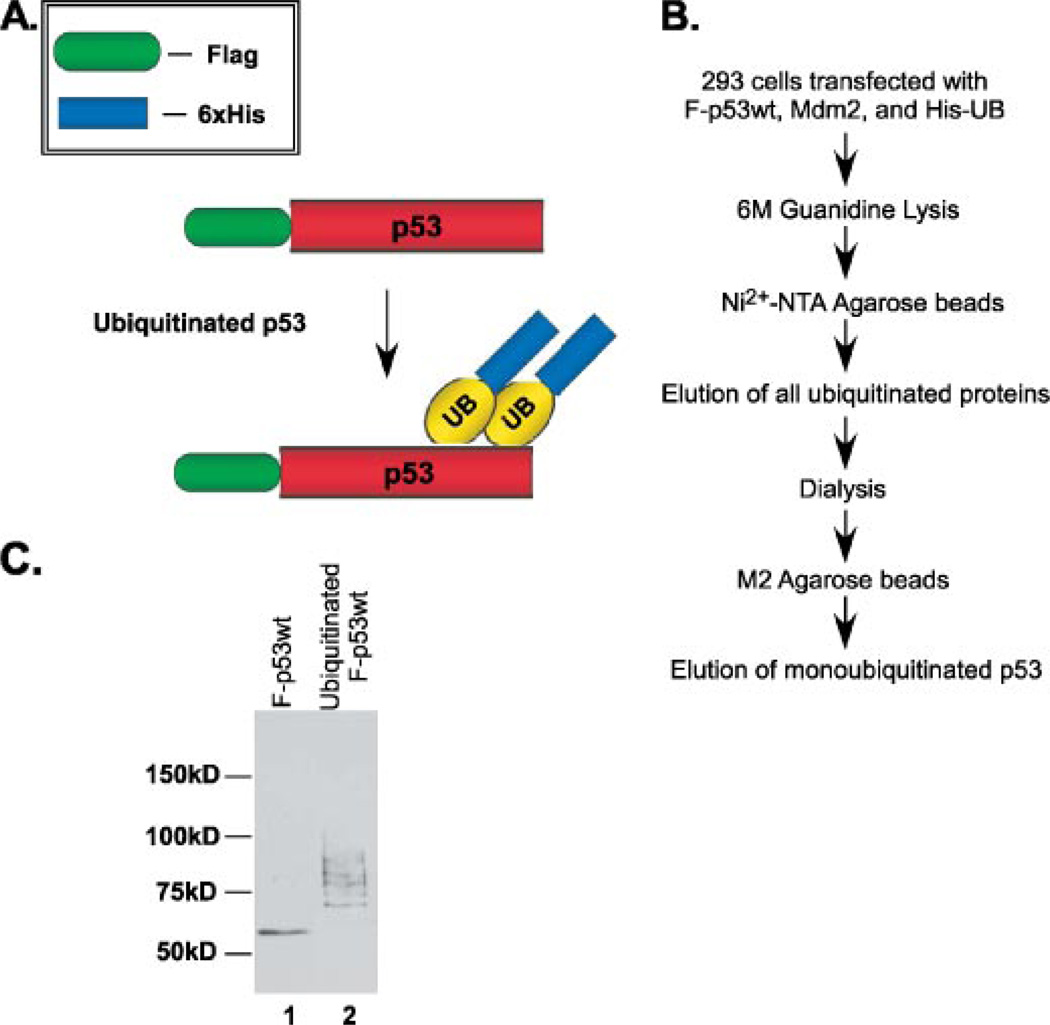
Monoubiquitination has been purported to be a key signal in the nuclear export of p53 and does not require Mdm2 to mediate the physical transfer from the nucleus to cytoplasm (8). It has also been reported that the nuclear export signal, a region located within the tetramerization domain of p53, needs to be “unmasked” in order for efficient nuclear export to occur (16). The unmasking of this domain would therefore occur when p53 was in the form of a monomer or dimer and not when in the tetramer formation. We wished to further explore this idea and the role that monoubiquitination might have in it. Particularly, we sought to isolate true monoubiquitinated p53 and assess its ability to tetramerize. To address this issue, we purified ubiquitinated p53 through a two-step purification method to enrich for monoubiquitinated p53. The first step isolated all ubiquitinated proteins and the second step refined the purification to p53-ubiquitinated protein. 293 cells were transiently transfected with FLAG-tagged p53wt, Mdm2, and His-tagged ubiquitin and subjected to two rounds of affinity purification (Fig. 1, A and B). First, all ubiquitinated protein was purified using Ni2+-nitrilotriacetic acid-agarose beads. The eluate was then dialyzed overnight against BC100. The dialysis procedure was critically important for removing all urea buffer and imidazole and to bring the ubiquitinated protein in balance with a buffer compatible with the M2 anti-FLAG beads. The dialyzed eluate was then subjected to a second round of affinity purification using anti-FLAG M2-agarose beads to purify ubiquitinated p53. Because the cells were not treated with 26 S proteosome inhibitors, the enriched population of purified, ubiquitinated p53 was true monoubiquitinated p53 because this particular form is not recognized and degraded by the 26 S proteasome (Fig. 1C, compare lane 1 versus lane 2). Using this method, polyubiquitinated protein would be freely degraded by the 26 S proteasome leaving an enriched population of monoubiquitinated p53. This double purification method using stringent conditions enables us to obtain a pure sample of the small population of monoubiquitinated p53 that exists among total ubiquitinated protein. Importantly, the purified, ubiquitinated p53 was 100% pure and did not contain any contamination from freeform p53 (Fig. 1C, compare lane 1 versus 2). The migration pattern on SDS-PAGE of the purified protein is similar to that of monoubiquitinated and multi-monoubiquitinated p53 as previously reported (Fig. 1C) (8).
FIGURE 1. Purification and fractionation of monoubiquitinated p53 from cells.
A, schematic representation of the method for purifying monoubiquitinated p53 from 293 cells. B, protocol for purifying monoubiquitinated p53. C, Western blot analysis of the purified monoubiquitinated p53. Cells were transfected with FLAG-p53, Mdm2, and His-UB and harvested 72 h post-transfection. Western blot analysis after 2 rounds of affinity purification (lane 2) was compared with wild type p53 control (lane 1) using anti-p53 (DO-1 antibody). The migration pattern seen (lane 2) represents monoubiquitination and multi-monoubiquitination.
Ubiquitinated p53 Proteins Can Still Form Tetramers
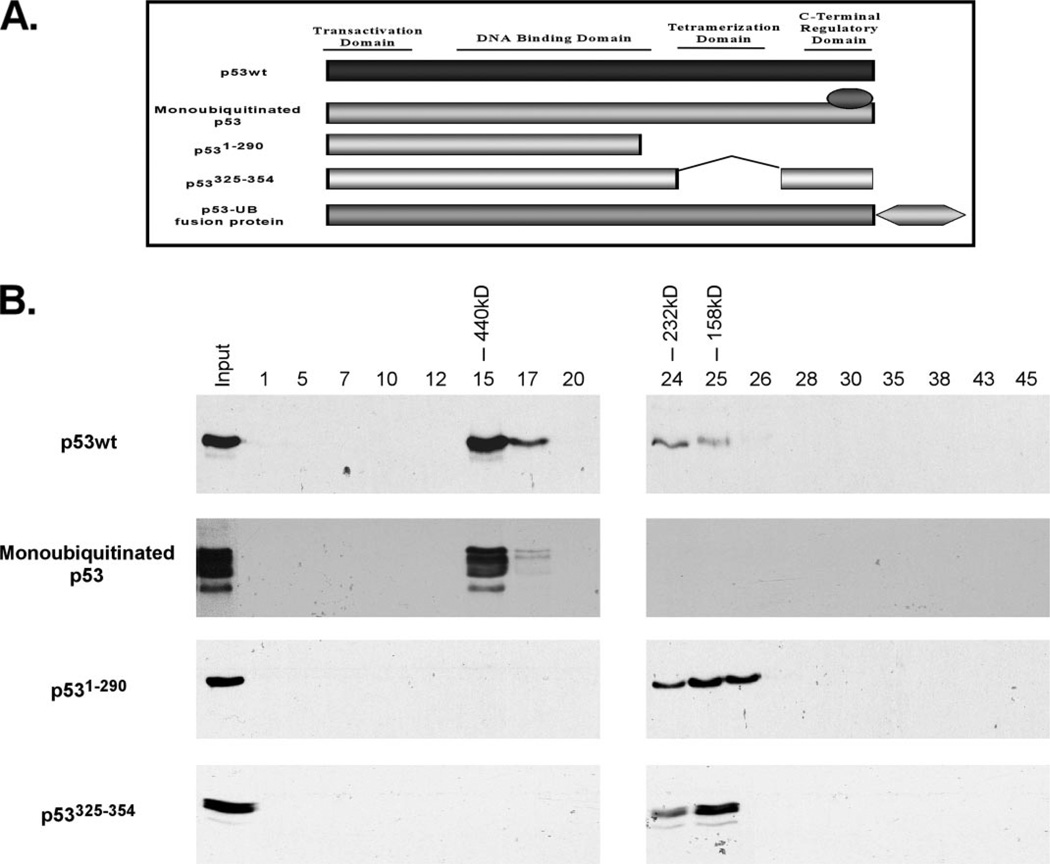
Once we had obtained a pure sample of monoubiquitinated p53, we wished to analyze its tetramerization properties. It has previously been reported that the tetramerization of p53, a requirement for its activity as a sequence-specific transcription factor, can be assessed using gel-filtration chromatography. When placed on a Superose-12 gel-filtration column using the SMART/HPLC system, wild type p53 in tetramer formation has been reported to elute in a fraction corresponding to 440 kDa when compared with known molecular mass standards including albumin, catalase, adolase, and ferritin (17).
