Abstract
Ribosomes are the nanomachines that synthesize all cellular proteins from mRNA templates. In eukaryotes, ribosomes, which are composed of ribosomal proteins and rRNA, are mainly assembled in the nucleus. Thus, ribosomal proteins require a nuclear transport step from their place of synthesis in the cytoplasm to their site of assembly in the nucleus. Recognition of import substrates is mediated by different types of nuclear localization signals, which are either directly recognized by import receptors or recruited to these via adaptor proteins. The novel transport adaptor Syo1 (Symportin), which is dedicated to the synchronous import of two functionally related ribosomal proteins, has recently been described. In this review, we highlight and discuss these findings in the context of our current knowledge of ribosome assembly and nucleocytoplasmic transport. We propose that nuclear co-import of functionally and topologically linked cargo could be a widespread strategy to streamline assembly of macromolecular complexes in the nucleus.
Keywords: nuclear import, transport adaptor, nuclear localization signal, ribosome assembly, ribosomal protein, chaperone
Ribosome Assembly in Eukaryotes
Proteins are major cellular constituents and play central roles in all processes of life. All proteins are synthesized by ribosomes—these are the nanomachines that catalyze peptide bond formation between amino acids using tRNA as the translation adaptor and mRNA as the blueprint. Rapidly dividing cells require the continuous synthesis of new ribosomes,1 a process that in eukaryotes primarily takes place in the nucleus. Ribosomes are complex structures, which consist of approximately 80 proteins (r-proteins) and four rRNAs (rRNA).2 Basically, the synthesis of the two ribosomal subunits consists in the ordered assembly of 33 r-proteins with the 18S rRNA and 46 r-proteins with the 25S, 5.8S and 5S rRNA, thus yielding mature 40S and 60S subunits, respectively.3 Since most r-proteins are incorporated into early pre-ribosomal particles, they need to traverse the nuclear pore complex (NPC) and be targeted to their site of assembly in the nucleolus or nucleoplasm (Fig. 1).4
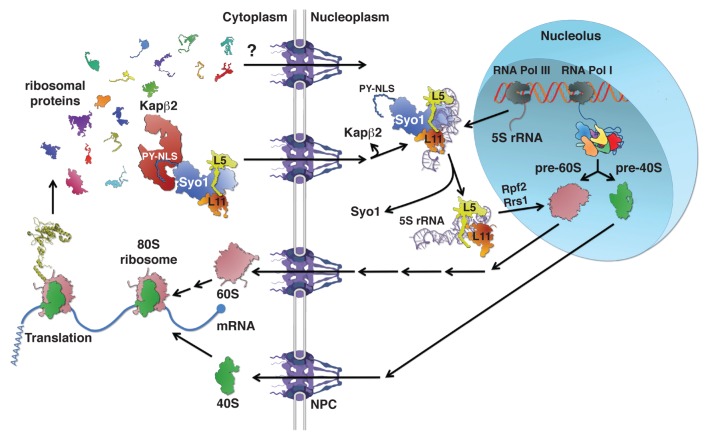
Figure 1. Symportin synchronizes nuclear import of the ribosomal proteins Rpl5 and Rpl11 with 5S RNP assembly. Ribosomal proteins (r-proteins) and ribosome biogenesis factors are synthesized in the cytoplasm and need to be imported into the nucleus before their assembly into pre-ribosomal particles. Passage across the nuclear pore complex (NPC) is facilitated by transport receptors of the importin-β family; however, in most cases the importin mediating nuclear import of a given r-protein or biogenesis factor is not yet known. Ribosome assembly starts in the nucleolus, a subnuclear compartment, by formation of a 90S pre-ribosomal particle upon transcription of the rDNA and the association of “early” r-proteins and biogenesis factors with the nascent 35S pre-rRNA. Concomitant to and/or after completion of transcription, endonucleolytic cleavage of the 35S pre-rRNA leads to the formation of pre-40S and pre-60S ribosomes. While pre-40S ribosomes are rapidly exported to the cytoplasm, the nuclear phase of pre-60S maturation is complex and involves several well-defined intermediates. The 5S RNP, composed of the 5S rRNA and the r-proteins Rpl5 and Rpl11, is incorporated into early pre-60S particles. The formation of the 5S RNP is already predetermined in the cytoplasm where Rpl5 and Rpl11 form an import complex with the transport adaptor Syo1. The trimeric Syo1-Rpl5-Rpl11 complex is recognized, via the N-terminal PY-NLS of Syo1, by the importin Kapβ2 (Kap104). Upon nuclear translocation and RanGTP-dependent cargo release, association of the 5S rRNA leads to the generation of a 5S RNP intermediate containing Syo1. Release of Syo1 is coupled to the formation of an assembly-competent 5S RNP that is most likely inserted with the aid of the biogenesis factors Rpf2 and Rrs1 into nucleolar pre-60S particles. Both pre-40S and pre-60S subunits undergo final maturation in the cytoplasm leading to acquisition of translation competence.
