Abstract
Drug addictions remain a substantial health issue, with limited treatment options currently available. Despite considerable advances in the understanding of our genetic architecture, the genetic underpinning of complex disorders remains elusive. Numerous candidate genes have been implicated in the etiology and response to treatment for different addictions based on our current understanding of the neurobiology. Genome-wide association studies have also provided novel targets. However, replication of these studies is often lacking which complicates interpretation; this will improve as issues such as phenotypic characterization, the apparent “missing heritability”, the identification of functional variants, and possible gene-environment interactions are addressed. In addition, there is growing evidence that genetic information can be useful for refining the choice of addiction treatment. As genetic testing becomes more common in the practice of medicine, a variety of ethical and practical challenges, some of which are unique to drug addiction, will also need to be considered.
Introduction
Drug addictions are a set of neurobiologically connected, chronic, and relapsing medical and psychiatric diseases characterized by persistent and compulsive use despite significant harmful consequences. Continued use of the addictive agent result in neuroadaptation, with these changes persisting long after use is discontinued. The World Health Organization estimates that there are currently 185 million users of illicit drugs, 1.3 billion tobacco smokers and 2 billion alcohol users worldwide (1). In addition to often immense damages to the individual, the economic costs of drug addiction are substantial. In the United States alone, this has been estimated annually at $181 billion for illicit drugs, $168 billion for tobacco and $185 billion for alcohol, and includes burdens on the health and criminal systems, as well as loss of productivity among the workforce (2).
The pathogenesis of addiction involves a series of complex interactions between biological (e.g., genetic vulnerability, gender, physiological and behavioral response to drug experimentation and use, drug-induced alterations in gene expression and resultant proteins), environmental (e.g., legality, acceptability, availability), psychological (e.g., novelty seeking or harm avoidance, personality traits), and drug factors (e.g., dose, pattern of use, and route of administration). The voluntary initiation and continuation of a behavior that is harmful to health is an important aspect of the etiology of many common diseases, including cardiovascular disease and metabolic syndrome. However, in addictions, the role of volition in initiation and later drug-induced impairments in judgment are most salient. Moreover, licit and illicit drug use is typically initiated in childhood, when the ability to balance the apparent short-term benefits of experimentation and use with the addictive potential and long-term physical and mental consequences of dependence is generally lacking. Very few adults initiate drug use voluntarily if they have remained drug naïve into adulthood, and tobacco and alcohol industries specifically target youth to recruit new users for this reason. However, abuse and addiction to prescription opiates is also becoming a problem among older adults. A strong genetic component in the etiologies of addictions has been identified, and several addictions are among the most heritable psychiatric disorders (3). Numerous family, twin and adoption studies have provided consistent evidence for the role of genetic factors, by estimating heritability as the fraction of inter-individual differences that can be attributed to genetic differences between individuals. Estimates of the heritability of smoking persistence/dependence vary at 0.4 to 0.8 (reviewed in (4, 5), table 1), while estimates for alcoholism typically range at 0.5 to 0.7 (reviewed in (3, 6), table 1). Similarly, heritability estimates for initiation and/or dependence of illicit drug use have been reported at 0.3 to 0.6, although fewer studies have focused on this (reviewed in (3, 7), table 1). It should be noted that heritability estimates are population- and time-specific, and other factors that can influence the risk of addiction indirectly (such as impulsivity) are also heritable themselves. Investigation of why heritability varies between populations and over time can provide insights into the role of novel environmental influences.
Table 1.
Heritability estimates for different drugs of abuse§
| Phenotype | Heritability estimates |
|---|---|
| Smoking | |
| Persistence | 28 – 84% |
| Cigarette consumption | 45 – 86% |
| Nicotine dependence | 31 – 75% |
| Nicotine withdrawal symptoms | 26 – 48% |
| Smoking cessation | 50 – 58% |
| Alcoholism | |
| Alcohol abuse/dependence | 50 – 70% |
| Consumption levels | 45 – 58% |
| Problem drinking | 8 – 50% |
| Opiates/Heroin | |
| Abuse and/or dependence | 43 – 60% |
| Sedatives | |
| Abuse and/or dependence | 29 – 58% |
| Psychostimulants | |
| Abuse and/or dependence | 42 – 74% |
Despite drug- and drug-class specific mechanisms of action and psychoactive effects, there is substantial overlap of genetic factors underlying addiction to most classes of drugs. For example, approximately 60% of genetic influences are shared between nicotine and alcohol dependence (7). The notion of a shared biological component underlying addiction to different drugs of abuse is reflected in the high rates of co-morbid dependence to different substances, similar patterns in the initiation and continuation of drug use, evidence of cross-tolerance and cross-dependence to different substances, and common mechanisms underlying drug reward in the brain. While the heritability of drug addictions has been determined by many twin and family studies, our understanding of the specific genes involved remains limited.
With the completion of a canonical sequence for the human genome in 2003, and very rapid advances in DNA sequencing technologies, the possibility that sequencing of all three billion DNA base pairs will become routine in medical practice is not farfetched. The canonical human genome sequence was a 13-year effort involving DNA fragment cloning, shotgun sequencing of random DNA fragments, advances in bioinformatic capabilities for sequence assembly, and required the efforts of hundreds of people and machines at a cost of $3 billion dollars. Using sequencing technologies in which up to a billion DNA fragments are simultaneously sequenced in a single run on one machine, a human genome can now be sequenced in several weeks for less than $50,000 (8). The $1,000 genome is thought to be a reasonable goal within the next few years, and large-scale sequencing of human genomes (such as the 1000 Genomes Project) is already underway. The International HapMap Project, which was also established in 2003, identifies and catalogs all common human genetic variants with the goal of using this information to find the genes that affect health, diseases, and individual responses to medications and environmental factors. To date, some 22 million polymorphisms are known and the allele frequencies and linkage disequilibrium relationships of many have been defined in HapMap which has a set of four world populations, and in the Human Genome Diversity Panel, a collection of 51 smaller population samples worldwide. However, despite efforts such as the ENCODE Project, our ability to identify variations in the human genome has vastly exceeded our understanding of their biological significance, a problem that is particularly important in unraveling the basis of common disorders such as drug addictions.
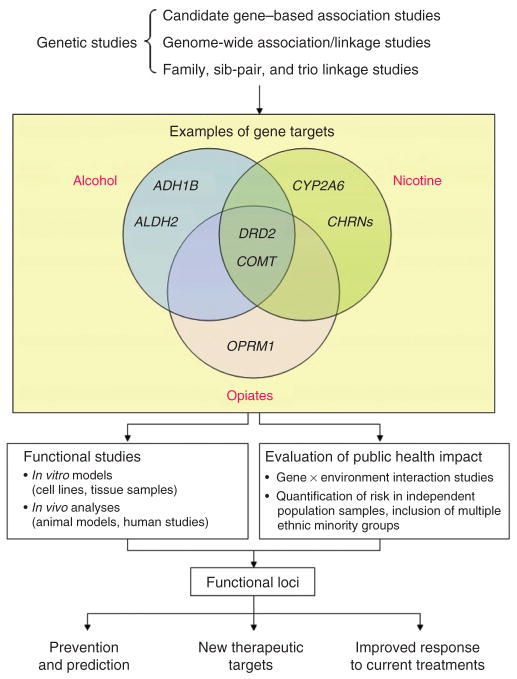
In this review, we will summarize the current knowledge of genes implicated in the etiology of addictions. We will explore the future of genetic research in this field, and the challenges that need to be addressed, such as a lack of replication of association studies, the somewhat disappointing results from genome-wide association studies conducted so far, and the importance of identifying functional predictors to help make sense of the information gained. Furthermore, gene-environment interactions have not been studied extensively in addiction despite their potential importance. Several examples of genetic testing in medicine are well known; we will consider how genetic findings might be translated into clinical practice for the prevention and treatment of addictions. The need for and importance of pharmacogenetic studies of functional variants in the understanding of disease and therapeutics is discussed. The use of genetic information presents a variety of ethical and practical challenges, several of which are unique or more salient in drug addictions. A comprehensive list of relevant publications for the sections discussed can be found in a supplementary file (available at http://www.nature.com/clpt/index.html).
Genes and addiction – what do we know?