Whereas the molecular mass of 440 kDa is not the expected size of a true p53 tetramer, which would be expected to be around 200 kDa, this validated observation has been reported to represent a multimeric species of p53 and occurs for both human and murine forms. Using these known standards for the p53 tetramer, we subjected our purified, ubiquitinated p53 protein to gel-filtration chromatography using a Superose-12 column. The eluted fractions were collected and then resolved by SDS-PAGE and Western blot using a p53-specific monoclonal antibody (DO-1) (Fig. 2B). The molecular weight pattern of purified, ubiquitinated p53 was compared with wild type FLAG-p53 that had been transfected into p53-null H1299 cells and purified by M2 anti-FLAG beads (Fig. 2B, second panel versus first panel). Our elution profile was compared with known size standards consisting of albumin, adolase, catalase, and ferritin (data not shown). The majority of purified, monoubiquitinated p53 protein eluted within fraction 15, which corresponds to 440 kDa based on the molecular size standard. Importantly, this pattern of size distribution matched that of p53wt, with the exception of a smaller amount of protein eluting with fractions 24–26, which corresponds to 232, 158, and 67 kDa, respectively. For monomeric p53 controls, we used two mutants that are unable to tetramerize due to a loss of the tetramerization domain: P531–290 and p53Δ325–354 (see Fig. 2A). These mutants were transfected into H1299 cells and purified using M2 beads. Equal amounts of purified protein were then placed on the column and subjected to SDS-PAGE and Western blot. The controls eluted within the fractions corresponding to the molecular size of monomeric p53, representing dimer and monomer fractions of p53 that collectively match the pattern of size distribution for p53wt previously reported (17) (Fig. 2B). Together, these results indicated that monoubiquitinated p53 is capable of forming tetramers in vivo. In addition, the molecular mass elution pattern of ubiquitinated p53 matched that of wild type p53 within the fraction corresponding to 440 kDa, indicating that monoubiquitinated p53 can also form multimeric species as previously reported for wild type 53.
FIGURE 2. Schematic representation of the proteins used in the SMART/HPLC chromatography.
A, p53 is divided into four functional domains: N-terminal transactivation domain (amino acid 1–42), DNA binding domain (amino acids 98–292), tetramerization domain (amino acids 325–254), and C-terminal regulatory domain (amino acids 363–393). The diagram indicates the following proteins used: wild type p53; purified, monoubiquitinated p53; p531–290 deletion mutant; p53Δ325–354 tetramerization domain mutant; monoubiquitination-mimicking p53 fusion protein; and the p53-GFP C-terminal fusion protein. B, monoubiquitinated p53 purified from cells can form tetramers. Western blot analysis of chromatographic fractions generated by the gel-filtration (Superose 12, SMART/FPLC system) of p53wt, monoubiquitinated p53, p531–290, and p53Δ325–354 immunoblotted with anti-p53 (DO-1) monoclonal antibody.
The Monoubiquitination-mimicking p53 Fusion Protein Can Form Tetramers
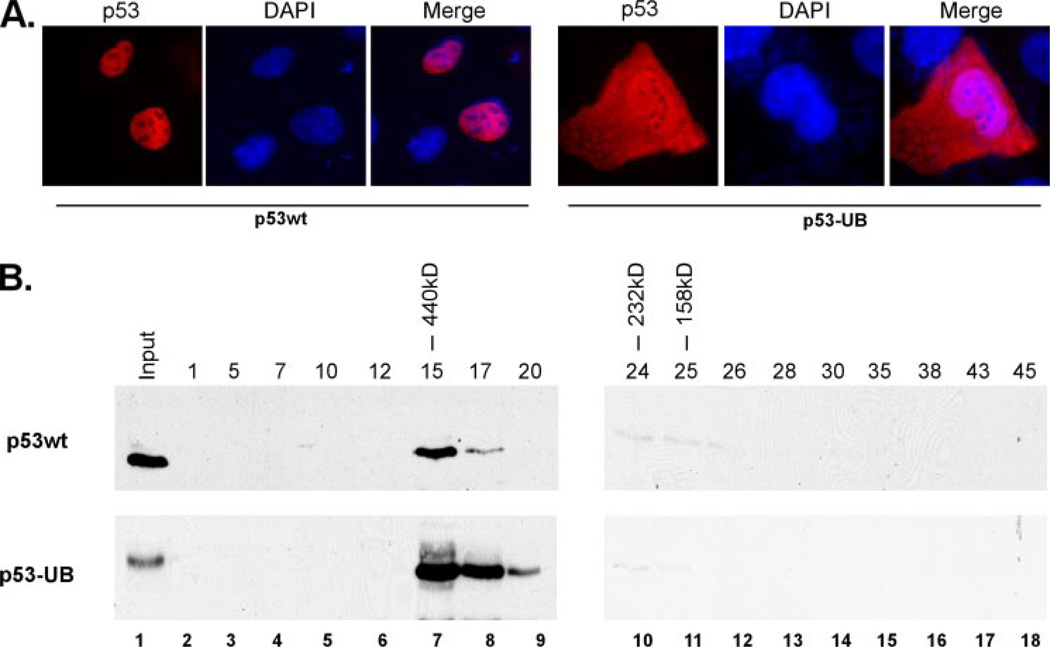
We next wanted to examine the tetramerization ability and function of monoubiquitinated p53 more closely. However, the purification of truly monoubiquitinated p53 protein from cells proved very challenging, and further the use of it in biochemical assays was difficult due to technical limitations. To help us better address these issues, we decided to use a monoubiquitination-mimicking p53 fusion protein that has been used in previous studies and shown to mimic the effects of post-translational ubiquitination (Fig. 2A) (8, 18). The major Mdm2-mediated ubiquitination sites of p53 are located at the C terminus (19–21), so we designed a molecule that mimics monoubiquitinated p53 though the C-terminal fusion of an ubiquitin moiety. This particular approach has been shown to alter the subcellular distribution of p53 and the yeast α-factor in a manner reminiscent of post-translational monoubiquitination (8, 18). Indeed, expression of this construct in H1299 cells showed a significant cytoplasmic distribution when compared with wild type p53, although a proportion was retained within the nucleus, similar to that previously reported (Fig. 3A) (8). Using this p53 fusion protein, we wished to validate our results with the purified, monoubiquitinated p53 by assessing the tetramerization ability of the fusion protein. To do this, p53-null H1299 cells were transiently transfected with constructs expressing the FLAG-tagged p53wt or the p53 ubiquitin fusion protein and affinity purified using anti-FLAG-agarose beads. The purified protein was then subjected to gel-filtration chromatography using a Superose-12 column in a similar fashion as described above and the eluted fractions resolved by SDS-PAGE and Western blot using the anti-p53 (DO-1) monoclonal antibody (Fig. 3B). When the monoubiquitination-mimicking p53 fusion protein was subjected to the column, the molecular weight elution pattern matched that of wild type p53, indicating that monoubiquitinated p53 has the ability of oligomerizing in vivo (Fig. 3B, compare lane 7 top panel versus bottom panel). These results indicate that the monoubiquitination-mimicking p53 fusion protein could readily form tetramers in vivo in a similar fashion to that of wild type p53. These data also support the findings of the tetramerization abilities of purified, monoubiquitinated p53 from cells and argue against the hypothesis that ubiquitination disrupts p53 tetramer formation.
FIGURE 3. The monoubiquitination-mimicking p53 fusion protein can form tetramers.
A, the monoubiquitinated p53-mimicking fusion protein (p53-UB) has approximately a 60:40% distribution between the cytoplasm and nucleus. Immunofluorescent staining of p53wt and p53-UB transfected in the p53-null H1299 cell line using anti-p53 (DO-1) antibody, counterstained with DAPI to visualize the nuclei. B, the monoubiquitinated-mimicking p53 fusion proteins can form tetramers. Western blot analysis of chromatographic fractions (lanes 2–18) generated by the gel filtration (Superose 12, SMART/FPLC system) of p53wt and p53-UB fusion protein transfected into H1299 cells and immunoblotted with anti-p53 (DO-1) monoclonal antibody.
