Abstract
We discuss recent progresses in computational studies of membrane proteins based on physical models with parameters derived from bioinformatics analysis. We describe computational identification of membrane proteins and prediction of their topology from sequence, discovery of sequence and spatial motifs, and implications of these discoveries. The detection of evolutionary signal for understanding the substitution pattern of residues in the TM segments and for sequence alignment are also discussed. We further discuss empirical potential functions for energetics of inserting residues in the TM domain, for interactions between TM helices or strands, and their applications in predicting lipid-facing surfaces of the TM domain. Recent progresses in structure predictions of membrane proteins are also reviewed, with further discussions on calculation of ensemble properties such as melting temperature based on simplified state space model. Additional topics include prediction of oligomerization state of membrane proteins, identification of the interfaces for protein-protein interactions, and design of membrane proteins.
Keywords: Membrane proteins, bioinformatics, empirical potential function, motifs, ensemble properties, melting temperature, protein-protein interaction, protein design
1. Introduction
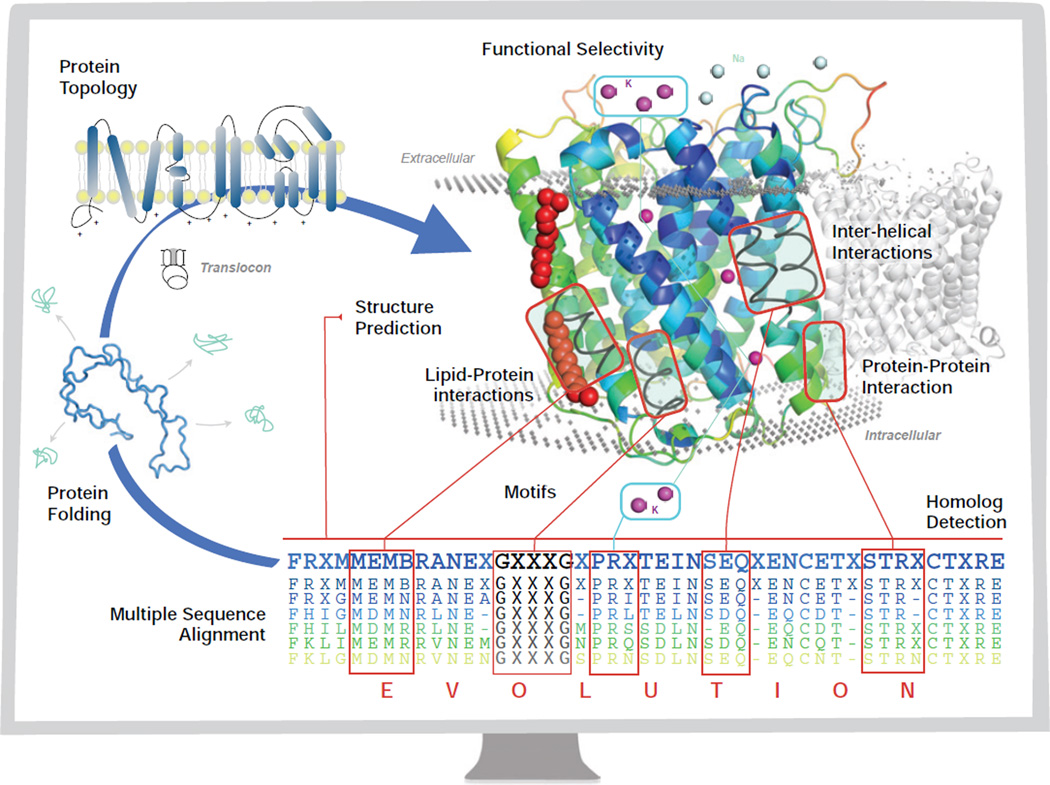
Membrane proteins account for about 20% to 30% of all proteins encoded in a typical genome (1; 2). They play central roles in transport of nutrients and metabolites, and in signaling of regulatory networks (3; 4; 5). A major obstacle in studying membrane proteins is the difficulty in experimental determination of their three dimensional structures. Computational studies of membrane proteins can compliment experimental studies and have made significant strides. In this review, we discuss recent work based on analysis of sequences and structures of membrane proteins, as well as important understandings gained from these studies on the physical processes of membrane protein assembly. An overview of the scope of studies surveyed in this review is shown in Fig 1, in the form of a diagram of the central dogma of molecular biology, in which different aspects where computational studies have made important contributions are depicted.
Figure 1.
The central dogma of molecular biology for membrane proteins (blue arrow) through the monitor of a computer. The chain of amino acids folds through the mediation of the translocon to a final stable low energy structure, with a specific topology (Section 2.2). The goal of structure prediction is to derive the three-dimensional structure of a membrane protein from its sequence (Section 7). The assembly of the helices in the transmembrane domains is facilitated by interhelical interactions (Section 6) via sequence and spatial motifs (Section 3), as well as protein lipid interactions (Sections 5.2 and 4). Membrane proteins also participate in protein-protein interactions (Section 8.2) for biological functions. The evolutionary relationship between membrane proteins can be detected through (multiple) sequence alignment, for which an evolutionary model of substitutions of residues in the transmembrane domain is essential in deriving specialized scoring matrices for alignment and for detection of homologs (Section 4). With significant understanding of the organizing principles of membrane proteins, computational studies can be carried out to design membrane proteins with desired properties such as functional selectivity (Section 8.3).
2. Identification of Membrane Proteins and Prediction of Their Topology
2.1. Predicting membrane proteins
It was discovered very early on that the presence of stretches of hydrophobic residues in a protein sequence is a good indicator that this sequence encodes a membrane protein (6). Because most transmembrane helices are hydrophobic, they appear as periodic stretches of non-polar amino acids of length 17–25 in the primary sequence. These stretches of hydrophobic residues cross the lipid membrane multiple times, and are connected by loops containing more polar residues. Such periodicity of hydrophobicity can be easily detected, and early methods for membrane protein prediction are based on calculation of a hydrophobicity index of residues within a window sliding along the protein sequence (6; 7).
A major source of misclassification with this approach is the existence of signal peptides important for targeting proteins for export. Signal peptide contains a hydrophobic region that can easily be mistaken for a transmembrane segment (8). Another source of difficulty is due to C-terminal peptides that are cleaved upon glycosylphosphatidylinositol (GPI)-anchoring, as these peptides are also often hydrophobic (9). An effective solution is to pre-process sequences by deleting signal peptides and cleaved peptides, both can be predicted accurately (10; 11; 12).
Predicting β-barrel membrane proteins is more challenging. Although residues facing the lipid membrane are predominantly hydrophobic, those facing the interior of the barrel can be quite polar (13). Unlike helical membrane proteins, there are no clear stretches of hydrophobic residues in their primary sequences.
2.2. Predicting Topology of Membrane Proteins
Many modern methods for identification of membrane proteins are based on techniques from machine learning and can also predict the topology of membrane proteins. The topology of a membrane protein refers to the number of transmembrane segments and the sidedness of the terminal ends of the protein, namely, whether the N- and C-end is on the non-translocated side or on the translocated side.
The topology of helical membrane protein can be predicted with high accuracy. Most prediction methods are based on processing multiple-sequence alignment data using machine-learning techniques such as neural networks (14; 15), Hidden Markov models (16; 17; 18; 19; 20), and support vector machines (21). The well-known “positive-inside” rule (22; 23; 24), namely, Arg and Lys residues are enriched in loops on the non-translocated side across the membrane compared to the translocated side, greatly aids in the development of these machine learning methods (22; 23; 24). For large scale prediction, recent experimentation using the consensus of many single-sequence based prediction methods also showed promise, which dispenses with time-consuming multiple-sequence alignments and are better suited for genome-scale predictions (25). For β-barrel membrane proteins, despite the lack of clear hydrophobic stretches of residues in the primary sequences, machine learning methods can now predict outer membrane proteins also very accurately (see (26) and (27)).
An approach alternative to machine learning is to predict membrane proteins and their topology based on physical considerations. This approach gives more mechanistic insight and is based on the fact that membrane protein folding and sorting are driven by physical processes. Using the scale of measured free energy contributions of inserting individual amino acids at different positions of the TM helices into the endoplasmic reticulum membrane (28), a simple additive free-energy model was used to identify putative TM helices. Combined with the positive-inside rule, this approach can predict the topology of α-helical membrane proteins accurately based on physical principles (29).
For β-barrel membrane protein, there are several characteristic observations that can help to determine their topology. First, the periplasmic loops are always short compared to extracellular loops (13), although this may not be true for mitochondrial and chloroplast outer membrane proteins. Second, there is a significant, albeit less dramatic bias in the topological sidedness of the distribution of charged residues. Different from the “positive-inside” rule for helical membrane proteins, there exists an overall “positive-outside” distribution. The extracellular cap region of the β-barrel membrane proteins are disproportionately enriched with positively charged Arg and Lys, which are disfavored in the periplasmic cap region (30). This is likely due to the asymmetric distribution of the two leaflets of the lipid bilayer, in which negatively charged lipopolysaccharides (LPS) is enriched in the outer-leaflet of the outer membrane (31). For gram’s negative bacteria, this “positive-out” rule for the outer membrane is consistent and complements the “positive-in” rule for the inner membrane, as both rules implies that positively charged residues are not favored in the periplasmic region.
Several computational methods based on machine learning techniques can predict the topology of β-barrel membrane proteins well (32; 33; 34). Built upon earlier results (35), a recent study based on measured physicochemical properties of residues and empirical statistical potential is also shown to have excellent performance in identifying β-barrel membrane proteins (36).
3. Motifs in Membrane Proteins
3.1. Sequence motifs
The GxxxG (or GG4) motif, in which two Gly are separated by three other residues, was the first sequence motif discovered (37). Originally observed in glycophorin A, this motif mediates close interaction of TM helices (38; 37). It is an example of the more general small-xxx-small motif forming helical dimer interface. Found in many biological systems, this class of motif provides a general framework for transmembrane helix association (39). Recent studies greatly broadened our view on the existence of different types of sequence motifs in membrane proteins, as well as their roles in providing structural stabilization and in regulating biological signaling (39; 40; 41; 42).
Computational discovery of sequence motifs of membrane proteins is a challenging task. Because the length of a transmembrane segment is short, there is strong coupling effects between the appearance of residues at one position and its consequential absence in another position (37; 43; 30; 44). The discovery of the GxxxG motif is the outcome of an important development, namely, the formulation of a rigorous statistical treatment of what would be the expected frequency of various patterns of residues for a given transmembrane helix (37). Prior to the study of Senes et al, widely used statistical models such as the Bernoulli/binomial model, the Markovian model, and the χ2 model do not account for this finite-size effect (45; 44). This model, subsequently termed as the permutation model (44), enables detection of very subtle signals even when only limited data are available. Similar permutation model was also later applied in studying spatial motifs in β-barrel membrane protein (30). Senes et al also introduced a dynamic programming method that makes it possible to compute efficiently the random distribution and p-values essential for identifying motifs using a database of membrane protein structures (37).
Subsequently, exact formulae for propensities of motifs with arbitrary number of residues under the permutation model were discovered, along with analytical formulae for p-value calculations for several types of sequence motifs (43; 44). An improved model, called positional null model that is based on exhaustive permutation but also account for bias of residue at certain positions was also developed (43; 44). Further studies showed that anti-motifs, which are sequence patterns that occur far less than would be expected, also reveal important biological information (30; 43; 44). Applications of these results have lead to the discovery of a large number of sequence motifs and antimotifs in β-barrel membrane proteins (43; 44). For example, the terminal motif YF2 was predicted to be important for recognition by periplasmic chaperon SurA for assisted folding (43), as mutations and deletion of the terminal F residue in PhoE from E. coli resulted in impairment of correct assembly of PhoE into the outer membrane (46). The MeMotif database contains many computationally derived sequence motifs for α-helical membrane proteins (47). A study of GPCRs using motifs of reduced alphabet of amino acids can be found in (48).
3.2. Spatial motifs
There are strong specific helix-helix and strand-strand interactions that can be detected through computational analysis. Interactions between TM helices and between strands as well as their overall assembly are the structural basis of sequence motifs. A global view on how TM helices interact spatially was obtained in a comprehensive study of interacting helical pairs, in which pairs of helices were clustered by their shape similarity (49). It was found that just five clusters accounts for about 74% of all observed interacting helical pairs. These clusters can be rationalized in simple principles of helix-helix packing that goes back to Crick (50). The recurring geometric patterns of helix-helix interactions were organized into a library of spatial motifs of interacting helical pairs (49). The classification of spatial motifs and the library of interacting helical pairs lead to important understanding of the structural organization rule of helix assembly (40). This approach also proved to be invaluable in predicting membrane protein structures (40).
Somewhat similar approach was adopted by Martin et al for β-barrel membrane proteins. From the decomposition of known structures of β-barrel membrane proteins, a library of four residue fragment were constructed (51). It was found that there are strong preferences for different fragments to be located at different regions, and there are also specific preferences for inter-strand contacts between these fragments (51).