The process of ribosome biogenesis is evolutionarily conserved among eukaryotes. Moreover, most of our current knowledge concerning this highly dynamic, multi-step process comes from studies with the yeast Saccharomyces cerevisiae, we will therefore briefly outline how ribosomes are assembled in this organism. Ribosome synthesis begins in the nucleolus, a specialized subnuclear compartment, with the transcription of the rDNA genes into precursors (pre-rRNAs), which undergo a series of covalent modifications and endo- and exonucleolytic cleavages.5,6 Three of the four rRNAs (18S, 5.8S and 25S) are transcribed as a single large 35S pre-rRNA by RNA polymerase I (RNA Pol I), whereas the fourth rRNA (5S) is transcribed as a pre-5S rRNA by RNA Pol III. Concomitant to transcription, small subunit r-proteins and early protein trans-acting factors associate with the nascent 35S pre-rRNA transcript to form 90S pre-particles (Fig. 1). A subsequent endonucleolytic cleavage step, either co- or post-transcriptional, generates the 43S and 66S pre-particles that are further matured to give rise to mature 40S and 60S r-subunits, respectively. Proteomic approaches revealed the protein and rRNA composition of several distinct and successive pre-40S and pre-60S ribosomal particles.3,4,7 Contrary to the relatively low compositional complexity of 43S pre-particles, the early pre-60S particles contain more than 40 associated protein trans-acting factors. As a consequence, the maturation of nuclear pre-60S particles is accompanied by major compositional changes that lead to a sequential reduction of complexity and to the acquisition of export competence. Export of pre-60S subunits, contrary to pre-40S export, is relatively well understood and relies on several factors, including the export adaptor Nmd3 and the exportin Crm1.3,8 Cytoplasmic maturation events then lead to the release of the last biogenesis factors and enable subunit joining;9,10 moreover, recent evidence indicates that pre-ribosomal subunits and/or 80S ribosomes are subjected to quality control prior to engaging in translation.11-13 Given the complexity of the process, it is not surprising that a multitude of protein trans-acting factors (> 200) are required for the assembly and maturation of pre-ribosomal particles as they travel from the nucleolus to the cytoplasm.3,14 Among these are, in agreement with the dynamic nature of the process, energy-consuming enzymes such as AAA-type ATPases (ATPases associated with various cellular activities), ATP-dependent RNA helicases, kinases and GTPases.3,15-18 This suggests that the energy derived from nucleotide hydrolysis or phosphorylation confers directionality to ribosome assembly and that such a large number of trans-acting factors is required to ensure accurate and efficient synthesis of ribosomal subunits.
While the structural and functional role of r-proteins within mature ribosomal subunits is well established, less is known about the contribution of r-proteins to the biogenesis of ribosomal subunits. A temporal assembly map of r-proteins was already established in the 1970s;19 however, only recently have their functional roles in ribosome assembly and nuclear export of pre-ribosomes started to be investigated, see for example.18,20-24 As can be seen in the crystal structures of ribosomes, most r-proteins fold into a globular domain, which is generally found on the surface of the ribosomal subunits, and long extensions that penetrate into the interior and stabilize the tertiary structure of the rRNA.25-28 Some r-proteins are even devoid of globular domains, and the extensions, which are rich in lysine and arginine residues, are likely disordered in the non-assembled r-proteins.26 In agreement with the higher regulatory complexity of eukaryotic translation, eukaryotic r-proteins are generally larger than their prokaryotic counterparts and eukaryotes also contain many additional, unique r-proteins. Moreover, the eukaryote-specific extensions within conserved r-proteins may be needed to confer nuclear targeting to those r-proteins that are assembled into nuclear pre-ribosomal subunits. Due to their capacity to bind to rRNA, mostly via their basic extensions, newly synthesized r-proteins must be prevented from binding non-specifically to cytoplasmic RNAs (e.g., tRNAs and mRNAs). In order to simultaneously promote nuclear targeting, the same basic stretches may be utilized for recognition by importins.29 Moreover, recent evidence suggests that, besides importins, r-proteins rely on specific binding partners or accessory domains that promote their soluble expression, nuclear targeting and/or assembly into pre-ribosomes.30-32
The Molecular Framework of Nucleo-Cytoplasmic Import
Nuclear import receptors: importin-β
With the acquisition of a nuclear compartment and the physical separation of the genetic material from the protein synthesis machinery, eukaryotic cells had to co-evolve sophisticated transport systems to ensure nuclear targeting of a large number of proteins essential to many nuclear processes, including ribosome biogenesis. Research over the past three decades, initiated by the discovery of a peptide segment mediating nuclear targeting,33 has unraveled the molecular framework of how proteins and ribonucleoproteins (RNPs) translocate across NPCs, which constitute the gates of the nucleo-cytoplasmic barrier.34-40 While proteins with a molecular mass below ~40 kDa can generally passively diffuse across NPCs, nuclear import of most proteins is dependent on an active transport process that is mediated by soluble transport factors, also referred to as importins, of the karyopherin-β protein family.41,42 The karyopherin-β protein family, which also includes exportins, is characterized by the occurrence of 19–20 HEAT (huntingtin, elongation factor 3, A subunit of PP2A and TOR) repeats arranged into an α-solenoid fold of concave or ring-like shape.42-45 While the genome of S. cerevisiae encodes ten importin-β proteins, 11 human karyopherin-β proteins are capable of transporting proteins into the nucleus.41 According to the available crystal structures, importin-β proteins can be divided into two functional domains, which are referred to as the N- and C-terminal arch.46,47 Despite their structural similarity and similar physico-chemical properties (isoelectric points ~4.5 / molecular weights 95–125 k Da), they only show ~20% of sequence identity.41 Nevertheless, importin-β proteins share several common features, such as the capacity to recognize their import cargo within the cytoplasm, to interact with hydrophobic phenylalanine-glycine (FG) repeats that line the transport channel of NPCs,48,49 and to release their cargo in the nucleus upon binding to RanGTP (see below). While the N-terminal arch, besides mediating the interaction with FG-repeats of nucleoporins, forms the RanGTP binding site, the C-terminal arch is the major contact site for recognition of cargo.
