Abstract
We have conducted a comprehensive case–control study of a nasopharyngeal carcinoma (NPC) population cohort from Guangxi Province of Southern China, a region with one of the highest NPC incidences on record. A total of 1407 individuals including NPC patients, healthy controls, and their adult children were examined for the human leukocyte antigen (HLA) association, which is so far the largest NPC cohort reported for such studies. Stratified analysis performed in this study clearly demonstrated that while NPC protection is associated with independent HLA alleles, most NPC susceptibility is strictly associated with HLA haplotypes. Our study also detected for the first time that A*0206, a unique A2 subtype to South and Southeast Asia is also associated with a high risk for NPC. HLA-A*0206, HLA-B*3802 alleles plus the A*0207–B*4601 and A*3303–B*5801 haplotypes conferred high risk for NPC showing a combined odds ratio (OR) of 2.6 (P<0.0001). HLA alleles that associate with low risk for NPC include HLA-A*1101, B*27, and B*55 with a combined OR of 0.42 (P<0.0001). The overall high frequency of NPC-susceptible HLA factors in the Guangxi population is likely to have contributed to the high-NPC incidence in this region.
Keywords: HLA, nasopharyngeal carcinoma, haplotype, stratified analysis
Introduction
Nasopharyngeal carcinoma (NPC) is an epithelial malignancy caused by a combination of Epstein–Barr virus (EBV) infection and environmental factors.1–3 Genetic predisposition also has a function in NPC susceptibility causing the observed familial aggregation4 and distinct racial and geographical distribution of the disease incidence.5–8 Southern China and Southeast Asia have a disproportionally high incidence of NPC. In these regions, NPC is one of the most common cancers,9,10 but in Caucasians, it is a rare disease.11
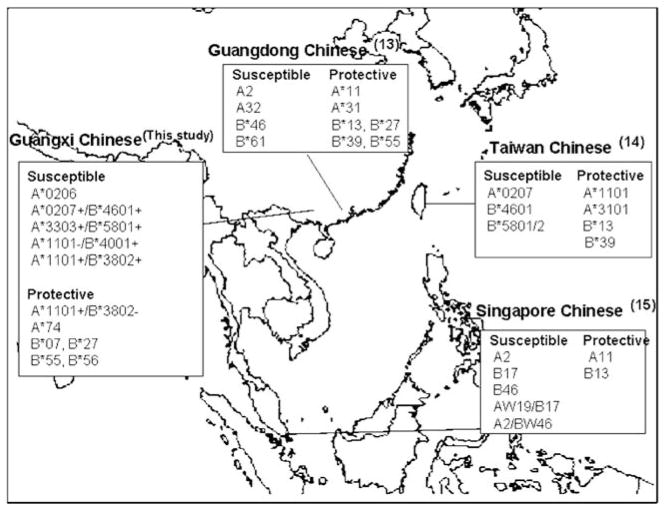
Among the host genetic markers that have been associated with NPC, the class I human leukocyte antigen (HLA) genes have shown a strong and consistent association with disease risk. As it was first reported by Simons et al.12 in a Singaporean Chinese cohort in the mid 1970s, the HLA association with NPC has been widely detected in different racial groups even though the exact HLA factors (alleles and haplotypes) associated with the disease sometimes vary among racial groups due to population-dependent HLA distributions. Populations of Southern China and Southeast Asia share high-NPC incidence as well as common HLA characteristics. Mainland Chinese,9,13 Taiwanese,14 and Singaporean Chinese15 NPC cohorts also show similar HLA associations, where HLA-A*11 and B*13 seem protective against NPC, and A*02 (A*0207), A*33, B*46, and B*58 associate with susceptibility to this disease (Figure 1). Other HLA loci including HLA-C and the class II loci, DR and DQ, have not shown independent associations with NPC even though certain class II alleles might be part of the extended HLA haplotypes associated with the disease as described by Hildesheim et al.14
Figure 1.
HLA associations with NPC in Chinese population.
Association of HLA polymorphism with human disease may indicate direct involvement of the HLA molecule in the disease pathogenesis. To date, however, there has been little in vitro or in vivo evidence that the NPC-associated HLA alleles affect differentially NPC-related EBV replication on pathogenesis. The role of HLA-A and -B alleles or haplotypes that include combinations of these alleles in NPC pathogenesis remains unknown. It is possible that the associated HLA class I alleles are simply marking by linkage disequilibrium (LD) the true NPC-causing gene(s). Large-scale cohort studies exploring combinations of associated HLA alleles may provide useful insights into the influence of HLA on NPC.
An early indicator of NPC development is the occurrence of immunoglobulin (Ig) A antibodies to EBV capsid antigens (EBV-IgA/VCA).16–18 Even though >95% of adults in the general population of all ethnic groups are healthy carriers of EBV, <2.5% are EBV-IgA/VCA antibody positive. In comparison, >95% of all NPC patients are EBV-IgA/VCA antibody positive.10 If HLA diversity is indeed directly responsible for the individually varied NPC risks, it is plausible that the development of the EBV-IgA/VCA antibodies in EBV-positive individuals may also be affected by the HLA polymorphism.
Here, we have conducted a case–control study of an NPC cohort recruited from Guangxi Province in Southern China where the NPC incidence is as high as 25–50 cases per 10000 individuals. DNA-based high-resolution HLA typing was performed on a total of 1407 individuals including NPC cases, matched controls, and offspring of the study subjects. This large study has allowed a comprehensive stratification of NPC-associated HLA factors and provided novel insights into the nature of the HLA association with the disease.
Results
HLA typing was informative for 356 NPC patients, 287 NPC free EBV-IgA/VCA antibody positive healthy individuals, and 342 NPC free EBV-IgA/VCA antibody negative healthy individuals. Comparative analyses between the two healthy groups failed to detect any significant deviation in the frequency distribution of HLA alleles and haplotypes (Supplementary Tables 1 and 2), indicating that HLA polymorphism does not affect the occurrence of the EBV-IgA/VCA antibody. Therefore, in subsequent analyses, the two NPC-free groups were combined as the control group (N = 629) for NPC cases.