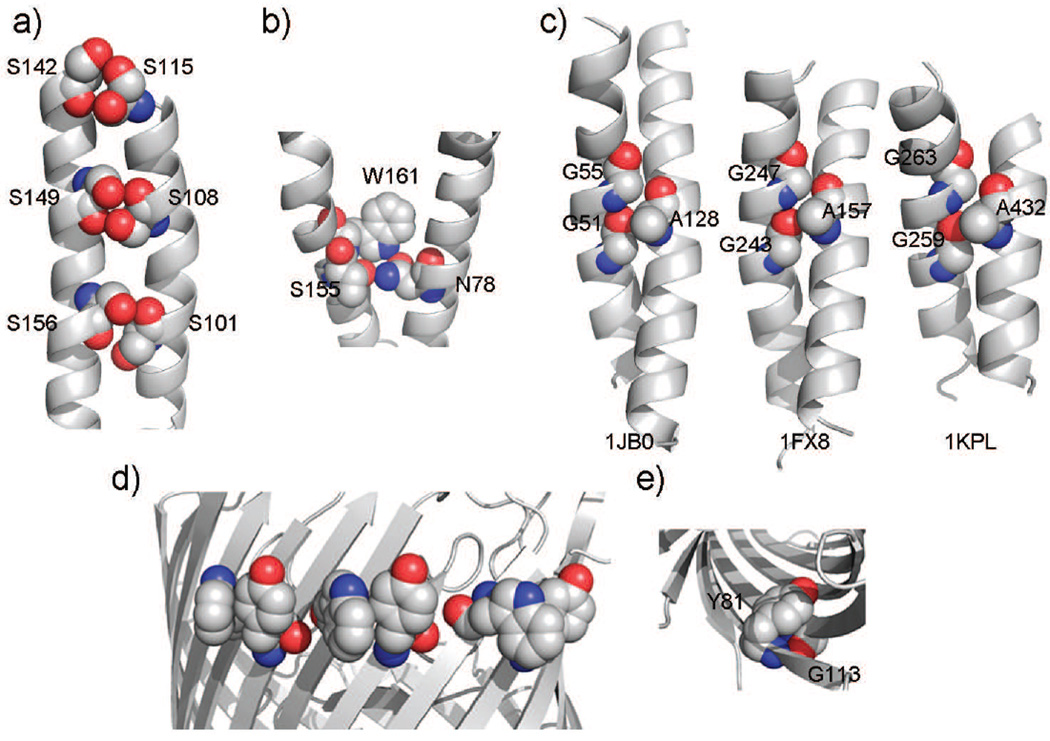
Another approach for discovery of spatial motifs of interacting residues is by comparing the frequency of observed appearance of certain spatial patterns of interacting residues with the frequency of what would be expected by random chance if there were no specific interhelical or interstrand interactions (52; 30; 44). The serine-zipper spatial motif (Fig 2a) was found in cytochrome c oxidase and in erythropoietin receptor (53; 54), where multiple repeated S-S interacting pairs form a large number of H-bonds (52). The placement of these small Ser ensure close packing between helices (55; 49). The polar clamps spatial motif (Fig 2b) involves three residues located on two helices, where a residue capable of forming two or more H-bonds is clamped by H-bonds formed with two residues (53). This motif is highly conserved among G-protein coupled receptors, and likely contributed to stability and specificity of the assembly of TM helices (53).
Figure 2.
Spatial motifs in α-helical and β-barrel membrane proteins: a) The serine zipper in bovine cytochrome c oxidase (helices III and IV), in which H-bonds are formed between S101-S156, S108-S149, and S115-S142. b) A polar clamp in bovine rhodopsin formed by residues W161 and T160 from helix IV and by N78 from helix II. The side chain of W161 is positioned such that its NE1 atom forms an H-bond with the OD1 atom from N78, while the OG1 oxygen from T160 is H-bonded to one of ND hydrogens of N78. c) The A-G-G triplet spatial motif interacting with GG4 sequential motif. Three helical pairs are from unrelated proteins (1jb0: photosystem I; 1fx8: glycerol conducting channel; 1jpl: Clc chloride channel), but all have similar parallel helical orientation with similar crossing angle values between −33 and −48 degrees. d) An instance of the WY non-H-bonded interaction motif in LamB. The aromatic side-chains of Trp and Tyr show considerable contact interaction. e) An instance of the GY strong H-bonded interaction motif in NspA. The protein has been tilted to show the motif on the internal side of the barrel. The aromatic side-chain of Tyr interacts with the Gly residue on the adjacent strand. This is an example of “aromatic rescue” (adapted from (30)).
A systematic analysis of triplet interactions involving three-residues revealed a number of additional spatial motifs, such as A-G-F and A-G-G (Fig 2c) (52). These well-defined spatial conformations exist on helices of unrelated proteins with similar parallel/antiparallel orientation and similar crossing angles (52). Often, well-known sequence motifs such as GG4 and AG4 participate in these higher order motifs of interaction (52).
In β-barrel membrane proteins, Trp and Tyr residues are found to form a frequently occurring motif through non-H-bonded interaction (Fig 2d). The spatial motif aromatic rescue consists of interacting G–Y residues and G–F residues across neighboring strands (30). The Tyr residue adopts an unusual rotamer and covers the backbone of Gly through H-bonding (Fig 2e). This motif stabilizes the protein structure by mitigates the instability Gly causes, as it prevents exposure of the backbone around Gly to solvent, at the same time minimizing exposure of aromatic ring to the solvent (56; 30). Experimental studies on similar motifs in soluble proteins showed that they contribute significantly to protein stability and affect folding dynamics (57). Other spatial motifs found in β-barrel membrane proteins are discussed in (30).
4. Patterns of Evolution in Membrane Proteins, Contact Prediction, and Functional Classification
Both sequence and spatial motifs are products of selection pressure on membrane proteins throughout evolution, either for structural integrity or for biological function. As evolution is a general driving force of biological machineries, we discuss how patterns of evolution of membrane proteins can be detected and how they can be used for biological predictions.
Scoring matrices and patterns of residue substitutions
An essential computational tool for membrane protein studies is sequence alignment (58; 59), which is used in database searches for homologous proteins. A key component of sequence alignment is the scoring matrix for quantification of sequence similarity.
Standard scoring matrices such as BLOSUM and PAM used in default NCBI sequence alignment were derived from soluble proteins (60; 61), and are inappropriate for membrane protein studies. Overall, membrane proteins are under unique physicochemical constraints, and experience selection pressure very different from that of soluble proteins. The patterns of allowed and forbidden substitutions at different positions of the transmembrane segments are different from that of soluble proteins. Scoring matrices therefore need to be specifically designed to capture the evolutionary pressure experienced by the TM segments.
A number of specialized scoring matrices have been developed for helical membrane proteins, including the SLIM and the PHAT matrices. Their applications result in significant improvement in identifying homologs of membrane protein (62; 63). These scoring matrices, however, are inappropriate for β-barrel membrane protein studies. As the lipid bilayer of bacterial outer membrane has different composition (eg., the presence of lipopolysac-charides, LPSs), there are significant differences in the selection pressure experienced between helical and barrel membrane proteins. Results of a rigorous test showed that scoring matrices SLIM and PHAT designed for helical membrane proteins misidentified soluble proteins and random sequences as β-barrel membrane proteins (64).
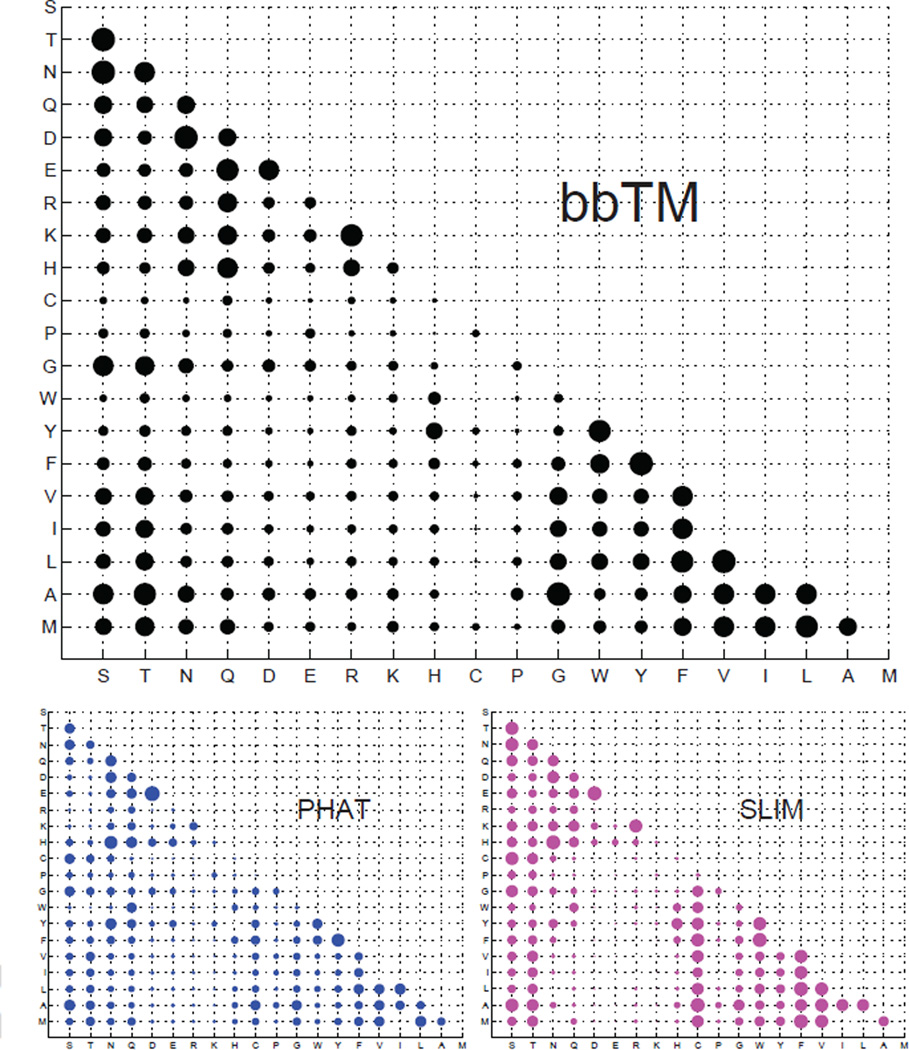
Customized specific scoring matrices can be derived based on a general framework for analyzing amino acid residue substitutions (65). Using a continuous-time Markov process to model amino acid substitution and a Bayesian Monte Carlo estimation algorithm (65), the instantaneous substitution rates of residues in the TM-segments of β-barrel membrane proteins were estimated (64). Scoring matrices specific for different evolutionary time were then derived from the estimated rates (Fig 3), and were shown to have significantly improved sensitivity and specificity in detecting remote homologs of β-barrel membrane proteins (64). As the estimated substitution rates encode probability of exchanges between different residue pairs, they can also be used to suggest design of mutagenesis studies.
Figure 3.
The scoring matrices representing the substitution probability between different residues in transmembrane segments of β-barrel membrane proteins (bbTM) and α-helical membrane proteins (according to the PHAT and Slim matrices) (62; 63). The size of a bubble is roughly proportional to the probability of substitution between the two corresponding residues (adapted from (64)).
A remaining open question is whether evolutionary patterns are sufficiently similar between bacterial and mitochondrial β-barrel membrane proteins, and whether the same scoring matrices would capture their common evolutionary selection pressure. As machineries and mechanisms involved in the assembly of both bacterial and mitochondrial β-barrel membrane protein are quite similar (66; 67; 68), their substitution patterns in the TM strands may be very similar. Further computational study is required to resolve this issue.
Another widely used empirical approach to extract evolutionary information from sequences is using the method of PSI-BLAST (69). Evolutionary information of the transmembrane segments are implicitly encoded in search results, and can be organized into a profile or a position specific weighted matrices (69). Such information can be effectively used to develop machine learning method, for example, as input data in the construction of a Hidden Markov Model or training a neural network for predicting the topology of transmembrane helices (70; 18).
Lipid binding sites are evolutionarily conserved
Phospholipid molecules are not only building blocks of membrane, they also play important roles in influencing the topology, folding, and assembly of membrane proteins, as well as in modulating their biological functions (71). By estimating the site-specific ratio of synonymous vs. non-synonymous substitution of the underlying DNA sequences, selection pressure experienced at individual amino acid positions can be measured (72). It was found that among lipid-facing residues, there are specific lipid binding sites that are evolutionarily conserved. These include the cholesterol-binding sites in β2-adrenergic receptor and in Na-K ATPase, the cardiolipin binding site in formate dehydrogenase-N, and the PG binding site in the KcsA potassium channel (72).
Discovery of packing interaction and predicting functional classes of membrane proteins from evolutionary analysis
If a particular mutation affects the stability or the function of the protein, another mutation might occur at a different position to compensate the effects of the original mutation (73; 74). This phenomenon of co-evolution of residues has been exploited for identification of packing interfaces between helices (75; 76) and for detection of residues that mediates gating in voltage-dependent potassium channels (77).
Evolutionary information can also help to understand the function and classes of poorly characterized membrane proteins, such as those obtained from large scale genome and meta genome sequencing projects (78; 79). For example, there are now a large number of new sequences homologous to ar-chaeal retinal-containing rhodopsin-like proteins found in marine bacteria, fungi, and unicellular algae (80). However, there is a lack of understanding of basic aspects of the biology of these sequences. Structures of known bacterial rhodopsins and evolutionary information contained in the homologous sequences helped to predict and delineate the functional relationship of these rhodopsin-like proteins (81). Although retinal-binding rhodopsins fold in similar structures, the residue make-up of the retinal-binding pockets may be tuned to adapt to different biological functions. Using residue fragments that form the retinal-binding pocket and amino acid substitution matrices derived specifically for the retinal-binding pockets, a relationship tree was obtained that groups rhodopsins by their biological function. This tree characterizes well rhodopsins with known functions, and predicts the functions of uncharacterized rhodpsin-like sequences (81). For example, Gloeobacter violaceus rhodopsin was grouped into the same branch as the xantorhodopsin from Salinibacter ruber, which uses carotenoids for light harvesting in the blue-green region of the light spectrum (82). Subsequent experimental studies showed that G. violaceus rhodopsin indeed binds specifically a carotenoid molecule, which functions as an antenna for light-harvesting (83).
5. Hydrophobicity scale from measurement and from calculation
The physical forces that hold membrane proteins together are of fundamental interests (84; 85; 86; 87; 88; 39; 89). Below we discuss experimentally measured hydrophobicity scale and how they are useful for computational studies. We also discuss equivalent scales derived from analysis of structures and sequences of membrane proteins, as well as their applications.
5.1. Insertion free energy and hydrophobicity scale
Extensive studies have been carried out to measure the free energy of inserting a residue into the lipid membrane. By measuring partitioning of a model helix-forming peptide between water and a reference state, the free energy of helix insertion into a membrane environment is obtained (90; 91; 85). As the environment of inserted helix is important, both membrane center and interface were taken as the reference state in measurements (90; 91; 92). Recently, the free energy contribution from individual amino acid for inserting a TM helix into the biological endoplasmic reticulum (ER) membrane via the Sec61 translocon were measured (93; 94; 28). The resulting insertion free energy scale, called the biological hydrophobicity scale or translocon scale, was the first free energy scale for insertion into a biological membrane. Very recently, the first water-to-bilayer transfer free energy scale measured in the context of a native membrane protein and lipid bilayer was reported (95).