Nuclear localization signals
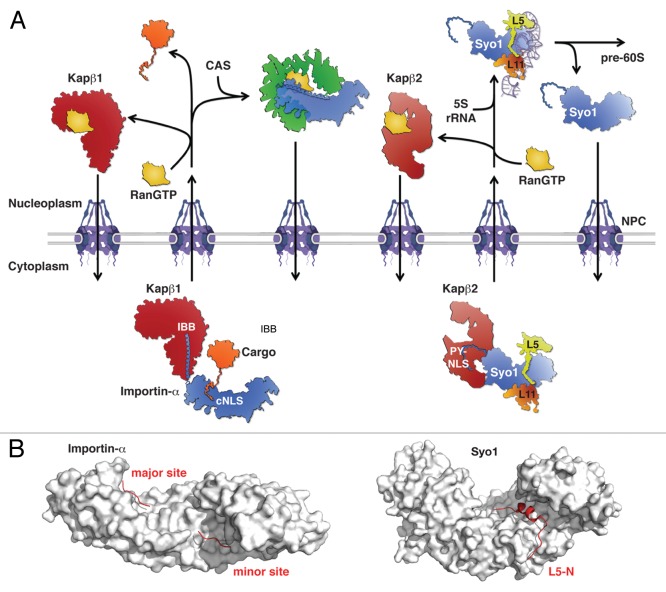
Recognition of import substrates is mediated by nuclear localization signals (NLSs), which are either directly recognized by importin-β or recruited to importin-β via transport adaptors, such as the co-importin importin-α.40,41,50,51 The classical NLSs (cNLS) can be subdivided into a monopartite- (consensus: K-K/R-X-K/R) and a bipartite-type (loose consensus of K/R-K/R-X(10–12)-(K/R)3/5).50,51 Both cNLS types are recognized by the importin-α subunit of the heterodimeric Kapβ1/importin-α (in S. cerevisiae Kap95/Kap60) transport receptor.40,41,50,51 Importin-α consists of ten armadillo (ARM) repeats whose inner α-helices form a concave surface containing a major (ARM-repeats 2–5) and a minor (ARM-repeats 6–8) binding site for basic residues in an extended conformation (Fig. 2). While monopartite cNLSs primarily bind to the major binding site, bipartite cNLSs bind to both sites with the C-terminal basic cluster being located in the major binding site.40,51-54 Upon cargo binding in the cytoplasm, importin-α interacts via its N-terminal importin-β1 binding (IBB) domain with Kapβ1 and the ternary cargo/importin-α/importin-β complex then traverses the NPC (Fig. 2).40,50 In the cargo-free form, importin-α binds its own IBB domain in the major cNLS binding site, thus ensuring that only cargo-loaded importin-α is recognized by Kapβ1 as a transport substrate. The IBB domain consists of ~40 amino acids composed of an N-terminal 310 helix connected via a short loop to a C-terminal basic α-helix of ~30 residues.50 The IBB domain binds to the inner concave surface formed by HEAT-repeats 7–19 of Kapβ1, thereby leading to a compaction of its elongated α-solenoid shape. The C-terminal α-helix of the IBB domain is the major determinant for the association with Kapβ1, and its contacts with HEAT-repeats 12–19 are mainly based on electrostatic interactions between basic residues of the IBB α-helix and conserved negatively charged side chains of Kapβ1. Besides the indirect recognition of import substrates via the IBB domain of cargo-loaded importin-α, both Kapβ1 and Kap95 can also directly bind to some of their cargo proteins.41,51,55,56
Figure 2.Modus operandi of the generic importin-α and the dedicated symportin transport adaptors. (A, left) Importin-α recognizes cargo proteins via classical nuclear localizations signals (cNLS). Subsequent binding, via its N-terminal IBB, to the Kapβ1 transport receptor leads to the generation of a Kapβ1/importin-α/cargo import complex. Upon nuclear translocation across the NPC, RanGTP binding to Kapβ1 releases cargo-loaded importin-α and the subsequent interaction of importin-α with the exportin CAS/Cse1 promotes cargo release. RanGTP-bound Kapβ1 and the CAS/RanGTP/importin-α export complex return to the cytoplasm where GTP hydrolysis releases RanGDP and allows Kapβ1 and its transport adaptor importin-α to engage in another round of nuclear protein import. (A, right) In contrast to the generic transport adaptor importin-α, the symportin Syo1 is solely dedicated to the synchronous nuclear import of the 5S rRNA binding r-proteins Rpl5 and Rpl11. Binding of the trimeric Syo1-Rpl5-Rpl11 complex to the transport receptor Kapβ2 is mediated by the N-terminal PY-NLS of Syo1. Upon nuclear translocation, RanGTP binding to Kapβ2 releases the trimeric Syo1-Rpl5-Rpl11 complex, which, together with the 5S rRNA, forms a Syo1-containing 5S RNP assembly intermediate. Conversion of this intermediate into an assembly-competent 5S RNP, which is incorporated into early pre-60S particles, leads to the dissociation of Syo1. Syo1 then returns to the cytoplasm by facilitated diffusion due to its affinity for FG-repeats of nucleoporins. (B) Surface representation of importin-α and Syo1 (gray) bound to the SV40-NLS and the N-terminal residues of Rpl5 (L5-N), respectively (red). The major and minor NLS-binding sites on importin-α are indicated. The shown structures are derived from the PDB entries 1BK6 (Kap60/SV40-NLS) and 4GMN (Syo1/L5-N).31,53
The remaining importin-β transport receptors do not rely on the recognition of cargo by co-importins.41 Functional and structural analyses revealed that the well-studied Kapβ2 (Kap104) binds via the inner concave surface of its C-terminal arch to the extended conformation of two types of linear sequences of ~15–30 amino acids, which have related but distinct consensus sequences and are accordingly referred to as hydrophobic or basic proline-tyrosine NLSs (PY-NLSs).41,47,57,58 The common denominator of both types of PY-NLSs is a C-terminal signature with the consensus sequence R/K/H-X(2–5)-P-Y/L. The distinguishing feature is the presence of either a hydrophobic- or basic-enriched N-terminal segment in the case of hydrophobic (hPY-NLS) or basic (bPY-NLS) PY-NLSs, respectively. While Kapβ2 recognizes both types, its yeast ortholog Kap104 is only capable of interacting with the bPY-NLS—this can be explained by the fact that residues within HEAT-repeat helices H16B and H17B that contact the hydrophobic motif are not conserved in Kap104.58 PY-NLSs, irrespective of the sub-type, contain three significant epitopes, consisting of (1) the N-terminal hydrophobic/basic motif, (2) the residue R/K/H and (3) the C-terminal P-Y/L signature.58
Ran GTPase drives nuclear transport
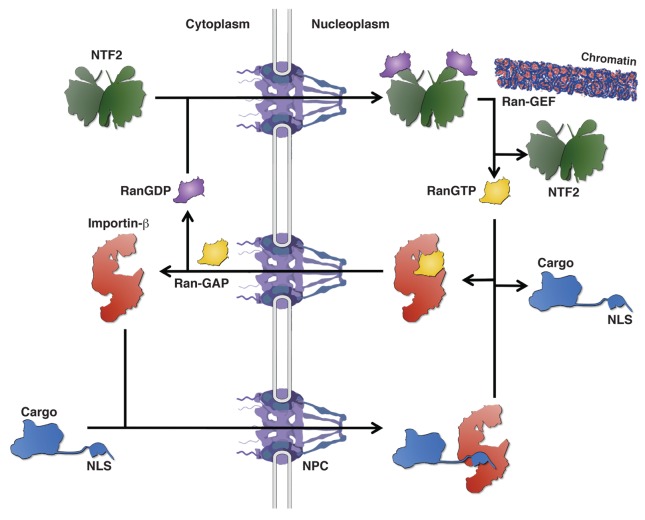