HLA alleles showing a significant difference in frequency distribution between cases and controls are listed in Table 1, and full analyses are presented in Supplementary Tables 3 and 4. For the reason of most of former NPC, HLA studies were based on serology typing, for better understanding the HLA influence in NPC, both HLA allotype and genotype were present in this table. For the HLA-A locus, 31 four-digit alleles were detected, 12 of which had an allele frequency >1% in either the case or control group. Five alleles, A*0206, A*0207, A*1101, A*3303, and A*7401/7402, showed a significant difference in frequency distribution between the case and control groups. After correction, however, only two alleles, A*1101 and A*3303, remained significant. Of the two detected A*11 alleles, A*1101 showed a reduced presence in patients compared with controls (P<0.0001), whereas the less common A*1102 showed no difference between these groups, but due to its low frequency, it need to be confirmed in even large study cohort. Four common A*02 subtypes, A*0201, A*0203, A*0206, and A*0207, were detected. Two of alleles, A*0206 and A*0207, showed an elevated frequency in patients. A*3303 also associated with increased risk of NPC (P = 0.0004).
Table 1.
Gene frequencies (%) of the two and four-digit HLA-A, -B, and -C alleles detected in NPC patients (N = 356) and controls (N = 629)
| Allele | Frequency
|
P-value | Pc-value* | ||
|---|---|---|---|---|---|
| NPC patients | Controls | ||||
| HLA-A | 02 | 38.62 (275) | 32.75 (412) | 0.01 | NS |
| 0206 | 5.2 (37) | 2.31 (29) | 0.0006 | NS | |
| 0207 | 14.89 (106) | 11.29 (143) | 0.02 | NS | |
| 11 | 23.74 (169) | 32.35 (407) | 0.0001 | 0.003 | |
| 1101 | 19.52 (139) | 29.09 (366) | <0.0001 | 0.0003 | |
| 33 | 20.51 (146) | 14.31 (180) | 0.0004 | 0.02 | |
| 3303 | 20.51 (146) | 14.31 (180) | 0.0004 | 0.04 | |
| 74 | 0.14 (1) | 1.11 (14) | 0.02 | NS | |
| HLA-B | 07 | 0.0 (0) | 1.03 (13) | 0.01 | NS |
| 13 | 8.43 (60) | 13.04 (164) | 0.002 | NS | |
| 1301 | 7.72 (55) | 12.08 (152) | 0.002 | NS | |
| 27 | 0.0 (0) | 1.59 (20) | 0.001 | NS | |
| 2704 | 0.0 (0) | 1.27 (16) | 0.005 | NS | |
| 38 | 12.78 (91) | 9.78 (123) | 0.04 | NS | |
| 3802 | 12.64 (90) | 9.30 (117) | 0.02 | NS | |
| 39 | 0.28 (2) | 1.19 (15) | 0.04 | NS | |
| 40 | 16.29 (116) | 12.32 (155) | 0.01 | NS | |
| 4001 | 14.75 (105) | 11.05 (139) | 0.02 | NS | |
| 55 | 0.56 (4) | 4.21 (53) | <0.0001 | 0.0002 | |
| 5502 | 0.56 (4) | 3.66 (46) | <0.0001 | 0.003 | |
| 56 | 0.84 (6) | 2.23 (28) | 0.02 | NS | |
| 5601 | 0.28 (2) | 1.67 (21) | 0.006 | NS | |
| 58 | 19.10 (136) | 14.07 (177) | 0.003 | NS | |
| 5801 | 19.10 (136) | 14.07 (177) | 0.003 | NS | |
| HLA-C | 0302 | 18.12 (129) | 13.43 (169) | 0.005 | NS |
| 0403 | 0.98 (7) | 2.23 (28) | 0.05 | NS | |
| 12 | 1.12 (8) | 4.29 (54) | 0.0001 | 0.006 | |
| 1202 | 0.56 (4) | 2.23 (28) | 0.005 | NS | |
| 1203 | 0.56 (4) | 2.07 (26) | 0.01 | NS | |
| 15 | 1.12 (8) | 2.62 (33) | 0.03 | NS | |
Abbreviations: HLA, human leukocyte antigen; NPC, nasopharyngeal carcinoma.
‘NS’ = P>0.05.
Fifty-five HLA-B alleles were detected, but only 14 had a frequency >1%. Among the 14 B alleles, seven showed a significantly different distribution between cases and controls, including B*1301, B*2704, B*3802, B*4001, B*5502, B*5601, and B*5801. After P-value correction, however, only the decrease of B*5502 in the patient group remained significant (P corrected = 0.003). Three B*27 subtypes, B*2704, B*2705, and B*2706 were detected only in the control group with a combined frequency of 1.59% (P = 0.0014). B*3802, B*4001, and B*5801 were each observed more frequently among the case group but the significance disappeared after correction.
Twenty-four alleles were detected at the HLA-C locus, 13 of which had a frequency >1%. Cw*0403, Cw*1202, and Cw*1203 were observed at a lower frequency in cases and Cw*0302 was the only allele showing an elevated frequency in cases. After correction, however, none of the four-digit HLA-C alleles remained significant.