These experimentally derived insertion free energy scales (or hydrophobic scale) have been used effectively in computational studies of membrane proteins. For example, both the Wimley-White whole residue octanol scale and interface scale can be used to accurately predict TM helices in membrane protein (96; 97). The biological hydrophobicity scale was also successfully used in predicting membrane protein topology, with the topology of 79% of a set of 123 membrane protein chains predicted correctly, which is better or comparable to hidden Markov model based methods (29).
Similar hydrophobic scales have also been developed computationally through statistical analysis of known structures of membrane proteins (7; 2; 98; 99; 30; 100; 36). The main idea is to estimate the ratio of the frequency of observing an amino acid residue in the TM segment vs what would be expected by random chance (101; 102). Similar to experimentally measured scale, the empirical hydrophobic scale can also be made dependent on the local helical position of the residue (103; 100), as well as the random model for expected frequency, which is equivalent to the reference states in experimental studies (102; 43; 44). For example, both computed and measured free energy costs of embedding Asn and Gln strongly depends on their location in the TM helix (103). An empirically derived statistical potential function has been successfully applied in genome wide prediction of membrane proteins, with test results indicating an accuracy of 99% (35; 36). Such potential function can also be used to estimate the tilt angle of a TM helix with respect to the bilayer normal, and to select amino acids in membrane protein design studies (100).
5.2. Predicting lipid facing regions of membrane proteins
Empirical hydrophobicity scale can be used to predict lipid-facing regions of membrane proteins. From structural analysis, it was found that there are strong preference for certain residues to face the headgroup and the hydrocarbon core regions of the lipid membrane (99). For example, Lys, Arg, Trp, Phe and Leu prefer to face the head-group region of the lipid bilayer instead of facing other helices, whereas Ile, Leu, Phe and Val prefer to face the hydrocarbon core region of the lipid bilayer. Small and polar residues are more likely to be buried inside the helical bundles and are lipophobic. In addition, it was found that Trp is frequently found in the hydrocarbon region, with its side-chain forming extensive interactions with residues on neighboring helices (99). This finding was consistent with subsequent experimental study in which it was found that Trp strongly supports self-assembly of TM helices, especially when placed on the g-position of the standard heptad. This position facilitates the side chain of Trp to interacting with neighboring helices (104). Overall, buried or interior-facing residues are significantly more polar and, hence, lipophobic, than the exterior residues (105). This lipophobic effect may play a general role in the folding and assembly of membrane proteins by encouraging the overall aggregation of TM helices, with the final structure determined through more specific interhelical H-bond, packing interactions, and loop constraints (105).
The lipid preferences of residues were quantified as a specialized empirical propensity scale called TMlip (for TransMembrane helix-LIPid) potential (99). TMlip was successfully used to predict the orientation of TM helices relative to the phospholipid bilayer (106). Based on a canonical model of the coiled-coil heptad repeat and the combination of the TM-lip propensity and evolutionary information, a computational method called LIPS (LIPid-facing Surface (105) can predict helical surface patches interfacing lipid molecules at 88% accuracy. Other studies based on surface propensity scale and evolutionary information also reported excellent results (77; 107; 108; 109). A recent study that integrates evolutionary profiles and propensities for both membrane exposed residues and solvent exposed residues reported excellent performance (110).
The Lips method is also useful in detecting inconsistencies in the structures of membrane proteins, such as the two structures of cytochrome b6f complex (105). It has also been used to aid in methods of template-free protein structure prediction (111), as well as in suggesting experimental studies (112). Further development based on TMlip potentials allowed the development of the Rants method (for RANKING of Transmembrane helices by Solvent accessibility) (106). Predictions made by Rants have been shown to be useful in designing experiments to identify interior facing residues and important polar interactions in the anion transporter SulP protein family (112).
6. Interactions between helices and between strands
6.1. Physical bases of interhelical and interstrand interactions
Physical forces beyond single body or insertion free energy are also at play in stabilizing membrane proteins. These include multibody interactions involving two or more helices or strands.
Polar interactions
Polar residues buried in the membrane environment likely contribute significantly for maintaining the stability of membrane proteins and their functions (85; 113; 114; 53; 103). Introduction of a single Asn, Asp, Glu or Gln in the TM segment can provide sufficiently strong driving force for helical self-association (113; 114). A survey of known membrane protein structures showed that polar and ionizable residues form extensive H-bond connections between TM helices, as virtually all TM helices form one or more interhelical H-bond (53).
Due to extreme experimental difficulty, quantitative assessment of the magnitude of the H-bond energy in the transmembrane environment became available only recently through elegant studies of double-mutant cycle analysis (115). The average energy of side-chain H-bond interactions is found to be modest (−0.6 kcal/mol). It is possible that the unfolded state of membrane proteins may already have H-bond largely satisfied through alternative interactions (116), as the polarity of the interior of both membrane and soluble proteins are quite similar (99). The apparent contribution of H-bond for specific helical interactions in membrane protein seems to vary significantly (115).
Other interactions
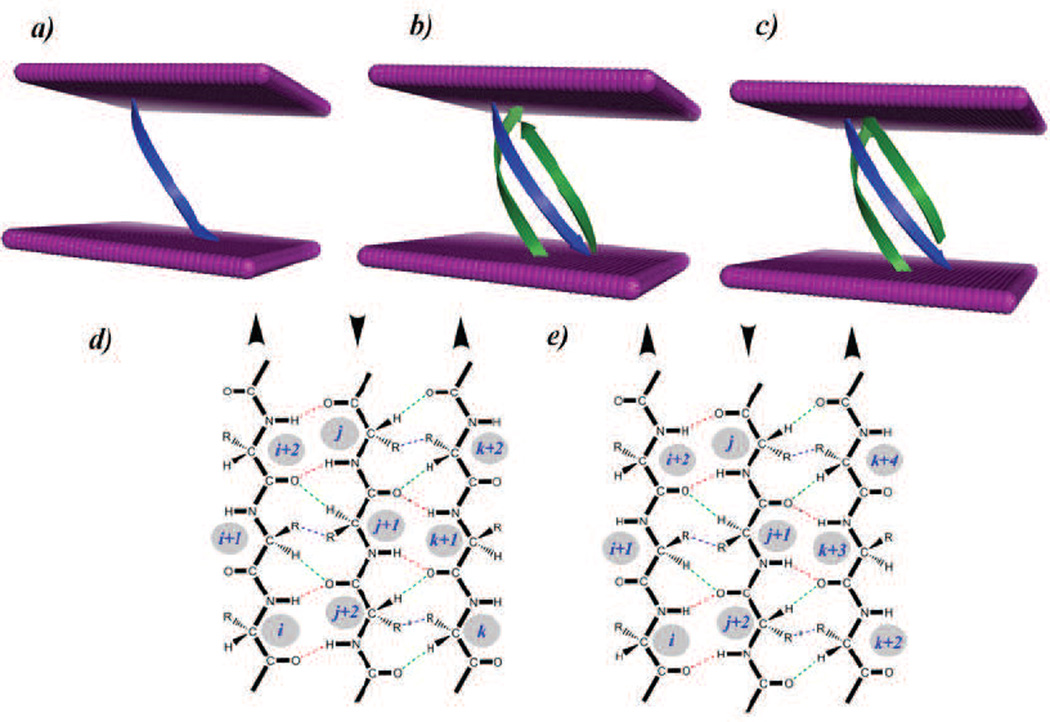
Other physical forces important for the assembly of TM helices include side chains packing, overall helical packing with small residues at helical-helical interfaces, aromatic interactions, and salt bridges (98; 117; 118; 119; 89; 120; 121). For β-barrel membrane proteins, the classical model of β-strand interactions of β-sheets, in which backbone H-bond, side-chain interactions, and weak H-bond stabilize neighboring strands, works well (122; 123; 30) (Fig 4a–c). The energetic contributions of H-bond and residues in the aromatic girdles of TM strands in the protein OmpA have been measured (124; 125). Recently, it was found that specific interactions between lipid and the TM strands of protein FhuA also provide significant stability to the TM domain (72). Based on the TMsip potential function and the reduced state space, it was found that strands 7–9 form the most unstable region in the protein FhuA, and strand 8, which runs through the middle of the LPS-binding site, has the highest energy. These strands are stabilized by biding a lipid molecule.
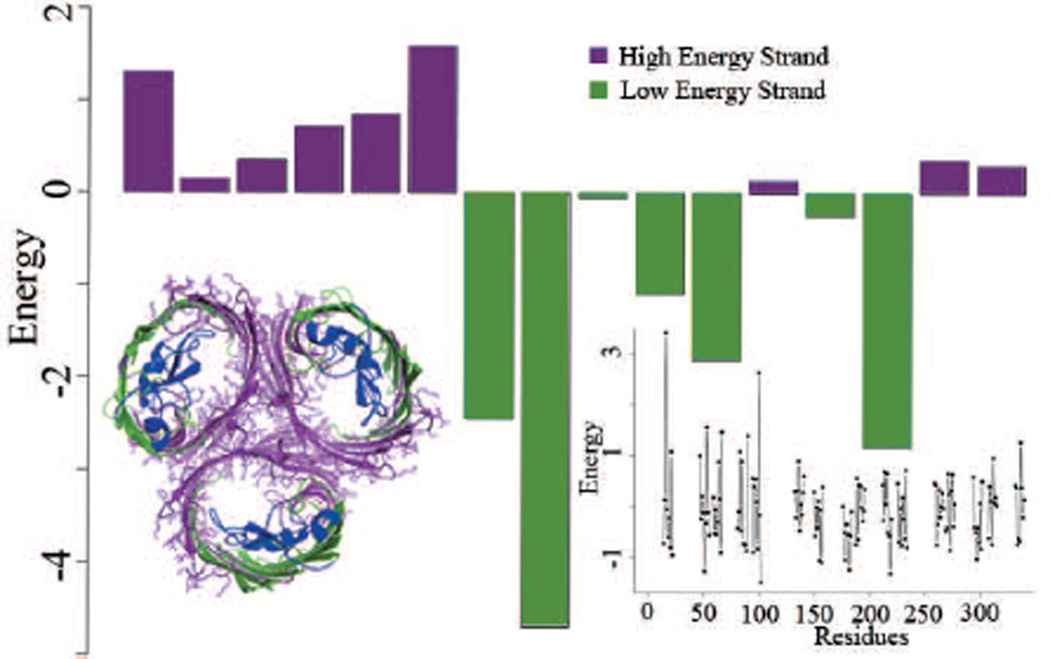
Figure 4.
Interactions of transmembrane strands in β-barrel membrane proteins and energetics of embedding Arg in the transmembrane domain. a) A single β-strand inserted in the membrane bilayer. Both experimental and computational potential functions show that the insertion energy of amino acid varies with their depth in the bilayer. b) Three strands inserted in the bilayer. Although the experimentally measured insertion scales derived from single helix experiments are insightful, a complete picture of the energetics requires considering interactions with neighboring strands/helices. c) An alternate conformation of the same three strands as shown in b). A good computational model can assess how prevalent each conformations is, and can estimate the associated probability. d) and e) show the pairwise interactions by the TMsip potential function according to the β-sheet canonical model (122; 123) for the strands shown in b) and c), respectively. Strong H-bonds between C-O…H-N, weak H-bonds between C-O…H-Cα, and side-chain interactions are shown.
Mechanical and thermal stability
Inter-helical and inter-strand interactions are also the major source of the mechanical stability of membrane protein, as shown by unfolding experiments of bacteriorhodopsin using atomic force microscopy and single-molecule force spectroscopy (126). In addition, these physical forces can be directly linked to protein stability as measured by the calculated melting temperatures of β-barrel membrane proteins (127) (Fig 4a–c and Fig 5).
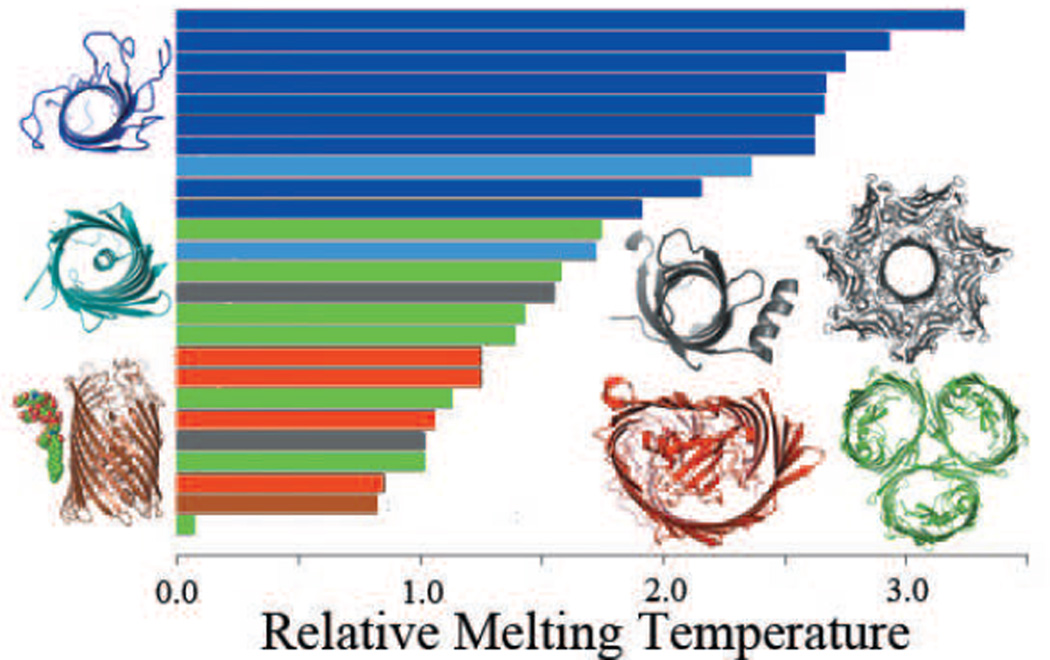
Figure 5.
The relative melting temperature of the transmembrane domains of 25 β-barrel membrane proteins can be calculated by enumerating all possible conformations in a reduced state space. Monomers that are stable without in-plugs and out-clamps, e.g., OmpA are shown in dark blue. Monomers stabilized by small in-plugs, e.g., NalP are shown in light blue. Monomers stabilized by out-clamps are represented by PagP and α-hemolysin (grey). β-barrels that require oligomerization for stability, e.g., ScrY are shown in green. Monomers stabilized by large in-plugs e.g FptA are shown in red. β-barrel membrane proteins can also have specific protein-lipid interactions, e.g., FhuA (brown) that increase protein stability. All stable monomers tend to have higher relative melting temperature and group towards the top of the graph (adapted from (127)).
