Summary
We have previously characterized several fungal-specific proteins from the human pathogen Candida albicans that either encode subunits of mitochondria Complex I (CI) of the electron transport chain (ETC) or regulate CI activity (Goa1p). Herein, the role of energy production and cell wall gene expression is investigated in the mitochondria mutant goa1Δ. We show that down regulation of cell wall-encoding genes in the goa1Δ results in sensitivity to cell wall inhibitors such as congo red and calcofluor white, reduced phagocytosis by a macrophage cell line, reduced recognition by macrophage receptors, and decreased expression of cytokines such as IL-6, IL-10, and IFN-γ. In spite of the reduced recognition by macrophages, the goa1Δ is still killed to the same extent as control strains. We also demonstrate that expression of the epithelial cell receptors E-cadherin and EGFR is also reduced in the presence of goa1Δ. Together, our data demonstrate the importance of mitochondria in the expression of cell wall biomolecules and the interaction of C. albicans with innate immune and epithelial cells. Our underlying premise is that mitochondrial proteins such as Goa1p and other fungal-specific mitochondrial proteins regulate critical functions in cell growth and in virulence. As such, they remain as valid drug targets for antifungal drug discovery.
Keywords: cell wall, mitochondria, immune recognition, macrophages, Candida albicans
Introduction
Candida albicans is a member of the commensal microbiota of mucosal surfaces. In HIV/AIDS patients, infections are usually limited to the oral, pharyngeal, and esophageal mucosa, while blood-borne invasive disease occurs in patients with risk factors such as surgery, neutropenia, and indwelling urinary track and central venous catheters (Rüping et al., 2008; Leventakos et al., 2010; Kriengkauykiat et al., 2011; Williams et al., 2011). Neutropenia occurs as a result of immune suppression previous to allogeneic bone marrow transplants or cancer chemotherapy. The reversible transition of the organism from a unicellular growth form to one that is filamentous has long been recognized as contributing to the invasiveness of the organism (Calderone et al., 2001). Also, cell surface adhesins of C. albicans are critical to the colonization of tissues. Thus, the ability to transform from a unicellular yeast into a multicellular, hyphal state, to adhere to, and invade a variety of tissues and immune cells, as well as utilizing strategies to evade immune protection are considered as the virulence factors for developing candidiasis (Brand et al., 2012).
The host protective response to commensal organisms such as Candida albicans relies on innate and cell-mediated immunity (Brown et al., 2012). Polymorphonuclear neutrophils (PMNs) and macrophages are both important for innate immune responses, and T-helper (Th) cell-mediated responses are also critical to protection against fungal infections (Romani et al., 2000; Lionakis et al., 2010). For fungal pathogens, the innate immune system recognizes conserved cell wall biomolecules such as mannan, β-glucan, and chitin (Porcaro et al., 2003). The recognition of fungal cells via receptors on these two types of immune cells is followed by the release of proinflammatory cytokines, such as interleukin (IL)-1, IL-6, interferon (IFN)-γ, and tumor necrosis factor (TNF)-α, and the activation of a respiratory burst following phagocytosis (Kim et al., 2005). Among the receptors for fungal cell wall oligosaccharides, dectin-1, toll-like receptors TLR-2 and TLR-4, seem to be most often studied. However, experiments on receptor-fungal cell wall ligands indicate that receptor interactions are required for recognition of a single ligand (Netea et al., 2006; Gow et al., 2007).
Endocytosis of C. albicans by epithelial and endothelial cells was suggested as a mechanism to provide access to the dermal basement membranes and blood vessels, which eventually results in systemic dissemination (Filler et al., 1995). To bind to a variety of host cells, C. albicans produces a number of cell surface adhesins. Among the most well-known of these is the ALS (agglutinin-like sequence) gene family, a group of glycoproteins that was first found to participate in a mating-associated adhesion event in S. cerevisiae (Draninis et al., 2007). In C. albicans, the roles of some of the ALS adhesins in oropharyngeal candidiasis and in the onset of endocytosis have been well established (Green et al., 2004; Cheng et al., 2005; Hoyer et al., 2007). For example, the Als3p of C. albicans mediates endocytosis by interacting with cadherins of epithelial and endothelial cells (Phan et al., 2007).