HLA typing was informative for 422 children of the study subjects, which enabled us to directly determine HLA haplotypes in 179 patients and 379 controls. For the remaining 177 patients and 250 controls, HLA haplotypes were assigned by population-based estimation methods. HLA-A/B haplotype frequencies were calculated on the basis of both methods of haplotype assignments. Stratification analyses were performed to determine the nature of the observed HLA association (Tables 2 and 3). Table 2 compares the frequency distribution of all HLA-A/B (where the HLA-A allele showed individual association with NPC) and -B/A (where the HLA-B allele showed individual association with NPC) haplotypes related to individual NPC-associated alleles between cases and controls to determine whether the observed allele association with NPC might be due to particular haplotypes or to an individual allele. Four of the five potentially NPC-related HLA-A alleles, A*0206, A*0207, A*1101, and A*3303, were each observed on HLA-A/B haplotypes with frequencies of >1% and were therefore included in Table 2A. Both the dominant A*0206–B*1502 haplotype (P = 0.0278) and all other A*0206-associated haplotypes combined (P = 0.0071) showed elevated frequencies in the patient group. A*0207 is predominantly associated with B*4601, and this was the only A*0207 haplotype associating with susceptibility to NPC (P = 0.0095). Similarly, A*3303–B*5801 was the only A*3303 haplotype associating with NPC susceptibility (P = 0.0003). The protective A*1101 allele was found on seven HLA-A/B haplotypes with frequencies >1% and these haplotypes showed inconsistent associations with NPC. A*1101–B*1301 (P = 0.0029), A*1101–B*4601 (P = 0.0198), and A*1101–B*others (P = 0.0009) showed reduced frequencies in the NPC group, whereas A*1101–B*3802 haplotype (P = 0.0228) showed an elevated frequency in the patient group.
Table 2.
HLA-A/B and B/A haplotype frequencies (%) in NPC patients (N = 356) and controls (N = 629)a
| Haplotype | Frequency
|
P-value* | Allele/haplotype effectb | ||
|---|---|---|---|---|---|
| NPC patients | Controls | ||||
| (A) | |||||
| HLA-A/B | |||||
| 0206 | 1502 | 1.26 | 0.4 | 0.03 | Allele A*0206 (S) |
| Others | 3.93 | 1.91 | 0.007 | ||
| 0207 | 4601 | 12.22 | 8.59 | 0.01 | Haplotype A*0207–B*4601 (S) |
| Others | 2.67 | 2.7 | NS | ||
| 1101 | 1301 | 3.79 | 7.07 | 0.003 | Allele A*1101 (P) |
| 1502 | 4.21 | 5.72 | NS | ||
| 3802 | 1.97 | 0.79 | 0.02 | Allele offset | |
| 4001 | 2.53 | 3.74 | NS | ||
| 4601 | 1.12 | 2.7 | 0.02 | Allele A*1101 (P) | |
| 5101 | 1.4 | 1.43 | NS | ||
| 5401 | 1.4 | 1.03 | NS | ||
| Others | 3.09 | 6.6 | 0.0009 | ||
| 3303 | 5801 | 17.13 | 11.29 | 0.0003 | Haplotype A*3303–B*5801 (S) |
| Others | 3.37 | 3.02 | NS | ||
| (B) | |||||
| HLA-B-A | |||||
| 1301 | 1101 | 3.79 | 7.07 | 0.003 | Allele A*1101 (P) |
| 2402 | 1.83 | 2.85 | NS | ||
| Others | 2.11 | 2.15 | NS | ||
| 3802 | 0203 | 9.41 | 7.07 | NS | |
| 1101 | 1.97 | 0.79 | 0.03 | Allele offset | |
| Others | 1.26 | 1.43 | NS | ||
| 4001 | 0203 | 3.23 | 1.59 | 0.02 | Allele B*4001 (S) |
| 1101 | 2.53 | 3.74 | NS | ||
| 1102 | 1.54 | 0.48 | 0.01 | Allele B*4001 (S) | |
| 2402 | 4.07 | 3.34 | NS | ||
| Others | 3.37 | 1.91 | NS | ||
| 5502 | 0203 | 0.42 | 2.07 | 0.004 | Allele B*5502 (P) |
| others | 0.14 | 1.59 | 0.003 | ||
| 5801 | 3303 | 17.13 | 11.29 | 0.0003 | Haplotype B*5801–A*3303 (S) |
| others | 1.97 | 2.78 | NS | ||
Abbreviations: HLA, human leukocyte antigen; NPC, nasopharyngeal carcinoma.
HLA-A/B haplotypes (Table 2A) are those containing HLA-A alleles that showed significant association with NPC in the analysis of individual alleles and HLA-B/A haplotypes (Table 2B) are those containing HLA-B alleles that showed significant association with NPC. Only the haplotypes with at least one allele showing significantly different gene frequencies between patients and controls are shown. Low-frequency haplotypes (<1.0% in both groups) associated with a particular A or B allele are combined as ‘others’. A*7400, which associated with protection (see Table 1), was not found on any haplotype with a frequency of >1%, and thus is not listed. This was also the case for B*2704 and B*5601.
‘P’ = Protective effect, ‘S’ = Susceptible effect.
‘NS’ = P>0.05.
Table 3.