6.2. Empirical potential function for biological understanding
A number of empirical potential functions have been developed for helix-helix interactions. In an early study, pairwise potential function based on statistics of known structures was successfully used to predict super-secondary structures of several packed small TM helices (128). Based on an atomic probabilistic model and packing contacts detected through Delaunay triangulation of membrane protein structures, a potential function for helix interaction called MHIP (for membrane helical interfacial pairwise propensity) was developed (98). By combining packing and helix contact analyses, Eilers et al developed an interfacial propensity scale for prediction of the relative orientation of TM helices (129). Dobbs et al developed a potential function for predicting inter-helical packing based on optimized discrimination of native helix-helix interactions from Monte Carlo generated decoy structures (130). Further development includes distance-based empirical potential that works well in predicting anchoring helix pairs (131). An interhelical contact potential was developed using a reduced alphabet of four amino acid types, which can discriminate native structures from many decoy conformations (132). Another empirical pairwise potential function for helical interaction was a major component of the force field used in the ROSETTA structure prediction method (111; 133; 134).
General mechanisms to stabilize β barrel membrane proteins
For β-barrel membrane proteins, an empirical potential function called the TMsip (for TransMembrane Strand Interaction Propensity) potential has been developed based on the canonical interaction model of β-sheet (122; 123; 30). Its null model is the rigorous permutation model discussed earlier. The TMsip potential function can be used to identify weakly stable region in the TM domain (127). Analysis of these weakly stable regions revealed four general mechanisms that β-barrel membrane proteins use to stabilize the TM domain (Fig 5): the well-known in-plug mechanism, as seen in FhuA (135), in which an inter-strand loop or a separate domain folds back and plug into the interior barrel to stabilize the TM barrel; the out-clamp mechanism as seen in PagP and hemolysin, in which a secondary structural element such as a helix outside the barrel stabilizes the TM barrel (136; 127), the newly discovered mechanism of specific lipid binding, in which the unstable region of the TM barrel is stabilized through specific strong binding with the LPS lipid molecule (72), as well as the mechanism of protein-protein interactions with which weakly stable regions are stabilized by another membrane protein (127).
Empirical potential function can also help to gain biological understanding. There are many examples where excellent agreement were reported between results obtained using experimentally derived free energy scale and those obtained using empirical potential function. For example, the measured free energy scales of inserting amino acid residues embedded in a helix into the endoplasmic reticulum agrees well with free-energy profiles derived from statistical analysis of membrane protein structures (28).
Understanding Arg in transmembrane segment from insertion energy and empirical potential function
Multiple Arg residues are found in the S4 trans-membrane helix of KvAP ion channel and other related channel proteins, and are likely to be important in sensing membrane depolarization and mediating channel gating (137; 138). Intuitively, these ionizable residues found in the hydrophobic core of lipid membrane would be energetically costly, and it is important to understand the physical basis of their locations. There is significant discrepancy in free energy of inserting Arg into the hydrophobic core when measured experimentally vs when calculated from molecular dynamics (MD) simulation (93; 139; 140; 141; 95). It was found that extra helices facilitate the retainment of hydration water molecules, which reduces solvation cost significantly (142). It was also suggested that part of the discrepancy may be because MD simulation does not account for the tendency of Arg side chain to snorkel towards the membrane-water interface (141). Although simulations are carried out using physics based force field, the large number of parameters involved and the difficulty in ensuring full sampling of an equilibrium ensemble of conformations may be sources of non-negligible errors (92; 141).
Hydrophobicity scale and empirical potential function can offer significant insight. According to the analysis of Hristova and Wimley using the experimentally derived Wimley-White scale (92), less than two Ala to Leu substitutions are required to compensate for one Ala to Arg substitution. It was found that it is easier to insert Arg in the interface region than the core of the bilayer (92).
The occurrence of Arg in hydrophobic core can also be understood through empirical potential function. Important favorable interactions across neighboring helices/strands often exist (98; 53; 103; 132), and such context dependent interactions will significantly modify the overall free energy of the protein. Since measured insertion free energy scales were mostly based on studies designed with single TM helices, the observed occurrence of Arg in the hydrophobic core of natural membrane proteins can be better interpreted with additional consideration incorporating inter-helical and inter-strand interactions.
This can be illustrated by analyzing the energetic consequence of embedding an Arg residue in the TM segment of a β-barrel membrane protein using the empirical potential function TMsip (Fig 4) (30). Arg in β-barrel membrane proteins facing inside the β-barrel pore is energetically favorable, but very unfavorable when facing the lipid membrane (30). Through interstrand interactions, there are three additional types of interactions that are major contributors to the stability of TM β-strands, namely, strong H-bond between main chain (C-O…H-N), side-chain interaction (R…R) including side chain H-bond, and weak H-bonds between C-O…H-Cα (122; 123; 30). According to the recently updated version of the TMsip scale incorporating additional structural data, Arg can be stabilized by main chain H-bond interactions with Ala, Trp, Val, and Thr, if they are located on appropriate positions of the neighboring strands (30). Since side chain H-bonds are known to contribute only modestly to the overall stability of membrane proteins (143), the context dependent main chain H-bond interactions are likely the main contributors that modifies the single-body energetics of Arg insertion.
According to TMsip, Arg is only slightly energetically unfavorable in the extracellular interfacial region, but is highly unfavorable in the hydrophobic core region and the periplasmic interfacial region (30). As Arg residue is inserted from the periplasmic side into the lipid bilayer, favorable main-chain H-bond interactions with Ala, Trp, Val, and Thr located on neighboring strands may compensate for the unfavorable insertion of Arg (Fig 4). This compensation effect would facilitate the translocation of Arg towards the more favorable extracellular interface in β-barrel membrane protein.
Experimentally measured insertion free energy derived from studies of single helix experiments can be regarded as one-body energetics, and the equivalent empirical potential function are hydrophobic scale involving only a single residue and its depth in the membrane environment. An accurate account of the full energetics of residues in the context of a wild type membrane protein needs also incorporate the effects of inter-helical or inter-strand interactions, namely, the two-body interactions. It is possible that higher order cooperative effects may also be relevant (52; 39).
In a recent study, the free energy changes in a wild type membrane protein were measured when an Ala was replaced with each of the 20 amino acids (95). This is the first time such measurements were made in wild type membrane protein placed in a lipid bilayer. Although Trp fluorescence was employed in experimental measurement, and Glu and Asp are mostly likely in the protonated state at the experimental condition, free energy changes of replacing Ala with the other residue types provide a wealth of quantitative information about membrane protein stability. It was found that Arg substitution incurs only a modest free energy cost (95). Although the analysis of this study was based on a simple one-body additive insertion energy model, interstrand interactions that is context dependent at the host position is likely to be non-negligible in wild type membrane proteins. The wealth of information provided in studies such as (95) can be used for alternative analyses using a statistical mechanical model (127) that considers context dependent interstrand interactions as well as non-native conformations, which works well to account for observed nonlinear and non-additive effects.
7. Predicting Structure of Membrane Protein
7.1. Irregular structures and their prediction
An idealized model of helical membrane proteins is that of an assembly of highly hydrophobic helices connected by loops, with orientations perpendicular to the membrane plane. This is the model upon which many successful hidden Markov model (HMM) methods for topology prediction were based (144). However, recent structures showed that there are many irregular structures. Transmembrane helices are often kinked at varying length and tilt angle (145; 146). In the water-membrane interfacial regions, there may exist amphipathic α-helices parallel to the membrane plane (147; 148). In addition, there exists re-entrant regions that enter and leave the membrane from the same side of the transmembrane region (149).
About 44% of TM helices have kinks, with 35% of which associated with Pro residue, and others with Ser and Gly at the center of the kink (150; 151). Kinks are likely to be important for membrane protein function, as they provide locations for movement such as hinge bending, and introduces structural diversity even among members of the same protein family. It was suggested that Pro in ancestral proteins may have initiated such kinks (152). TM helices subsequently were stabilized through evolution to an extent that the maintenance of the kinked conformation no longer required the presence of Pro residues (152). Molecular dynamics simulation of single TM helix has been successful in identifying many kinks (151). In a study of 405 TM helices, it was found that 79% of the proline kinks, 59% of the vestigial proline kinks, and 18% of the non-proline helical kinks can be reproduced from 1 ns of MD simulation (151).
A study of the re-entrant regions using the technique of principal component analysis for dimension reduction revealed that these regions have distinct amino acid composition (149). As many re-entrant regions are found in transporters, Gly and Ala are abundantly found in this region (149). In addition, Ser and Thr are also enriched (153). Hidden Markov models developed based on these patterns can now predict the re-entrant regions successfully at 70–75% accuracy (149; 153).
7.2. Comparative three-dimensional model of membrane protein structure
If the structure of a homologous membrane protein exists, comparative or homology structural model can be built based on the template structure (154; 155). This technique has been applied fruitfully to study the G-protein coupled receptors (GPCRs), an important receptor for cellular signal transduction (155; 156). When a template structure is identified and a quality alignment is obtained, a specialized comparative modeling method Medeller can identify a reliable core structure, and build a structural model by extending the core to other TM region and to the loop region (157). This approach showed higher accuracy in modeled structure than generic homology modeling methods. For β-barrel membrane proteins, the TMBPro method takes predicted secondary structures and evaluate their overall energy to each structural template containing the same number of strands (158). Combined with conformational search via simulated annealing for the lowest energy alignment of the sequence to the structural template, the conformation with the lowest overall energy can be taken as the predicted structure (158). It is expected that improvement in alignment and detection of remote homologs can be obtained through usage of customized scoring matrices (64; 159). This will allow further leverage of current knowledge of existing membrane protein structures, at a rate of about 130 proteins per template structure (159). Furthermore, these scoring matrices are found to be useful for identifying mitochondria outer membrane proteins in eukaryotes (159).
7.3. Template-free prediction of membrane protein structure
A more challenging task in structure prediction is when there is no known structures that can serve as the template structure. That is, none of the homologous proteins have known structures. The Rosetta de novo protein structure prediction method has been extended to predict structures of helical membrane proteins, without the need of a template structure (111; 133; 134), although no template-free methods currently exist that can predict structures of β-barrel membrane proteins.
Using an empirical potential function that combines van der Waals interaction, backbone torsional force, electrostatic interaction, and orientation dependent H-bond interaction, Barth et al developed a method based on Rosetta Monte Carlo sampling that can successfully recover the side chain conformations of membrane proteins, can model distorted TM helices, and can predict the conformation of glycophorin A interface (133). Further prediction of likely interacting helical pairs with a large sequence separation was obtained from a carefully constructed library of interacting helical pairs and the evolutionary profiles of the two helices. With such predicted inter-helical geometry and co-factor coordination when available to restrict the conformational space, Barth et al successfully predicted three dimensional structures of a divers set of membrane proteins with different size, topologies, and biological functions, with excellent results at the level of about 4 Å in RMSD (134).
7.4. Structure prediction through combined experimental and computational studies
Partial experimental information that is insufficient on its own right for structure determination can be very effective in guiding computational prediction methods towards a much smaller feasible space for conformational search. An important form of experimental data is coarse grained density map of cryo-electron microscopy at medium-resolution (7–10 Å), in which helices are better resolved as rods than strands and loops. By placing predicted helices into the density rods for helices and adding modeled loops, the overall structures of helical membrane proteins can be predicted in some cases with much improved resolution, although this method hinges upon the correct prediction of helices (160). Combining CryoEM data with evolutionary information, the Cα-trace model of the transmembrane domain of human copper transporter 1 was also successfully constructed (119; 161).
Another approach is to integrate experimental mutagenesis data into the structure prediction protocol by biasing the selection of the final model towards those that are consistent with the experimental mutagenesis results. This approach has been applied successfully to predict the structure of the transmembrane domain of the homodimeric BNIP3 (162) and the heterodimeric structure of complete αIIb and β3 complex (163). However, significant amount of experimental data are required, and therefore this approach is best-suited for well studied membrane proteins. A general theoretical framework to generate protein structures that satisfy different experimentally derived restraints described in (164) may be useful for such tasks.
8. Beyond Structure Prediction: Ensemble Properties, Protein-Protein Interactions, and Protein Design
Great progresses have been made in predicting structures of membrane proteins. However, many important problems in membrane protein studies require information beyond that of a single native structure. Below we first discuss studies on the ensemble nature of conformations of membrane proteins, which is the basis of their thermodynamics properties. We also discuss prediction of oligomerization state and protein-protein interactions. In addition, we discuss future development in protein design, in which computational studies will likely make significant contributions.
8.1. Ensemble Nature of Membrane Protein Structure and Their Thermodynamic Properties
There are many important questions beyond the knowledge of a single predicted structure. For example, do membrane proteins exist in multiple conformations (Fig 4)? What are their associated probabilities? How thermodynamic properties can be calculated from ensemble properties of conformations? How do dynamic transitions occur among these conformations and how such changes may contribute to the observed biological functions?
In the study of β-barrel membrane proteins, progresses have been made in addressing some of these questions (127). Because of the relatively regular pattern in strand interactions, the conformational space of TM strands can be effectively modeled using a simplified state-space model (30). By assuming a reduced conformational space in which each strand can slide up or down for a total of 7 positions, one can enumerate all possible conformations and calculate the energy value for each conformation. Thermodynamic properties of the transmembrane domain can then be computed (127). Fig 5 depicts one such thermodynamic property, namely, the relative melting temperature calculated for the TM domains of a number of β-barre membrane proteins.
Role of nonnative and alternative conformations
It is important to consider non-native conformations in computing thermodynamic properties of membrane proteins. Although it was not immediately obvious why TM strands would not always adopt the ground state conformation, as it would be very costly to break all the H-bonds to move up or down to a different register, experimental results on PagP showed that there can be significant conformational change when different detergent is present (165). In fact, alternative conformations with low energy may serve as obligate on-pathway transient states (166).