Confirmation of the Tetramerization Ability of the Monoubiquitination-mimicking p53 Fusion Protein Using an Oligomerization Assay
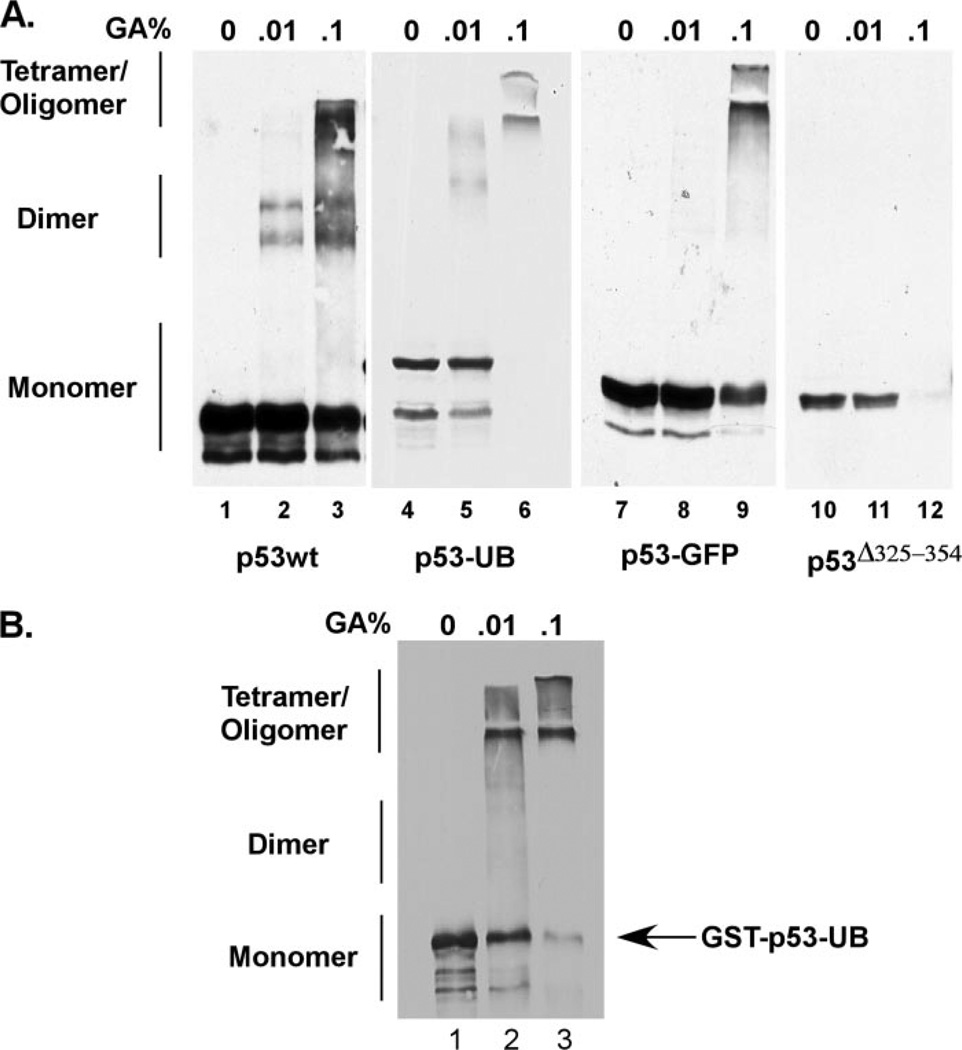
We next wanted to expand our tetramerization analysis further for monoubiquitinated p53. To do this, we took an in vitro approach to assess the ability of the monoubiquitination-mimicking p53 fusion protein to form oligomers in an in vitro oligomerization assay. p53-null H1299 cells were transiently transfected with constructs expressing p53 wild type, the monoubiquitination-mimicking p53 fusion protein, p53 protein fused to GFP, or p53Δ325–254. 24 h post-transfection, the cells were harvested on ice and lysed with the FLAG lysis buffer. The lysates were then incubated with an increasing percentage of gluteraldehyde to covalently cross-link any dimers or tetramers that formed. After incubation, equal protein amounts were resolved by SDS-PAGE and Western blot using the DO-1 antibody (Fig. 5A). Interestingly, the monoubiquitinated p53 fusion protein formed both dimer and tetramer/oligomer populations in a similar pattern as that of p53wt (Fig. 4A, compare lanes 1–3 to 4–6). We used a p53 protein fused with GFP at the C terminus as a control (Fig. 2) to ensure that the addition of a nonspecific bulky adduct did not disrupt tetramer formation (Fig. 4A, lanes 7–9). A mutant p53 protein that lacks the tetramerization domain was included as a negative control and showed no slower migrating species on Western blot (Fig. 4A, lanes 10–12). To further confirm these findings, purified GST-p53-UB protein was also used in the in vitro oligomerization assay (Fig. 4B). This protein also showed a gel migration shift with increasing amounts of gluteraldehyde representing dimers and tetramers/oligomers of the protein. Taken together, the in vitro oligomerization assay indicated that the monoubiquitination-mimicking p53 fusion protein was capable of forming dimers and oligomers in vitro in a similar fashion to that of wild type p53.
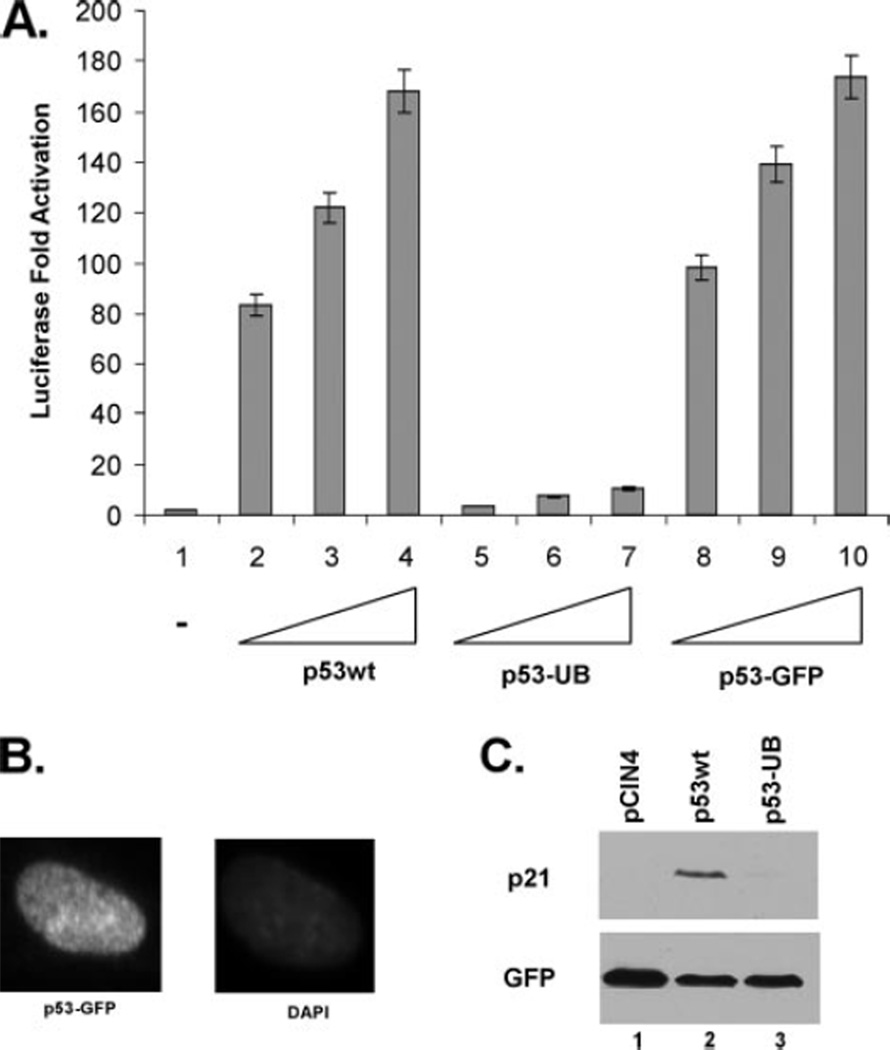
FIGURE 5. Monoubiquitination of p53 inhibits its transactivation activity and DNA binding in vivo.
A, monoubiquitinated p53 impairs its transactivation ability for p21 induction. Dual luciferase reporter assay using the p21 reporter construct in H1299 cells transfected with p53wt (lanes 2–4), p53-UB (lanes 5–7), or p53-GFP (lanes 8–10). Results are shown as -fold activation over the control (lane 1) averaging three independent experiments. B, the control p53-GFP exclusively localizes to the nucleus. The p53-null H1 299 cell line was transfected with p53-GFP and immunofluorescence was done 24 h post-transfection showing GFP signal exclusively within the nucleus. C, p53 monoubiquitination impairs endogenous p21 induction. Western blot analysis of p21 expression in H1299 cells with pCIN4 control (lane 1), p53wt (lane 2), or p53-UB fusion protein (lane 3) immunoblotted withanti-p21 (C19) polyclonal antibody and anti-GFP monoclonal antibody as a transfection control.
FIGURE 4. The monoubiquitination-mimicking p53 fusion protein can form tetramers in an oligomerization assay.
A equal amounts of total protein were incubated with 0, 0.01 %, and 0.1% gluteraldehyde on ice for 30 min and resolved on a 7% SDS-PAGEgel followed by Western blot using anti-p53 (DO-1) antibody. B, the in vitro oligomerization assay was repeated using purified GST-p53-UB protein incubated with increasing amounts of gluteraldehyde and resolved on a 7% SDS-PAGE followed by Western blot using DO-1.