Effect of allelic combinatioNS of NPC-associated HLA alleles on the disease development
| Allele combinationa A/B | NPC patients (N = 356) % (n) | Controls (N = 629) % (n) | OR (95% CI) | P-value* | Allele/haplotype effect |
|---|---|---|---|---|---|
| Protective | |||||
| 1101/4001 | Allele A*1101 | ||||
| −/+ | 19.38 (69) | 8.59 (54) | 2.56 (1.76–3.72) | <0.0001 | |
| +/− | 28.37 (101) | 38.95 (254) | 0.62 (0.47–0.82) | 0.0008 | |
| +/+ | 8.15 (29) | 11.92 (75) | 0.66 (0.42–1.02) | NS | |
| 0203/5502 | Allele B*5502 | ||||
| −/+ | 0.28 (1) | 2.54 (16) | 0.11 (0.02–0.57) | 0.009 | |
| +/− | 27.53 (98) | 23.37 (147) | 1.25 (0.93–1.68) | NS | |
| +/+ | 0.84 (3) | 4.45 (28) | 0.18 (0.06–0.53) | 0.002 | |
| 1101/1301 | Allele A*1101 | ||||
| −/+ | 5.9 (21) | 6.52 (41) | 0.9 (0.52–1.55) | NS | |
| +/− | 27.25 (97) | 34.02 (214) | 0.73 (0.55–0.97) | 0.03 | |
| +/+ | 9.27 (33) | 16.85 (106) | 0.5 (0.33–0.76) | 0.001 | |
| Susceptible | |||||
| 0206/1502 | Allele A*0206 | ||||
| −/+ | 13.2 (47) | 15.74 (99) | 0.81 (0.56–1.18) | NS | |
| +/− | 7.02 (25) | 3.66 (23) | 1.99 (1.12–3.53) | 0.02 | |
| +/+ | 3.37 (12) | 0.95 (6) | 3.62 (1.43–9.15) | 0.007 | |
| 0207/4601 | Haplotype A*0207–B*4601 | ||||
| −/+ | 7.87 (28) | 12.24 (77) | 0.61 (0.39–0.96) | 0.03b | |
| +/− | 3.93 (14) | 4.93 (31) | 0.79 (0.41–1.50) | NS | |
| +/+ | 23.31 (83) | 16.38 (103) | 1.55 (1.12–2.14) | 0.008 | |
| 3303/5801 | Haplotype A*3303–B*5801 | ||||
| −/+ | 3.37 (12) | 4.45 (28) | 0.75 (0.28–1.49) | NS | |
| +/− | 5.06 (18) | 5.41 (34) | 0.93 (0.52–1.68) | NS | |
| +/+ | 32.87 (117) | 21.46 (135) | 1.79 (1.34–2.39) | 0.0001 | |
Abbreviations: CI, confidence interval; HLA, human leukocyte antigen; NPC, nasopharyngeal carcinoma; OR, odds ratio.
‘+’ = positive for the allele; ‘−’ = negative for the allele.
The A0207−/B4601+ signal (P = 0.0325) is a weak ‘protective’ confirmed, the opposite of the strong susceptible haplotype A0207/B4601.
‘NS’ = P>0.05.
Five of the seven NPC-associated HLA-B alleles composed haplotypes with frequencies >1%. The B*5801–A*3303 haplotype showed an elevated frequency in NPC patients, whereas the other B*5801 haplotypes did not. The other NPC-associated B alleles were all found on at least two common HLA-B/A haplotypes. Both B*1301 and B*3802 were significantly more common in the control group only when A*1101 was also present on these haplotypes (P = 0.0029 and P = 0.0228, respectively). B*4001 was associated with four haplotypes with a frequency >1%. All haplotypes containing this allele were observed more frequently in the patients (B*4001–A*0203 and B*4001–A*1102 reaching significance; P = 0.0167 and P = 0.0138, respectively), except for the A*1101-associated B*4001 haplotype, which was more common in the controls albeit not significantly.
Table 3 stratifies six common NPC-associated HLA-A/B allelic combinations to examine independent as well as collective effects of HLA alleles on NPC risk analyzed in Table 2. For each tested HLA-A/B phenotypic combination, the study subjects were divided into three groups: those having both of the relevant HLA-A and HLA-B alleles and those that had one, but not the other. The protective A*1101 and the susceptible B*4001 allele combination is included first in Table 3 to show the collective effect of these two offsetting NPC-associated alleles. The protective B*5502 is in LD with A*0203, and stratification for this allele combination showed that the protective effect of B*5502 could be detected in the presence (odds ratio (OR) = 0.18, P = 0.0018) or absence (OR = 0.11, P = 0.0088) of A*0203. A*1101, which is commonly linked to the NPC-related B alleles B*1301 and B*4001, is the most common protective allele in the Guangxi NPC cohort. Stratified analysis showed that the A*1101 protection was stronger when B*1301 was also present (OR = 0.5, P = 0.0010; without B*1301; OR = 0.73, P = 0.0280). However, B*1301 had no effect (OR = 0.9, P>0.05) in the absence of A*1101. Remarkably, B*4001 actually associated strongly with risk of developing NPC if A*1101 was missing (OR = 2.56, P<0.0001), but in the presence of A*1101, the susceptibility effect of B*4001 was completely abrogated (OR = 0.66, P>0.05). The susceptibility effect of A*0206 seems to be largely independent of HLA-B, though the A*0206 and B*1502 combination was somewhat stronger (OR = 3.63, P = 0.0065) than that in the presence of other HLA-B alleles (OR = 1.99, P = 0.0184). Both A*0207 (Table 1) and B*460114,19 were thought to be independent high-risk factors for NPC in Southern Chinese and Southeast Asia. The two alleles are in strong LD in the Guangxi cohort, as in most Southern Chinese populations, and the A*0207–B*4601 haplotype associated with susceptibility (Table 3). However, A*0207 showed no effect in the absence of B*4601, whereas B*4601 had a weak protective effect in the absence of A*0207 (OR = 0.61, P = 0.0325). Finally, A*3303 and B*5801 forms the most common A–B haplotype in the Guangxi cohort and both alleles were associated with an elevated NPC risk (Table 1). Stratification of the A*3303 and B*5801 combination showed that the NPC effect was more strongly associated with the presence of both alleles (OR = 1.79, P = 0.0001) as one allele without the other had no effect.
Table 4 summarizes the HLA alleles and the allele combinations associated with the risk of developing NPC. HLA protection against NPC mainly involves independent allele influence except that the A*1101 protection may be overridden by the presence of the high-risk B*3802. The presence of one or more of the six protective factors has a frequency of 58.98% within the control group and 34.83% within the NPC patients. Together, these protective factors delivered a combined OR of 0.37 (P<0.0001). NPC susceptibility, on the other hand, associates most strongly with certain allele combinations, as opposed to single alleles, with the exception of the A*0206 effect. Individual positive for any of the five high-risk factors accounted for 50.08 and 73.03% of the controls and cases, respectively. These high-risk factors showed individual ORs between 2.05 and 3.56 and a collective OR of 2.70 (P<0.0001).
Table 4.