Recent studies in helical membrane protein demonstrated the flexible nature of transmembrane helices, which contain many kinks, bulges, and re-entrant loops (149; 167; 150). Furthermore, the spatial close proximity among newly synthesized TM helices during co-tranlational insertion to membrane suggests that there may exist interhelical interactions even in the early stage of membrane protein folding (168). For example, several experimentally determined TM helices in Gltph glutamate transporter were found not to have lowest free energy of insertion in wild type protein, and the segment with both measured and predicted lowest free energy has significant position displacement compared to the wild type protein (168). These findings suggest that TM helices may shift positions dramatically during the folding and oligomerization process, which may be important for bringing functionally important polar residues into places.
Overall, the population of alternative conformational states may play important roles in determining the final native structure and function of membrane protein, and in ensuring the overall stability and robustness of the cell machineries in which membrane proteins are important components.
8.2. Protein-Protein Interactions
A genomic scale survey of domain combinations of helical membrane proteins suggested that membrane proteins exist mostly as single domains, and oligomerization within the membrane may be the general mechanism for membrane proteins to gain new biological functions (169; 170). For GPCRs, characterizing their oligomerization state is of considerable importance (171). Computational docking and molecular dynamics simulations have been applied in gaining insight into the oligomerization state and in delineating the protein-protein interface (see ref (171) for a recent review).
The oligomerization state of β-barrel membrane proteins can be accurately predicted computationally (127). Based on the TMsip empirical potential function and the reduced conformational state model, it was found that the average deviation in energy of the unstable strands from the mean of all strands serves as an excellent predictor of the overall oligomerization state of the membrane protein. In a leave-one out blind test of 25 non-homologous β-membrane proteins, in which each of the protein is taken in turn for testing, while the remaining 24 proteins used for model construction, excellent results are obtained in predicting the oligomeric state. As subsequently realized that protein FhuA can exist in dimeric form, the predictions of the oligomerization state for these 25 β-barrel membrane proteins are 100% accurate with 100% specificity (172; 127). These predictions are robust, as the outcome does not depend on specific choice of structures used in the construction of the energy function. Furthermore, as structural information is not essential for such predictions, the oligomerization state can also be predicted quite successful even when only sequence information is employed (127): The accuracy and specificity are 96% and 94% when only sequence information is used (127), respectively, with the consideration that protein FhuA indeed form a dimer (172).
The interface of protein-protein interaction for β-barrel membrane proteins can also be predicted (127). Based on the observation that the protein-protein interface is enriched with weakly stable strands, interfaces can be predicted either with the knowledge of the structure where high accuracy can be achieved, or with sequence information only where accuracy is slightly degraded (Fig 6). Another approach based on the machine learning method of random forest can also predict residues located in the protein-protein interface accurately (173).
Figure 6.
The β-barrel membrane protein OmpF exists as a trimer, with strands 1–5 and 16 forming the protein-protein interaction (PPI) interface. The expected energy of the trans-membrane domains of each of the β-strands is calculated using the TMsip statistical potential function. The consecutive strands 1–6 and 15–16 have high expected energy and coincide with the real PPI interface of the protein. Here high energy strands are also termed as weakly stable. The accuracy of identifying the β-strands located in the PPI interface in a data set of 25 non-redundant β-barrel membrane proteins is 78% using structural information and 66% using sequence information only. The right inset plots the contribution of each residue to the stability of the protein. This can be used to suggest mutagenesis studies that aim to change the stability of the protein (adapted from (127)).
Success in predicting the oligomerization state and in identifying protein-protein interaction interface in the TM domain will likely reveal novel insight into the mechanism of many membrane proteins. For β-barrel membrane proteins, mutations can be suggested that would strongly affect the oligomerization state (Fig 6, inlet). It is conceivable that protein-protein interface for eukaryotic membrane proteins can also be predicted, and mutants with different oligomerization behavior can be engineered. For example, the eukaryotic proteins VDAC found in mitochondria oligomerizes during the induction of apoptosis (174). Predicted oligomerization site on VDAC can aid in experimental design of studies to identify key residues involved in VDAC oligomerization. Such investigations will be important for studying the underlying mechanism of apoptosis (174; 175).
8.3. Design and engineering of membrane proteins
De novo design of membrane proteins and inhibitors
De novo protein design and protein engineering aim to produce proteins with new or enhanced activity and stability. Although significant progress has been made in recent years (176), there are only a limited number of reported successes in de novo membrane protein design. The most promising approach is to extend computational methods used for the design of globular proteins. This approach lead to the successful design of a four helix bundle membrane protein engineered to bind two Fe(II/III) diphenylporphyrins in a bis-His geometry. This designed membrane protein forms a channel capable of transmembrane electron transfer (177). There has also been significant progress in the design of small peptides that target the transmembrane proteins and inhibit protein-protein interactions in the TM domain (178). Anti-αIIb peptide that targets the transmembrane domain of the α subunit of the integrin αIIbβ3 disrupts the heteromeric helix-helix interactions. The specificity of the designed anti-αIIb was validated both in vitro and in vivo (178; 179).
Engineering stability and oligomerization state of membrane proteins
As more structures of membrane proteins become available, improved understanding of their organizational principles has led to efforts in engineering membrane proteins with improved protein stability (180). For example, a metal binding site was engineered in the mastoparan X protein, an am-phiphilic α-helix that is too short to form a stable helix in water. This newly acquired metal binding ability stabilizes the helical structure of the protein, and increased the binding and lysis ability of the protein to the membrane (181). Longer transmembrane regions were also engineered for the β-barrel membrane protein FhuA to match the hydrophobic cores of thick polymeric membranes, with the goal for targeted drug delivery (182).
There have also been successes in engineering stability of oligomerized membrane proteins. Using a statistical potential function, mutations that would stabilize or destabilize the dimeric interface of GPCRs were predicted based on a de novo designed rhodopsin homodimer model (183). These predictions compared favorably with experimental studies (183). Computational study on β-barrel membrane protein has also suggested that oligomers form primarily due to the instability of monomers. Such oligomerization can be altered by mutations that stabilize or destabilize the monomeric form of the β-barrel membrane protein (127).
Geometry and selectivity
Success has also been reported on engineering the geometry of β-barrel membrane protein. Most β-barrel membrane proteins consist of an even number of strands, and β-hairpins are often thought as the basic repeating unit (184; 185). It is plausible that the evolution of β-barrel membrane proteins are based on the modularity of hairpin duplication and oligomeric assembly of these hairpins (185). Indeed, bacterial toxin α-hemolysin and the multidrug efflux system TolC forms β-barrel membrane protein upon oligomerization once multiple hairpins are inserted into the lipid membrane (186; 187). Arnold et al constructed an artificial β-barrel membrane protein by duplicating the sequence of 8-strand OmpX. The resulting protein has a pore size of that of a 16-strand porin based on single-channel conductance measurements (185).
Pores with specially constructed filters have been successfully engineered to control the flow of ions and metabolites through the membrane bilayer. β-barrel membrane protein OmpF, which is slightly cation-selective due to the −1 net charge in the filter region, has been converted into Ca2+-selective channel by carefully mutating two Args located in the constriction zone to Glus (188). Similarly, aquaporin-1 filter was engineered to enhance proton conductance computationally and the results were subsequently confirmed by experiments (189). Dynamics of reconstituted native plugged FhuA channels in an ion-conducting state have been studied by adding 4M urea on the cis side, which reversibly unfolds the plug domain and open an ion-conducting pathway that mimics the TonB dependent channel (190). Mutants of OmpF whose extracellular loops were deleted one at a time were also engineered to be pH insensitive (191).
It is likely that the pace of designing membrane protein will accelerate, and many more novel membrane proteins with desirable biophysical properties and novel or enhanced functions will be made.
9. Conclusion
We have summarized key aspects of computational studies of membrane proteins, including bioinformatics prediction of membrane proteins and their topology, the discovery and implication of sequence and spatial motifs, membrane protein evolution and the substitution patterns of amino acids in the TM domain, as well as the modeling of the underlying physical forces through empirical potential function. We have also discussed recent successes in structure prediction and in protein-protein interactions prediction, as well as progress in characterization of ensemble properties of membrane proteins. We believe that computational studies based on both the underlying physical forces as well as bioinformatics analysis of evolutionary signal will continue to make important contributions in understanding and manipulating membrane proteins that compliments experimental investigations.
Highlights.
-
>
Prediction of membrane proteins and topology,
-
>
Discovery of sequence and spatial motifs, detection of evolutionary signal,
-
>
Empirical potential functions,
-
>
Structure predictions and ensemble properties,
-
>
Prediction of protein-protein interactions
Acknowledgment
We thank Drs. Patrick Barth, Ann Delcour, Robert Eisenberg, Alan Finkelstein, Dennis Gessman, Nathan Joh, Linda Kenney, Renhao Li, Stephan Nussberger, Varda Shoshan-Barmatz, Alessandro Senes, Ann Stanley, and Bill Wimley for very helpful discussions. This work was supported by NIH grants GM079804 and GM086145, NSF grants DBI 1062328 and DMS-0800257, and ONR grant N00014-09-1-0028.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms, Protein Science: A Publication of the Protein Society. 1998;7(4):1029–1038. doi: 10.1002/pro.5560070420. PMID: 9568909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arkin I, Brunger A. Statistical analysis of predicted transmembrane alpha-helices. Biochim Biophys Acta. 1998;1429(1):113–128. doi: 10.1016/s0167-4838(98)00225-8. [DOI] [PubMed] [Google Scholar]
- 3.DeFelice L. Transporter structure and mechanism. Trends Neurosci. 2004;27(6):352–359. doi: 10.1016/j.tins.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Cho W, Stahelin R. Membrane-protein interactions in cell signaling and membrane trafficking. Annu Rev Biophys Biomol Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- 5.von Heijne G. The membrane protein universe: what’s out there and why bother? J Intern Med. 2007;261(6):543–557. doi: 10.1111/j.1365-2796.2007.01792.x. [DOI] [PubMed] [Google Scholar]
- 6.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. Journal of Molecular Biology. 1982;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. PMID: 7108955. URL http://www.ncbi.nlm.nih.gov/pubmed/7108955. [DOI] [PubMed] [Google Scholar]
- 7.von Heijne G. Membrane protein structure prediction, hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Engineering. 1997;10(1):1–6. doi: 10.1093/protein/10.1.1. PMID: 9051728. URL http://www.ncbi.nlm.nih.gov/pubmed/9051728. [DOI] [PubMed] [Google Scholar]
- 9.Orlean P, Menon AK. Thematic review series: lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. Journal of Lipid Research. 2007;48(5):993–1011. doi: 10.1194/jlr.R700002-JLR200. PMID: 17361015. doi:10.1194/jlr.R700002-JLR200. URL http://www.ncbi.nlm.nih.gov/pubmed/17361015. [DOI] [PubMed] [Google Scholar]
- 10.Bendtsen J, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340(4):783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 11.Poisson G, Chauve C, Chen X, Bergeron A. FragAnchor: a large-scale predictor of glycosylphosphatidylinositol anchors in eukaryote protein sequences by qualitative scoring, Genomics. Proteomics Bioinformatics / Beijing Genomics Institute. 2007;5(2):121–130. doi: 10.1016/S1672-0229(07)60022-9. PMID: 17893077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pierleoni A, Martelli PL, Casadio R. PredGPI: a GPI-anchor predictor. BMC Bioinformatics. 2008;9:392. doi: 10.1186/1471-2105-9-392. PMID: 18811934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulz GE. beta-Barrel membrane proteins. Current Opinion in Structural Biology. 2000;10(4):443–447. doi: 10.1016/s0959-440x(00)00120-2. PMID: 10981633. URL http://www.ncbi.nlm.nih.gov.proxy.cc.uic.edu/pubmed/10981633. [DOI] [PubMed] [Google Scholar]