A Fraction of the Monoubiquitination-mimicking p53 Fusion Proteins Are in the Nucleus but Remain Transcriptionally Inactive
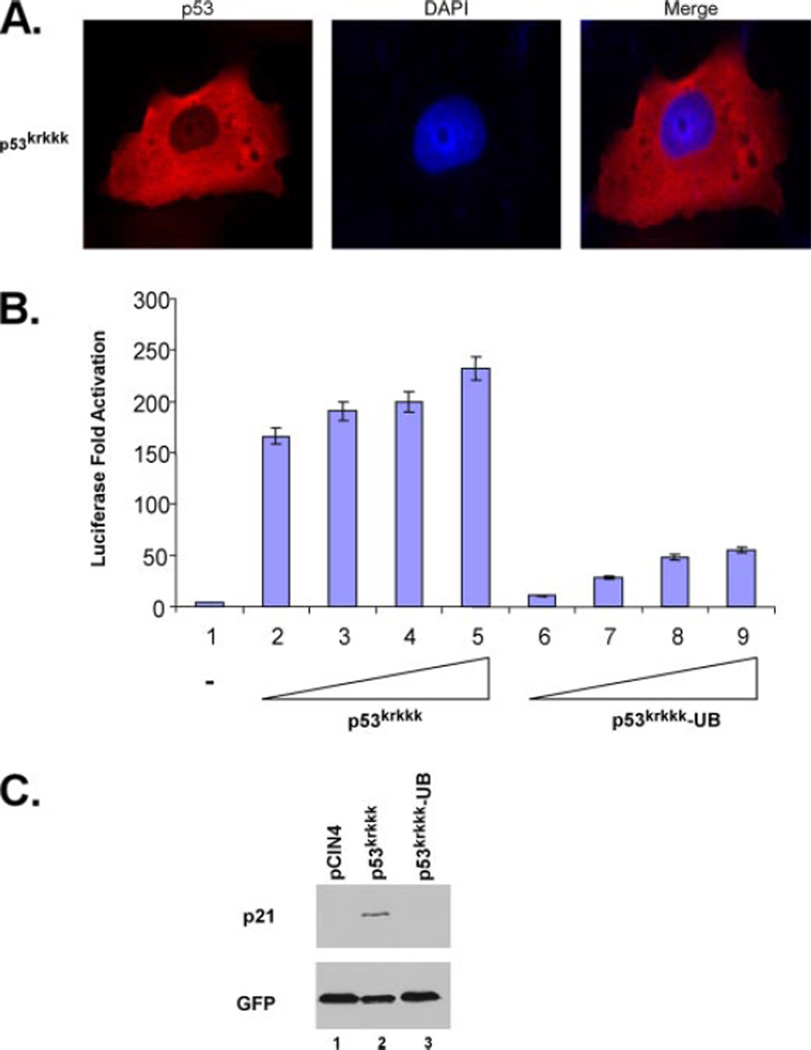
The majority of monoubiquitinated p53 is effectively transported out of the nucleus into the cytoplasm and away from its genetic targets (8).3 However, we have consistently seen a significant amount of the monoubiquitination-mimicking p53 fusion protein retained in the nucleus despite a shift in the subcellular localization for a majority of the protein (Fig. 3A, compare left panel with right panel). It has recently been reported that E4F1-mediated ubiquitination of p53 at lysine 320 positively promotes transcriptional activation (22). Because of this and the significant amount of the monoubiquitination-mimicking p53 fusion protein remaining in the nucleus, we were interested in assessing the transcriptional activity of monoubiquitinated p53. As p53 is a unique transcription factor in that it can exist in the cell as a tetramer without the presence of DNA, the possibility exists that despite it having no effect on the tetramerization ability, monoubiquitination may be an important regulatory step in p53 function through as yet undefined mechanisms. To assess this, p53-null H1299 cells were transfected with a luciferase reporter construct (p21-Luc) that contains a p53 responsive element from within the p21 5′ short promoter, and luciferase activity was measured as -fold activation relative to control (Fig. 6A). Surprisingly, comparison of p53wt transcriptional activity on the p21 promoter to that of the monoubiquitination-mimicking p53 fusion protein showed a significant inhibition of p21-Luc transcription with the fusion protein (Fig. 5A, compare lanes 2–4 with 5 and 6). Importantly, p53 protein fused to GFP remained transcriptionally active at comparative levels to p53 wild type, indicating that the observations seen were specific to ubiquitin and not due to the addition of a nonspecific bulky adduct (Fig. 5A, lanes 8–10). As shown in Fig. 5B, the p53-GFP protein is exclusively within the nucleus based on immunofluorescence. To confirm the validity of these reporter assays, in vivo endogenous gene expression was also analyzed. Overexpression of p53 in H1299 cells induces the expression of the p53-responsive gene p21 that can be readily assessed by Western blot. These cells were transfected with pCIN4, p53wt, or the p53-ubiquitin fusion construct and harvested 24 h post-transfection. SDS-PAGE and Western blot were then performed using the DO-1 antibody and showed robust induction of the p21 gene when p53wt is expressed, but no induction when the p53 fusion protein is expressed (Fig. 5C, compare lanes 2 and 3 to lane 1). These data indicate that the fusion of ubiquitin to the C terminus of p53 in a manner to mimic monoubiquitinated p53 significantly inhibits the transactivation activity of p53 both in a luciferase reporter assay and in the induction of the endogenous p21 gene.
FIGURE 6. The p53krkkk construct mimics the subcellular localization of the monoubiquitination-mimicking p53 fusion protein and retains transcriptional activity.
A, immunofluorescent staining of p53wt and p53krkkk expressed in H1299 cells using anti-p53 (DO-1) antibody and DAPI as a counterstain to visualize the nuclei. B, mimicking monoubiquitination of p53krkkk impairs its transcriptional activity. Dual luciferase reporter assay showing -fold activation of the p21 reporter construct using p53krkkk (lanes 2–5) or p53krkkk-UB fusion protein (lanes 6–9) compared with control (lane 1). Three independent experiments are indicated here. C, fusion of a C-terminal ubiquitin monomer to the p53krkkk mutant impairs endogenous p21 induction. Western blot analysis of p21 expression in H1299 cells with pCIN4 control (lane 1), p53wt (lane 2), or p53krkkk-UB fusion protein (lane 3) immunoblotted with anti-p21 (C19) polyclonal antibody and anti-GFP monoclonal antibody as a transfection control.
Nuclear Export Is Not Sufficient for Inhibiting p53 Transcriptional Activity
Our results using the p53-UB C-terminal fusion suggested that that addition of ubiquitin to p53 induced nuclear export and inhibited its transactivation activity, considering that a significant proportion remained nuclear. However, the observation that a proportion of expressed p53-UB protein localized to the cytoplasm prevented us from excluding the possibility that the transcriptional inhibition could be a result of its localization within the cytoplasm. Despite the retention of a significant proportion of p53-UB within the nucleus, the lack of transcriptional functions of monoubiquitinated p53 could be due to its altered subcellular localization within the cytoplasm and away from its genetic targets. To address this issue, we searched for a p53 mutant that mimicked the subcellular localization of p53-UB but still retained at least some transcriptional activity. One particular mutant, p53krkkk, contains 5 mutations within the bipartite NLS sequence of p53 (K305A, R306A, K319A, K320A, K321A) and localizes within the cytoplasm when expressed in H1299 cells at a similar percentage as p53-UB (Fig. 6A). To assess the transcriptional activity of this mutant, H1299 cells were transfected with the p21-Luc luciferase reporter construct and p53krkkk (Fig. 6B). The luciferase activity of this mutant had similar dose-dependent activity as that of p53wt. These results indicate that the nuclear export of p53 is not sufficient for inhibiting its transcriptional activity. This is most likely due to the homeostatic shuttling balance of p53 between the nucleus and cytoplasm. To then address transcriptional activity of monoubiquitinated p53krkkk, a C-terminal fusion construct was designed in a similar fashion as p53-UB. Surprisingly, addition of one ubiquitin moiety also greatly reduced the transcriptional activity of p53krkkk when compared with the mutant without ubiquitin (Fig. 6B). To validate these data, the in vivo induction of p21 was assessed using both p53krkkk and p53krkkk-UB (Fig. 6C). p53krkkk was able to induce the expression of p21 after overexpression at a similar level to that of p53wt, whereas monoubiquitinated p53krkkk failed to induce p21, again similar to the findings with monoubiquitinated p53wt. Taken together, these data suggest that nuclear export is not sufficient for inhibition of p53 transcriptional activity. Furthermore, the addition of a single ubiquitin monomer greatly inhibits the transactivation ability of p53.