Oppositely, C. albicans has evolved ways to escape the host immune system and progress to a pathogenic status following initial host cell interactions. One such mechanism is the hyphal transition, during which the organism converts to a filamentous growth post-phagocytosis in macrophages to exit these cells, and invade tissues (Sudbery et al., 2004).
The morphological switch from yeast to filamentous forms can be introduced by a variety of environmental conditions that require signal transduction pathways. In addition, phagocytosis and outgrowth by hyphae from macrophages has been studied in the context of metabolic changes occurring during the intracellular phase of C. albicans (Lorenz et al., 2004). Coordination of intracellular events following its phagocytosis with nutrient availability and oxygen radical levels in the macrophage occurs. Once internalized by macrophages, during early stage events, the organism begins a low nutrient growth program, characterized by gluconeogenesis, β-oxidation of fatty acids, and the conservation of carbon via the glyoxylate cycle. These events require crosstalking of peroxisomes and mitochondria in C. albicans. As hyphal transition follows and the organism initiates outgrowth from the macrophage, its metabolism changes to a glycolytic metabolism (Lorenz et al., 2004). Therefore, and not surprisingly, the switch to a hyphal morphology in C. albicans depends upon regulated metabolic events that include carbon metabolism.
In this study, we focus upon mitochondrial events that influence interactions with macrophages and epithelial cells. Using a well-characterized mitochondrial mutant lacking the gene GOA1, we have previously shown its role in maintaining membrane potential, ATP synthesis, reduction of toxic levels of cell ROS, and virulence. We also know that during oxidant stress, Goa1p translocates to the mitochondria and regulates Complex I (CI) of the electron transport chain (ETC) (Bambach et al., 2009; Li, et al., 2011; Chen et al., 2012). Herein, we report the cell surface changes in goa1Δ and the consequences of those changes on the recognition by macrophage and epithelial cell receptors.
Results
Mitochondria of C. albicans are required for cell wall maintenance and expression of adherence genes
The cell wall of Candida species consists of an inner layer adjacent to the plasma membrane composed mostly β-glucans and chitin, which provide a rigid cell structure and an outer fibrillar layer that is mainly composed of mannan and glycoproteins such as those of the ALS adherence family. Covalent attachment to either β-1,3 or β-1,6 glucan anchors glycoproteins to the wall matrix. We previously reported a significant down regulation of 86 genes (5.38% of total) associated with cell wall functions in the goa1Δ compared to its parental strain (Sun et al., 2013). Of these, 40 genes are associated with cell wall biogenesis and integrity, 26 genes encode adhesin proteins that included 7 of the ALS gene family, and 14 genes that are required for the yeast-hyphal switch (summary, Table 2). Also available for analysis was the ndh51Δ and reintegrant strain. NDH51 encodes a subunit of the CI ETC. The total number of down regulated genes associated with cell wall functions in this mutant is approximately the same as with the goa1Δ. Of these, both mutants display similar wall-associated gene changes, indicating a crucial role for mitochondria in these processes (data not shown).
Table 2.