- 14.Rost B, Fariselli P, Casadio R. Topology prediction for helical trans-membrane proteins at 86% accuracy. Protein Sci. 1996;5(8):1704–1718. doi: 10.1002/pro.5560050824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martelli PL, Fariselli P, Casadio R. Bioinformatics. Suppl 1. Vol. 19. England: Oxford; 2003. An ENSEMBLE machine learning approach for the prediction of all-alpha membrane proteins; pp. 205–211. PMID: 12855459. URL http://www.ncbi.nlm.nih.gov/pubmed/12855459. [DOI] [PubMed] [Google Scholar]
- 16.Tusnády GE, Simon I. Bioinformatics. 9. Vol. 17. England: Oxford; 2001. The HMMTOP transmembrane topology prediction server; pp. 849–850. PMID: 11590105. URL http://www.ncbi.nlm.nih.gov/pubmed/11590105. [DOI] [PubMed] [Google Scholar]
- 17.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. Journal of Molecular Biology. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. PMID: 11152613. doi:10.1006/jmbi.2000.4315. URL http://www.ncbi.nlm.nih.gov/pubmed/11152613. [DOI] [PubMed] [Google Scholar]
- 18.Viklund H, Elofsson A. Best alpha-helical transmembrane protein topology predictions are achieved using hidden markov models and evolutionary information. Protein Science: A Publication of the Protein Society. 2004;13(7):1908–1917. doi: 10.1110/ps.04625404. PMID: 15215532. doi:10.1110/ps.04625404. URL http://www.ncbi.nlm.nih.gov/pubmed/15215532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Käll L, Krogh A, Sonnhammer ELL. A combined trans-membrane topology and signal peptide prediction method. Journal of Molecular Biology. 2004;338(5):1027–1036. doi: 10.1016/j.jmb.2004.03.016. PMID: 15111065. doi:10.1016/j.jmb.2004.03.016. URL http://www.ncbi.nlm.nih.gov/pubmed/15111065. [DOI] [PubMed] [Google Scholar]
- 20.Käll L, Krogh A, Sonnhammer ELL. Bioinformatics. Suppl 1. Vol. 21. Oxford; England: 2005. An HMM posterior decoder for sequence feature prediction that includes homology information; pp. 251–257. PMID: 15961464. doi:10.1093/bioinformatics/bti1014. URL http://www.ncbi.nlm.nih.gov/pubmed/15961464. [DOI] [PubMed] [Google Scholar]
- 21.Nugent T, Jones DT. Transmembrane protein topology prediction using support vector machines. BMC Bioinformatics. 2009;10:159. doi: 10.1186/1471-2105-10-159. PMID: 19470175. doi:10.1186/1471-2105-10-159. URL http://www.ncbi.nlm.nih.gov/pubmed/19470175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Heijne G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 1986;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Heijne G, Gavel Y. Topogenic signals in integral membrane proteins. Eur J Biochem. 1988;174(4):671–678. doi: 10.1111/j.1432-1033.1988.tb14150.x. [DOI] [PubMed] [Google Scholar]
- 24.Gavel Y, Steppuhn J, Herrmann R, von Heijne G. The ‘positive-inside rule’ applies to thylakoid membrane proteins. FEBS Lett. 1991;282(1):41–46. doi: 10.1016/0014-5793(91)80440-e. [DOI] [PubMed] [Google Scholar]
- 25.Hennerdal A, Elofsson A. Bioinformatics. 9. Vol. 27. England: Oxford; 2011. Rapid membrane protein topology prediction; pp. 1322–1323. PMID: 21493661. doi:10.1093/bioinformatics/btr119. URL http://www.ncbi.nlm.nih.gov/pubmed/21493661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gromiha MM, Suwa M. Discrimination of outer membrane proteins using machine learning algorithms. Proteins. 2006;63(4):1031–1037. doi: 10.1002/prot.20929. PMID: 16493651. doi:10.1002/prot.20929. URL http://www.ncbi.nlm.nih.gov/pubmed/16493651. [DOI] [PubMed] [Google Scholar]
- 27.Hu J, Yan C. A method for discovering transmembrane beta-barrel proteins in gram-negative bacterial proteomes. Computational Biology and Chemistry. 2008;32(4):298–301. doi: 10.1016/j.compbiolchem.2008.03.010. PMID: 18467177. doi:10.1016/j.compbiolchem.2008.03.010. URL http://www.ncbi.nlm.nih.gov/pubmed/18467177. [DOI] [PubMed] [Google Scholar]
- 28.Hessa T, Meindl-Beinker N, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White S, von Heijne G. Molecular code for transmembrane-helix recognition by the sec61 translocon. Nature. 2007;450(7172):1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- 29.Bernsel A, Viklund H, Falk J, Lindahl E, von Heijne G, Elofsson A. Prediction of membrane-protein topology from first principles. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(20):7177–7181. doi: 10.1073/pnas.0711151105. PMID: 18477697. doi:10.1073/pnas.0711151105. URL http://www.ncbi.nlm.nih.gov/pubmed/18477697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackups R, Jr., Liang J. Interstrand pairing patterns in beta-barrel membrane proteins: the positive-outside rule, aromatic rescue, and strand registration prediction. J Mol Biol. 2005;354(4):979–993. doi: 10.1016/j.jmb.2005.09.094. [DOI] [PubMed] [Google Scholar]
- 31.Nikaido H, Nakae T. The outer membrane of Gram-negative bacteria. Adv Microb Physiol. 1979;20:163–250. doi: 10.1016/s0065-2911(08)60208-8. [DOI] [PubMed] [Google Scholar]
- 32.Bagos PG, Liakopoulos TD, Spyropoulos IC, Hamodrakas SJ. PRED-TMBB: a web server for predicting the topology of beta-barrel outer membrane proteins. Nucleic Acids Research. 2004;32(Web Server issue):W400–W404. doi: 10.1093/nar/gkh417. PMID: 15215419 PMCID: 441555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Randall A, Cheng J, Sweredoski M, Baldi P. Bioinformatics. 4. Vol. 24. England: Oxford; 2008. TMBpro: secondary structure, beta-contact and tertiary structure prediction of transmembrane beta-barrel proteins; pp. 513–520. PMID: 18006547. [DOI] [PubMed] [Google Scholar]
- 34.Singh NK, Goodman A, Walter P, Helms V, Hayat S. TMBHMM: a frequency profile based HMM for predicting the topology of transmembrane beta barrel proteins and the exposure status of transmembrane residues. Biochimica Et Biophysica Acta. 2011;1814(5):664–670. doi: 10.1016/j.bbapap.2011.03.004. PMID: 21426944. doi:10.1016/j.bbapap.2011.03.004. URL http://www.ncbi.nlm.nih.gov/pubmed/21426944. [DOI] [PubMed] [Google Scholar]
- 35.Wimley W. Toward genomic identification of beta-barrel membrane proteins: composition and architecture of known structures. Protein Sci. 2002;11(2):301–312. doi: 10.1110/ps.29402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas C JF, Wimley WC. Bioinformatics. 16. Vol. 26. England: Oxford; 2010. Ahighly accurate statistical approach for the prediction of transmembrane beta-barrels; pp. 1965–1974. PMID: 20538726. doi:10.1093/bioinformatics/btq308. URL http://www.ncbi.nlm.nih.gov/pubmed/20538726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Senes A, Gerstein M, Engelman D. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J Mol Biol. 2000;296(3):921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- 38.Lemmon M, Flanagan J, Treutlein H, Zhang J, Engelman D. Sequence specificity in the dimerization of transmembrane alpha-helices. Biochemistry. 1992;31(51):12719–12725. doi: 10.1021/bi00166a002. [DOI] [PubMed] [Google Scholar]
- 39.Senes A, Engel DE, DeGrado WF. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol. 2004;14:465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Moore D, Berger B, DeGrado W. Protein-protein interactions in the membrane: sequence, structural, and biological motifs. Structure. 2008;16(7):991–1001. doi: 10.1016/j.str.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herrmann J, Panitz J, Unterreitmeier S, Fuchs A, Frish-man D, Langosch D. Complex patterns of histidine, hydroxylated amino acids and the GxxxG motif mediate high-affinity transmembrane domain interactions. J Mol Biol. 2009;385(3):912–923. doi: 10.1016/j.jmb.2008.10.058. [DOI] [PubMed] [Google Scholar]
- 42.Berger B, Kulp D, Span L, DeGrado J, Billings P, Senes A, Bennett J, DeGrado W. Consensus motif for integrin transmembrane helix association. Proc Natl Acad Sci U S A. 2010;107(2):703–708. doi: 10.1073/pnas.0910873107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jackups R, Jr., Cheng S, Liang J. Sequence motifs and antimotifs in beta-barrel membrane proteins from a genome-wide analysis: the Ala-Tyr dichotomy and chaperone binding motifs. J Mol Biol. 2006;363(2):611–623. doi: 10.1016/j.jmb.2006.07.095. [DOI] [PubMed] [Google Scholar]
- 44.Jackups R, Jr., Liang J. Combinatorial analysis for sequence and spatial motif discovery in short sequence fragments. IEEE/ACM Trans Comput Biol Bioinform. 2010;7(3):524–536. doi: 10.1109/TCBB.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robin S, Rodophe F, Schbath S. DNA, Words and Models: Statistics of Exceptional Words. Cambridge University Press; 2005. [Google Scholar]
- 46.Struyvé M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol. 1991;218:141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- 47.Marsico A, Scheubert K, Tuukkanen A, Henschel A, Winter C, Winnenburg R, Schroeder M. MeMotif: a database of linear motifs in alpha-helical transmembrane proteins. Nucleic Acids Res. 2010;38(Database issue):D181–D189. doi: 10.1093/nar/gkp1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gangal R, Kumar K. Reduced alphabet motif methodology for GPCR annotation. J Biomol Struct Dyn. 2007;25(3):299–310. doi: 10.1080/07391102.2007.10507178. [DOI] [PubMed] [Google Scholar]
- 49.Walters R, DeGrado W. Helix-packing motifs in membrane proteins. Proc Natl Acad Sci U S A. 2006;103(37):13658–13663. doi: 10.1073/pnas.0605878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crick FHC. The packing of α-helices: simple coiled-coils. Acta Crystallographica. 1953;6(8-9):689–697. URL http://dx.doi.org/10.1107/S0365110X53001964. [Google Scholar]
- 51.Martin J, de Brevern A, Camproux A. In silico local structure approach: a case study on outer membrane proteins. Proteins. 2008;71(1):92–109. doi: 10.1002/prot.21659. [DOI] [PubMed] [Google Scholar]
- 52.Adamian L, Jackups R, Binkowski TA, Liang J. Higher-order interhelical spatial interactions in membrane proteins. J Mol Biol. 2003;327:251–272. doi: 10.1016/s0022-2836(03)00041-x. [DOI] [PubMed] [Google Scholar]
- 53.Adamian L, Liang J. Interhelical hydrogen bonds and spatial motifs in membrane proteins: polar clamps and serine zippers. Proteins. 2002;47:209–218. doi: 10.1002/prot.10071. [DOI] [PubMed] [Google Scholar]
- 54.Ruan W, Becker V, Klingmuller U, Langosch D. The interface between self-assembling erythropoietin receptor transmembrane segments corresponds to a membrane-spanning leucine zipper. J Biol Chem. 2004;279(5):3273–3279. doi: 10.1074/jbc.M309311200. [DOI] [PubMed] [Google Scholar]
- 55.Ruan W, Lindner E, Langosch D. The interface of a membrane-spanning leucine zipper mapped by asparagine-scanning mutagenesis. Protein Sci. 2004;13(2):555–559. doi: 10.1110/ps.03357404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hutchinson E, Sessions R, Thornton J, Woolfson D. Determinants of strand register in antiparallel β-sheets of proteins. Protein Sci. 1998;7(11):2287–2300. doi: 10.1002/pro.5560071106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Merkel JS, Regan L. Aromatic rescue of glycine in β sheets. Fold Des. 1998;3:449–455. doi: 10.1016/s1359-0278(98)00062-5. [DOI] [PubMed] [Google Scholar]
- 58.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 59.Larkin M, Blackshields G, Brown N, Chenna R, McGet-tigan P, McWilliam H, Valentin F, Wallace I, Wilm A, Lopez R, Thompson J, Gibson T, Higgins D. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 60.Dayhoff M, Schwartz R, Orcutt B. Ch. A model of evolutionary change in proteins. suppl. 3. vol.5. Washington, D.C: National Biomedical Research Foundation; 1978. Atlas of Protei Sequence and Structure; pp. 345–352. [Google Scholar]
- 61.Henikoff S, Henikoff J. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ng P, Henikoff J, Henikoff S. PHAT: a transmembrane-specific substitution matrix. predicted hydrophobic and transmembrane. Bioinformatics. 2000;16(9):760–766. doi: 10.1093/bioinformatics/16.9.760. [DOI] [PubMed] [Google Scholar]
- 63.Muller T, Rahmann S, Rehmsmeier M. Non-symmetric score matrices and the detection of homologous transmembrane proteins. Bioinformatics. 2001;17(Suppl 1):S182–S189. doi: 10.1093/bioinformatics/17.suppl_1.s182. [DOI] [PubMed] [Google Scholar]
- 64.Jimenez-Morales D, Adamian L, Liang J. Detecting remote homologues using scoring matrices calculated from the estimation of amino acid substitution rates of beta-barrel membrane proteins, Conference Proceedings: … Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. 2008:1347–1350. doi: 10.1109/IEMBS.2008.4649414. Conference 2008. PMID: 19162917. doi:10.1109/IEMBS.2008.4649414. URL http://www.ncbi.nlm.nih.gov.proxy.cc.uic.edu/pubmed/19162917. [DOI] [PMC free article] [PubMed]
- 65.Tseng Y, Liang J. Estimation of amino acid residue substitution rates at local spatial regions and application in protein function inference: A Bayesian Monte Carlo approach. Mol. Biol. Evol. 2006;23(2):421–436. doi: 10.1093/molbev/msj048. [DOI] [PubMed] [Google Scholar]
- 66.Walther D, Papic D, Bos M, Tommassen J, Rapaport D. Signals in bacterial beta-barrel proteins are functional in eukaryotic cells for targeting to and assembly in mitochondria. Proc Natl Acad Sci U S A. 2009;106(8):2531–2536. doi: 10.1073/pnas.0807830106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walther D, Bos M, Rapaport D, Tommassen J. The mitochondrial porin, VDAC, has retained the ability to be assembled in the bacterial outer membrane. Mol Biol Evol. 2010;27(4):887–895. doi: 10.1093/molbev/msp294. [DOI] [PubMed] [Google Scholar]
- 68.Tommassen J. Assembly of outer-membrane proteins in bacteria and mitochondria. Microbiology. 2010;156(Pt 9):2587–2596. doi: 10.1099/mic.0.042689-0. [DOI] [PubMed] [Google Scholar]
- 69.Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rost B, Casadio R, Fariselli P, Sander C. Transmembrane helices predicted at 95% accuracy. Protein Sci. 1995;4(3):521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dowhan W, Bogdanov M. Lipid-dependent membrane protein topogenesis. Annu Rev Biochem. 2009;78:515–540. doi: 10.1146/annurev.biochem.77.060806.091251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adamian L, Naveed H, Liang J. Lipid-binding surfaces of membrane proteins: evidence from evolutionary and structural analysis. Biochim Biophys Acta. 2011;1808(4):1092–1102. doi: 10.1016/j.bbamem.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Altschuh D, Lesk AM, Bloomer AC, Klug A. Correlation of coordinated amino acid substitutions with function in viruses related to tobacco mosaic virus. Journal of Molecular Biology. 1987;193(4):693–707. doi: 10.1016/0022-2836(87)90352-4. PMID: 3612789. [DOI] [PubMed] [Google Scholar]