The driving force behind nuclear import is provided by the predominant nuclear localization of a small GTPase, called Ran (Gsp1), in its GTP-bound state.37,40 The gradient of RanGTP across the NPC is established by the exclusive nuclear or cytoplasmic localization of its guanine-exchange factor (Ran-GEF) and its GTPase activating protein (Ran-GAP), respectively. This RanGTP gradient is then employed to specifically dissociate the import substrate from cargo-loaded importin-β in the nuclear compartment. The complete protein import cycle can be summarized as follows (Fig. 3): (i) Importin-β proteins bind their import substrates in the cytoplasm, then, the affinity of transport receptors for the FG-repeat meshwork allows facilitated diffusion of importin-cargo complexes across the NPC. (ii) Upon nuclear entry, RanGTP binds to the N-terminal arch of cargo-loaded importin-β and thereby induces release of the import substrate in the nucleus. (iii) The newly formed RanGTP/importin-β complex traverses again the NPC and dissociates upon GTP hydrolysis, catalyzed by Ran-GAP, in the cytoplasm due to the low binding affinity of RanGDP for importin-β proteins. (iv) To replenish the nuclear pool of Ran, RanGDP is re-imported by the transport factor NTF2 (Ntf2) into the nucleus, where it is again loaded with GTP by its GEF. This cycle is slightly more complex in case of the heterodimeric Kapβ1/importin-α transport receptor—upon its nuclear entry, RanGTP binding leads to the dissociation of importin-α from Kapβ1 and concomitantly, with the aid of the nucleoporin NUP50 (Nup2), the cNLS-cargo is released from importin-α, which is then recycled to the cytoplasm by the exportin CAS (Cse1) (Fig. 3).40
Figure 3. The GTPase Ran drives the nuclear transport cycle. Importin-β transport receptors recognize their NLS-containing cargo proteins in the cytoplasm and promote nuclear translocation of the importin-cargo complex across the NPC. Binding of RanGTP to the importin-β transport receptor in the nucleoplasm leads to the dissociation of the cargo protein. The RanGTP-bound transport receptor returns to the cytoplasm where Ran-GAP stimulation of GTP hydrolysis releases RanGDP. To replenish the pool of nuclear RanGTP, NTF2 re-imports RanGDP into the nucleus where the chromatin-associated Ran-GEF promotes GTP loading of Ran.
Even though different models have been put forward, the exact mechanism of how the transport barrier, devised by FG-repeat containing nucleoporins, is traversed by import and export cargo complexes is still controversially debated.39,40,59,60 Nevertheless, it seems clear that transport receptors are required for the “solubilisation” and the equilibration of the cargo complex within/across the transport channel. Directionality of the transport process is then brought about by the dissociation of the cargo from the transport receptor in an energy-consuming manner, thus preventing the released cargo from passing again through the NPC channel. Therefore, the process of transport across the NPC can be viewed as a rectified Brownian motion.40,61 The energy input in the case of karyopherin-β-dependent transport is provided by the RanGTP gradient and the GTP hydrolysis on the cytoplasmic side of the transport channel.
Symportin—A Novel Transport Adaptor for Co-Import of Rpl5 and Rpl11
Mechanism of Syo1-mediated co-import
Aside from the general task to import r-proteins for ribosome assembly to the nucleus, cells face another important logistic problem: a number of r-proteins form functional clusters on the surface of the ribosomal subunits or they need to be incorporated into nascent pre-ribosomes at distinct temporal or spatial entry points; thus, suggesting a coordination between nuclear import and assembly of r-proteins in order to streamline ribosome assembly. A recent study showed that functionally related r-proteins are indeed co-imported.31 Specifically, the newly discovered protein Syo1 (for synchronized import or symportin) (i) simultaneously imports the two r-proteins, Rpl5 and Rpl11, which together with the 5S rRNA form the 5S RNP that constitutes the central protuberance on mature 60S subunits;25 and (ii) coordinates the nuclear import of Rpl5 and Rpl11 with their assembly into the 5S RNP and hence early pre-60S ribosomes (Fig. 1).31 Eukaryotic Rpl5 consists of a globular domain flanked by N- and C-terminal extensions of ~45 amino acids that clamp the 5S rRNA.25 While the C-terminal extension of Rpl5 is eukaryote specific, only the first ~15 amino acids of the N-terminal extension are unique to eukaryotes, as indicated by their absence from the archaeal Rpl5 ortholog L18P. On the other hand, eukaryotic Rpl11 and its archaeal ortholog L5P are relatively well conserved over their entire lengths and are structurally very similar in their ribosome-bound state.25,62 Both the N- and C-terminal extensions of Rpl5 contain predicted bipartite NLSs that are sufficient to target human r-protein L5 to the nucle(ol)us.63 Rpl11 also harbors a predicted monopartite NLS within its long C-terminal loop, which forms extensive contacts with the 5S rRNA.25 However, the importins that mediate nuclear import of Rpl5 and Rpl11 via these predicted NLSs have not yet been identified. Interestingly, our study unveiled an additional and unexpected mechanism for the import of Rpl5 and Rpl11. Structural analyses showed that Syo1 recognizes the very N-terminal region of Rpl5 (amino acids 2–20),31 which also contributes to 5S rRNA binding and notably includes the eukaryote-specific extension. This finding indicates that Syo1 binding to the N-terminal extension of Rpl5 will preclude the formation of an interaction between Rpl5 and importin-α or an importin-β transport receptor. In vitro binding assays revealed that Syo1 can bind Rpl5 and Rpl11 simultaneously, but also individually. In line with a physiological relevance of these interactions, Syo1 affinity purification from yeast cells yielded both Rpl5 and Rpl11. Moreover, low levels of Kap104 could be detected; thus, suggesting a possible role for Syo1 in nuclear import of Rpl5 and Rpl11 via the importin-β Kap104. Subsequent experiments corroborated this model: Syo1 contains an N-terminally located bPY-NLS that is required and sufficient to mediate the interaction with Kap104. Furthermore, in vitro, the trimeric Syo1-Rpl5-Rpl11 complex forms a stoichiometric tetrameric complex with Kap104—this interaction is sensitive to the presence of RanGTP, thus establishing the trimeric Syo1-Rpl5-Rpl11 complex as a bona fide transport cargo of Kap104 (Fig. 1 and 2). Further proof for this scenario was provided by