Summary of down regulated genes encoding cell wall functions in goa1Δ
| Gene | Fold | Function |
|---|---|---|
| Cell wall biogenesis/integrity (n=40) | ||
| orf19.1160, orf19.5412, | 0.28-0.29 | fungal-type cell wall organization |
| orf19.719 | 0.37 | |
| orf19.4984 | 0.47 | chitin catabolic process |
| orf19.1196 | 0.44 | role in MAPKKK cascade involved in cell wall biogenesis |
| Orf19.4581 | 0.48 | glycosylphosphatidylinositol-alpha 1,4 mannosyltransferase I |
| ADA2 | 0.24 | transcriptional coactivator; cell wall integrity role |
| CHK1 | 0.21 | histidine kinase, regulates cell wall biosynthesis |
| CHT1,3 | 0.39, 0.27 | chitinase |
| CRH11 | 0.33 | GPI-anchored cell wall transglycosylase |
| ECM1,4,7,15,17,21,331 | 0.16-0.48 | cell wall organization or biogenesis |
| ENG1 | 0.43 | endo-1,3-beta-glucanase |
| FKS1 | 0.49 | beta-1,3-glucan synthase subunit |
| MNN4, orf19.5557 | 0.27-0.49 | mannosyltransferase |
| MNT4 | 0.45 | mannosyltransferase |
| PHR1,3 | 0.46, 0.20 | beta-1,3-glucanosyltransferase |
| PIR1 | 0.34 | 1,3-beta-glucan-linked structural cell wall protein |
| IRS4 | 0.28 | roles in cell wall integrity |
| KIC1 | 0.47 | Ste20p kinases; in RAM cell wall integrity signaling network |
| PGA10,31,52 | 0.25-0.47 | GPI anchored cell surface protein |
| SLK19 | 0.39 | alkaline-induced membrane protein; affects cell aggregation |
| SMI1 | 0.48 | cell wall biosynthesis protein |
| SUR7 | 0.38 | required for normal cell wall, plasma membrane |
| XOG1 | 0.18 | exo-1,3-beta-glucanase |
| BGL22 | 0.21 | glucanase; induced during cell wall regeneration |
| BMT3 | 0.08 | beta-mannosyltransferase |
| EXG2 | 0.44 | GPI-anchored cell wall protein |
| Orf19.5070 | 0.42 | cell-wall mannoproteins |
| VPS28 | 0.49 | proteolytic activation of Rim101p, role in echinocandin sensitivity |
| Cell membrane associated genes (n=6) | ||
| PLB5 | 0.42 | GPI-linked phospholipase B |
| YPT31 | 0.50 | required for resistance to toxic ergostero analog |
| ERG27 | 0.42 | 3-Keto sterol reductase of ergosterol biosynthesis |
| Orf19.3226 | 0.28 | intracellular sterol transport |
| orf19.4096 | 0.06 | acylglycerophosphocholine O-acyltransferase |
| orf19.7547 | 0.27 | phosphatidylinositol-3-phosphate binding |
| Adhesin/antigenic genes (n=26) | ||
| ALS1, 2,3,4,5,6,7 | 0.08-0.49 | ALS family protein, cell-surface glycoprotein, adhesion |
| IFF4,5,8,11 | 0.11-0.43 | GPI-anchored protein , adhesion-like protein |
| HSP70 | 0.15 | antigenic, role in entry into host cells |
| HYR3 | 0.45 | adhesion-like protein |
| orf19.251 | 0.30 | binds human immunoglobulin E |
| AHP1 | 0.42 | alkyl hydroperoxide reductase; immunogenic in mouse |
| CSH1 | 0.38 | aldo-keto reductase family member, role in fibronectin adhesion |
| CYC3 | 0.50 | cytochrome c heme lyase, antigenic cell-wall protein |
| DDR48 | 0.05 | immunogenic stress-associated protein |
| MET15, 6 | 0.04, 0.23 | antigenic during murine or human systemic infection |