- 74.Altschuh D, Vernet T, Berti P, Moras D, Nagai K. Coordinated amino acid changes in homologous protein families. Protein Engineering. 1988;2(3):193–199. doi: 10.1093/protein/2.3.193. [DOI] [PubMed] [Google Scholar]
- 75.Fleishman SJ, Unger VM, Yeager M, Ben-Tal N. A calpha model for the transmembrane alpha helices of gap junction intercellular channels. Molecular Cell. 2004;15(6):879–888. doi: 10.1016/j.molcel.2004.08.016. PMID: 15383278. [DOI] [PubMed] [Google Scholar]
- 76.Fuchs A, Martin-Galiano AJ, Kalman M, Fleishman S, Ben-Tal N, Frishman D. Bioinformatics. 24. Vol. 23. England: Oxford; 2007. Co-evolving residues in membrane proteins; pp. 3312–3319. [DOI] [PubMed] [Google Scholar]
- 77.Fleishman SJ, Yifrach O, Ben-Tal N. An evolutionarily conserved network of amino acids mediates gating in voltage-dependent potassium channels. Journal of Molecular Biology. 2004;340(2):307–318. doi: 10.1016/j.jmb.2004.04.064. PMID: 15201054. doi:10.1016/j.jmb.2004.04.064. URL http://www.ncbi.nlm.nih.gov/pubmed/15201054. [DOI] [PubMed] [Google Scholar]
- 78.Beja O, Aravind L, Koonin E, Suzuki M, Hadd A, Nguyen L, Jovanovich S, Gates C, Feldman R, Spudich J, Spudich E, DeLong E. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289(5486):1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- 79.Gilbert J, Dupont C. Microbial metagenomics: beyond the genome. Ann Rev Mar Sci. 2011;3:347–371. doi: 10.1146/annurev-marine-120709-142811. [DOI] [PubMed] [Google Scholar]
- 80.Brown L, Jung K. Bacteriorhodopsin-like proteins of eubacteria and fungi: the extent of conservation of the haloarchaeal proton-pumping mechanism. Photochem Photobiol Sci. 2006;5(6):538–546. doi: 10.1039/b514537f. [DOI] [PubMed] [Google Scholar]
- 81.Adamian L, Ouyang Z, Tseng Y, Liang J. Evolutionary patterns of retinal-binding pockets of type I rhodopsins and their functions. Photochem Photobiol. 2006;82(6):1426–1435. doi: 10.1562/2006-02-14-RA-802. [DOI] [PubMed] [Google Scholar]
- 82.Balashov S, Imasheva E, Boichenko V, Anton J, Wang J, Lanyi J. Xanthorhodopsin: a proton pump with a light-harvesting carotenoid antenna. Science. 2005;309(5743):2061–2064. doi: 10.1126/science.1118046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Imasheva E, Balashov S, Choi A, Jung K, Lanyi J. Reconstitution of gloeobacter violaceus rhodopsin with a light-harvesting carotenoid antenna. Biochemistry. 2009;48(46):10948–10955. doi: 10.1021/bi901552x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.White S, Wimley W. Hydrophobic interactions of peptides with membrane interfaces. Biochim Biophys Acta. 1998;1376(3):339–352. doi: 10.1016/s0304-4157(98)00021-5. [DOI] [PubMed] [Google Scholar]
- 85.White S, Wimley W. Membrane protein folding and stability: physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 86.Popot J, Engelman D. Helical membrane protein folding, stability, and evolution. Annu Rev Biochem. 2000;69:881–922. doi: 10.1146/annurev.biochem.69.1.881. [DOI] [PubMed] [Google Scholar]
- 87.Liang J. Experimental and computational studies of the determinants of membrane protein folding. Curr. Opin. Chem. Biol. 2002;6:878–884. doi: 10.1016/s1367-5931(02)00393-9. [DOI] [PubMed] [Google Scholar]
- 88.Engelman D, Chen Y, Chin C, Curran A, Dixon A, Dupuy A, Lee A, Lehnert U, Matthews E, Reshetnyak Y, Senes A, Popot J. Membrane protein folding: beyond the two stage model. FEBS Lett. 2003;555(1):122–125. doi: 10.1016/s0014-5793(03)01106-2. [DOI] [PubMed] [Google Scholar]
- 89.Bowie J. Solving the membrane protein folding problem. Nature. 2005;438(7068):581–589. doi: 10.1038/nature04395. [DOI] [PubMed] [Google Scholar]
- 90.Wimley W, Creamer T, White S. Solvation energies of amino acid side chains and backbone in a family of host-guest pentapeptides. Biochemistry. 1996;35(16):5109–5124. doi: 10.1021/bi9600153. [DOI] [PubMed] [Google Scholar]
- 91.Wimley W, White S. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol. 1996;3(10):842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 92.Hristova K, Wimley W. A look at arginine in membranes. J Membr Biol. 2011;239(1-2):49–56. doi: 10.1007/s00232-010-9323-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White S, von Heijne G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433(7024):377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 94.Hessa T, White S, von Heijne G. Membrane insertion of a potassium-channel voltage sensor. Science. 2005;307(5714):1427. doi: 10.1126/science.1109176. [DOI] [PubMed] [Google Scholar]
- 95.Moon C, Fleming K. From the cover: Side-chain hydrophobicity scale derived from transmembrane protein folding into lipid bi-layers. Proc Natl Acad Sci U S A. 2011;108(25):10174–10177. doi: 10.1073/pnas.1103979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jayasinghe S, Hristova K, White S. Energetics, stability, and prediction of transmembrane helices. J Mol Biol. 2001;312(5):927–934. doi: 10.1006/jmbi.2001.5008. [DOI] [PubMed] [Google Scholar]
- 97.Snider C, Jayasinghe S, Hristova K, White SH. MPEx: a tool for exploring membrane proteins. Protein Science. 2009;18(12):2624–2628. doi: 10.1002/pro.256. URL http://onlinelibrary.wiley.com/doi/10.1002/pro.256/abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Adamian L, Liang J. Helix-helix packing and interfacial pairwise interactions of residues in membrane proteins. J Mol Biol. 2001;311:891–907. doi: 10.1006/jmbi.2001.4908. [DOI] [PubMed] [Google Scholar]
- 99.Adamian L, Nanda V, DeGrado W, Liang J. Empirical lipid propensities of amino acid residues in multispan alpha helical membrane proteins. Proteins. 2005;59(3):496–509. doi: 10.1002/prot.20456. [DOI] [PubMed] [Google Scholar]
- 100.Senes A, Chadi D, Law P, Walters R, Nanda V, DeGrado W. E(z), a depth-dependent potential for assessing the energies of insertion of amino acid side-chains into membranes: derivation and applications to determining the orientation of transmembrane and interfacial helices. J Mol Biol. 2007;366(2):436–448. doi: 10.1016/j.jmb.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 101.Miyazawa S, Jernigan R. Residue-residue potentials with a favorable contact pair term and an unfavorable high packing density term, for simulation and threading. J Mol Biol. 1996;256(3):623–644. doi: 10.1006/jmbi.1996.0114. [DOI] [PubMed] [Google Scholar]
- 102.Li X, Liang J. Ch. Knowledge-based energy functions for computational studies of proteins. Springer; 2006. Computational algorithms for protein structure prediction. [Google Scholar]
- 103.Lear J, Gratkowski H, Adamian L, Liang J, DeGrado W. Position-dependence of stabilizing polar interactions of asparagine in transmembrane helical bundles. Biochemistry. 2003;42(21):6400–6407. doi: 10.1021/bi020573j. [DOI] [PubMed] [Google Scholar]
- 104.Ridder A, Skupjen P, Unterreitmeier S, Langosch D. Tryptophan supports interaction of transmembrane helices. J Mol Biol. 2005;354(4):894–902. doi: 10.1016/j.jmb.2005.09.084. [DOI] [PubMed] [Google Scholar]
- 105.Adamian L, Liang J. Prediction of transmembrane helix orientation in polytopic membrane proteins. BMC Struct Biol. 2006;6:13. doi: 10.1186/1472-6807-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Adamian L, Liang J. Prediction of buried helices in multispan alpha helical membrane proteins. Proteins. 2006;63(1):1–5. doi: 10.1002/prot.20874. [DOI] [PubMed] [Google Scholar]
- 107.Beuming T, Weinstein H. A knowledge-based scale for the analysis and prediction of buried and exposed faces of transmembrane domain proteins. Bioinformatics. 2004;20(12):1822–1835. doi: 10.1093/bioinformatics/bth143. [DOI] [PubMed] [Google Scholar]
- 108.Yuan Z, Zhang F, Davis M, Boden M, Teasdale R. Predicting the solvent accessibility of transmembrane residues from protein sequence. J Proteome Res. 2006;5(5):1063–1070. doi: 10.1021/pr050397b. [DOI] [PubMed] [Google Scholar]
- 109.Park Y, Hayat S, Helms V. Prediction of the burial status of trans-membrane residues of helical membrane proteins. BMC Bioinformatics. 2007;8:302. doi: 10.1186/1471-2105-8-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Illergard K, Callegari S, Elofsson A. MPRAP: an accessibility predictor for a-helical transmembrane proteins that performs well inside and outside the membrane. BMC Bioinformatics. 2010;11:333. doi: 10.1186/1471-2105-11-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yarov-Yarovoy V, Schonbrun J, Baker D. Multipass membrane protein structure prediction using rosetta. Proteins. 2006;62(4):1010–1025. doi: 10.1002/prot.20817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Leves F, Tierney M, Howitt S. Polar residues in a conserved motif spanning helices 1 and 2 are functionally important in the SulP transporter family. Int J Biochem Cell Biol. 2008;40(11):2596–2605. doi: 10.1016/j.biocel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 113.Zhou F, Cocco M, Russ W, Brunger A, Engelman D. Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nat Struct Biol. 2000;7(2):154–160. doi: 10.1038/72430. [DOI] [PubMed] [Google Scholar]
- 114.Choma C, Gratkowski H, Lear J, DeGrado W. Asparagine-mediated self-association of a model transmembrane helix. Nat Struct Biol. 2000;7(2):161–166. doi: 10.1038/72440. [DOI] [PubMed] [Google Scholar]
- 115.Joh N, Min A, Faham S, Whitelegge J, Yang D, Woods V, Bowie J. Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins. Nature. 2008;453(7199):1266–1270. doi: 10.1038/nature06977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bowie J. Membrane protein folding: how important are hydrogen bonds? Curr Opin Struct Biol. 2011;21(1):42–49. doi: 10.1016/j.sbi.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Smith S, Eilers M, Song D, Crocker E, Ying W, Groesbeek M, Metz G, Ziliox M, Aimoto S. Implications of threonine hydrogen bonding in the glycophorin a transmembrane helix dimer. Biophys J. 2002;82(5):2476–2486. doi: 10.1016/S0006-3495(02)75590-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Faham S, Yang D, Bare E, Yohannan S, Whitelegge J, Bowie J. Side-chain contributions to membrane protein structure and stability. J Mol Biol. 2004;335(1):297–305. doi: 10.1016/j.jmb.2003.10.041. [DOI] [PubMed] [Google Scholar]
- 119.Fleishman S, Harrington S, Friesner R, Honig B, Ben-Tal N. An automatic method for predicting transmembrane protein structures using cryo-em and evolutionary data. Biophys J. 2004;87(5):3448–3459. doi: 10.1529/biophysj.104.046417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.North B, Cristian L, Fu Stowell X, Lear J, Saven J, DeGrado W. Characterization of a membrane protein folding motif, the Ser zipper, using designed peptides. J Mol Biol. 2006;359(4):930–939. doi: 10.1016/j.jmb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 121.Zhang Y, Kulp D, Lear J, DeGrado W. Experimental and computational evaluation of forces directing the association of trans-membrane helices. J Am Chem Soc. 2009;131(32):11341–11343. doi: 10.1021/ja904625b. [DOI] [PubMed] [Google Scholar]
- 122.Wouters M, Curmi P. An analysis of side chain interactions and pair correlations within antiparallel β-sheets: the differences between backbone hydrogen-bonded and non-hydrogen-bonded residue pairs. Proteins. 1995;22(2):119–131. doi: 10.1002/prot.340220205. [DOI] [PubMed] [Google Scholar]
- 123.Ho B, Curmi P. Twist and shear in β-sheets and β-ribbons. J Mol Biol. 2002;317(2):291–308. doi: 10.1006/jmbi.2001.5385. [DOI] [PubMed] [Google Scholar]
- 124.Hong H, Szabo G, Tamm L. Electrostatic couplings in OmpA ion-channel gating suggest a mechanism for pore opening. Nat Chem Biol. 2006;2(11):627–635. doi: 10.1038/nchembio827. [DOI] [PubMed] [Google Scholar]
- 125.Hong H, Park S, Jimenez R, Rinehart D, Tamm L. Role of aromatic side chains in the folding and thermodynamic stability of integral membrane proteins. J Am Chem Soc. 2007;129(26):8320–8327. doi: 10.1021/ja068849o. [DOI] [PubMed] [Google Scholar]
- 126.Oesterhelt F, Oesterhelt D, Pfeiffer M, Engel A, Gaub H, Muller D. Unfolding pathways of individual bacteriorhodopsins. Science. 2000;288(5463):143–146. doi: 10.1126/science.288.5463.143. [DOI] [PubMed] [Google Scholar]
- 127.Naveed H, Jackups R, Jr., Liang J. Predicting weakly stable regions, oligomerization state, and protein-protein interfaces in transmembrane domains of outer membrane proteins. Proc Nat Aca Sci USA In press. 2009 doi: 10.1073/pnas.0902169106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jones D. Successful ab initio prediction of the tertiary structure of NK-lysin using multiple sequences and recognized supersecondary structural motifs. Proteins Suppl. 1997;1:185–191. doi: 10.1002/(sici)1097-0134(1997)1+<185::aid-prot24>3.3.co;2-t. [DOI] [PubMed] [Google Scholar]
- 129.Eilers M, Patel AB, Liu W, Smith SO. Comparison of helix interactions in membrane and soluble α-bundle proteins. Biophys J. 2002;82:2720–2736. doi: 10.1016/S0006-3495(02)75613-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dobbs H, Orlandini E, Bonaccini R, Seno F. Optimal potentials for predicting inter-helical packing in transmembrane proteins. Proteins. 2002;49(3):342–349. doi: 10.1002/prot.10229. [DOI] [PubMed] [Google Scholar]
- 131.Wendel C, Gohlke H. Predicting transmembrane helix pair configurations with knowledge-based distance-dependent pair potentials. Proteins. 2008;70(3):984–999. doi: 10.1002/prot.21574. [DOI] [PubMed] [Google Scholar]