in vitro import assays using permeabilized HeLa cells, showing that nuclear accumulation of the trimeric Syo1-Rpl5-Rpl11 is dependent on an active, importin-dependent transport process. Following transport across the NPC, the RanGTP-released trimeric Syo1-Rpl5-Rpl11 complex can, as shown in vitro, bind the 5S rRNA; thus, possibly forming a transient 5S RNP intermediate that will later be converted into an assembly-competent 5S RNP lacking Syo1. Up to now, it is not clear how exactly the 5S RNP is assembled into pre-60S ribosomes. Data from the Woolford laboratory indicate that the biogenesis factors Rpf2 and Rrs1, which form a sub-complex with 5S rRNA, Rpl5 and Rpl11 when maturation of nuclear pre-60S subunits is blocked by mutational inactivation of Rrp1, may be responsible for the recruitment of the 5S RNP into pre-60S ribosomes.64 Future experiments are required to elucidate whether and how the Rpf2-Rrs1 hetero-dimer replaces Syo1 from a Syo1-containing 5S RNP intermediate in order to promote assembly of the 5S RNP. Moreover, it will be of interest to see how exactly correct 3′ processing of the pre-5S rRNA is sensed and linked to 5S RNP formation and assembly.65
Structure of Syo1 and cargo recognition
The crystal structure, solved for the orthologous protein from the thermophilic ascomycete Chaetomium thermophilum,66 reveals that Syo1 is an unusual chimera composed of four complete N-terminal ARM repeats followed by six HEAT repeats.31 Such a combination of an ARM- and HEAT-repeat domain has so far not been observed in other proteins; however, the evolutionary and functional implications of this arrangement are not yet clear. The ARM- and HEAT repeats of Syo1 form an all α-helical elongated superhelix or α-solenoid, which is specifically reminiscent of the overall length and shape of the transport adaptor importin-α (Fig. 2).31,53 Syo1 recognizes the N-terminal residues of Rpl5 (amino acids 2–20) via side chains emanating from the inner α-helices of the concave surface formed by the HEAT-repeat domain. While the key residues of Rpl5 that mediate the interaction with Syo1 are essentially a succession of properly spaced and conserved aromatic (Phe/Tyr) and basic (Arg/Lys) amino acids, the major (ARM-repeats 2–5) and minor (ARM-repeats 6–8) binding sites of importin-α have evolved to accommodate 3–5 and 1–2 basic amino acids, respectively.52-54,67 This narrow amino acid recognition bias is most notably reflected by the occurrence of a conserved W-X3-N signature within the inner α-helices of ARM-repeats 2–4, 7 and 8 of importin-α; these tryptophan and asparagine residues, together with conserved acidic side chains, are the main determinants for cNLS binding. Notably, this signature is absent from the inner ARM-repeat α-helices of Syo1, which should therefore not be competent to bind monopartite or bipartite cNLSs. So far, there is no structural information available on how Rpl11 is recognized by Syo1; however, binding studies revealed that Rpl11 binding to Syo1 is more complex and is not restricted to a linear motif as in the case of Rpl5.31 Rpl11 recognition by Syo1 mainly involves the C-terminal end of the acidic loop of Syo1.
Formation of the Kap104-Syo1-Rpl5-Rpl11 import complex
In analogy to importin-α, which utilizes its N-terminal IBB to interact with Kapβ1,46 Syo1 is recruited to Kapβ2/Kap104 via its N-terminally located bPY-NLS (Fig. 2).31 However, unlike the IBB, which folds back to occupy the major binding site in the cargo-free form of importin-α,68 there is so far no evidence for such a self-binding mechanism to the concave ARM-repeat surface in the case of Syo1. Elegant experiments from the Stewart laboratory have shown that binding of NUP50 (Nup2) to the minor site and C-terminal region of importin-α accelerates cargo release at the nucleoplasmic face of the NPC.67,69 Concomitantly, the interaction with its RanGTP-loaded export receptor CAS (Cse1) displaces NUP50 (Nup2), thus promoting recycling of importin-α to the cytoplasm. Moreover, the nucleoporin-assisted cargo release also ensures retention of importin-α at the nuclear basket and therefore increases the efficiency of export complex formation.70-72 In the case of Syo1, release of Rpl5 and Rpl11 is coupled to their transfer onto the 5S rRNA, likely via a Syo1-containing 5S RNP intermediate (see above), and accordingly, Syo1 localizes predominantly to the nucleus.31 It will be interesting to learn whether a similar mechanism of nucleoporin-assisted cargo release at the nucleoplasmic side of the NPC also occurs in the case of importin-β transport receptors such as Kapβ2/Kap104. While importin-α is recycled to the cytoplasm by an active transport mechanism, free nucleoplasmic Syo1 crosses the NPC by facilitated diffusion due to its affinity for FG-repeats of certain nucleoporins (Fig. 3).31 It must be assumed that the directionality of diffusion is provided by encountering Rpl5 and Rpl11 in the cytoplasm, because cargo binding severely reduces FG-repeat binding and nuclear translocation in the HeLa cell import assay.31 However, Syo1’s export could also be mediated by an export receptor, such as CAS or Crm1. Future experiments are required to understand how Syo1 interacts with FG-repeats and how cargo binding regulates this interaction. An interesting but yet unresolved issue is how cargo loading of Syo1, and hence import competence, is sensed by Kapβ2/Kap104. Intuitively, in order to avoid a futile cycle of Syo1 exchange across the NPC, it would only make sense to transport cargo-bound Syo1 into the nucleus. Therefore, it is very likely that binding of Rpl5 and Rpl11 contribute to the interaction with Kapβ2/Kap104, either by rendering the bPY-NLS of Syo1 accessible or by directly contacting Kapβ2/Kap104. Alternatively, the relatively high abundance of the newly synthesized r-proteins Rpl5 and Rpl11, coupled with their high affinity for Syo1, may ensure a rapid loading of free Syo1 in the cytoplasm before encountering the transport receptor.