| PGA62 | 0.44 | adhesin-like cell wall protein |
| RBR3 | 0.37 | cell wall adhesin-like protein |
| PST2 | 0.07 | NADH:quinone oxidoreductase, immunogenic in mice |
| SAP10, 9 | 0.22, 0.24 | secreted aspartyl protease; roles in adhesion |
| XYL2 | 0.16 | D-xylulose reductase; immunogenic in mice |
| Yeast-hyphal morphological transition genes (n= 14) | ||
| CEK1 | 0.44 | ERK-family protein kinase, yeast-hyphal switching |
| CHO1 | 0.33 | phosphatidylserine synthase, required for filamentous growth |
| CSP37 | 0.25 | hyphal cell wall protein |
| DFG5 | 0.28 | N-linked mannoprotein, role in hyphal growth |
| GPH1 | 0.28 | glycogen phosphorylase, localizes to cell surface of hyphae |
| MID1 | 0.38 | high affinity calcium uptake system, role in thigmotropism |
| PHO85,111 | 0.50,0.50 | acid phosphatase, negatively regulated by Rim101p |
| RBT5 | 0.09 | GPI-anchored cell wall protein |
| RHD3 | 0.32 | GPI-anchored cell wall protein; yeast-associated protein |
| RVS167 | 0.40 | involved in endocytosis; hyphal growth required |
| SAM2 | 0.50 | S-adenosylmethionine synthetase, localizes to surface of hyphal cells |
| YDC1 | 0.21 | Mob2p-dependent hyphal regulatio |
| SNZ1 | 0.12 | induced on yeast to hyphal switch |
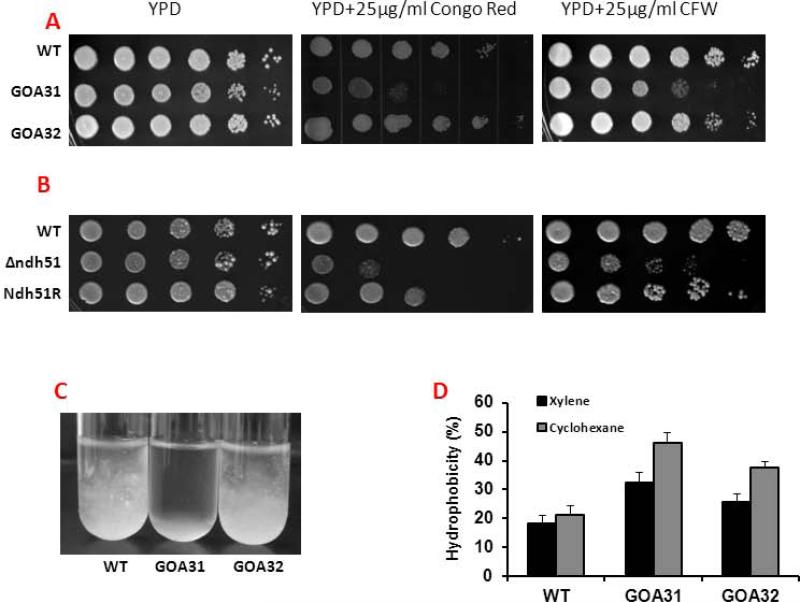
Based upon these data, we compared the sensitivities of strains to Congo red (CR) and calcofluor white (CFW), cell flocculation, and cell wall hydrophobicity. As shown in Table 2, a decrease in genes encoding a mannosyltransferase (MNN4. MNT4) and glucan synthases (FKS1, ENG1, PHR1, PHR3) correlates with the sensitivity of goa1Δ to CR and CFW (Fig. 1A) in contrast to the wild type (WT) and reconstituted strain (GOA32). Similar growth inhibition by CR and CFW was noted with ndh51Δ compared to its integrant strain (Ndh51R) (Fig. 1B). The goa1Δ also flocculated much more extensively in RPMI after 6 h (Fig1.C) and had increased hydrophobicity (Fig. 1D). The sensitivity to cell wall inhibitors, increased hydrophobicity (p<0.01), and increased flocculation demonstrate that major cell wall changes have occurred in the mutants.
Figure 1.
Deletion of GOA1 and NDH51 of C. albicans causes changes in its cell wall. (A) A cell suspension of WT (SC5314), goa1Δ (GOA31) and its gene-reconstituted strain (GOA32) were serially diluted and applied to YPD agar, or YPD agar supplemented with 25 μg/ml of calcoflour white (CFW), or 25 μg/ml of Congo red (CR). Images were taken 48 h after plating cells. The mutant strain was more sensitive than WT and GOA32 to each cell wall inhibitor. (B) Similarly, the ndh51Δ was also more sensitive to both CR and CFW than control strains (C) Flocculation of goa1Δ and control strains is compared in RPMI medium. The null mutant flocculated more than control strains. (D) Differences in cell surface hydrophobicity with cyclohexane or xylene among strains are represented.