Protective and susceptible HLA alleles, haplotypes, and genotypes for the development of NPC
| HLA factor | NPC cases (N = 356) % (n) | Controls (N = 629) % (n) | OR (95% CI) | P-value |
|---|---|---|---|---|
| Protective | ||||
| A*1101+/B*3802 | 32.58 (116) | 49.76 (313) | 0.49 (0.37–0.64) | <0.0001 |
| A*74 | 0.28 (1) | 2.07 (13) | 0.13 (0.05–0.32) | 0.03 |
| B*07 | 0.0 (0) | 2.07 (13) | 0.07 (0.01–0.56) | 0.01 |
| B*27 | 0.0 (0) | 3.18 (20) | 0.04 (0.01–0.29) | 0.001 |
| B*55 | 1.12 (4) | 7.95 (50) | 0.13 (0.05–0.32) | <0.0001 |
| B*56 | 1.69 (6) | 4.45 (28) | 0.37 (0.16–0.87) | 0.02 |
| Combined | 34.83 (124) | 58.98 (371) | 0.37 (0.28–0.49) | <0.0001 |
| Susceptible | ||||
| A*0206 | 10.39 (37) | 4.61 (29) | 2.40 (1.47–3.92) | 0.0005 |
| A*0207+/B*4601+ | 23.03 (82) | 16.38 (103) | 1.53 (1.11–2.11) | 0.01 |
| A*3303+/B*5801+ | 32.87 (117) | 21.14 (133) | 1.83 (1.37–2.44) | <0.0001 |
| A*1101−/B*4001+ | 23.03 (82) | 14.15 (89) | 1.82 (1.30–2.53) | 0.0004 |
| A*1101+/B*3802+ | 3.93 (14) | 1.59 (10) | 2.53 (1.14–5.61) | 0.02 |
| Combined | 73.03 (260) | 50.08 (315) | 2.70 (2.05–3.56) | <0.0001 |
Abbreviations: CI, confidence interval; HLA, human leukocyte antigen; NPC, nasopharyngeal carcinoma; OR, odds ratio.
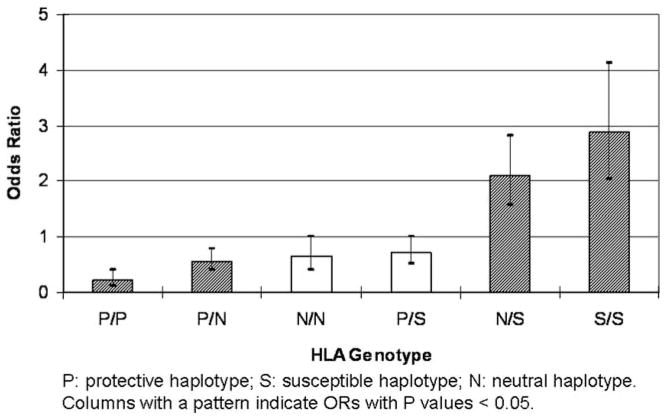
On the basis of analyses shown, HLA haplotypes were classified as NPC protective (P), susceptible (S), and neutral (N), generating six genotypes. Figure 2 shows the ORs of the six genotypes for NPC development. Interestingly, neither the protective nor the susceptible genotypes seemed dominant over the other. The exact values are presented in Supplementary Table 5. The ORs of the remaining genotypes were distributed in a manner that was expected given that the protective and susceptible genotypes were defined in this same cohort. The validity of this scheme will be particularly interesting to test in an independent cohort.
Figure 2.
ORs of HLA genotypes for NPC development.
The influence of HLA genotypes on NPC risk was analyzed by comparing all individual genotypes in HLA-A and -B loci separately and the compound genotypes of HLA-A and -B. Two HLA-A, six HLA-B genotypes and six HLA-A-B compound genotypes were significantly associated with either elevated NPC risk or protection (Supplementary Table 6). These genotypes all involve at least one allele showing a higher risk or protection for the disease.
Discussion
Southern China has a disproportionally high incidence of NPC compared with other parts of the world. The study population from Wuzhou City of Guangxi Province in Southern China holds, perhaps, the highest recorded NPC incidence.9,10 Apart from unique environmental factors (mainly traditional diet), our data support a role for genetic predisposition contributing to the high-disease incidence. The results from this study confirm and extend previously reported HLA and NPC associations in Southern Chinese populations,12–14,19 such as the protective effect associated with HLA-A*1101 and susceptibility effects associated with B*0207, A*3303, and B*5801. This study provides further insights into the nature of the HLA association with NPC, such as the observation that the dominant A*11 protection can be entirely attributed to the major subtype A*1101. The other A*11 subtype detected in this population, A*1102, did not show any protective effect even though the two A*11 subtypes differ by only a single amino acid in position 19 outside of the peptide-binding groove.
Our stratified analyses demonstrated that a proportion of the HLA-associated NPC susceptibility is likely to be haplotype-dependent in this cohort (Table 3). In earlier cohort studies from Southern China13 and Taiwan,14 the A*0207–B*4601 allele combination was consistently associated with a high-NPC risk, but there are contradictory reports on whether the two alleles have independent effects. In our study cohort, two major high-risk HLA-A/B allele combinations, A*0207–B*4601 and A*3303–B*5801, were identified. Patients with either of these two allele combinations accounted for half of the NPC cohort and susceptibility conferred by these HLA-A/B combinations was strictly dependent on the presence of both alleles. The alleles comprising both pairs are in strong LD, forming the two most common HLA-A/B haplotypes in the Guangxi population.