Diverging roads for Rpl5 and Rpl11 to the nucleus?
Another pertinent question concerns the loading state of Syo1—does Syo1 exclusively operate as a transport adaptor that simultaneously carries Rpl5 and Rpl11, or can each of these be an individual cargo? Genetic data suggest that import of Rpl5 is more dependent on the Syo1 pathway than Rpl11 since only overexpression of Rpl5, but not of Rpl11, suppresses the cold-sensitive growth phenotype associated with the absence of Syo1.31 Most strikingly, Syo1 overexpression completely restores growth of yeast cells expressing dysfunctional and even lethal Rpl5 variants, which notably harbor single mutations in the globular domain.31 This finding strongly suggests that Syo1 can be viewed as a chaperone that (i) prevents the globular Rpl5 domain from misfolding and aggregation and (ii) escorts Rpl5 from the site of its cytoplasmic synthesis until its incorporation, as part of the 5S RNP, into nuclear pre-60S ribosomes. Once assembled into the ribosome, these faulty Rpl5 variants permit mature 60S subunits to properly operate. Since Syo1 is not an essential protein, it seems evident that alternative import routes for Rpl5 and Rpl11 exist. What is then the advantage of a specialized, Syo1-dependent import route for Rpl5 and Rpl11? One obvious benefit consists in the optimization of 5S RNP assembly by the simultaneous co-import of the two 5S rRNA binding r-proteins Rpl5 and Rpl11—thereby establishing the correct stoichiometry and geometry for their efficient transfer onto the 5S rRNA already in the cytoplasm. In that sense, the Syo1-dependent import of Rpl5 and Rpl11 may represent the preferred route, especially at low temperatures where macromolecular assembly processes are generally less thermodynamically favored. Moreover, the utilization of a dedicated transport adaptor may also ensure that the cargo proteins do not stand in competition with other import substrates that utilize the same generic transport receptor. However, this scenario also implies that the trimeric Syo1-Rpl5-Rpl11 complex should represent one of the preferred Kapβ2/Kap104 substrates. Alternatively, the additional flexibility provided by specialized transport adaptors may in general be harnessed to specifically import selected cargoes under certain growth conditions or according to the cellular needs. In this respect it is noteworthy that there are five importin-α isoforms in mammalian cells; while all isoforms are capable of recognizing cNLSs, some isoforms show striking substrate specificity for non-classical cargoes.50
Synchronized Nuclear Import of Functionally Related Proteins—A Widespread Scenario?
The discovery of the specialized transport adaptor Syo1 for the synchronized nuclear import of the topologically linked and functionally related r-proteins, Rpl5 and Rpl11, suggests that further cases of synchronized import may exist. We therefore anticipate that other r-proteins may as well be imported in a synchronized manner in order to coordinate their nuclear import with ribosome assembly. Analysis of the ribosomal crystal structures now allows defining structurally and functionally related r-protein pairs that might be subject to synchronized co-import. Testing this hypothesis might unravel further—yet unrecognized—symportins. However, it might also show that the importin-βs and/or their co-importins have the capacity to transport simultaneously more than one cargo. Syo1 binds a linear motif, which is present at the N-terminus of Rpl5. In contrast, Rpl11 binding to Syo1 is more complex and is not restricted to a linear motif.31 Rpl11 binding involves mainly the C-terminal part of the acidic loop of Syo1. The well-studied Kapβ2 also contains an acidic loop insertion of ~60 amino acids between its HEAT-repeat α-helices H8A and H8B. The function of this acidic loop, however, remains widely enigmatic to date, and might be part of a yet unrecognized binding platform for further cargoes. However, synchronization of nuclear import might not only be important for assembly of ribosomes, also other macromolecular assembly processes may rely on nuclear import of assembly modules. Evidence suggests that nucleosome assembly may also be streamlined by co-import of histones, as shown for H5 and the H2A/B heterodimer, with histone chaperones, such as nucleoplasmin and Nap1.73,74 While the nucleocytoplasmic shuttling protein Nap1 binds directly to Kap114 and increases the affinity of Kap114 for the H2A/B NLSs,74 the nucleoplasmin-H5 complex can be recruited to the importin-α/β transport receptor via the bipartite cNLS of nucleoplasmin.73 A scenario, where importin-α or a dedicated transport adaptor ensures synchronized nuclear import, provides an attractive principle for the transport of functional units or pre-assembled (sub-)complexes into the nucleus and may represent a general strategy to streamline the assembly of macromolecular complexes.
Acknowledgments
Research in the authors’ laboratories is supported by the LOEWE program of the State of Hesse (to G.B.), the German Research Council (Hu363/9–4 to E.H. and SFB638 to I.S.) and the Swiss National Science Foundation (PP00P3_123341 to D.K.). G.B. thanks Florian Altegoer and Jan Schuhmacher for critical reading of the manuscript.