Mitochondrial mutant goa1Δ is phagocytosed poorly by mouse phagocytes
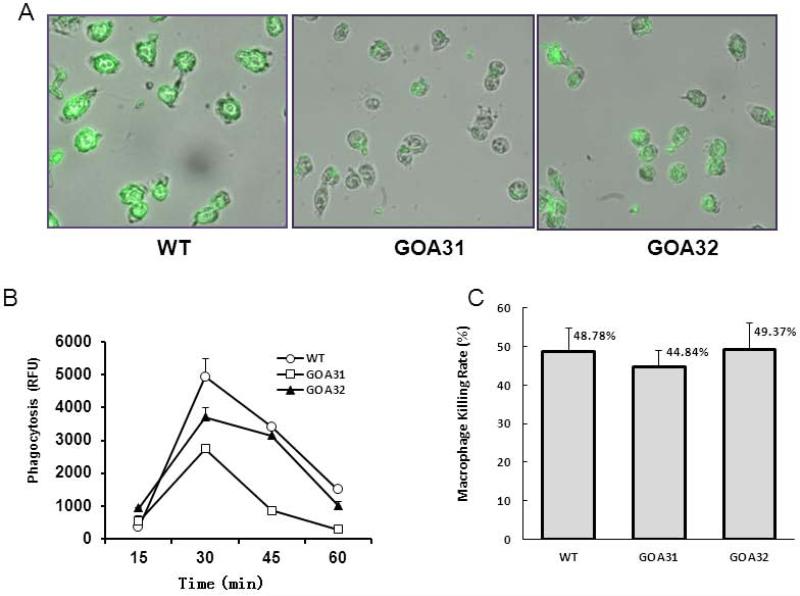
From previous published data, Goa1p is required for mitochondrial complex I (CI) activity, cristae integrity, and oxidative phosphorylation (Li, et al., 2011; Chen et al., 2012). The goa1Δ was also avirulent in a blood-borne, invasive candidiasis murine model (Bambach et al., 2009). In vitro studies indicated an increased killing of the mutant by polymorphonuclear neutrophils even though it was phagocytosed less than control strains (Bambach et al., 2009). To continue our studies of the goa1Δ and interactions with host cells, we used a mouse macrophage cell line to measure phagocytosis (Fig. 2A and 2B). In agreement with reduced gene expression of cell wall components, phagocytosis by the macrophage cell line RAW264.7 was reduced by approximately 50% after 30 min for the goa1Δ mutant compared with WT and GOA32 strains, p <0.001 (Fig. 2B). We also measured macrophage killing of goa1Δ, WT and the GOA1-reconstituted strain (GOA32) after 24 h of incubation. Unlike our previous studies with PMN, we found no significant differences in killing of all strains by macrophages (Fig. 2C). These results indicate that goa1Δ is defective in its ability to induce phagocytosis by phagocytes, although its killing by phagocytes was not impaired. Our interpretation of these data is that extracellular killing by macrophages of goa1Δ accounts for the similar levels of killing in all strains. The anti-Candida killing by PMNs but not equally by macrophages is well-known (Miramón, et al., 2012).
Figure 2.
Phagocytosis of all strains is shown following incubation with macrophages (RAW264.7) for the times indicated. All strains were pre-labeled with FITC. Fluorescence by un-labeled yeast cells was quenched with trypan blue prior to measuring the fluorescence of ingested yeasts. (A) Images are displayed from cells following an incubation of 30 min; (B) The fluorescence of internalized yeast was followed for 1 h at 15 min intervals by spectrofluorimetrics (Ex/Em=495/525). A reduction in the phagocytosis of goa1Δ (GOA31) compared to WT and GOA32 was noted at 30 min (P< 0.01). (C) Viability of all strains is shown after an overnight incubation with macrophages (RAW264.7). Viable colonies of each strain were counted under microscopy. Data are averages of two separate experiments. A Student's unpaired t test was used to determine P values, P> 0.05.
Cytokine production by macrophages is reduced in the presence of goa1Δ
In addition to the innate immune response against C. albicans infections, cellular immune responses that require the activation of CD4+ (Th-1) cells are also critical. The outer mannan polysaccharides, glycoproteins of the C. albicans cell wall, and inner layer of β-glucan are recognized by innate immune cells via their cell surface receptors. Since many genes encoding these polymers in goa1Δ are down regulated (Table 2), we measured TLR-2, TLR-4 and dectin-1 expression after incubation of all strains with the same macrophage cell line. Although synergized responses of cytokine release have been reported (Ferwerda, et al., 2008), the Toll-like receptors TLR-2 and TLR-4 bind mannan while dectin-1 recognizes β-glucan mostly.