Haplotype-dependent disease associations may indicate one of the two possibilities: (1) the two alleles on the haplotype are behaving in an epistatic manner to reach functional synergy or (2) the disease-associated HLA haplotype is tracking an unidentified disease locus present on that specific haplotype. An example of the former is that the structural stability of the HLA-DQ αβ dimer is affected by the type of DQA and DQB alleles carried on HLA haplotypes, which in turn influences the risk of insulin-dependent diabetes mellitus (IDDM).20 Functional synergy between HLA-A and -B molecules and any potential clinical relevance such synergy may have, however, are yet to be established. In terms of the alternative HLA marker model, there are no data that unequivocally support or refute the hypothesis in NPC studies, despite efforts to identify non-HLA disease genes in the extended MHC region.21,22 Still, direct involvement of HLA molecules in NPC pathogenesis, including that involving the causative EBV infection, also lacks functional data support. Interpretations of the present results tend to support the locus tracking hypothesis in that there is evidence for (1) a lack of HLA association with the occurrence of IgA antibodies to EBV-IgA/VCA, a well-known precursor of NPC, (2) haplotype-dependent but allele-independent disease susceptibility, and (3) the lack of association of A*1102 despite its identical peptide-binding structure shared with the well-documented protective A*1101 allele.14 Furthermore, distinct population-specific HLA alleles associate with susceptibility to NPC, which is unexpected if HLA is directly involved in disease pathogenesis.
In contrast to the haplotype-dependent but allele-independent susceptibility exhibited by the two major high-risk haplotypes A*0207–B*4601 and A*3303–B*5801, the dominant protective effect conferred by A*1101 did not show a clear haplotypic dependence. In fact, most A*1101-linked A/B haplotypes showed a trend of under-representation in the case group except the one linked with the susceptible B*3802. A*1101 seems to protect against other viral infections as well. For example, A*1101 may confer protection against AIDS through restriction of immunodominant HIV peptides,23,24 but the structural and functional basis for its direct involvement in NPC pathogenesis has not been thoroughly explored.
The detected allele and haplotype associations with NPC seem to be independent of HLA genotypes as genotypes showing significant ORs are essentially those with least two copies of NPC-associated alleles or haplotypes. Also, no particular genotypes showed stronger association than individual alleles or haplotypes.
Another noteworthy HLA association observed in the Guangxi NPC cohort is an opportune ‘complete’ protection of B*27 against disease development. B*27 was detected with a typical frequency in the control group (1.59%), but was completely absent in the patient group (OR = 0.04, P = 0.001). Earlier cohort studies from Southern China and Taiwan have also detected B*27 as a low-risk allele though the protection was never ‘complete.’14,19,25 B*27 is well known for its high-risk association with the inflammatory autoimmune disease ankylosing spondylitis and it confers protection against AIDS progression apparently through its peptide-binding properties for immunodominant HIV epitopes.26 Perhaps B*27 is also involved in EBV-related pathogenesis, impacting the risk of NPC development. Given the low frequency of B*27 in the study population, however, the absence of B*27 in the patient group should be confirmed in larger replication cohort studies.
The overall HLA profile of a population is likely to influence the incidence of HLA-associated diseases. An example is the correlation between the rates of IDDM incidence and the frequency distribution of the IDDM-associated HLA-DQ and DR alleles in world populations.27 The NPC incidence in a population may also correlate positively with the frequencies of NPC-susceptible HLA alleles and haplotypes of the population. The two dominant susceptible HLA haplotypes, A*0207–B*4601 and A*3303–B*5801, are the two most common HLA-A/B haplotypes in Guangxi and together they account for 20% of the total haplotype frequency of the study Wuzhou population, which is one of the highest in Far East Asian populations. Overall, the susceptible HLA factors were found in about half of the individuals in the Wuzhou population, which is also one of the highest. Therefore, the unusually high presence of NPC-associated HLA alleles and haplotypes may partly explain the disproportionally high-NPC incidence in Southern China and Southeast Asia.
Materials and methods
Study cohorts
NPC cases and controls were recruited from Wuzhou City and Cangwu County of Guangxi Province.10 All study subjects were of Han ethnic origin. Informed consent was obtained from all study participants. An effort was made to enroll triads consisting of a proband (either NPC patient or NPC-free but EBV-IgA/VCA antibody positive), an unaffected spouse and an adult child. The case group included 356 unrelated patients with biopsy-confirmed NPC. The mean age was 50.1 years (range 19–80), 95.5% of them being EBV-IgA/VCA antibody positive. Two groups of control were the case’s spouse or geographically matched residents who were NPC free at the time of study enrollment. An antibody to EBV capsid antigen (EBV-IgA/VCA) were confirmed by serologic testing at the time of study enrollment. One group was positive (N = 287) and the other negative (N = 342) for the EBV-IgA/VCA antibody. The mean age was 45.7 and 46.9, respectively, for the antibody positive and negative groups. The controls were matched to the cases by age, ethnicity, and geographic residence (Table 5). In addition, 422 adult children of the study subjects in the case and control groups were recruited to allow elucidation of HLA haplotypes, but they were excluded in all other analyses.
Table 5.
Gender and age of NPC patients and EBV-IgA/VCA positive and negative controls
| NPC patients | Controls
|
||
|---|---|---|---|
| EBV-IgA/ VCA positive | EBV-IgA/ VCA negative | ||
| N = 356 | N = 287 | N = 342 | |
| Male/female (% male) | 237/119 (67%) | 141/146 (49%) | 129/213 (37%) |
| Age | 50.09±10.83 | 45.69±8.90 | 46.87±10.24 |
| EBV-IgA/VCA | 95.50% | 100% | 0% |
Abbreviations: EBV, Epstein–Barr virus; NPC, nasopharyngeal carcinoma.
Detection of EBV-IgA/VCA antibody
The presence of the EBV-IgA/VCA antibody was detected using the immunoperoxidase assay as described earlier.28 EBV positive B95-8 cells fixed on slides were incubated with multiple dilutions of the testing serum followed by incubation with antihuman IgA horseradish peroxidase and staining with diaminobenzidine. Testing sera with a staining titer of 1:10 or higher dilution were considered positive for the EBV-IgA/VCA antibody.