Glossary
Abbreviations:
- r-protein
ribosomal protein
- rRNA
ribosomal RNA
- pre-rRNA
precursor rRNA
- RNA Pol
RNA polymerase
- NPC
nuclear pore complex
- NLS
nuclear localization signal
- cNLS
classical NLS
- PY-NLS
proline-tyrosine NLS
- IBB
importin-β1 binding domain
- RNP
ribonucleoprotein
- FG repeats
phenylalanine-glycine repeats
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/24792
References
- 1.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–40. doi: 10.1016/S0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 2.Jenner L, Melnikov S, de Loubresse NG, Ben-Shem A, Iskakova M, Urzhumtsev A, et al. Crystal structure of the 80S yeast ribosome. Curr Opin Struct Biol. 2012;22:759–67. doi: 10.1016/j.sbi.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 3.Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim Biophys Acta. 2010;1803:673–83. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Tschochner H, Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–63. doi: 10.1016/S0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- 5.Kressler D, Linder P, de La Cruz J. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7897–912. doi: 10.1128/mcb.19.12.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 7.Dez C, Tollervey D. Ribosome synthesis meets the cell cycle. Curr Opin Microbiol. 2004;7:631–7. doi: 10.1016/j.mib.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Faza MB, Chang Y, Occhipinti L, Kemmler S, Panse VG. Role of Mex67-Mtr2 in the nuclear export of 40S pre-ribosomes. PLoS Genet. 2012;8:e1002915. doi: 10.1371/journal.pgen.1002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lo KY, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell. 2010;39:196–208. doi: 10.1016/j.molcel.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panse VG, Johnson AW. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem Sci. 2010;35:260–6. doi: 10.1016/j.tibs.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bussiere C, Hashem Y, Arora S, Frank J, Johnson AW. Integrity of the P-site is probed during maturation of the 60S ribosomal subunit. J Cell Biol. 2012;197:747–59. doi: 10.1083/jcb.201112131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lebaron S, Schneider C, van Nues RW, Swiatkowska A, Walsh D, Böttcher B, et al. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat Struct Mol Biol. 2012;19:744–53. doi: 10.1038/nsmb.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strunk BS, Novak MN, Young CL, Karbstein K. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell. 2012;150:111–21. doi: 10.1016/j.cell.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henras AK, Soudet J, Gérus M, Lebaron S, Caizergues-Ferrer M, Mougin A, et al. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci. 2008;65:2334–59. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kressler D, Hurt E, Bergler H, Bassler J. The power of AAA-ATPases on the road of pre-60S ribosome maturation--molecular machines that strip pre-ribosomal particles. Biochim Biophys Acta. 2012;1823:92–100. doi: 10.1016/j.bbamcr.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strunk BS, Karbstein K. Powering through ribosome assembly. RNA. 2009;15:2083–104. doi: 10.1261/rna.1792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferreira-Cerca S, Sagar V, Schäfer T, Diop M, Wesseling AM, Lu H, et al. ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit. Nat Struct Mol Biol. 2012;19:1316–23. doi: 10.1038/nsmb.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schäfer T, Maco B, Petfalski E, Tollervey D, Böttcher B, Aebi U, et al. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 2006;441:651–5. doi: 10.1038/nature04840. [DOI] [PubMed] [Google Scholar]
- 19.Kruiswijk T, Planta RJ, Krop JM. The course of the assembly of ribosomal subunits in yeast. Biochim Biophys Acta. 1978;517:378–89. doi: 10.1016/0005-2787(78)90204-6. [DOI] [PubMed] [Google Scholar]
- 20.Fernández-Pevida A, Rodríguez-Galán O, Díaz-Quintana A, Kressler D, de la Cruz J. Yeast ribosomal protein L40 assembles late into precursor 60 S ribosomes and is required for their cytoplasmic maturation. J Biol Chem. 2012;287:38390–407. doi: 10.1074/jbc.M112.400564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferreira-Cerca S, Pöll G, Gleizes PE, Tschochner H, Milkereit P. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol Cell. 2005;20:263–75. doi: 10.1016/j.molcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 22.Ferreira-Cerca S, Pöll G, Kühn H, Neueder A, Jakob S, Tschochner H, et al. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol Cell. 2007;28:446–57. doi: 10.1016/j.molcel.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 23.Pöll G, Braun T, Jakovljevic J, Neueder A, Jakob S, Woolford JL, Jr., et al. rRNA maturation in yeast cells depleted of large ribosomal subunit proteins. PLoS ONE. 2009;4:e8249. doi: 10.1371/journal.pone.0008249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosado IV, Kressler D, de la Cruz J. Functional analysis of Saccharomyces cerevisiae ribosomal protein Rpl3p in ribosome synthesis. Nucleic Acids Res. 2007;35:4203–13. doi: 10.1093/nar/gkm388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524–9. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 26.Klein DJ, Moore PB, Steitz TA. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J Mol Biol. 2004;340:141–77. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 27.Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334:941–8. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- 28.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–6. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 29.Jäkel S, Mingot JM, Schwarzmaier P, Hartmann E, Görlich D. Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 2002;21:377–86. doi: 10.1093/emboj/21.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koch B, Mitterer V, Niederhauser J, Stanborough T, Murat G, Rechberger G, et al. Yar1 protects the ribosomal protein Rps3 from aggregation. J Biol Chem. 2012;287:21806–15. doi: 10.1074/jbc.M112.365791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kressler D, Bange G, Ogawa Y, Stjepanovic G, Bradatsch B, Pratte D, et al. Synchronizing nuclear import of ribosomal proteins with ribosome assembly. Science. 2012;338:666–71. doi: 10.1126/science.1226960. [DOI] [PubMed] [Google Scholar]
- 32.Lacombe T, García-Gómez JJ, de la Cruz J, Roser D, Hurt E, Linder P, et al. Linear ubiquitin fusion to Rps31 and its subsequent cleavage are required for the efficient production and functional integrity of 40S ribosomal subunits. Mol Microbiol. 2009;72:69–84. doi: 10.1111/j.1365-2958.2009.06622.x. [DOI] [PubMed] [Google Scholar]
- 33.Dingwall C, Sharnick SV, Laskey RA. A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell. 1982;30:449–58. doi: 10.1016/0092-8674(82)90242-2. [DOI] [PubMed] [Google Scholar]