A common feature of all drugs with addiction liability is their ability to activate the mesolimbic brain reward pathway and increase dopamine levels in the nucleus accumbens (9). This can occur by facilitation of dopamine release from presynaptic neurons or inhibition of its reuptake (such as cocaine and amphetamines), or by increasing the activity of dopaminergic neurons (such as alcohol, nicotine, opioids and cannabis) (9). While dopamine plays an important role in mediating the rewarding properties of drugs of abuse, other neurotransmitters including serotonin, opioid peptides, γ-aminobutyric acid, acetylcholine, endocannabinoids and glutamate also contribute (9). It is notable that learning processes are crucial to the neurobiology of addiction. In both addicted humans and laboratory animals, drug-associated conditioned stimuli can evoke dopamine release alone.
In general, genes implicated in addiction can be categorized into those that influence the liability to experiment with the drug in the first place, and those that are directly involved in the biological processes underlying addiction once the individual has been exposed to the drug. As such, genes that are related to personality traits (such as impulsivity, risk-taking or response to stress) may predispose one to drug experimentation, while others may be involved in the initial, immediate subjective and physiological reaction to drug use, helping to determine whether drug use will continue and escalate. Genes that encode proteins involved in the brain reward system are also important in the development of dependence to various classes of drugs. Variability in the function of receptors, transporters and metabolic enzymes of the neurotransmitter systems listed above may modify the risk of drug dependence. For example, variability in the gene(s) encoding the dopamine D2 receptor (DRD2) and/or its adjacent ankyrin repeat and protein kinase domain-containing protein 1 (ANKK1) has been implicated in dependence to nicotine, heroin and cocaine, as well as alcoholism and abuse of psychostimulants (7, 10). In addition, genes that influence dependence in a drug-specific manner have also been implicated. For example, variability in cytochrome P450 2A6 (CYP2A6), the main enzyme involved in the metabolism of nicotine, has been associated with smoking behaviors (4). Table 2 summarizes the different candidate genes implicated in drug addiction and the literature where they have been reviewed in greater detail.
Table 2.
Selected examples of genetic variation that have been implicated in drug addiction phenotypes§
| Target | Gene | Phenotype |
|---|---|---|
| Brain reward pathways | ||
| Dopamine | ||
| Receptors | DRD2 | Smoking initiation, persistence, cigarette consumption, cessation |
| Heroin use, cocaine dependence | ||
| Psychostimulant polysubstance abuse | ||
| Alcoholism | ||
| DRD4 | Smoking risk, time to first cigarette, craving and response to smoking cues, nicotine dependence | |
| Heroin, cocaine dependence | ||
| Methamphetamine use | ||
| Transporters | SLC6A3 (DAT1) | Smoking risk |
| Cocaine dependence | ||
| Metabolism Monoamine oxidase A | MAO-A | Cigarette consumption, smoking risk, nicotine dependence |
| Tyrosine hydroxylase | TH | Smoking risk |
| Dopamine β-hydroxylase | DBH | Smoking risk, nicotine dependence |
| Catechol-O-methyltransferase | COMT | Smoking risk, treatment response to nicotine spray and patch, nicotine dependence |
| Heroin, cocaine dependence | ||
| Alcohol dependence | ||
| Methamphetamine use | ||
| Serotonin | ||
| Transporters | SLC6A4 (5HTT, SERT) | Smoking risk, cigarette consumption, dependence |
| Alcohol dependence | ||
| Heroin dependence | ||
| Metabolism Tryptophan hydroxylase | TPH1, 2 | Age of smoking initiation, risk of smoking |
| Heroin dependence | ||
| Alcohol dependence | ||
| Pharmacodynamic targets | ||
| Opioid receptor (μ, δ, κ) | OPRM1 | Heroin, opiate dependence |
| Alcohol dependence | ||
| OPRK1 | Heroin, opiate dependence | |
| Alcohol dependence | ||
| OPRD1 | Heroin, cocaine dependence | |
| Nicotinic acetylcholine receptor, α4 subunit | CHRNA4 | Nicotine dependence |
| Nicotinic acetylcholine receptor, α5, α3, β4 subunit | CHRNA5, A3, B4 gene cluster on chr. 15 | Nicotine dependence, cigarette consumption, lung cancer and COPD risk |
| Alcohol, cocaine dependence | ||
| Cannabinoid receptor | ||
| GABAA receptor, γ2 subunit | GABRG2 | Heroin dependence, methamphetamine use |
| GABAA receptor, α2 subunit | GABRA2 | Alcohol dependence |
| GABAB receptor, subunit 2 | GABBR2 | Nicotine dependence |
| Pharmacokinetic targets | ||
| Cytochrome P450 2A6 | CYP2A6 | Smoking risk, cigarette consumption, smoking cessation |
| Alcohol dehydrogenase 2 | ADH2, ADH4, ADH1B, ADH1C | Alcohol dependence |
| Aldehyde dehydrogenase 2 | ALDH2 | Alcohol dependence |
| Cytochrome P450 2E1 | CYP2E1 | Alcohol dependence |
For a comprehensive list of primary reviews, see Supplementary Table 3.
In addition to more traditional genotyping approaches, research on addiction and other complex diseases have been extended to genome-wide association (GWA) studies during the past few years, where large numbers of SNPs (ranging from several hundred thousand to more than one million) spread across the genome are assessed in affected individuals and controls, and associated with disease outcome. SNPs that are in high linkage disequilibrium (i.e., tend to be inherited together more commonly than expected due to chance) can serve as proxies for each other, and as such not all of the markers within a given region need to be genotyped, as long as the marker panels can capture the variation at those loci that have not been genotyped (11). GWA studies have provided new insights into our understanding of the genetics of addiction as they are conducted without a prior hypothesis based on gene function or disease pathways as described above. As a result, a number of novel targets with potential biological relevance have been discovered. For instance, GWA studies have found associations for genes involved in cell adhesion such as neurexin 1 (NRXN1) with nicotine dependence, while neurexin 3 (NRXN3) has been implicated with alcohol, opioid and polysubstance abuse (reviewed in (7), Supplementary Table 1). Furthermore, the association between the CHRNA5-A3-B4 gene cluster with smoking-related illnesses such as lung cancer and COPD was initially revealed by GWA studies that reported significant associations with the chromosomal regions 15q24-25 (reviewed in (12), Supplementary Table 1). These novel targets provide clues to the neurobiology of addiction, but require more detailed assessment of their functionality as discussed below.
Conversely, as our understanding of the neurobiology of addiction improves, we have novel candidate genetic targets to consider. One such example are microRNAs (miRNA) that interact with the 3′-untranslated region (3′-UTR) of the target mRNA and can mediate their degradation or repress translation (13). Upon development of drug dependence, the brain undergoes significant remodeling and adaptation, and the expression profile of a number of genes in the brain are known to change following acute and repeated administration of psychostimulants and other drugs of abuse (14). These changes are thought to represent a compensatory mechanism to maintain homeostasis in response to drug-induced effects, and are manifested as drug-seeking behaviors, withdrawal and tendency to relapse. With the discovery of miRNAs and their role in the regulation of gene expression it has been proposed that drug-induced gene expression changes may be mediated through this intermediate pathway. One study has shown that nicotine treatment up- and down-regulates a number of miRNAs, in particular miR-140*, which regulates the expression of a number of genes including dynamin 1 (Dmn1) that may be involved in endocytosis and be important in drug-induced neural plasticity (15) and nicotine dependence (16). Similarly, miR-504* has been implicated in the regulation of allele-specific differential expression of a functional SNP in the 3′ UTR of DRD1 that has been associated with nicotine addiction (17).
Recent evidence also suggests that epigenetic regulation of gene expression may contribute to the pathogenesis of drug addiction. In the nucleus accumbens of rodents, cocaine increased histone acetylation of promoters in a number of genes (cfos, fos-b, bdnf, cdk5, npy) that are known to be important in the addiction process (reviewed in (18), Supplementary Table 1). Also, higher methylation levels in the promoter region of OPRM1 have been found in the lymphocytes of former long-term heroin addicts undergoing methadone maintenance treatment (19). Alcohol withdrawal increased expression of histone deacetylases, and decreased expression of CREB and NPY in the amygdala in rodent models, while inhibition of histone deacetylases reduced the anxiety resulting from alcohol withdrawal (20). Thus, variability in these gene regulators may be additional important determinants of drug addiction phenotypes.