Mdm2-mediated Ubiquitination Can Inhibit p53 DNA Binding in Vivo
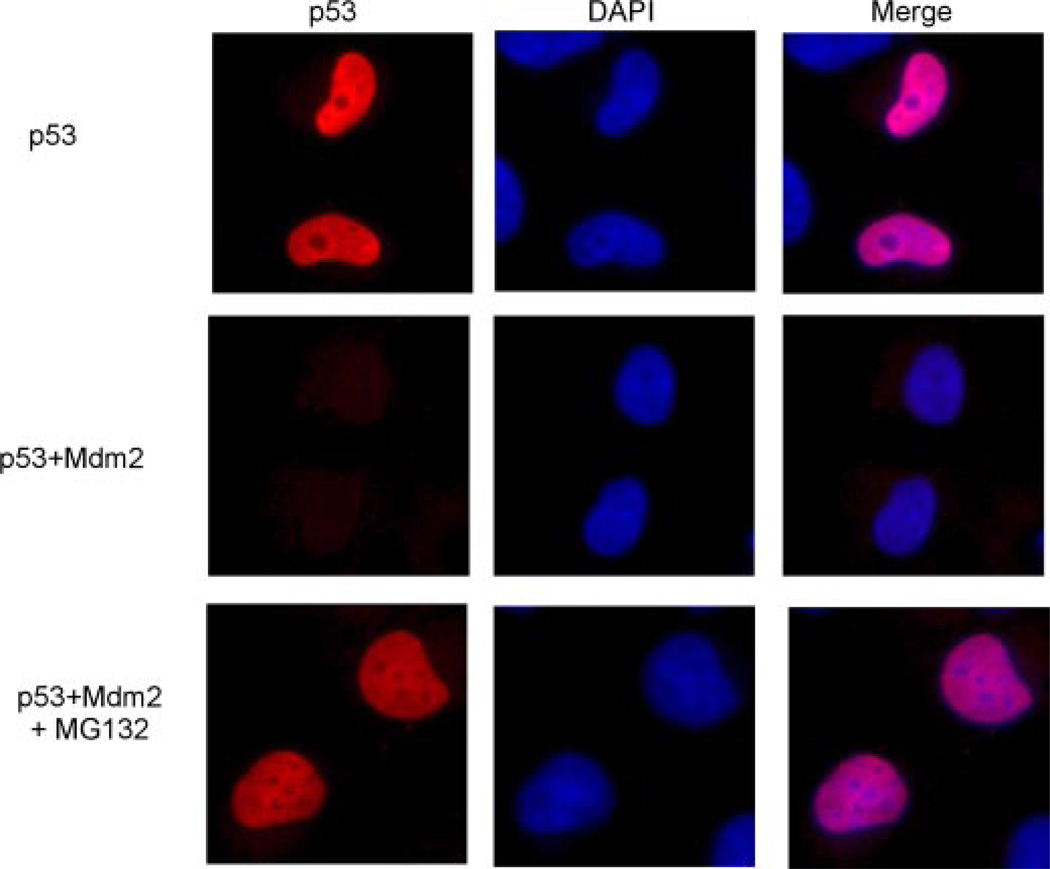
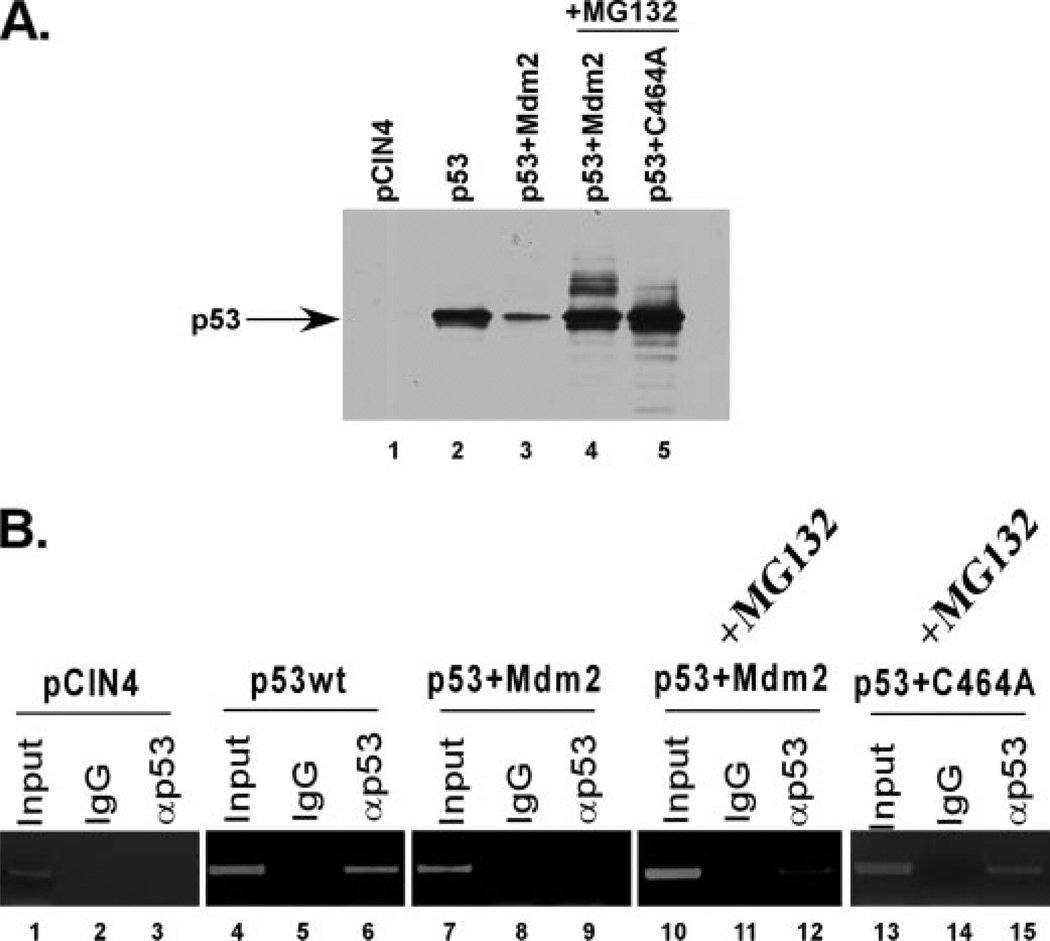
We next wanted to assess the observed transcriptional repression under more physiological conditions. Because the monoubiquitination-mimicking p53 fusion protein was transcriptionally inactive, and because it was not a direct result of nuclear exclusion as shown by the p53krkk-UB mutant, we hypothesized that Mdm2-mediated monoubiquitination of p53 in vivo may also inhibit the transcriptional activity possibly through the inhibition of DNA binding. To first confirm the localization of p53 when co-expressed with Mdm2, we performed a series of immunofluorescence assays (Fig. 7). H1299 cells were transfected with p53, p53 and Mdm2, or p53 and Mdm2 in the presence of the proteasome inhibitor MG132. As shown in the top panel, p53 localizes exclusively to the nucleus. However, when expressed with Mdm2, p53 is completely degraded and undetectable (Fig. 7, middle panel). Upon treatment with MG132, p53 protein is restored and remains in the nucleus, suggesting that Mdm2-mediated ubiquitination of p53 occurs predominantly within the nucleus. Because of this, we next used ChIP experiments to assess whether Mdm2-dependent ubiquitination of p53 had an effect on its DNA binding ability of the 5′ p21 short promoter region. ChIP was performed using p53, p53 and Mdm2, or p53 and an inactive form of Mdm2 (C464A) with or without proteasome inhibitor treatment (Fig. 8A). We hypothesized that without proteasome inhibitor treatment, p53 would be degraded to a protein level insufficient for promoter binding in the ChIP (Fig. 8A, compare lanes 2, 3, and 4). However, treatment with MG132 would allow for the accumulation of a sufficient amount of ubiquitinated p53 protein for the assay. As shown in Fig. 8B, in the presence of Mdm2, p53 fails to bind to the endogenous p21 promoter at equivalent protein levels as wild type (Fig. 8, B, compare lanes 6 and 9, and compare lanes 6 and 12, respectively). Importantly, this inhibition is Mdm2 and ubiquitination dependent, as the catalytically inactive Mdm2 mutant (C464A) does not inhibit the binding (Fig. 8B, lane 15). These results indicate that Mdm2-mediated ubiquitination of p53 inhibits in vivo binding to the p21 target promoter.
FIGURE 7. p53 localizes to the nucleus and is degraded by Mdm2 within this cellular compartment.
H1299 cells were transfected with p53, p53 + Mdm2, or p53 + Mdm2 in the presence of the proteasome inhibitor MG132. Immunofluorescent staining is shown of p53 using DO-1 and DAPI as a counterstain to visualize the nucleus.
FIGURE 8. Mdm2 can inhibit p53 DNA binding in vivo.
A, Western blot analysis of p53 inputs taken from the ChIP experiment in Fig. 9B. 8% SDS-PAGE was performed with pCIN4 (lane 1), p53 (lane 2), p53 and Mdm2 (lane 3), p53 and Mdm2 in the presence of the proteasome inhibitor MG132 (lane 4), or p53 and Mdm2(C464A) (lane 5) immunoblotted with DO-1. B, ChIP assay using anti-p53 (FL)(lanes 3, 6, 9, 12, and 15) or IgG (lanes 2, 5, 8, 11, and 14) compared with input DNA (lanes 1, 4, 7, 10, and 13).