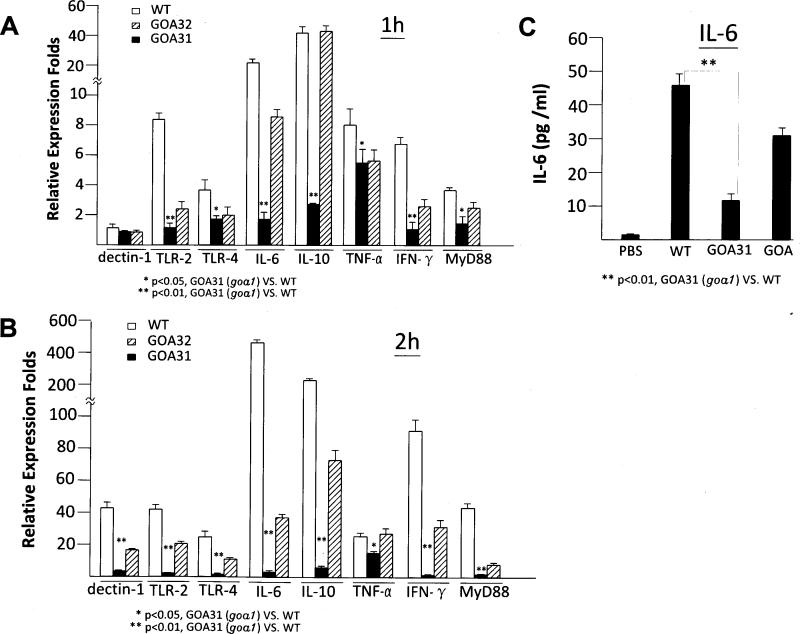
In addition to the three macrophage receptors that recognize fungal biomolecules, we also measured downstream pro-inflammatory cytokines such as TNF-α, IL-10, IL-6, IFN-γ, and MyD88 in the presence of goa1Δ and control strains after 1 and 2h of incubation (Fig. 3A,B). Expression of TLR-2 and TLR4, was significantly reduced after 1 h in the presence of goa1Δ compared to WT and GOA32 (p <0.001). No differences were observed in dectin-1 expression among strains after 1 h of incubation. After 2 h of incubation, the expression of TLR-2 and TLR-4 remained lower in goa1Δ, and while expression of dectin-1 was low for all strains, less was observed in goa1Δ (Fig. 3A,B).
Figure 3.
Expression (qPCR) of cell surface receptors and cytokines of macrophages infected with strains of C. albicans at 1 and 2 h (A and B, respectively). The macrophage receptors, Dectin-1, TLR-2 and TLR-4 usually synergize to trigger downstream cytokine and adaptor protein expression after binding to ligands such as β-glucan, phospholipomannan and O-linked mannosyl residues. Less expression of receptors occurred in co-culture with goa1Δ, while the cytokines, IL-6, IL-10, IFN-γ and adaptor protein MyD88 were significantly decreased at 1h. (C) The concentration of IL-6 was also determined by ELASA in the supernatants of RAW264.7 cells infected with all yeast strains for 3h to verify the qPCR expression profile. (*P< 0.05; **P< 0.01).
Cytokines IL-10 and IL-6 were significantly reduced in goa1Δ compared to control strains, but TNF-α remained at comparable levels in all strains (p = 0.10 at 1h and p = 0.02 at 2h) (Fig.3A,B). A significant reduction of IL-6 was also confirmed by ELASA (p <0.005) (Fig. 3C). IFN-γ, released by the activation of Th-1 cells and which also plays a major role in activating phagocytic cells against Candida, was reduced in goa1Δ as was the adapter protein MyD88 (Marr et al., 2003), which mediates a signaling pathway that results in translocation of NF-κB and subsequent cytokine genes (Fig 3A,B).
The most dramatic differences in cytokine profiles among strains were that of IL-6 and IL-10. Both cytokines were decreased by 20-fold at 1 h in goa1Δ. IL-6 is one of the cytokines necessary for the development of Th17, a T-cell subset essential for immunity to C. albicans (Ghosh, et al., 2010). IL-6 and IL-10 are both anti-inflammatory cytokines that are secreted by T-cells and macrophages whose activation, in general, inhibits macrophage pro-inflammatory cytokine production, such as IFN-γ, TNF-α, and also blocks NF-κB activity. Correspondingly, we observed less expression but still differences in IFN-γ, TNF-α and MyD88 expression profiles between mutant and control strains.