Genes and addiction – where do we go from here?
A) Need for better replication of results
Despite enormous efforts over the past few decades, the progress in finding the genes and causal variants underlying drug addiction has been slow. The variants examined in candidate gene association studies so far have been based on a rather imperfect understanding of biological pathways, and studies have often yielded inconsistent results. For example, while case-control association studies have associated the Taq1 A1 allele with a number of smoking and alcoholic phenotypes, several studies have also been negative and meta-analyses have generally failed to support an association (4, 21, 22), (Table 2, Supplementary Table 3).
Similarly, given that GWA studies necessarily involve multiple statistical tests, stringent levels of significance are required which may hinder the replication of results. At a nominal p-value of 0.05, a GWA study examining 500,000 SNPs may potentially result in 25,000 false positives. For this reason, a genome-wide statistical significance of p < 1 × 10−7 is typically used; however, this results in very large sample sizes required in order to have sufficient statistical power. Thus, to address the potentially high number of false positive results, it is particularly vital to replicate early results in independent samples, and this is now often included in the same GWA study as part of a multi-stage design. However, replication of initial findings demonstrating similar magnitude and direction of effect within the same or similar phenotype and population is often not observed, complicating interpretation of results (11). It is notable however that the associations can be quite robust and replicable, such as the association of the CHRNA5-A3-B4 gene cluster with smoking phenotypes (12).
There are several possible explanations for the lack of replication. Addiction is a complex behavioral trait, and substantial heterogeneity exists between studies as there are a number of variables that may differ or may not have been controlled for across studies. Many studies have been underpowered, and different study methodologies (e.g. prospective vs. retrospective, population-based vs. clinical trial samples) may result in different sample populations. There is also a lack of consistency in phenotypes and outcome measures, such as definition of an appropriate control group (ever vs. former vs. never users). Inclusion of individuals of different ethnic backgrounds, while important for understanding predictors of diseases in these populations, may result in erroneous conclusions due to population stratification as allele frequencies and cultural acceptability of drug use may differ. GWA studies have indicated potential population stratification even within geographical regions previously considered to be genetically homogeneous. As such, the use of better selected controls and examination of ancestry informative markers may address these issues to an extent. The use of intermediate phenotypes, as discussed below, may also help improve reproducibility of studies by reducing the heterogeneity between studies. Publication bias, where positive results are more likely to be published, may inflate the apparent effect of a genetic variant and its association to addiction outcomes. Care must also be taken to ensure that claims of replication of data are made with sound justification to avoid further confusion in the literature. These issues occur widely in genetic research, including studies in drug addiction, and need to be considered in the design of future genetic studies.
B) Need for more consistent definitions of phenotypes
One of the biggest challenges of genetic research in addiction is the heterogeneity of the phenotype studied and a lack of consistent measurements for outcomes across studies. For instance, alcoholics vary greatly in their age of onset of problem drinking, alcohol symptoms, drinking history and co-morbid disorders. A number of scales also exist to measure nicotine dependence; the two most commonly used being the Fagerström Test for Nicotine Dependence (FTND) and Diagnostic and Statistical Manual – IV (DSM-IV). However, these two measures correlate only weakly (23), and they are likely capturing different aspects of dependence. The FTND may be a stronger measure of physical dependence, while the DSM-IV emphasizes the awareness of dependence, such as recognition of adverse consequences of smoking, a desire to reduce use, and mood changes that occurs during withdrawal. Thus, there is a need for consistent instrumentation so data from different studies can be combined and/or compared, and development of measures that better capture the multi-dimensional nature of dependence and its evolution over time in the individual’s addiction history. Limiting the use of retrospective, self-report data may also help reduce inconsistencies in phenotype outcomes; biomarkers should be incorporated where possible to confirm self-report. In addition, proxy (e.g. sibling or spouse) reports can be used to provide supporting information as medical records rarely comprehensively record substance use and not all dependent subjects seek treatment. Funding agencies and scientific societies should make a major effort to ensure that all large scale studies in drug addiction incorporate a core set of measures that are fully comparable, starting with definitions of use, quantity, frequency as well as aspects of abuse and dependence.
In recent years, intermediate phenotypes have been proposed as an alternative to traditional phenotype measures. These include neurophysiological, biochemical, endocrinological, neuroanatomical, cognitive and neuropsychological correlates that are heritable and quantifiable and are thought to more closely manifest etiologies of addictions. The relationship between genes and clinically observable symptomologies of addiction is highly complex, and intermediate phenotypes are thought to capture information on mediating variables within this chain of events. Because they are objective measures, and are potentially less complex compared to the diagnostic criteria for addiction, with wide-ranging symptomologies and clinical courses and a dependency on environmental exposures, intermediate phenotypes may be more representative of gene action. Electrophysiological measures, as well as structural and functional imaging data in so-called “imaging genetics” studies, have been used to investigate brain alterations associated with alcohol dependence in relation to genetic variations in GABRA2 and CHRM2 (reviewed in (24), Supplementary Table 1), COMT and mGluR3 (25), and DRD4 and OPRM1 (26).
C) Where is the missing heritability?
When GWA studies were first introduced in the field of genetic research of complex diseases their premise and method were simple; screen for association using markers spaced evenly across the genome to capture the effects of most, if not all, the common genetic variation in any individual, determine their associations with phenotypes of interest, and assess these loci for their functional effects in relation to disease process. However, some major challenges rapidly arose. The common alleles examined in these studies account for relatively small increments in risk and explain only a minor proportion of the phenotypic variance observed, with the odd ratios of associated SNPs in the range of 1.1 ~ 2.0 (27). Much of the genetic variance for a number of complex traits has not been accounted for; human height is one example, with an estimated heritability of ~80%, but only ~6% of the phenotypic variance accounted for by GWA studies (27). A similar observation was made for nicotine addiction; the proportion of variance in cigarette consumption accounted for by markers identified in GWA studies only ranges from ~1% (for cigarettes per day) to ~4% (for cotinine level) (28), even though the heritability of this trait has been estimated at ~50%.
Failure of GWA studies to account for this apparent missing heritability has stimulated debate on the utility of these studies, particularly given the enormous financial and scientific investments dedicated to them. The relative merit of this approach has been debated and discussed in greater detail elsewhere (11, 27). The common disease/common variant hypothesis, which states that common complex diseases are attributable to relatively few common genetic variants of moderate effect, has not been supported. Instead, the genetic underpinning of addictions and other complex diseases may be attributed to either multiple common variants, each contributing a very minor role, or multiple rare variants with intermediate to larger effect sizes, for which the resulting odds ratios fail to reach genome-wide statistical significance using attainable sample sizes. In the past few years, GWA studies have been performed on a very large scale, with tens of thousands of samples from multiple independent studies in order to identify the effects of common variants with small effect sizes (29). However, caution needs to be taken to ensure that these large sample sets are not collected at the expense of detailed environmental and behavioral information, or do not limit the use of intermediate phenotypes, and that internal heterogeneity within studies does not offset the advantage of increased sample size. These meta-analytic studies are designed to detect the residual main genotype effects, and likely identify only those variants whose main effects are large enough to be detectable against the backdrop of the myriad different environments from which these samples are taken.
Targeted re-sequencing of genes previously found to have common variants associated with complex diseases, or of functionally related genes, may be another method by which new sources of variation can be discovered. For example, four rare genetic variants (each with ~1% allele frequency) were found through re-sequencing that together accounted for a greater proportion of the variance in the risk of type I diabetes than a single common variant located in the same gene as detected by GWA studies (30). In addition, while SNPs represent alterations at single nucleotides which may only reduce function to a modest extent, other large types of structural changes to genetic architecture, such as copy number variations (CNVs), may have larger effects on gene function and thus have a greater phenotypic impact. The 1000 Genomes Project (http://www.1000genomes.org), an international consortium with the goal of creating a complete detailed catalogue of all genetic variation (including rare SNPs, CNVs and insertions/deletions) in at least 1000 genomes from across the world, will serve as a useful reference for future studies of addictive disorders. It is notable that although samples will be drawn from various world populations, some populations will likely be underrepresented and thus detection of variants present in these groups may be missed.