The Monoubiquitination-mimicking p53 Fusion Protein Fails to Bind p53 Target Promoters in Vivo
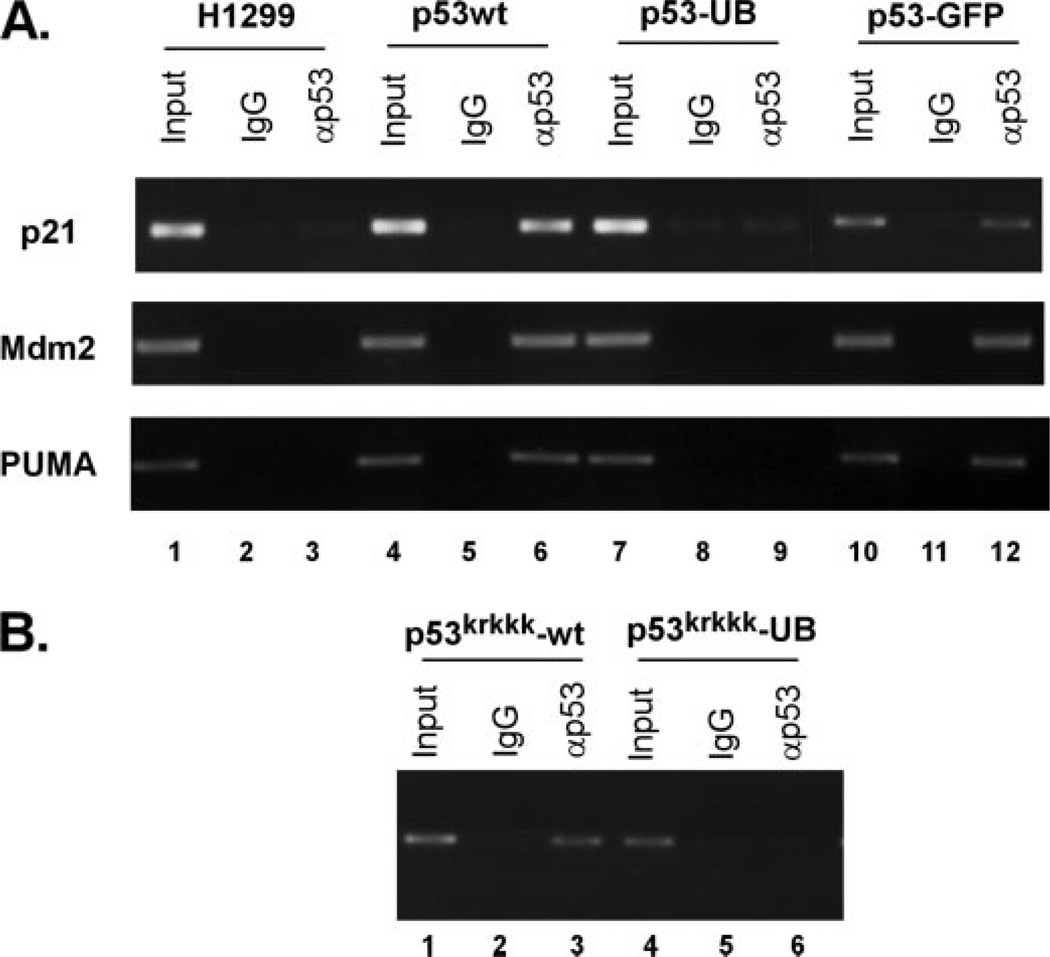
The fusion of a C-terminal ubiquitin moiety on p53 had a profound effect on its activity as a sequence-specific transcription factor. Because of this, we wished to assess whether the monoubiquitination-mimicking p53 fusion protein had a similar defect in DNA binding as when p53 and Mdm2 were co-expressed above. To do this, we performed ChIP assays to answer the question of what effect monoubiquitination had on the DNA binding ability of p53 in vivo. Using the 5′ p21 short promoter region as a target, ChIP was performed by using either wild type p53 or the p53-ubiquitin fusion protein and performing the immunoprecipitation with full-length p53 polyclonal antibody (Fig. 9A). Although p53 wild type efficiently bound the endogenous p21 promoter, the monoubiquitination-mimicking p53 fusion protein failed to do so when compared with wild type p53 (Fig. 9A, top panel, compare lanes 4–6 to 7–9). The use of p53 fused with GFP at the C terminus as a control indicates that the observations seen using the monoubiquitination-mimicking p53 protein are specific for ubiquitination and not other nonspecific additions (Fig. 9A, top panel, lanes 10–12). We also performed ChIP assays using other p53-responsive promoters such as mdm2 and PUMA to assess whether these observations were gene specific or applied generally to p53-responsive genes (Fig. 9A, middle and bottom panels). The monoubiquitination-mimicking p53 fusion protein also failed to bind to the endogenous promoters of mdm2 and PUMA when compared with wild type p53 (Fig. 9A, middle and bottom panels, compare lanes 4–6 to 7–9). The ChIP assays therefore indicate that the monoubiquitination-mimicking p53 fusion protein fails to bind p53-responsive promoters in vivo and supports the transcriptional repression as well as the Mdm2-mediated DNA binding defect for p53 seen above. Use of this fusion protein as a way of imitating the effects of monoubiquitinated p53 in cells suggests that monoubiquitination of p53 represses its transcriptional activity by inhibiting site-specific DNA binding.
FIGURE 9. Monoubiquitination of p53 impairs its site-specific DNA binding in vivo.
A, ChIP assay using anti-p53 (FL) (lanes 3, 6, 9, and 12) or IgG (lanes 2, 5, 8, and 11) compared with input DNA (lanes 1, 4, 7, and 10). The p53-GFP C-terminal fusion protein was included as a control. ChIP was performed on the promoters of three p53 responsive genes including p21, mdm2, and PUMA. B, monoubiquitination of p53krkkk impairs DNA binding in vivo. ChIP assay using anti-p53 (FL) antibody (lanes 3 and 6) or IgG (lanes 2 and 5) compared with total DNA input (lanes 1 and 4).
We then wished to confirm that the inhibition of transactivator activity observed for p53krkkk-UB was indeed do to a loss of DNA binding similar to that of p53-UB. A ChIP assay was performed again using the 5′ p21 short promoter region as a target. H1299 cells were transfected with p53krkkk and p53krkkk-UB constructs and after cross-linking, subjected to immunoprecipitation using the full-length p53 antibody (Fig. 9B). In a manner similar to the p53-UB fusion protein, monoubiquitination blocked the DNA binding ability of p53krkkk when compared with p53wt control (Fig. 9B, compare lanes 1–3 and 4–6). The ChIP assays therefore indicate that monoubiquitination of p53 blocks its ability to bind to the p21 short promoter and therefore suggested that monoubiquitinated p53 may have an effect on its transactivation activities.
Ubiquitination of p53 Inhibits Site-specific DNA Binding
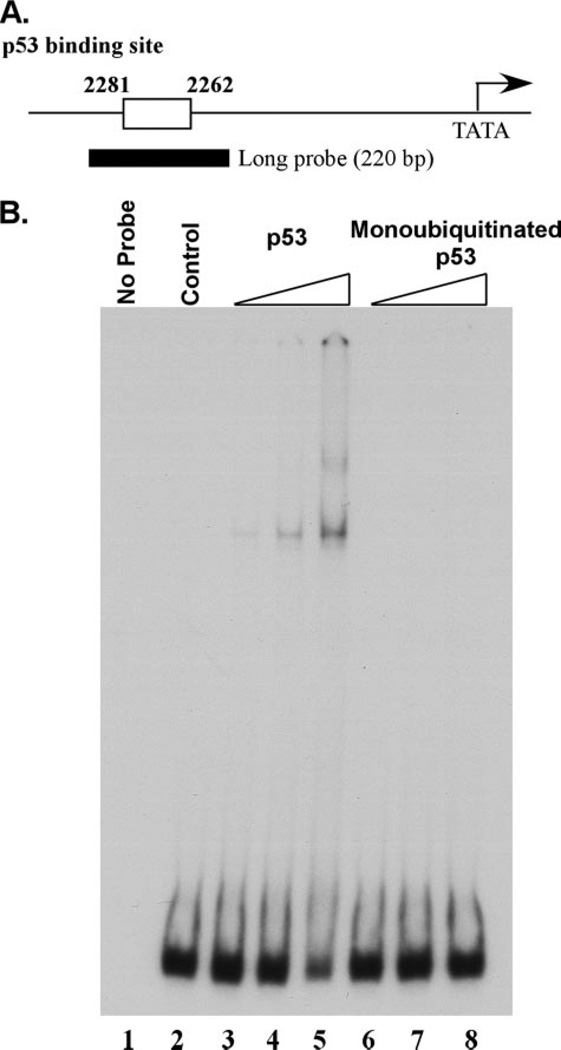
Use of the monoubiquitination-mimicking p53 fusion protein suggested that monoubiquitination repressed site-specific DNA binding in vivo. To further address this, we wished to use purified, monoubiquitinated p53 from cells in a DNA binding assay in vitro. The technical challenges of using true monoubiquitinated protein in an in vivo assay, as stated above, lead us to use purified, monoubiquitinated p53 in an in vitro system. To examine this, we used an established EMSA for p53 that has been previously used in assessing p53 site-specific binding (14, 23, 24). We tested the DNA binding ability of purified, monoubiquitinated p53 on a 220-bp long DNA fragment derived from the p21 promoter region (Fig. 10A (23). Equal protein amounts of wild type p53 and monoubiquitinated p53 protein were incubated with probe and resolved on a 4% PAGE gel followed by autoradiography (Fig. 10B). The purified, wild type p53 protein showed site-specific DNA binding by EMSA in a dose-dependent manner (Fig. 10B, lanes 3–5). However, purified monoubiquitinated p53 protein failed to bind to the same DNA fragment when incubated at equivalent amounts (Fig. 10B, lanes 6–8). These data together with the ChIP observations suggest that monoubiquitination of p53 causes transcriptional repression through the inhibition of DNA binding. The use of truly monoubiquitinated p53 purified from cells in this in vitro DNA binding assay supports the observations seen with the monoubiquitination-mimicking p53 fusion protein in vivo and provides significant support for the effects of p53 monoubiquitination, despite the technical challenges encountered when addressing these questions.
FIGURE 10. Monoubiquitination of p53 inhibits its DNA binding activity in vitro.
A, schematic representation of the long probe in the p21 promoter region used for EMSA. B, EMSA of wild type p53 (lanes 2–5) and purified, monoubiquitinated p53 (lanes 6–8) using the long probe with increasing amounts of protein.