Decreased recognition and endocytosis of goa1Δ by human epithelial cells
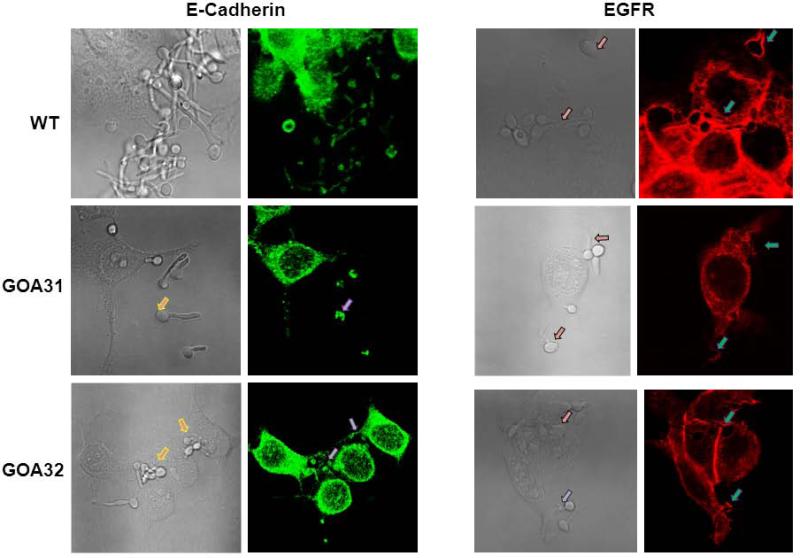
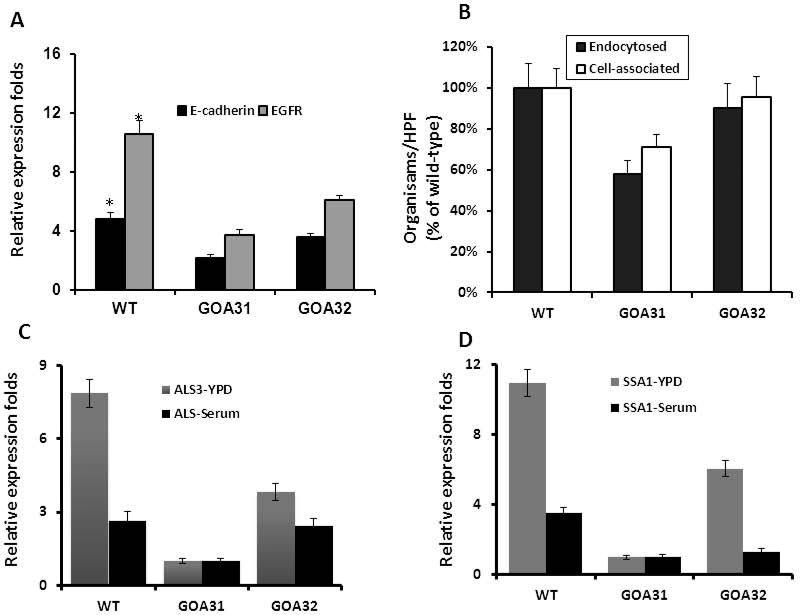
Invasion of epithelial cells follows colonization by C. albicans in candidiasis patients. C. albicans binds to the epithelial receptors E-cadherin (calcium-dependent adhesion) and EGFR (epidermal growth factor receptor) via its adherence proteins Als3 and Ssa1p (Zhu et al., 2012) and induces endocytosis. To determine if reduced expression of adherence glycoproteins in goa1Δ (Table 2) affected epithelial endocytosis, we examined the distribution of epithelial receptor E-cadherin and EGFR by confocal microscopy after infection of the FaDu oral epithelial cell line with goa1Δ and control strains. As shown at Fig. 4, both receptors were more highly expressed in WT or GOA32 than when infected with goa1Δ. Fluorescence was more intense on yeast buds and germinating hyphae of control strains, whereas fluorescence was less on mother cells, or partially covered the germ tubes of the goa1Δ mutant. This observation was also confirmed by a significant reduction in the expression of receptors by the epithelial cell line to goa1Δ, as determined by qPCR after infection for 1 h (Fig. 5A). Accordingly, less endocytosis of goa1 was observed compared to control strains (Fig. 5B). As Als3p and Ssa1p are important adhesin glycoproteins that promote binding to epithelial cells, we also measured the expression of both in goa1Δ compared to control strains (Fig. 5C, D). Both genes were down regulated in the goa1Δ in YPD (Table 2) or 10% BSA.