D) Discovery of functional predictors
Another limitation of GWA studies is that the markers identified are often not the causal variants themselves, but rather, are in linkage disequilibrium with them. This may contribute to the lack of reproducibility between studies as these markers may be in linkage disequilibrium in one population but not another. Furthermore, causal variants may not be well tagged by SNPs used in commercial genotyping arrays. This highlights the importance of identifying functional genetic predictors involved in complex diseases such as drug addiction.
Genetic manipulations in cell or animal models, as well as studies using tissue samples or cell lines, can be used to elucidate the functional impact of genetic variants. One example is the case of the CHRNA5-A3-B4 cluster on chromosome 15, encoding nicotinic acetylcholine receptor (nAChRs) subunits, which has been implicated as a susceptibility locus for nicotine addiction and lung cancer in numerous GWA studies. Even though a number of variants within the CHRNA5-A3-B4 cluster have been implicated in nicotine addiction, a rationale for how these receptor subunits may be mediating these phenotypes is currently lacking. For example, it is not known whether these genes are involved in initiation, progression or maintenance of nicotine dependence. Furthermore, whether the associations of this gene cluster with lung cancer and COPD is a direct or indirect effect via an influence on cigarette consumption remains to be clarified. Understanding the biological significance of the CHRNA5-A3-B4 receptor subunits and the functional significance of the variants implicated in this gene cluster will help interpret the observed associations.
The nAChRs are pentameric ligand-gated cation channels consisting of some combination of nine α (α2–α10) and three β (β2–β4) subunits. Mouse knockout models have demonstrated that the α4 and β2 subunits, the most widely expressed forms in the brain, are critical for nicotine self-administration and nicotine-induced dopamine release in the VTA (31, 32). Mouse knockout models for α5, α3 and β4 subunits have also been created, although only in heterozygous form for α3 as the complete knockouts suffer severe physical abnormalities and die within weeks of birth (33). Their responses to nicotine have not been extensively analyzed yet, though in general these animals have reduced sensitivity to nicotine-induced seizures and the locomotor suppressant effects of nicotine (reviewed in (34), Supplementary Table 1). Animal models will also be useful in determining whether these nAChR subunits have a direct role in mediating lung cancer or COPD independently of smoking behaviors.
Functional tests have also shown that rs16969968, a nonsynonymous SNP (Asp398Asn) in CHRNA5 implicated in GWA studies, lies in the M3-M4 intracellular loop of the receptor and may be involved in receptor trafficking but not receptor expression levels (35). While such studies are encouraging, the functional significance of genetic variation in the CHRNA5-A3-B4 gene cluster is not yet clear.
It is also notable that SNPs that have been implicated in this region also lie within genes encoding an iron regulatory protein (IREB2), an α4 proteosome subunit protein (PSMA4) and a putative protein of unknown function (LOC123688) that may also be important in cell proliferation and apoptosis (12). Further research, such as behavioral tests in animal models and targeted re-sequencing of this gene cluster, will help clarify the role of these genes and their variants in mediating smoking phenotypes. This also provides the basis for the hypothesis-driven testing of gene-gene interactions, for example, by conditioning analyses on a known functional SNP.
E) Genes and Environment interactions
The genetic influence on any behavioral outcome likely depends on exposure to certain environments; for example, an individual cannot become addicted to a drug, regardless of their genetic liability, if they were never given the opportunity to experiment with it in the first place. It is becoming clear that research on addiction should not be restricted to study of environmental or genetic effects in isolation, but rather that their interactions (G x E) should be considered, including the tendency of people to non-randomly assort to particular environments (G - E correlation). However, the majority of studies in drug addiction do not account for G x E interactions, though a few examples in alcoholism have been reported. Several studies have reported interactions of 5-HTTLPR, MAO-A-LPR and DRD2 TaqA1 genotypes with family relations, maltreatment, or negative life events on alcohol use, intoxication, and dependence (reviewed in (36), Supplementary Table 1). Additional instances where the influence of genetic variation on function may depend on gene-environment interactions have been studied. The association between increased 5-HTT expression and low levels of response to alcohol as well as increased alcohol intake has been reported in individuals with two copies of the L-allele of 5-HTTLPR, and a similar effect was also observed among carriers of the S-allele that have been exposed to early developmental stress (37). As such, the presence of gene-environment interactions may potentially confound the interpretation of genetic association studies, account for part of the ‘missing heritability’, and contribute to the lack of reproducibility between studies. However, in the majority of cases the main genotype effect should still be observed regardless of environmental influences if the study is sufficiently powered.
A number of challenges underlie G x E studies and their application remains controversial to some. One of the main issues is that just as the genetic factors underlying drug addiction are not well known, the set of relevant environmental factors are also not clearly understood. As such, the discovery of G x E interactions depends on having appropriate subgroups (particularly those with a sound theoretical or biological basis) in the analyses where the genetic factors are having an effect. Inappropriate subgrouping may lead to erroneous conclusion of a lack of genetic, environmental or G x E effect, and failure to replicate a G x E results may be due to the effect acting upon further subgroups compared to those examined in the original study. However, particular care must be taken in selecting subgroups as unguided exploration through the endless permutations of possible subpopulations increases the risk of Type I errors, making the criteria for replication more stringent. Well-validated and consistent measures of environmental factors are needed, as some G x E interactions has been shown to be artifacts caused by scaling of environmental measures. A lack of clear distinction between environmental and genetic influences is also an issue. For example, bad parenting as an aversive environmental risk factor is in part related to genetics as manifested by individual differences in personality traits. Other contextual variables, such as social cohesion in the local community, may also be needed in the analysis. For example, certain G x E interactions (such as gene-parenting interactions) may appear in certain cultures but not others due to the different context of drug use and abuse. Multi-level analyses may be one method by which to model these effects. Finally, the nature of genetic, environmental factors and their interactions is likely transient and their relative importance will vary according to the life stage of the individual. Thus, longitudinal and age-specific models may be important, particularly for drug addiction.
It is notable that in spite of the potential importance of gene-environment interactions, only a portion of the studies of complex diseases have attempted to account for these effects to date. While there are methodological issues in the design and analysis of gene-environment effects, recently methods have been proposed to account for these interactions in GWA studies (38). Furthermore, inclusion of environmental covariates into analyses of data generated by GWA studies revealed associations with new genetic loci and their levels of statistical significance were increased compared to unadjusted analyses (39). Thus, accounting for genetic and environmental factors and their interactions may increase statistical power to detect true risk factors.
F) Re-emergence of family linkage studies
Family linkage studies examine genetically related individuals exposed to a similar familial and to some extent community environment; as such, they have been of great utility in genetic epidemiology and have proven effective in identifying genes of simple Mendelian diseases such as cystic fibrosis. Related individuals affected with the disease are recruited along with their unaffected family members and the inheritance pattern of the disease is examined via genomic markers. Differential transmission of alleles to the affected individuals indicates linkage of the marker with the phenotype measured. However, obtaining samples for family-based studies, particularly for psychiatric illnesses such as drug addiction, can be difficult given the associated stigma (40). Genotyping costs are also greater because of the higher density of markers required compared to studies with population-based samples, and the large size of chromosomal regions shared between family members makes it more difficult to isolate the signal associated with disease outcomes. Nevertheless, in comparison with GWA studies, which use population-based samples and are powered to detect common variants with modest effects, traditional family linkage studies may have greater power to detect rare variants associated with drug addiction phenotypes. To some extent the same argument applies to the use of founder populations and genetic isolates in which some variants that are rare on a world-wide basis can be expected to be far more common in these samples.
How can genetic information be applied to drug addiction?
A) Significant pharmacogenetic impacts on drug addiction
There are several studies where functional genetic variants were found to have a clinically relevant pharmacogenetic impact on drug addiction. A brief summary of these important examples is provided here as they have been reviewed more extensively elsewhere (41–45).