- 132.Sodt A, Head-Gordon T. Driving forces for transmembrane alpha-helix oligomerization. Biophys J. 2010;99(1):227–237. doi: 10.1016/j.bpj.2010.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Barth P, Schonbrun J, Baker D. Toward high-resolution prediction and design of transmembrane helical protein structures. Proc Natl Acad Sci U S A. 2007;104(40):15682–15687. doi: 10.1073/pnas.0702515104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Barth P, Wallner B, Baker D. Prediction of membrane protein structures with complex topologies using limited constraints. Proc Natl Acad Sci U S A. 2009;106(5):1409–1414. doi: 10.1073/pnas.0808323106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.van den Berg B. The FadL family: unusual transporters for unusual substrates. Curr Opin Struct Biol. 2005;15(4):401–407. doi: 10.1016/j.sbi.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 136.Huysmans G, Radford S, Brockwell D, Baldwin S. The N-terminal helix is a post-assembly clamp in the bacterial outer membrane protein pagp. J Mol Biol. 2007;373(3):529–540. doi: 10.1016/j.jmb.2007.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait B, MacKinnon R. X-ray structure of a voltage-dependent k+ channel. Nature. 2003;423(6935):33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 138.Ruta V, Chen J, MacKinnon R. Calibrated measurement of gating-charge arginine displacement in the KvAP voltage-dependent k+ channel. Cell. 2005;123(3):463–475. doi: 10.1016/j.cell.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 139.Dorairaj S, Allen T. On the thermodynamic stability of a charged arginine side chain in a transmembrane helix. Proc Natl Acad Sci U S A. 2007;104(12):4943–4948. doi: 10.1073/pnas.0610470104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.MacCallum J, Bennett W, Tieleman D. Partitioning of amino acid side chains into lipid bilayers: results from computer simulations and comparison to experiment. J Gen Physiol. 2007;129(5):371–377. doi: 10.1085/jgp.200709745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schow E, Freites J, Cheng P, Bernsel A, von Heijne G, White S, Tobias D. Arginine in membranes: the connection between molecular dynamics simulations and translocon-mediated insertion experiments. J Membr Biol. 2011;239(1–2):35–48. doi: 10.1007/s00232-010-9330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Johansson A, Lindahl E. Protein contents in biological membranes can explain abnormal solvation of charged and polar residues. Proc Natl Acad Sci U S A. 2009;106(37):15684–15689. doi: 10.1073/pnas.0905394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Joh N, Min A, Faham S, Whitelegge J, Yang D, Woods V, Bowie J. Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins. Nature. 2008;453(7199):1266–1270. doi: 10.1038/nature06977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sonnhammer E, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 145.von Heijne G. Proline kinks in transmembrane alpha-helices. J Mol Biol. 1991;218(3):499–503. doi: 10.1016/0022-2836(91)90695-3. [DOI] [PubMed] [Google Scholar]
- 146.Papaloukas C, Granseth E, Viklund H, Elofsson A. Estimating the length of transmembrane helices using z-coordinate predictions. Protein Sci. 2008;17(2):271–278. doi: 10.1110/ps.073036108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hristova K, Wimley W, Mishra V, Anantharamiah G, Segrest J, White S. An amphipathic alpha-helix at a membrane interface: a structural study using a novel X-ray diffraction method. J Mol Biol. 1999;290(1):99–117. doi: 10.1006/jmbi.1999.2840. [DOI] [PubMed] [Google Scholar]
- 148.Granseth E, von Heijne G, Elofsson A. A study of the membrane-water interface region of membrane proteins. J Mol Biol. 2005;346:377–385. doi: 10.1016/j.jmb.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 149.Viklund H, Granseth E, Elofsson A. Structural classification and prediction of reentrant regions in alpha-helical transmembrane proteins: application to complete genomes. J Mol Biol. 2006;361(3):591–603. doi: 10.1016/j.jmb.2006.06.037. [DOI] [PubMed] [Google Scholar]
- 150.Kauko A, Illergard K, Elofsson A. Coils in the membrane core are conserved and functionally important. J Mol Biol. 2008;380(1):170–180. doi: 10.1016/j.jmb.2008.04.052. [DOI] [PubMed] [Google Scholar]
- 151.Hall S, Roberts K, Vaidehi N. Position of helical kinks in membrane protein crystal structures and the accuracy of computational prediction. J Mol Graph Model. 2009;27(8):944–950. doi: 10.1016/j.jmgm.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 152.Yohannan S, Faham S, Yang D, Grosfeld D, Chamberlain A, Bowie J. A C alpha-H...o hydrogen bond in a membrane protein is not stabilizing. J Am Chem Soc. 2004;126(8):2284–2285. doi: 10.1021/ja0317574. [DOI] [PubMed] [Google Scholar]
- 153.Yan C, Luo J. An analysis of reentrant loops. Protein J. 2010;29(5):350–354. doi: 10.1007/s10930-010-9259-z. [DOI] [PubMed] [Google Scholar]
- 154.Forrest L, Tang C, Honig B. On the accuracy of homology modeling and sequence alignment methods applied to membrane proteins. Biophys J. 2006;91(2):508–517. doi: 10.1529/biophysj.106.082313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhang Y, Devries M, Skolnick J. Structure modeling of all identified G protein-coupled receptors in the human genome. PLoS Comput Biol. 2006;2(2):e13. doi: 10.1371/journal.pcbi.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Goldfeld D, Zhu K, Beuming T, Friesner R. Successful prediction of the intra- and extracellular loops of four G-protein-coupled receptors. Proc Natl Acad Sci U S A. 2011;108(20):8275–8280. doi: 10.1073/pnas.1016951108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kelm S, Shi J, Deane C. MEDELLER: homology-based coordinate generation for membrane proteins. Bioinformatics. 2010;26(22):2833–2840. doi: 10.1093/bioinformatics/btq554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Randall A, Cheng J, Sweredoski M, Baldi P. TMBpro: secondary structure, beta-contact and tertiary structure prediction of transmembrane beta-barrel proteins. Bioinformatics. 2008;24(4):513–520. doi: 10.1093/bioinformatics/btm548. [DOI] [PubMed] [Google Scholar]
- 159.Jimenez-Morales D, Liang J. Pattern of amino acid substitutions in transmembrane domains of -barrel membrane proteins for detecting remote homologs in bacteria and mitochondria. PLoS ONE. doi: 10.1371/journal.pone.0026400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lindert S, Staritzbichler R, Wotzel N, Karakas M, Stewart P, Meiler J. EM-fold: De novo folding of alpha-helical proteins guided by intermediate-resolution electron microscopy density maps. Structure. 2009;17(7):990–1003. doi: 10.1016/j.str.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Schushan M, Barkan Y, Haliloglu T, Ben-Tal N. C(alpha) -trace model of the transmembrane domain of human copper transporter 1, motion and functional implications. Proc Natl Acad Sci U S A. 2010;107(24):10908–10913. doi: 10.1073/pnas.0914717107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Metcalf D, Law P, DeGrado W. Mutagenesis data in the automated prediction of transmembrane helix dimers. Proteins. 2007;67(2):375–384. doi: 10.1002/prot.21265. [DOI] [PubMed] [Google Scholar]
- 163.Metcalf D, Kulp D, Bennett J, DeGrado W. Multiple approaches converge on the structure of the integrin alphaIIb/beta3 transmembrane heterodimer. J Mol Biol. 2009;392(4):1087–1101. doi: 10.1016/j.jmb.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lin M, Lu H, Chen R, Liang J. Generating properly weighted ensemble of conformations of proteins from sparse or indirect distance constraints. J Chem Phys. 2008;129(9):094101. doi: 10.1063/1.2968605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cuesta-Seijo J, Neale C, Khan M, Moktar J, Tran C, Bishop R, Pomes R, Prive G. PagP crystallized from SDS/cosolvent reveals the route for phospholipid access to the hydrocarbon ruler. Structure. 2010;18(9):1210–1219. doi: 10.1016/j.str.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lin M, Zhang J, Lu H, Chen R, Liang J. Constrained proper sampling of conformations of transition state ensemble of protein folding. J Chem Phys. 2011;134(7):075103. doi: 10.1063/1.3519056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Elofsson A, von Heijne G. Membrane protein structure: prediction versus reality. Annu Rev Biochem. 2007;76:125–140. doi: 10.1146/annurev.biochem.76.052705.163539. [DOI] [PubMed] [Google Scholar]
- 168.Kauko A, Hedin L, Thebaud E, Cristobal S, Elofsson A, von Heijne G. Repositioning of transmembrane alpha-helices during membrane protein folding. J Mol Biol. 2010;397(1):190–201. doi: 10.1016/j.jmb.2010.01.042. [DOI] [PubMed] [Google Scholar]
- 169.Lehnert U, Xia Y, Royce T, Goh C, Liu Y, Senes A, Yu H, Zhang Z, Engelman D, Gerstein M. Computational analysis of membrane proteins: genomic occurrence, structure prediction and helix interactions. Q Rev Biophys. 2004;37(2):121–146. doi: 10.1017/s003358350400397x. [DOI] [PubMed] [Google Scholar]
- 170.Liu Y, Gerstein M, Engelman D. Transmembrane protein domains rarely use covalent domain recombination as an evolutionary mechanism. Proc Natl Acad Sci U S A. 2004;101(10):3495–3497. doi: 10.1073/pnas.0307330101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Simpson L, Taddese B, Wall I, Reynolds C. Bioinformatics and molecular modelling approaches to GPCR oligomerization. Curr Opin Pharmacol. 2010;10(1):30–37. doi: 10.1016/j.coph.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 172.Locher K, Rosenbusch J. Oligomeric states and siderophore binding of the ligand-gated FhuA protein that forms channels across escherichia coli outer membranes. Eur J Biochem. 1997;247(3):770–775. doi: 10.1111/j.1432-1033.1997.t01-1-00770.x. [DOI] [PubMed] [Google Scholar]
- 173.Bordner A. Predicting protein-protein binding sites in membrane proteins. BMC Bioinformatics. 2009;10:312. doi: 10.1186/1471-2105-10-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Keinan N, Tyomkin D, Shoshan-Barmatz V. Oligomerization of the mitochondrial protein voltage-dependent anion channel is coupled to the induction of apoptosis. Mol Cell Biol. 2010;30(24):5698–5709. doi: 10.1128/MCB.00165-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Shoshan-Barmatz V, Keinan N, Abu-Hamad S, Tyomkin D, Aram L. Apoptosis is regulated by the VDAC1 n-terminal region and by VDAC oligomerization: release of cytochrome c, AIF and Smac/Diablo. Biochim Biophys Acta. 2010;1797(6-7):1281–1291. doi: 10.1016/j.bbabio.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 176.Samish I, MacDermaid C, Perez-Aguilar J, Saven J. Theoretical and computational protein design. Annu Rev Phys Chem. 2011;62:129–149. doi: 10.1146/annurev-physchem-032210-103509. [DOI] [PubMed] [Google Scholar]
- 177.Korendovych I, Senes A, Kim Y, Lear J, Fry H, Therien M, Blasie J, Walker F, DeGrado W. De novo design and molecular assembly of a transmembrane diporphyrin-binding protein complex. J Am Chem Soc. 2010;132(44):15516–15518. doi: 10.1021/ja107487b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yin H, Slusky J, Berger B, Walters R, Vilaire G, Litvinov R, Lear J, Caputo G, Bennett J, DeGrado W. Computational design of peptides that target transmembrane helices. Science. 2007;315(5820):1817–1822. doi: 10.1126/science.1136782. [DOI] [PubMed] [Google Scholar]
- 179.Caputo G, Litvinov R, Li W, Bennett J, DeGrado W, Yin H. Computationally designed peptide inhibitors of protein-protein interactions in membranes. Biochemistry. 2008;47(33):8600–8606. doi: 10.1021/bi800687h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sarkar C, Dodevski I, Kenig M, Dudli S, Mohr A, Hermans E, Pluckthun A. Directed evolution of a G protein-coupled receptor for expression, stability, and binding selectivity. Proc Natl Acad Sci U S A. 2008;105(39):14808–14813. doi: 10.1073/pnas.0803103105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Signarvic R, DeGrado W. Metal-binding dependent disruption of membranes by designed helices. J Am Chem Soc. 2009;131(9):3377–3384. doi: 10.1021/ja809580b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Muhammad N, Dworeck T, Fioroni M, Schwaneberg U. Engineering of the E. coli Outer Membrane Protein FhuA to overcome the Hydrophobic Mismatch in Thick Polymeric Membranes. J Nanobiotechnology. 2011;9(1):8. doi: 10.1186/1477-3155-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Taylor M, Fung H, Rajgaria R, Filizola M, Weinstein H, Floudas C. Mutations affecting the oligomerization interface of G-protein-coupled receptors revealed by a novel de novo protein design framework. Biophys J. 2008;94(7):2470–2481. doi: 10.1529/biophysj.107.117622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Schulz G. The structure of bacterial outer membrane proteins. Biochim Biophys Acta. 2002;1565(2):308–317. doi: 10.1016/s0005-2736(02)00577-1. [DOI] [PubMed] [Google Scholar]
- 185.Arnold T, Poynor M, Nussberger S, Lupas A, Linke D. Gene duplication of the eight-stranded beta-barrel OmpX produces a functional pore: a scenario for the evolution of transmembrane beta-barrels. J Mol Biol. 2007;366(4):1174–1184. doi: 10.1016/j.jmb.2006.12.029. [DOI] [PubMed] [Google Scholar]
- 186.Song L, Hobaugh M, Shustak C, Cheley S, Bayley H, Gouaux J. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274(5294):1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 187.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405(6789):914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 188.Miedema H, Meter-Arkema A, Wierenga J, Tang J, Eisenberg B, Nonner W, Hektor H, Gillespie D, Meijberg W. Permeation properties of an engineered bacterial OmpF porin containing the EEEE-locus of Ca2+ channels. Biophys J. 2004;87(5):3137–3147. doi: 10.1529/biophysj.104.041384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Li H, Chen H, Steinbronn C, Wu B, Beitz E, Zeuthen T, Voth G. Enhancement of proton conductance by mutations of the selectivity filter of aquaporin-1. J Mol Biol. 2011;407(4):607–620. doi: 10.1016/j.jmb.2011.01.036. [DOI] [PubMed] [Google Scholar]
- 190.Udho E, Jakes K, Buchanan S, James K, Jiang X, Klebba P, Finkelstein A. Reconstitution of bacterial outer membrane tonb-dependent transporters in planar lipid bilayer membranes. Proc Natl Acad Sci U S A. 2009;106(51):21990–21995. doi: 10.1073/pnas.0910023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Basle A, Qutub R, Mehrazin M, Wibbenmeyer J, Delcour A. Deletions of single extracellular loops affect ph sensitivity, but not voltage dependence, of the escherichia coli porin ompf. Protein Eng Des Sel. 2004;17(9):665–672. doi: 10.1093/protein/gzh078. [DOI] [PubMed] [Google Scholar]