- 34.Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–43. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 35.Köhler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol. 2007;8:761–73. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 36.Brohawn SG, Partridge JR, Whittle JR, Schwartz TU. The nuclear pore complex has entered the atomic age. Structure. 2009;17:1156–68. doi: 10.1016/j.str.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–71. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 38.Köhler A, Hurt E. Gene regulation by nucleoporins and links to cancer. Mol Cell. 2010;38:6–15. doi: 10.1016/j.molcel.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 39.Rout MP, Aitchison JD, Magnasco MO, Chait BT. Virtual gating and nuclear transport: the hole picture. Trends Cell Biol. 2003;13:622–8. doi: 10.1016/j.tcb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 40.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 41.Chook YM, Süel KE. Nuclear import by karyopherin-βs: recognition and inhibition. Biochim Biophys Acta. 2011;1813:1593–606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu D, Farmer A, Chook YM. Recognition of nuclear targeting signals by Karyopherin-β proteins. Curr Opin Struct Biol. 2010;20:782–90. doi: 10.1016/j.sbi.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andrade MA, Petosa C, O’Donoghue SI, Müller CW, Bork P. Comparison of ARM and HEAT protein repeats. J Mol Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- 44.Cook AG, Conti E. Nuclear export complexes in the frame. Curr Opin Struct Biol. 2010;20:247–52. doi: 10.1016/j.sbi.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 45.Güttler T, Görlich D. Ran-dependent nuclear export mediators: a structural perspective. EMBO J. 2011;30:3457–74. doi: 10.1038/emboj.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cingolani G, Petosa C, Weis K, Müller CW. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399:221–9. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- 47.Lee BJ, Cansizoglu AE, Süel KE, Louis TH, Zhang Z, Chook YM. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126:543–58. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bayliss R, Littlewood T, Strawn LA, Wente SR, Stewart M. GLFG and FxFG nucleoporins bind to overlapping sites on importin-β. J Biol Chem. 2002;277:50597–606. doi: 10.1074/jbc.M209037200. [DOI] [PubMed] [Google Scholar]
- 49.Terry LJ, Wente SR. Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot Cell. 2009;8:1814–27. doi: 10.1128/EC.00225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lott K, Cingolani G. The importin β binding domain as a master regulator of nucleocytoplasmic transport. Biochim Biophys Acta. 2011;1813:1578–92. doi: 10.1016/j.bbamcr.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marfori M, Mynott A, Ellis JJ, Mehdi AM, Saunders NF, Curmi PM, et al. Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim Biophys Acta. 2011;1813:1562–77. doi: 10.1016/j.bbamcr.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 52.Conti E, Kuriyan J. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure. 2000;8:329–38. doi: 10.1016/S0969-2126(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 53.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/S0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 54.Fontes MR, Teh T, Kobe B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J Mol Biol. 2000;297:1183–94. doi: 10.1006/jmbi.2000.3642. [DOI] [PubMed] [Google Scholar]
- 55.Fernández-Cid A, Vega M, Herrero P, Moreno F. Yeast importin-β is required for nuclear import of the Mig2 repressor. BMC Cell Biol. 2012;13:31. doi: 10.1186/1471-2121-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strahl T, Hama H, DeWald DB, Thorner J. Yeast phosphatidylinositol 4-kinase, Pik1, has essential roles at the Golgi and in the nucleus. J Cell Biol. 2005;171:967–79. doi: 10.1083/jcb.200504104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cansizoglu AE, Lee BJ, Zhang ZC, Fontoura BM, Chook YM. Structure-based design of a pathway-specific nuclear import inhibitor. Nat Struct Mol Biol. 2007;14:452–4. doi: 10.1038/nsmb1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Süel KE, Gu H, Chook YM. Modular organization and combinatorial energetics of proline-tyrosine nuclear localization signals. PLoS Biol. 2008;6:e137. doi: 10.1371/journal.pbio.0060137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frey S, Richter RP, Görlich D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006;314:815–7. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- 60.Weis K. The nuclear pore complex: oily spaghetti or gummy bear? Cell. 2007;130:405–7. doi: 10.1016/j.cell.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 61.Stewart M. Ratcheting mRNA out of the nucleus. Mol Cell. 2007;25:327–30. doi: 10.1016/j.molcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 62.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–20. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 63.Rosorius O, Fries B, Stauber RH, Hirschmann N, Bevec D, Hauber J. Human ribosomal protein L5 contains defined nuclear localization and export signals. J Biol Chem. 2000;275:12061–8. doi: 10.1074/jbc.275.16.12061. [DOI] [PubMed] [Google Scholar]
- 64.Zhang J, Harnpicharnchai P, Jakovljevic J, Tang L, Guo Y, Oeffinger M, et al. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 2007;21:2580–92. doi: 10.1101/gad.1569307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nariai M, Tanaka T, Okada T, Shirai C, Horigome C, Mizuta K. Synergistic defect in 60S ribosomal subunit assembly caused by a mutation of Rrs1p, a ribosomal protein L11-binding protein, and 3′-extension of 5S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2005;33:4553–62. doi: 10.1093/nar/gki772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, et al. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146:277–89. doi: 10.1016/j.cell.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 67.Matsuura Y, Lange A, Harreman MT, Corbett AH, Stewart M. Structural basis for Nup2p function in cargo release and karyopherin recycling in nuclear import. EMBO J. 2003;22:5358–69. doi: 10.1093/emboj/cdg538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kobe B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat Struct Biol. 1999;6:388–97. doi: 10.1038/7625. [DOI] [PubMed] [Google Scholar]
- 69.Matsuura Y, Stewart M. Nup50/Npap60 function in nuclear protein import complex disassembly and importin recycling. EMBO J. 2005;24:3681–9. doi: 10.1038/sj.emboj.7600843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Booth JW, Belanger KD, Sannella MI, Davis LI. The yeast nucleoporin Nup2p is involved in nuclear export of importin alpha/Srp1p. J Biol Chem. 1999;274:32360–7. doi: 10.1074/jbc.274.45.32360. [DOI] [PubMed] [Google Scholar]
- 71.Denning D, Mykytka B, Allen NP, Huang L, Al Burlingame, Rexach M. The nucleoporin Nup60p functions as a Gsp1p-GTP-sensitive tether for Nup2p at the nuclear pore complex. J Cell Biol. 2001;154:937–50. doi: 10.1083/jcb.200101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dilworth DJ, Suprapto A, Padovan JC, Chait BT, Wozniak RW, Rout MP, et al. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J Cell Biol. 2001;153:1465–78. doi: 10.1083/jcb.153.7.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arregi I, Falces J, Bañuelos S, Urbaneja MA, Taneva SG. The nuclear transport machinery recognizes nucleoplasmin-histone complexes. Biochemistry. 2011;50:7104–10. doi: 10.1021/bi2008867. [DOI] [PubMed] [Google Scholar]
- 74.Mosammaparast N, Ewart CS, Pemberton LF. A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. EMBO J. 2002;21:6527–38. doi: 10.1093/emboj/cdf647. [DOI] [PMC free article] [PubMed] [Google Scholar]