The discovery of functional polymorphisms in alcohol dehydrogenase IB (ADH1B) and aldehyde dehydrogenase 2 (ALDH2) represents one of the earliest and still most successful examples of pharmacogenetics as applied to drug addictions. These enzymes catalyze consecutive steps in alcohol metabolism (see Fig. 1); both have functional polymorphisms common in East Asians that additively alter the risk of alcoholism, with protective effects of 4–10 fold depending on the population (41). The two most common genetic variants in these enzymes are ADH1B His47Arg in which Arg47 is an increase-of-function allele, and ALDH2 Glu487Lys in which Lys487 is an inactive allele. Accumulation of acetaldehyde, the intermediate from ADH metabolism, potently releases histamine, triggering an aversive skin flushing reaction as well as headaches, nausea and palpitations that are thought to deter heavy alcohol use and development of dependence. Indeed, a number of studies have shown that genetic polymorphisms in ADH1B and ALDH2 that have functional impacts on enzyme function alter the risk of alcohol dependence (reviewed in (10), Supplementary Table 1, 3). Acetaldehyde is also a mutagen that can react with a variety of biomolecules and individuals with the Glu487Lys loss-of-function variant in ALDH2 should refrain from drinking large quantities of alcohol as they have a substantially elevated risk of upper GI cancer compared to individuals with the fully active enzyme (42).
Figure 1.
Similarly, genetic variants in the mu-opioid receptor have significant implications in drug addiction. One SNP (118A>G) corresponding to an amino acid change of Asn40Asp in the N-terminus of the receptor is of particular interest. Receptors with this variant have a three-fold greater binding of beta-endorphin, with a corresponding three-fold greater activation of the G-protein coupled inwardly rectifying potassium channels, although no differences in binding or activation by other endogenous opioid peptides nor exogenous opiates were observed (46). In specific cell lines morphine, methadone and DAMGO are all less potent at inhibiting adenylyl cyclase activity for receptors with the Asp40 variant. The Asp40 SNP also lowers receptor levels in expression cell systems compared to the wildtype Asn40 allele, which may be due to differences in glycosylation (46)). The presence of this variant influences the diverse physiological functions under modulation by the mu-opioid receptor, such as stress responsivity and pain perception. Processes related to abnormal stress responsivity, such as alcohol and opioid addictions, were found to be associated with genetic variation at the mu-opioid receptor even though these are very distinct disorders (reviewed in (44, 45), Supplementary Table 1, 3). Several studies, including those in the relatively non-admixed populations of central Sweden and the Han Chinese, have shown that the Asn40Asp variant is associated with opiate addiction, and one study has shown that it is also associated with alcoholism, although negative studies have also been reported ((44), Supplementary Table 1, 3). Of particular pharmacogenetic and clinical significance is that the presence of one or two copies of this variant predicts favorable outcome to treatment of alcoholism with naltrexone, a selective opioid antagonist (47).
It has been demonstrated that smokers titrate levels of smoking in order to maintain a particular level of nicotine in their system, and manipulation of the rates of nicotine clearance alter smoking behaviors (reviewed in (43)). As such, variability in CYP2A6, the main metabolic inactivating enzyme for nicotine (Fig. 1), can influence smoking behaviors, dependence and cessation. There are currently 38 CYP2A6 alleles identified (including SNPs, gene conversions, deletions and duplications, http://www.cypalleles.ki.se/cyp2a6.htm), and much progress has been made to understand their functional impact. Many of these genetic variants significantly alter the rate of nicotine metabolism among a variety of populations (reviewed in (43), Supplementary Table 1). Consistent with this, many studies (of primarily heavy-smoking Caucasians or Japanese ethnicity) have shown that smokers with genetic variants that impair CYP2A6 function, reducing nicotine metabolism, smoke fewer cigarettes, are less likely to be adult current smokers, and have a lower risk of lung cancer (reviewed in (4, 43), Supplementary Table 1, 3). Some studies have also indicated that reduced CYP2A6 activity may also alter the rate of acquisition of nicotine dependence and the rate of the escalation of dependence (48, 49).
B) Genetic testing in the prevention of drug addiction
Genetic testing already has important clinical applications in the prevention of diseases. Screening for deleterious variants in BRCA1/2 associated with increased risk of hereditary breast and ovarian cancer in women is becoming more common, and individuals with deleterious variants of BRCA1/2 can increase surveillance (such as increasing frequency of mammograms), undergo prophylactic surgery and take steps to reduce other risk factors. However, implementation of genetic testing in prevention programs for drug addiction has additional complexity as experimentation and initiation of drug use is more or less a choice made by the individual. It is unclear whether targeted prevention programs for individuals, in particular youths and adolescents who are the prime targets of such interventions, based on certain genetic vulnerabilities to developing drug dependence will be effective or socially acceptable. While we are still far from having sufficiently powerful genetic predictors for addictions, one example of genetic prevention that could be useful today is the Glu487Lys variant of aldehyde dehydrogenase-2 (ALDH2), which is found in approximately 500 million people as described above. Physicians should inform patients with the loss-of-function variant of their elevated risk of upper GI cancer and suggest they refrain from consuming large amounts of alcohol (42).
While it may be difficult to incorporate genetic testing into drug prevention programs, genetic research of drug addictions can assist in understanding the mechanisms underlying its etiology, and the prospects of using genetics to tailor medical treatment for drug addiction are encouraging.
C) Genetic testing in the treatment of drug addiction
Once an individual is addicted, the current clinical options for treatment are rather limited and only partially effective. Current pharmacotherapies and behavioral counseling improves the likelihood of smoking cessation by approximately 1.5 to 2.0 fold, with high rates of relapse and an average of 4–5 quit attempts needed before success (50). Personalized medication choices can be facilitated to a great extent by genetic studies, and provide novel insights into pharmacodynamics, pharmacokinetics and other aspects of the disposition of medications. Pharmacogenetic studies have already identified individuals that respond better to certain types of therapies for drug addiction based on their genetic makeup (Table 3). For example, functional variation in DBH, DRD2, OPRM1, CYP2A6 and CYP2B6 has been associated with smoking abstinence rates in clinical trials, either in response to pharmacotherapy or placebo (reviewed in (4), Supplementary table 4), although not always consistently. Other pharmacogenetic studies have examined treatment response for alcohol or opiate dependence. Variability in genes that are involved in methadone metabolism, such as CYP3A4 and CYP2D6, as well as genetic variation in P-glycoprotein (ABCB1, MDR1), for which methadone is a substrate, and DRD2 may alter the methadone dosage required for maintenance in heroin addicts (51–53). However, methadone is a complex drug for which the pharmacokinetic and pharmacodynamic responses are not well understood, and the genetic factors that underlie the variability in its response are not well known.
Table 3.
Common pharmacotherapies for drug dependence and the genetic variation that has been implicated in treatment response*
| Treatment | Mechanism of action | Genes | Effect |
|---|---|---|---|
| Nicotine dependence | |||
| Nicotine replacement therapy (gum, transdermal patch, lozenges, nasal spray) | nAChR agonist | CYP2A6 | CYP2A6 slow metabolizers, as indicated by phenotype indicator (trans-3′hydroxycotinine/cotinine ratio), were found to have higher quit rates |
| DRD2/ANKK1 | Taq1A A1 allele is associated with higher quit rates compared to the A2 allele | ||
| DRD2 -141C Del genotype was associated with higher quit rates compared to DRD2 -141C Ins genotype | |||
| DRD2 957T allele was associated with higher quit rates compared to the C allele | |||
| DBH | DBH 1368A allele was associated with higher quit rates compared to the G allele | ||
| COMT | For the COMT Val108/158Met polymorphism, individuals with the low-activity Met allele was associated with higher quit rates | ||
| OPRM1 | For the Asn40Asp polymorphism, the Asp40 variant was associated with higher quit rates compared to those homozygous for Asn40 | ||
| Bupropion | Dopamine/norepinephrine reuptake inhibitor | CYP2B6 | effect primarily driven by low quit rates of CY2B6*6 in placebo arm |
| Noncompetitive inhibitor of nAChR | DRD2/ANKK1 | Taq1A A2 allele was associated with higher quit rates | |
| Individuals homozygous for the DRD2 -141C Ins allele had significantly higher quit rates | |||
| CHRNB2 | GG genotype for 3′ UTR SNP (rs2072661) was associated with higher quit rates | ||
| Varenicline | α4β2 nAChR partial agonist | None examined to date | |
| Alcohol dependence | |||
| Disulfiram | aldehyde dehydrogenase inhibitor dopamine β-hydroxylase inhibitor |
None examined to date | |
| Naltrexone | non-selective opioid receptor antagonist | OPRM1 | Individuals with the Asp40 variant had significantly lower rates of relapse and longer time to return to heavy drinking compared to those homozygous for Asn40, though no differences were seen in overall abstinence rates |
| Acamprosate | unclear, may be metabotropic glutamate receptor subtype 5 (mGluR5) antagonist | None examined to date | |
| Opiate dependence | |||
| Methadone | opiate receptor agonist | ABCB1 (MDR1) encoding P- glycoprotein | Those with TT genotype for rs1128503 C>T SNP were more likely to receive higher methadone doses. |
| DRD2/ANKK1 | Individuals with the T allele of DRD2 rs6275C > T SNP required higher doses of methadone for maintenance therapy | ||
| Levo-α-acetylmethadol | μ-opioid receptor agonist | None examined to date | |
| Buprenorphine | μ-opioid receptor partial agonist κ-opioid receptor antagonist |
None examined to date | |
| Naltrexone | non-selective opioid receptor antagonist | None examined to date | |
Reviewed in (4, 10, 51). A complete list of references can be found in Supplementary Table 3.