HLA typing
HLA class I alleles were characterized using a PCR-SSOP (sequence-specific oligonucleotide probe) typing protocol developed by the 13th International histocompatibility Workshop.29 Briefly, the gene fragment spanning exon 2, intron 2, and exon 3 was amplified using locus-specific primers for HLA-A, -B, and -C separately. The PCR products were immobilized on nylon membranes and hybridized with a panel of P32-labeled oligonucleotide (19 mers) matching all known sequence variations of the HLA genes. Typing results were interpreted by SSOP hybridization patterns based on sequences of known HLA alleles. Typing ambiguities were resolved by sequencing exons 2 and 3 completely. For sequencing analysis, the PCR product of HLA-A, -B, or -C was used as the template for the sequencing reaction. For each of the HLA genes, two sequencing reactions (one for exon 2 and one for exon 3) were performed using exon-specific sequencing primers. The sequencing analysis was performed using the ABI Big Dye Terminator Cycle Sequencing Kit and ABI3730xl DNA analyzer (Applied Biosystems, Foster City, CA, USA). HLA alleles were assigned on the basis of the sequence database of known alleles with the help of the ASSIGN software developed by Conexio Genomics (Conexio Genomics, Perth City, WA, Australia). Ambiguous heterozygous genotypes were resolved by additional PCR and sequencing procedures using allele-specific PCR primers to selectively amplify only one of the two alleles.
Statistical analyses
HLA allele frequencies were calculated based on observed genotypes, and HLA-A and -B haplotype frequencies were estimated using one of the two methods: (1) unambiguous assignment based on familial segregation for the cases and controls using recruited spouse and child genotype data or (2) indirect assignment based on maximum likelihood estimation for the study subjects without recruited family members. For the latter, the haplotypic analysis was performed using the BLOCKHEAD genetic analysis software developed by George Nelson in the Laboratory of Genomic Diversity, National Cancer Institute.
The effect of HLA alleles on the development of NPC and EBV-IgA/VCA antibody was evaluated by computing ORs and 95% confidence intervals as well as exact P-values using the FREQ procedure of the SAS 9.1 software (The SAS Institute, NC, USA). A correction of 0.5 was applied on every cell of the 2 × 2 table that contains a zero. The analyses were performed at four-digit and two-digit resolution levels separately. P-value was calculated by χ2-test for each allele and was corrected by multiplying the number of all detected alleles. Significance was considered at P<0.05. Both uncorrected and corrected P-values were presented in our tables. Stratified analyses were applied to evaluate the effect of haplotypes of HLA-A and -B that contained alleles showing individual significant associations with NPC risk.
Supplementary Material
Acknowledgments
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Conflict of interest
The authors declare no conflict of interest
Supplementary Information accompanies the paper on Genes and Immunity website (http://www.nature.com/gene)
References
- 1.Brown TM, Heath CW, Lang RM, Lee SK, Whalley BW. Nasopharyngeal cancer in Bermuda. Cancer. 1976;37:1464–1468. doi: 10.1002/1097-0142(197603)37:3<1464::aid-cncr2820370331>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 2.Yu MC, Garabrant DH, Huang TB, Henderson BE. Occupational and other non-dietary risk factors for nasopharyngeal carcinoma in Guangzhou, China. Int J Cancer. 1990;45:1033–1039. doi: 10.1002/ijc.2910450609. [DOI] [PubMed] [Google Scholar]
- 3.Guo X, Johnson RC, Deng H, Liao J, Guan L, Nelson GW, et al. Evaluation of non-viral risk factors for nasopharyngeal carcinoma in a high-risk population of Southern China. Int J Cancer. 2009;124:2942–2947. doi: 10.1002/ijc.24293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coffin CM, Rich SS, Dehner LP. Familial aggregation of nasopharyngeal carcinoma and other malignancies. A clinicopathologic description. Cancer. 1991;68:1323–1328. doi: 10.1002/1097-0142(19910915)68:6<1323::aid-cncr2820680623>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 5.Jeannel D, Hubert A, de Vathaire F, Ellouz R, Camoun M, Ben Salem M, et al. Diet, living conditions and nasopharyngeal carcinoma in Tunisia–a case-control study. Int J Cancer. 1990;46:421–425. doi: 10.1002/ijc.2910460316. [DOI] [PubMed] [Google Scholar]
- 6.Laramore GE, Clubb B, Quick C, Amer MH, Ali M, Greer W, et al. Nasopharyngeal carcinoma in Saudi Arabia: a retrospective study of 166 cases treated with curative intent. Int J Radiat Oncol Biol Phys. 1988;15:1119–1127. doi: 10.1016/0360-3016(88)90193-9. [DOI] [PubMed] [Google Scholar]
- 7.Johansen LV, Mestre M, Overgaard J. Carcinoma of the nasopharynx: analysis of treatment results in 167 consecutively admitted patients. Head Neck. 1992;14:200–207. doi: 10.1002/hed.2880140307. [DOI] [PubMed] [Google Scholar]
- 8.Lee AW, Foo W, Mang O, Sze WM, Chappell R, Lau WH, et al. Changing epidemiology of nasopharyngeal carcinoma in Hong Kong over a 20-year period (1980–99): an encouraging reduction in both incidence and mortality. Int J Cancer. 2003;103:680–685. doi: 10.1002/ijc.10894. [DOI] [PubMed] [Google Scholar]