DISCUSSION
The mechanism controlling p53 nuclear export has been the focus of intense study for some time and has resulted in the solidification of three main hypotheses: Mdm2-dependent p53 export (25, 26), Mdm2-independent autonomous p53 export (16, 27), and ubiquitination-dependent p53 export (28, 29). Consistent with several studies from others, our recent study provides the direct evidence that monoubiquitination act as a key signal in the nuclear export of p53 (8). The question that remains is what the precise mechanism is by which monoubiquitination mediates this event. Molecules that are meant for export contain a nuclear export signal (NES) and are recognized by the export receptor CRM1. CRM1 is a member of the karyo-pherin-β family of receptor proteins and has been proposed to be the key factor that mediates p53 export (30). p53 contains two nuclear export sequences, one in the N terminus and one in the C terminus of the protein (31, 32). It has been shown that nuclear export of p53 is augmented by ectopic expression of the export protein CRM1 (19); conversely, leptomycin B inhibits the function of CRM1 and also blocks nuclear export of p53 (16, 31). Interestingly, oligomerization of p53, which occurs through the C-terminal domain, may obscure the C-terminal NES and inhibit nuclear export (16). Our previous study demonstrates that monoubiquitination is a key signal in the nuclear export of p53 and does not require Mdm2 to mediate the physical transfer from the nucleus to cytoplasm (8). It has also been proposed that the nuclear export signal, a region located within the tetramerization domain of p53, needs to be unmasked for efficient nuclear export to occur (16, 32). The unmasking of this domain would therefore occur when p53 was in the form of a monomer or dimer but not when in the tetramer formation. It is therefore possible that the monoubiquitination of p53 may affect its oligomerization, thus revealing the NES for CRM1 binding. However, to test this hypothesis, it is crucial to obtain pure ubiquitinated p53 protein. Purification of this protein population is technically challenging, making it a very difficult task. Here, we have established a new method for isolating pure monoubiquitinated forms of p53 and have used them to assess the ability of ubiquitinated p53 protein to tetramerize. Surprisingly, however, we found that ubiquitinated p53 protein apparently forms stable tetramers/oligomers. This was confirmed using several different approaches and shows that monoubiquitination has no effect on p53 tetramerization. Furthermore, the striking change in subcellular localization of monoubiquitinated p53 suggests that dissociation of the p53 tetramer for efficient nuclear export is not required. Thus, we demonstrate that this seemly well accepted model may not be the case and that ubiquitination-mediated effects on nuclear export are more complicated than once thought. Nevertheless, it is still possible that ubiquitination of p53 directly modulates the interactions with the nuclear export machinery protein CRM1 to facilitate nuclear export of p53. It is also possible that monoubiquitination causes a change in the structural conformation of the p53 tetramer resulting in exposure of the NES for CRM1 binding or recruitment of other factors without tetramer dissociation. Indeed, recent findings suggest that dissociation of the p53 tetramer is not required for efficient nuclear export and that ubiquitination contributes to the exposure of the C-terminal NES, enhanced sumoylation of the C terminus of p53, and dissociation of Mdm2 (33). These data in combination with our findings suggest a mechanism for nuclear export whereby monoubiquitination of p53 leads to the exposure of the C-terminal NES and further C-terminal modifications that result of p53 nuclear export without the need for tetramer dissociation.
Mdm2 is a critical and specific repressor of p53 function and controls precise regulation of the protein during times of cellular stress as well as resting homeostasis. Whereas p53 regulation is at least, in part, through the degradation-dependent mechanisms of Mdm2, degradation-independent mechanisms also exist for controlling the transcriptional activation abilities of p53 (22, 34, 35). For example, nuclear export of p53-mediated but Mdm2-dependent ubiquitination is a significant and well accepted degradation-independent mechanism for regulating p53 as it moves the protein out of the nucleus and away from critical transcriptional targets (8, 16, 25–29, 31). Still, other mechanism must exist because a significant proportion of p53 remains in the nucleus but is transcriptionally repressed by Mdm2. Our results here show that the inhibition of p53 activity not only occurs as a result of nuclear export, but also occurs through the direct inhibition of its DNA binding ability in an ubiquitination-dependent but degradation-independent manner. This was further confirmed with the use of the p53krkkk mutant that mimics the subcellular distribution of the p53-UB C-terminal fusion protein. These results suggest that nuclear export is not sufficient for inhibiting p53 transcriptional activity as this mutant retains transcriptional activity despite the fact that a majority of the protein is localized to the cytoplasm. Importantly, addition of an ubiquitin monomer to this mutant severely inhibits its transcriptional activity, further supporting the notion that the transcriptional repression seen with the addition of ubiquitin to p53 is not a result of nuclear exclusion; rather, monoubiquitination has a direct inhibition on the DNA binding and transcriptional activity of p53. This novel mechanism may be very important for repressing the transcriptional activity of p53 when p53 cannot be exported out of the nucleus under certain cellular conditions. Movement into the cytoplasm is a critical step for separating p53 from its genetic targets, and the inhibition of its activity may represent an additional layer of functional regulation to ensure that it remains inactive when not needed.
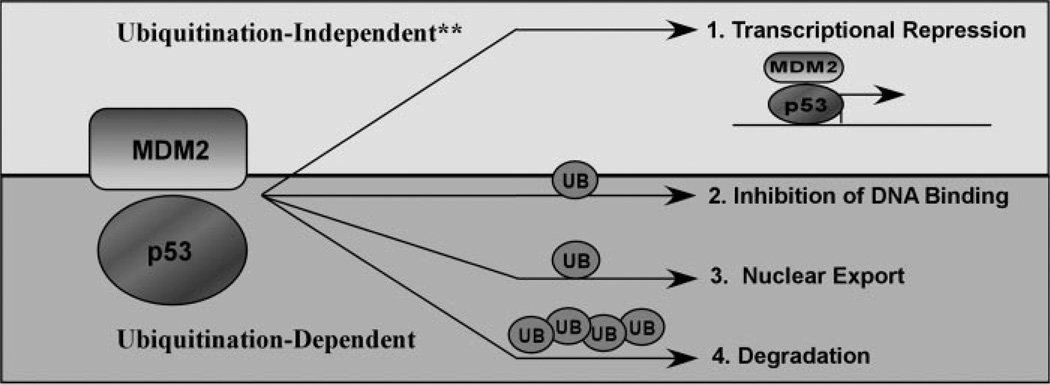
It is well accepted that Mdm2 not only induces p53 degradation but also acts as a repressor of p53-mediated transcription, at least in part through the nuclear export of p53 (8). Interestingly, based on this study, we have found that Mdm2-mediated ubiquitination of p53 also inhibits its site-specific DNA binding. Other known E3 ubiquitin ligases of p53, namely COP1, Pirh2, and ARF-BP1, have only been shown to facilitate p53 degradation but not transcriptional repression (5). Another p53 E3 ligase, E4F1, has even been shown to positively promote p53-mediated transcription through the ubiquitination of lysine 320 (22). Therefore, ubiquitination-mediated transcriptional repression shown here may be specific for Mdm2-mediated effects on p53. Our in vivo data using ChIP assays indicate that Mdm2-mediated ubiquitination of p53 does severely inhibit its DNA binding ability. Importantly, stability of p53 protein in the presence of Mdm2 through the use of the proteasome inhibitor MG132 still does not rescue the DNA binding defect, indicating that the observations seen are Mdm2-mediated effects and not due to degradation of p53. Use of the catalytically inactive mutant of Mdm2 confirms that the DNA binding deficiency seen is a result of Mdm2-mediated ubiquitination and not just the interaction with Mdm2. It is interesting to note that Mdm2 and p53 form a complex on the promoters of specific p53-responsive genes and facilitates transcriptional repression (34, 35). However, it is unlikely that this form of p53 is ubiquitinated, as Mdm2-mediated ubiquitination of p53 would block the DNA binding ability and be released from the promoter. Mdm2 has been shown to inhibit p53 function through both ubiquitination-dependent and -independent mechanisms (Fig. 11) (36, 37). Further studies will elucidate the precise mechanisms involved.
FIGURE 11. Model of Mdm2 functions on p53.
The predominant mechanisms for p53 regulation by Mdm2 include monoubiquitination-dependent inhibition of DNA binding (2), nuclear export (3), and polyubiquitination dependent degradation (4). Asterisks, in addition, ubiquitination-independent transcriptional repression can occur (1) and this may also result from ubiquitination of other substrates such as H2A and H2B (37).