A few genetic tests to optimize medical treatments in psychiatry, either by enhancing therapeutic efficacy or reducing adverse effects, are already in place. The FDA has added genetic testing to the prescription of carbamazepine in the treatment of bipolar disorder and neuropathic pain, as variation in the human leukocyte antigen gene (HLA-B*1502) has been associated with a potentially fatal skin reaction (reviewed in (53)). The FDA also approved the first diagnostic pharmacogenetic test in 2005. The AmpliChip CYP450 Genotyping Platform assesses variants in CYP2D6 and CYP2C19, the enzymes that metabolize numerous drugs including antidepressants, antipsychotics and opiates, and is intended to help physicians prescribe the type and dosage of medications based on an individual’s genotype (53). The next step is to determine whether pharmacogenetic testing can improve treatment outcomes for drug dependence in prospective studies, and to determine whether such procedures are cost-effective compared to standard care. Such economic analyses have already been performed for smoking cessation treatments, and suggest that genetic testing can be beneficial under certain assumptions (54, 55). These include the allele frequency of the genetic variant examined being neither too common nor rare, and the treatment response effect size of one genotype group being sufficiently larger than the other. Even though genetic testing may not necessarily be more cost-efficient in some cases (55), a demonstration that individuals with certain genetic variants will respond better to this treatment may encourage use among those who would otherwise be reluctant to do so. There is already evidence that genetic feedback can result in behavioral modifications related to drug addiction. For example, Marteau and Munafò et al. (personal communications) have also demonstrated that disclosure of genetic information can alter behavior and treatment compliance for smoking cessation.
Challenges & barriers to genetic research in drug addiction
A) Informatics
A plethora of data can now be generated via GWA studies, RNA expression studies based on microarrays and RNA sequencing, DNA sequencing and methylation studies, studies of epigenetic changes in histones, and re-sequencing studies targeting both SNPs and CNVs. However, many research laboratories are understaffed and ill-equipped to face the informatics technology (IT) challenges inherent in these high throughput methodologies. Also, research is constrained by software and hardware capabilities to handle extensive clinical genetic databases. It should be noted that these issues are applicable to all genetic studies of complex diseases and have been covered in greater detail elsewhere; a brief discussion will be provided here.
The IT challenge exists at several levels: planning (such as the informatics required to generate DNA capture arrays for sequencing), data capture, transfer, processing and storage (data files can be as large as 5 terabytes), primary analyses including mapping of hundreds of millions of DNA fragments to reference genomes, secondary analyses including identification of SNPs from sequence, tertiary analyses including gene ontology and clustering of differences in expression and epigenetic status of genes, and quaternary analyses integrating across modalities. There are several potential solutions to these problems. Primary analyses are usually performed on dedicated servers acquired with machines. For secondary analyses, fiber-optic connections to cluster servers and cloud computing networks with adequate storage (presently 30–100 terabytes) and automated data backup is mandatory. Tertiary analyses may be accomplished on cluster servers, cloud computing networks, and petaflop supercomputers.
Every individual’s DNA is unique and can be potentially used as an identifier; thus measures need to be taken to ensure the privacy of the data collected from genetic studies. However, firewalls that are necessary for the security and confidentiality of patient data can impede access and sharing of data by researchers. Data Enclaves where information is not collected, banked or shared, but allow investigators to work with specific datasets who have filed a research protocol regarding what can be printed, saved and removed from the site is one method of sharing datasets while protecting the identity of participants. The transfer of large files is another frequent problem but can be addressed by high speed links to nearby supercomputers and cloud computing systems. Availability of cheap and effective Laboratory Information Management Systems (LIMS) would also accelerate progress.
Clearly, collaborations between biologists, statisticians and IT specialists will be needed to best obtain, store and interpret the massive amounts of data created. Courses in advanced software development and system management can be developed and would be integral to professional development. An important focus could be algorithm development in specialized areas, for example Hidden-Markov Chain, simulations, E-M algorithms, and statistical genetics.
B) Practical issues of implementing genetic testing
The use of genetic testing in medicine has been the subject of much debate. As the day of whole genome sequencing for every individual draws near, legislation is required to protect individuals from potential misuse of the information. Recently, the United States Senate unanimously passed the Genetic Information Nondiscrimination Act which prohibits employers from inquiring about genetic testing or using one’s genetics as the basis for hiring, firing or promoting (56). The act also applies to health insurance plans, and prohibits the setting of eligibility, premium or contribution amount based on genetics, or requiring an individual to take a genetic test (56), and a similar voluntary moratorium also exists in the United Kingdom. Other considerations include the disclosure of genetic information when it can benefit third parties, such as sharing an individual’s genetic test result with their children, and as discussed above, the security of stored genetic data needs to be assured.
Another issue entails provision of funding and resources for genetic research in addictions. Scientific funding agencies vary across countries, and drug addiction is often not a high research priority given the general belief that drug use can be controlled by the law (even though prohibition has not proven successful in the past), and many in the general public still tend to view it as an issue of willpower. Thus, there needs to be better education of the general public, health care providers, and policy makers on how to interpret genetic information. The general public tends to view genes as deterministic, and it is important to improve the ability to convey risk, as opposed to absolutes, when disseminating information on diseases such as drug addiction where many other factors also contribute. Individuals also need to be protected against companies that attempt to profit from unsound or presently un-validated genetic tests. The commercialization of genetic testing is well underway with companies offering sequencing services alongside a list of diseases to which one is susceptible for a price. There are also companies that claim to be able to optimize the treatment of addictions based on genetics (e.g. Salugen, http://www.salugen.com; NicoTest™, http://www.nicotest.com, accessed February 2010), and predict the risk of developing lung cancer (http://www.synergenz.com, accessed February 2010). Such ventures need to be regarded with caution, especially given the predictive power of genetic tests is still highly limited at best.
Conclusions
In spite of the substantial consequences to the individual and society, genetic research on addictions has remained a relatively low priority. Recent technological advances have provided the tools to detect variation in the genetic architecture between individuals; however, much work still needs to be done to determine the biological relevance and to interpret the associations between these genetic variants with addiction-related phenotypes. An improvement in our understanding of the genetic and environmental factors underlying drug addiction has the potential to increase our understanding of the etiology and neurobiology of addictions, leading to greatly reduced morbidity and mortality by providing novel treatments and by improving the success rates of existing treatments.
Supplementary Material
Acknowledgments
This paper originated from an Esteve Foundation Discussion Group meeting and the authors are grateful for their support. Additional support from the Centre for Addictions and Mental Health, NSERC CGS-D Scholarship, and a SPICE Scholarship from Interdisciplinary Capacity Enhancement Team for MKH are acknowledged. Dr. A. Heinz receives support from the German Federal Ministry of Education and Research (National Genome Research Network, NGFNplus: 01GS08159). Dr. J. Kaprio receives funding from the Academy of Finland Centre of Excellence in Complex Disease Genetics, Academy of Finland grant 118555 and DA012854. Dr. M.J. Kreek is in part supported by NIH-NIDA P60-DA005130, NIH-NIMH R01-MH79880, NIH UL1 RR024143 and R01 MH076537 (H. Crystal, PI),. Dr. M.D. Li is in part supported by DA012844 and DA013783. Dr. R.F. Tyndale receives support from the Centre for Addictions and Mental Health, a Canada Research Chair in Pharmacogenetics, CIHR MOP86471, and NIH DA 0220830.
Abbreviations
- Heritability
The proportion of phenotypic variation that can be attributed to genetic variation between individuals.