- 9.Lu SJ, Day NE, Degos L, Lepage V, Wang PC, Chan SH, et al. Linkage of a nasopharyngeal carcinoma susceptibility locus to the HLA region. Nature. 1990;346:470–471. doi: 10.1038/346470a0. [DOI] [PubMed] [Google Scholar]
- 10.Guo XC, Scott K, Liu Y, Dean M, David V, Nelson GW, et al. Genetic factors leading to chronic Epstein-Barr virus infection and nasopharyngeal carcinoma in South East China: study design, methods and feasibility. Hum Genomics. 2006;2:365–375. doi: 10.1186/1479-7364-2-6-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burt RD, Vaughan TL, McKnight B, Davis S, Beckmann AM, Smith AG, et al. Associations between human leukocyte antigen type and nasopharyngeal carcinoma in Caucasians in the United States. Cancer Epidemiol Biomarkers Prev. 1996;5:879–887. [PubMed] [Google Scholar]
- 12.Simons MJ, Wee GB, Chan SH, Shanmugaratnam K, Day NE, de-The G. Immunogenetic aspects of nasopharyngeal carcinoma (NPC) III. HL-a type as a genetic marker of NPC predisposition to test the hypothesis that Epstein-Barr virus is an etiological factor in NPC. IARC Sci Publ. 1975:249–258. [PubMed] [Google Scholar]
- 13.Hu SP, Day NE, Li DR, Luben RN, Cai KL, Ou-Yang T, et al. Further evidence for an HLA-related recessive mutation in nasopharyngeal carcinoma among the Chinese. Br J Cancer. 2005;92:967–970. doi: 10.1038/sj.bjc.6602347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hildesheim A, Apple RJ, Chen CJ, Wang SS, Cheng YJ, Klitz W, et al. Association of HLA class I and II alleles and extended haplotypes with nasopharyngeal carcinoma in Taiwan. J Natl Cancer Inst. 2002;94:1780–1789. doi: 10.1093/jnci/94.23.1780. [DOI] [PubMed] [Google Scholar]
- 15.Chan SH, Day NE, Kunaratnam N, Chia KB, Simons MJ. HLA and nasopharyngeal carcinoma in Chinese—a further study. Int J Cancer. 1983;32:171–176. doi: 10.1002/ijc.2910320206. [DOI] [PubMed] [Google Scholar]
- 16.Henle G, Henle W. Epstein-Barr virus-specific IgA serum antibodies as an outstanding feature of nasopharyngeal carcinoma. Int J Cancer. 1976;17:1–7. doi: 10.1002/ijc.2910170102. [DOI] [PubMed] [Google Scholar]
- 17.Zeng Y, Zhang LG, Li HY, Jan MG, Zhang Q, Wu YC, et al. Serological mass survey for early detection of nasopharyngeal carcinoma in Wuzhou City, China. Int J Cancer. 1982;29:139–141. doi: 10.1002/ijc.2910290204. [DOI] [PubMed] [Google Scholar]
- 18.Zeng Y, Zhang LG, Wu YC, Huang YS, Huang NQ, Li JY, et al. Prospective studies on nasopharyngeal carcinoma in Epstein-Barr virus IgA/VCA antibody-positive persons in Wuzhou City, China. Int J Cancer. 1985;36:545–547. doi: 10.1002/ijc.2910360505. [DOI] [PubMed] [Google Scholar]
- 19.Wu SB, Hwang SJ, Chang AS, Hsieh T, Hsu MM, Hsieh RP, et al. Human leukocyte antigen (HLA) frequency among patients with nasopharyngeal carcinoma in Taiwan. Anticancer Res. 1989;9:1649–1653. [PubMed] [Google Scholar]
- 20.Kwok WW, Schwarz D, Nepom BS, Hock RA, Thurtle PS, Nepom GT. HLA-DQ molecules form alpha-beta heterodimers of mixed allotype. J Immunol. 1988;141:3123–3127. [PubMed] [Google Scholar]
- 21.Lu CC, Chen JC, Tsai ST, Jin YT, Tsai JC, Chan SH, et al. Nasopharyngeal carcinoma-susceptibility locus is localized to a 132 kb segment containing HLA-A using high-resolution microsatellite mapping. Int J Cancer. 2005;115:742–746. doi: 10.1002/ijc.20946. [DOI] [PubMed] [Google Scholar]
- 22.Ooi EE, Ren EC, Chan SH. Association between microsatellites within the human MHC and nasopharyngeal carcinoma. Int J Cancer. 1997;74:229–232. doi: 10.1002/(sici)1097-0215(19970422)74:2<229::aid-ijc16>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 23.Beyrer C, Artenstein AW, Rugpao S, Stephens H, VanCott TC, Robb ML, et al. Epidemiologic and biologic characterization of a cohort of human immunodeficiency virus type 1 highly exposed, persistently seronegative female sex workers in northern Thailand. Chiang Mai HEPS Working Group. J Infect Dis. 1999;179:59–67. doi: 10.1086/314556. [DOI] [PubMed] [Google Scholar]
- 24.Fukada K, Tomiyama H, Chujoh Y, Miwa K, Kaneko Y, Oka S, et al. HLA-A*1101-restricted cytotoxic T lymphocyte recognition for a novel epitope derived from the HIV-1 Env protein. Aids. 1999;13:2597–2599. doi: 10.1097/00002030-199912240-00017. [DOI] [PubMed] [Google Scholar]
- 25.Yu MC, Huang TB, Henderson BE. Diet and nasopharyngeal carcinoma: a case-control study in Guangzhou, China. Int J Cancer. 1989;43:1077–1082. doi: 10.1002/ijc.2910430621. [DOI] [PubMed] [Google Scholar]
- 26.Gao X, Bashirova A, Iversen AK, Phair J, Goedert JJ, Buchbinder S, et al. AIDS restriction HLA allotypes target distinct intervals of HIV-1 pathogenesis. Nat Med. 2005;11:1290–1292. doi: 10.1038/nm1333. [DOI] [PubMed] [Google Scholar]
- 27.Bao MZ, Wang JX, Dorman JS, Trucco M. HLA-DQ beta non-ASP-57 allele and incidence of diabetes in China and the USA. Lancet. 1989;2:497–498. doi: 10.1016/s0140-6736(89)92102-8. [DOI] [PubMed] [Google Scholar]
- 28.Cevenini R, Donati M, Rumpianesi F, Moroni A, Paolucci P. An immunoperoxidase assay for the detection of specific IgA antibody in Epstein-Barr virus infections. J Clin Pathol. 1984;37:440–443. doi: 10.1136/jcp.37.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao X, Nelson GW, Karacki P, Martin MP, Phair J, Kaslow R, et al. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N Engl J Med. 2001;344:1668–1675. doi: 10.1056/NEJM200105313442203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.