The importance of monoubiquitination for post-translational regulation is becoming more apparent for varying cellular functions (38, 39). Monoubiquitination is an important regulatory step for diverse mechanisms such as protein-protein interactions, transcriptional activation, and protein internalization. It has also been shown to regulate the functions of other transcription factors such as FOXO4 and DNA repair proteins and FANCD2 and PCNA (40–44). It is clear that 26 S proteasome-mediated degradation requires a polyubiquitin chain signal consisting of at least 4 ubiquitin molecules, and therefore the striking effects of the addition of one ubiquitin moiety on p53 indicates a potentially important layer of regulatory complexity in the p53 pathway (45). Monoubiquitination of p53 not only has drastic effects on its subcellular localization, but our data indicate that it also has important consequences for its function as well. The addition of ubiquitin blocks the transactivation activity of p53 but does not alter its ability to tetramerize. As p53 is a unique transcription factor in that it can exist in the cell as a tetramer without the presence of DNA, the possibility exists that monoubiquitination is an important regulatory step through as yet undefined mechanisms. If p53 does indeed exist as a balance of forms between tetramers and monomers/dimers, a regulatory mechanism would need to exist to prevent p53 tetramers from binding DNA and activating p53 responsive genes. Monoubiquitination provides an avenue for regulating these forms of p53 within the cell. And, because these observations are specific for monoubiquitination and not from the addition of other bulky adducts, monoubiquitination could be acting as the specific signal for the inhibition of p53 activity. p53 is a key transcription factor for inducing cell growth arrest and apoptosis, and there have been several E3 ubiquitin ligases described for p53 to date (46–49). Therefore, it is not unexpected that there are multiple layers of pathway regulation. Monoubiquitination could represent a critical mechanism within the ubiquitin-proteasome pathway, independent of protein degradation, for the inhibition of p53 activity. Importantly, monoubiquitination is not a mechanism for blocking the tetramerization of p53 nor does it block the nuclear export of p53 as a tetramer. The blockage of DNA binding indicates monoubiquitination may have important roles in regulating p53 that are independent of the ubiquitin-proteasome pathway.
In summary, we have developed a novel two-step method for isolating pure, ubiquitinated p53 protein for the purpose of analyzing its functional abilities. The well accepted model for nuclear export suggests that dissociation of the p53 tetramer and exposure of the NES is an obligate step for efficient nuclear export to occur; however, our data challenges this notion by showing not only that Mdm2-mediated monoubiquitination is a key signal for nuclear export, but that it also has no effect on p53 tetramerization. Importantly, this degradation independent function of Mdm2-mediated ubiquitination causes p53 transcriptional repression for the population of the protein that remains in the nucleus. Because controlling p53 transcriptional activity is of vital importance for cell cycle regulation, this degradation-independent function of ubiquitin provides a novel mechanism for repressing p53 in the nucleus under conditions where it cannot be exported out. Elucidating the functional consequences of p53 monoubiquitination in vivo is technically challenging due to the difficulty in isolating the protein. To aid this approach we have used a molecule that mimics p53 monoubiquitination to support these findings in vivo. Although this fusion protein is not an exact representation of in vivo monoubiquitinated p53, it functionally behaves in a similar manner as purified monoubiquitinated p53 protein and has been previously reported to mimic post-translational monoubiquitination of the yeast α-factor (18). Our data indicates that monoubiquitination of p53 in this regard inhibits its sequence-specific DNA binding and leads to transcriptional repression. Further studies are needed to analyze the exact mechanism for this observed repression.
Footnotes
The abbreviations used are: E3, ubiquitin-protein isopeptide ligase; El, ubiquitin-activating enzyme; E2, ubiquitin-conjugating protein; EMSA, electrophoretic mobility shift assay; NES, nuclear export signal; UB, ubiquitin; BSA, bovine serum albumin; DPBS, Dulbecco’s phosphate-buffered salt; DAPI, 4’,6-diamidino-2-phenylindole;GFP, green fluorescent protein; ChIP, chromatin immunoprecipitation.
C. L Brooks and W. Gu, unpublished data.
REFERENCES
- 1.Vogelstein B, Lane D, Levine AJ. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Caron de Fromentel C, Soussi T. Genes Chromosomes Cancer. 1992;4:1–15. doi: 10.1002/gcc.2870040102. [DOI] [PubMed] [Google Scholar]
- 3.Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 4.Chao C, Hergenhahn M, Kaeser MD, Wu Z, Saito S, Iggo R, Hollstein M, Appella E, Xu Y. J. Biol. Chem. 2003;278:41028–41033. doi: 10.1074/jbc.M306938200. [DOI] [PubMed] [Google Scholar]
- 5.Brooks CL, Gu W. Mol. Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stommel JM, Wahl GM. EMBO J. 2004;23:1547–1556. doi: 10.1038/sj.emboj.7600145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laney JD, Hochstrasser M. Cell. 1999;97:427–430. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- 8.Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 9.Sigismund S, Polo S, Di Fiore PP. Curr. Top. Microbiol. Immunol. 2004;286:149–185. doi: 10.1007/978-3-540-69494-6_6. [DOI] [PubMed] [Google Scholar]
- 10.Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR. Science. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 11.Leu JI, Dumont P, Hafey M, Murphy ME, George DL. Nat. Cell Biol. 2004;6:443–450. doi: 10.1038/ncb1123. [DOI] [PubMed] [Google Scholar]
- 12.Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. Mol. Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 13.Xirodimas DP, Stephen CW, Lane DP. Exp. Cell Res. 2001;270:66–77. doi: 10.1006/excr.2001.5314. [DOI] [PubMed] [Google Scholar]
- 14.Gu W, Roeder RG. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 15.Weinmann AS, Bartley SM, Zhang T, Zhang MQ, Farnham PJ. Mol. Cell. Biol. 2001;21:6820–6832. doi: 10.1128/MCB.21.20.6820-6832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman PN, Chen X, Bargonetti J, Prives C. Proc. Natl. Acad. Sci. U. S.A. 1993;90:3319–3323. doi: 10.1073/pnas.90.8.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terrell J, Shih S, Dunn R, Hicke L. Mol. Cell. 1998;1:193–202. doi: 10.1016/s1097-2765(00)80020-9. [DOI] [PubMed] [Google Scholar]
- 19.Lohrum MA, Woods DB, Ludwig RL, Balint E, Vousden KH. Mol. Cell. Biol. 2001;21:8521–8532. doi: 10.1128/MCB.21.24.8521-8532.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura S, Roth JA, Mukhopadhyay T. Mol. Cell. Biol. 2000;20:9391–9398. doi: 10.1128/mcb.20.24.9391-9398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez MS, Desterro JM, Lain S, Lane DP, Hay RT. Mol. Cell. Biol. 2000;20:8458–8467. doi: 10.1128/mcb.20.22.8458-8467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Cam L, Linares Lk, Paul C, Julien E, Lacroix M, Hatchi E, Triboulet R, Bossis G, Shmueli A, Rodriguez MS, Coux O, Sardet C. Cell. 2006;127:775–788. doi: 10.1016/j.cell.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 23.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 24.Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, Gu W. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2259–2264. doi: 10.1073/pnas.0308762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roth J, Dobbelstein M, Freedman DA, Shenk T, Levine AJ. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tao W, Levine AJ. Proc. Natl. Acad. Sci. U.S.A. 1999;96:3077–3080. doi: 10.1073/pnas.96.6.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Middeler G, Zerf k, Jenovai S, Thulig A, Tschodrich-Rotter M, Ku-bitscheck U, Peters R. Oncogene. 1997;14:1407–1417. doi: 10.1038/sj.onc.1200949. [DOI] [PubMed] [Google Scholar]
- 28.Boyd SD, Tsai KY, Jacks T. Nat. Cell Biol. 2000;2:563–568. doi: 10.1038/35023500. [DOI] [PubMed] [Google Scholar]
- 29.Geyer RIC, Yu ZK, Maki CG. Nat. Cell Biol. 2000;2:569–573. doi: 10.1038/35023507. [DOI] [PubMed] [Google Scholar]
- 30.Kau TR, Way JC, Silver PA. Nat. Rev. Cancer. 2004;4:106–117. doi: 10.1038/nrc1274. [DOI] [PubMed] [Google Scholar]
- 31.Freedman DA, Wu L, Levine AJ. Cell Mol. Life Sci. 1999;55:96–107. doi: 10.1007/s000180050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Xiong Y. Cell Growth & Differ. 2001;12:175–186. [PubMed] [Google Scholar]
- 33.Carter S, Bischof O, Dejean A, Vousden ICH. Nat. Cell Biol. 2007;9:428–435. doi: 10.1038/ncb1562. [DOI] [PubMed] [Google Scholar]
- 34.Arva NC, Gopen TR, Talbott ICE, Campbell LE, Chicas A, White DE, Bond GL, Levine AJ, Bargonetti J. JBiol. Chem. 2005;280:26776–26787. doi: 10.1074/jbc.M505203200. [DOI] [PubMed] [Google Scholar]
- 35.White DE, Talbott ICE, Arva NC, Bargonetti J. Cancer Res. 2006;66:3463–3470. doi: 10.1158/0008-5472.CAN-05-1381. [DOI] [PubMed] [Google Scholar]
- 36.Ohkubo S, Tanaka T, Taya Y, Kitazato IC, Prives C. J. Biol. Chem. 2006;281:16943–16950. doi: 10.1074/jbc.M601388200. [DOI] [PubMed] [Google Scholar]
- 37.Minsky N, Oren M. Mol. Cell. 2004;16:631–639. doi: 10.1016/j.molcel.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 38.Hicke L. Nat. Rev. Mol. Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 39.Sun L, Chen ZJ. Curr. Opin. Cell Biol. 2004;16:119–126. doi: 10.1016/j.ceb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, DAndrea AD. Mol. Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 41.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 42.Kannouche PL, Wing J, Lehmann AR. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 43.van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM. Nat. Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 44.Watanabe IC, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pickart CM. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 46.Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 47.Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, O’Rourke IC, Koeppen H, Dixit VM. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 48.Honda R, Tanaka H, Yasuda H. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 49.Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, Parant JM, Lozano G, Hakem R, Benchimol S. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]