- Genome-wide association study
Study of common genetic variation across the entire human genome to identify genetic associations with observable traits. It is an approach of searching for genes underlying diseases without any a priori hypotheses.
- Allele
Alternative forms of a gene that occurs at a given locus on a specific chromosome.
- Linkage disequilibrium
When different genetic loci are inherited together more commonly than expected due to chance alone.
- Haplotype
A combination of alleles found at different genetic loci, but within the same chromosome.
Footnotes
Conflicts of interests
Dr. J. Kaprio has served as a consultant to Pfizer. To receive compensation from the University of Virginia, Dr. M.D. Li has served as a consultant to NIH, DeCODE genetics, University of Pennsylvania, Reckitt Benckiser Pharmaceuticals, Pennsylvania Department of Health, and Informational Managements Consulting. Dr. M.D. Li also serves as a scientific advisor to ADial Pharmaceuticals. Dr. R.F. Tyndale hold shares and is a CSO in Nicogen Research Inc., a company that is focused on novel smoking cessation treatment approaches. None of the data contained in this manuscript alters or improves any commercial aspect of Nicogen, no Nicogen funds were used in this work, and the manuscript was not reviewed by others affiliated with Nicogen. Dr. R.F. Tyndale has also been a paid consultant for Novartis. MKH and Drs. D. Goldman, A. Heinz, M.J. Kreek and M.R. Munafò report no conflicts of interest.
References
- 1.The World Health Organization. Substance Abuse Facts and Figures: The Global Burden. 2002 Available from: http://www.who.int/substance_abuse/facts/global_burden/en/index.html.
- 2.Office of National Drug Control Policy. The Economic Costs of Drug Abuse in the United States, 1992–2002. Washington, D.C: 2004. [Google Scholar]
- 3.Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nat Rev Genet. 2005;6(7):521. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- 4.Ho MK, Tyndale RF. Overview of the pharmacogenomics of cigarette smoking. Pharmacogenomics J. 2007;7(2):81–98. doi: 10.1038/sj.tpj.6500436. [DOI] [PubMed] [Google Scholar]
- 5.Rose RJ, Broms U, Korhonen T, Dick DM, Kaprio J. Handbook of Behavior Genetics. Springers; 2009. Genetics of Smoking Behavior; pp. 411–32. [Google Scholar]
- 6.Agrawal A, Lynskey MT. Are there genetic influences on addiction: evidence from family, adoption and twin studies. Addiction. 2008;103(7):1069–81. doi: 10.1111/j.1360-0443.2008.02213.x. [DOI] [PubMed] [Google Scholar]
- 7.Li MD, Burmeister M. New insights into the genetics of addiction. Nat Rev Genet. 2009;10(4):225. doi: 10.1038/nrg2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pushkarev D, Neff NF, Quake SR. Single-molecule sequencing of an individual human genome. Nat Biotech. 2009;27(9):847. doi: 10.1038/nbt.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross S, Peselow E. The Neurobiology of Addictive Disorders. Clinical Neuropharmacology. 2009;32(5):269–76. doi: 10.1097/wnf.0b013e3181a9163c. [DOI] [PubMed] [Google Scholar]
- 10.Khokhar JY, Ferguson CS, Zhu AZX, Tyndale RF. Pharmacogenetics of Drug Dependence: Role of Gene Variations in Susceptibility and Treatment. Annual Review of Pharmacology and Toxicology. 2010;50:39–61. doi: 10.1146/annurev.pharmtox.010909.105826. [DOI] [PubMed] [Google Scholar]
- 11.Pearson TA, Manolio TA. How to Interpret a Genome-wide Association Study. JAMA. 2008 Mar 19;299(11):1335–44. doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- 12.Bierut LJ. Convergence of genetic findings for nicotine dependence and smoking related diseases with chromosome 15q24-25. Trends in Pharmacological Sciences. 2009;31(1):46–51. doi: 10.1016/j.tips.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meltzer PS. Cancer genomics: Small RNAs with big impacts. Nature. 2005;435(7043):745. doi: 10.1038/435745a. [DOI] [PubMed] [Google Scholar]
- 14.Yuferov V, Nielsen D, Butelman E, Kreek MJ. Microarray Studies of Psychostimulant-Induced Changes in Gene Expression. Addiction Biology. 2005;10(1):101–18. doi: 10.1080/13556210412331308976. [DOI] [PubMed] [Google Scholar]
- 15.Huang W, Li MD. Nicotine modulates expression of miR-140*, which targets the 3?-untranslated region of dynamin 1 gene ( Dnm1) The International Journal of Neuropsychopharmacology. 2009;12(04):537–46. doi: 10.1017/S1461145708009528. [DOI] [PubMed] [Google Scholar]
- 16.Xu Q, Huang W, Payne TJ, Ma JZ, Li MD. Detection of Genetic Association and a Functional Polymorphism of Dynamin 1 Gene with Nicotine Dependence in European and African Americans. Neuropsychopharmacology. 2008;34(5):1351–9. doi: 10.1038/npp.2008.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang W, Li MD. Differential Allelic Expression of Dopamine D1 Receptor Gene (DRD1) Is Modulated by microRNA miR-504. Biological Psychiatry. 2009;65(8):702. doi: 10.1016/j.biopsych.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renthal W, Nestler EJ. Epigenetic mechanisms in drug addiction. Trends in Molecular Medicine. 2008;14(8):341. doi: 10.1016/j.molmed.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nielsen DA, Yuferov V, Hamon S, Jackson C, Ho A, Ott J, et al. Increased OPRM1 DNA Methylation in Lymphocytes of Methadone-Maintained Former Heroin Addicts. Neuropsychopharmacology. 2008;34(4):867–73. doi: 10.1038/npp.2008.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandey SC, Ugale R, Zhang H, Tang L, Prakash A. Brain Chromatin Remodeling: A Novel Mechanism of Alcoholism. J Neurosci. 2008 Apr 2;28(14):3729–37. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munafo MR, Matheson IJ, Flint J. Association of the DRD2 gene Taq1A polymorphism and alcoholism: a meta-analysis of case-control studies and evidence of publication bias. Mol Psychiatry. 2007;12(5):454. doi: 10.1038/sj.mp.4001938. [DOI] [PubMed] [Google Scholar]
- 22.Munafo MR, Timpson NJ, David SP, Ebrahim S, Lawlor DA. Association of the DRD2 gene Taq1A polymorphism and smoking behavior: A meta-analysis and new data. Nicotine Tob Res. 2009 Jan 1;11(1):64–76. doi: 10.1093/ntr/ntn012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes JR, Oliveto AH, Riggs R, Kenny M, Liguori A, Pillitteri JL, et al. Concordance of different measures of nicotine dependence: Two pilot studies. Addictive Behaviors. 2004;29(8):1527. doi: 10.1016/j.addbeh.2004.02.031. [DOI] [PubMed] [Google Scholar]
- 24.Strat YL, Ramoz N, Schumann G, Gorwood P. Molecular Genetics of Alcohol Dependence and Related Endophenotypes. Curr Genomics. 2008;9(7):444–51. doi: 10.2174/138920208786241252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puls I, Mohr J, Wrase J, Priller J, Behr J, Kitzrow W, et al. Synergistic effects of the dopaminergic and glutamatergic system on hippocampal volume in alcohol-dependent patients. Biological Psychology. 2008;79(1):126. doi: 10.1016/j.biopsycho.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Filbey FM, Ray L, Smolen A, Claus ED, Audette A, Hutchison KE. Differential Neural Response to Alcohol Priming and Alcohol Taste Cues Is Associated With DRD4 VNTR and OPRM1 Genotypes. Alcoholism: Clinical and Experimental Research. 2008;32(7):1113–23. doi: 10.1111/j.1530-0277.2008.00692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Visscher PM, Montgomery GW. Genome-wide Association Studies and Human Disease: From Trickle to Flood. JAMA 2009. 2009 Nov 11;302(18):2028–9. doi: 10.1001/jama.2009.1643. [DOI] [PubMed] [Google Scholar]
- 28.Keskitalo K, Broms U, Heliovaara M, Ripatti S, Surakka I, Perola M, et al. Association of serum cotinine level with a cluster of three nicotinic acetylcholine receptor genes (CHRNA3/CHRNA5/CHRNB4) on chromosome 15. Hum Mol Genet 2009. 2009 Oct 15;18(20):4007–12. doi: 10.1093/hmg/ddp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106(23):9326–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare Variants of IFIH1, a Gene Implicated in Antiviral Responses, Protect Against Type 1 Diabetes. Science 2009. 2009 Apr 17;324(5925):387–9. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tapper AR, McKinney SL, Nashmi R, Schwarz J, Deshpande P, Labarca C, et al. Nicotine activation of alpha4* receptors: sufficient for reward, tolerance, and sensitization. Science. 2004;306(5698):1029–32. doi: 10.1126/science.1099420. [DOI] [PubMed] [Google Scholar]
- 32.Picciotto MR, Zoli M, Rimondini R, Lena C, Marubio LM, Pich EM, et al. Acetylcholine receptors containing the [beta]2 subunit are involved in the reinforcing properties of nicotine. Nature. 1998;391(6663):173. doi: 10.1038/34413. [DOI] [PubMed] [Google Scholar]
- 33.Xu W, Gelber S, Orr-Urtreger A, Armstrong D, Lewis RA, Ou C-N, et al. Megacystis, mydriasis, and ion channel defect in mice lacking the α3 neuronal nicotinic acetylcholine receptor. Proceedings of the National Academy of Sciences of the United States of America. 1999 May 11;96(10):5746–51. doi: 10.1073/pnas.96.10.5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Portugal GS, Gould TJ. Genetic variability in nicotinic acetylcholine receptors and nicotine addiction: Converging evidence from human and animal research. Behavioural Brain Research. 2008;193(1):1. doi: 10.1016/j.bbr.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Grucza RA, Xuei X, et al. Variants in Nicotinic Receptors and Risk for Nicotine Dependence. Am J Psychiatry. 2008 Sep 1;165(9):1163–71. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carmen SvdZ, Rutger CMEE. Gene-environment interactions and alcohol use and dependence: current status and future challenges. Addiction. 2009;104(6):907–14. doi: 10.1111/j.1360-0443.2009.02563.x. [DOI] [PubMed] [Google Scholar]
- 37.Heinz A, Mann K, Weinberger DR, Goldman D. Serotonergic Dysfunction, Negative Mood States, and Response to Alcohol. Alcoholism: Clinical and Experimental Research. 2001;25(4):487–95. [PubMed] [Google Scholar]
- 38.Chatterjee N, Wacholder S. Invited Commentary: Efficient Testing of Gene-Environment Interaction. Am J Epidemiol 2009. 2009 Jan 15;169(2):231–3. doi: 10.1093/aje/kwn352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Igl W, Johansson Ãs, Wilson JF, Wild SH, Polasek O, Hayward C, et al. Modeling of Environmental Effects in Genome-Wide Association Studies Identifies SLC2A2 and HP as Novel Loci Influencing Serum Cholesterol Levels. PLoS Genet. 6(1):e1000798. doi: 10.1371/journal.pgen.1000798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kathleen AB. The Art of Recruitment: The Foundation of Family and Linkage Studies of Psychiatric Illness. Family Process. 1998;37(2):153–65. doi: 10.1111/j.1545-5300.1998.00153.x. [DOI] [PubMed] [Google Scholar]
- 41.Goldman D, Ducci F. The genetics of alcoholism and other addictive disorders. In: Speicher MR, Antonarakis SE, Motulsky AG, editors. Human Genetics. 4. Springer-Verlag; Berlin Heidelberg: 2010. [Google Scholar]
- 42.Brooks PJ, Enoch M-A, Goldman D, Li T-K, Yokoyama A. The Alcohol Flushing Response: An Unrecognized Risk Factor for Esophageal Cancer from Alcohol Consumption. PLoS Med. 2009;6(3):e1000050. doi: 10.1371/journal.pmed.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mwenifumbo JC, Tyndale RF. Genetic variability in CYP2A6 and the pharmacokinetics of nicotine. Pharmacogenomics. 2007;8(10):1385–402. doi: 10.2217/14622416.8.10.1385. [DOI] [PubMed] [Google Scholar]
- 44.Kreek MJ, LaForge KS. Stress Responsivity, Addiction, and a Functional Variant of the Human Mu-Opioid Receptor Gene. Molecular Interventions 2007. 2007 Apr;7(2):74–8. doi: 10.1124/mi.7.2.7. [DOI] [PubMed] [Google Scholar]
- 45.Kreek MJ. Role of a Functional Human Gene Polymorphism in Stress Responsivity and Addictions. Clin Pharmacol Ther. 2008;83(4):615–8. doi: 10.1038/clpt.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bond C, LaForge KS, Tian M, Melia D, Zhang S, Borg L, et al. Single-nucleotide polymorphism in the human mu opioid receptor gene alters Î2-endorphin binding and activity: Possible implications for opiate addiction. Proceedings of the National Academy of Sciences of the United States of America. 1998 Aug 4;95(16):9608–13. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oslin DW, Berrettini W, Kranzler HR, Pettinati H, Gelernter J, Volpicelli JR, et al. A Functional Polymorphism of the [mu]-Opioid Receptor Gene is Associated with Naltrexone Response in Alcohol-Dependent Patients. Neuropsychopharmacology. 2003;28(8):1546–52. doi: 10.1038/sj.npp.1300219. [DOI] [PubMed] [Google Scholar]
- 48.O’Loughlin J, Paradis G, Kim W, DiFranza J, Meshefedjian G, McMillan-Davey E, et al. Genetically decreased CYP2A6 and the risk of tobacco dependence: a prospective study of novice smokers. Tob Control. 2004 Dec 1;13(4):422–8. doi: 10.1136/tc.2003.007070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Audrain-McGovern J, Koudsi NA, Rodriguez D, Wileyto EP, Shields PG, Tyndale RF. The Role of CYP2A6 in the Emergence of Nicotine Dependence in Adolescents. Pediatrics. 2007 Jan 1;119(1):e264–74. doi: 10.1542/peds.2006-1583. [DOI] [PubMed] [Google Scholar]
- 50.Maseeh A, Kwatra G. A Review of Smoking Cessation Interventions. MedGenMed. 2005;7(2):24. [PMC free article] [PubMed] [Google Scholar]
- 51.Kreek MJ, Zhou Y, Butelman ER, Levran O. Opiate and cocaine addiction: from bench to clinic and back to the bench. Current Opinion in Pharmacology. 2009;9(1):74. doi: 10.1016/j.coph.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doehring A, Hentig Nv, Graff J, Salamat S, Schmidt M, Geisslinger G, et al. Genetic variants altering dopamine D2 receptor expression or function modulate the risk of opiate addiction and the dosage requirements of methadone substitution. Pharmacogenetics & Genomics. 2009;19(6):407–14. doi: 10.1097/FPC.0b013e328320a3fd. [DOI] [PubMed] [Google Scholar]
- 53.Haile CN, Kosten TA, Kosten TR. Pharmacogenetic Treatments for Drug Addiction: Alcohol and Opiates. The American Journal of Drug and Alcohol Abuse. 2008;34(4):355–81. doi: 10.1080/00952990802122564. [DOI] [PubMed] [Google Scholar]
- 54.Heitjan DF, Asch DA, Ray R, Rukstalis M, Patterson F, Lerman C. Cost-effectiveness of pharmacogenetic testing to tailor smoking-cessation treatment. Pharmacogenomics J. 2008;8(6):391. doi: 10.1038/sj.tpj.6500492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Welton NJJE, David SP, Munafo MR. A cost-effectiveness analysis of genetic testing of the DRD2 Taq1A polymorphism to aid treatment choice for smoking cessation. Nicotine & Tobacco Research. 2008;10(1):231–40. doi: 10.1080/14622200701767761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dressler LG, Terry SF. How Will GiNa influence Participation in Pharmacogenomics research and clinical testing? Clin Pharmacol Ther. 2009;86(5):472– 5. doi: 10.1038/clpt.2009.146. [DOI] [PubMed] [Google Scholar]
- 57.Heath AC, Martin NG. Genetic Influences on Alcohol Consumption Patterns and Problem Drinking: Results from the Australian NH&MRC Twin Panel Follow-up Survey. Annals of the New York Academy of Sciences. 1994;708:72–85. doi: 10.1111/j.1749-6632.1994.tb24699.x. Types of Alcoholics: Evidence from Clinical, Experimental, and Genetic Research. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.