Figure 4.
Confocal microscopic images of the FaDu epithelial cell line E-Cadherin and EGFR. Yeast cells were incubated with FaDu cells for 90 min in MEM Earl's salts plus 10% FBS, then stained with either the anti-ECadherin antibody conjugated with Alexa 488 (left panel) or anti-EGFR antibody conjugated with Alexa 555 (right panel). Images (left panel of each stain) are compared to bright-field images (right panel). The goa1Δ null was poorly or partially stained with both antibodies compared to control strains. The arrows indicate the accumulation of fluorescent stain around the hyphae of control strains but not the goa1Δ (GOA31).
Figure 5.
(A) Expression levels of E-Cadherin and EGFR in FaDu epithelial cells are shown by real-time PCR after infection with each strain post-1h. The endocytosis and cell associated strains are shown after an incubation of 3 h. (B) Data are presented for each strain as the percentage of endocytosed or cell-associated cells per 200 epithelial cells. The expression of the Candida albicans cell adhesins ALS3 (C) and SSA1 (D) was significantly lower than control strains in both YPD and serum broth.
Discussion
Previously, we have shown that GOA1 from C. albicans is required for normal mitochondrial functions. The lack of GOA1 caused a major reduction in respiration associated with a decrease in both CI activity of the ETC and oxidative phosphorylation and an increase in cell ROS. As a consequence, the mutant had a shortened chronological aging and cell apoptosis, changes that were probably of consequence to a reduction in virulence and colonization of the kidney in a murine model of blood-borne candidiasis (Bambach et al, 2009). In vitro experiments demonstrated a greater killing by human neutrophils of goa1Δ. We have also noted a down regulation of mitochondria and peroxisomal genes that likely indicate an inability of these two organelles to cross-talk in shared pathways such as gluconeogenesis, fatty acid β-oxidation, and the glyoxylate bypass pathway (Li, et al., 2011; Chen et al., 2012; Sun et al., 2013). The ability of Candida albicans to adapt to carbohydrate starvation, nitrosative stress, and oxidative stress during phagocytosis is essential for its virulence (Miramón, et al., 2012).
Our current data indicate a failure by macrophages and epithelial cells to fully recognize the mutant. Our explanation is that decreased expression of genes associated with cell wall biosynthesis and integrity, including adhesins, are contributory to reduced recognition of the mutant. Cell surface changes were initially shown as an increased sensitivity of the mutant to cell wall perturbing compounds such as Congo red and calcoflour white. Heightened sensitivity of the mutant compared to control strains was associated with reduced expression of 72 genes encoding proteins associated with cell wall biogenesis, organization, and adherence proteins such as the ALS family of glycoproteins. Cell surface changes should have an effect on host-fungus interactions, now clearly observed in our current report. Our data indicate that mitochondria/peroxisomes play an important role in the biosynthesis and organization of the fungal cell wall that also has implications in host recognition (Chen, et al., 2012; Sun, et al., 2013).
The fungal cell wall is a highly dynamic structure that changes in regard to environmental conditions such as nutrient limitation. Fungal cell wall ligands for dectin-1, TLR-2 and TLR-4 are β-glucan, phospholipomannan, and O-linked mannans, respectively (Lionakis, et al., 2013). Dectin-1 expression in macrophages was lower for all strains at 1h due perhaps to its inner cell wall localization, while TLR-2 expression was significantly reduced in macrophages at 1-2 h post-infection. Among the three receptors, down regulation of TLR-2 leads to a significant reduction in IL-6 and IL-10 production in goa1Δ since TLR-2 is responsible for pro-inflammatory and anti-inflammatory cytokine expression. However, TNFα, a downstream cytokine of TLR-2, was maintained at sufficient levels in goa1Δ by 1h. Cell wall mannosylation is a key determinant in phagocytosis, since glycosylation mutants have delayed endocytosis when encountering macrophages (Lewis, et al., 2012). Therefore, with regard to the deficiencies in mitochondria in goa1Δ, cell surface phospholipomannan for TLR-2 may be more important to mediate host-organism interaction and recognition by macrophages. We cannot determine precisely as yet how the loss of GOA1 affects the proportion of cell wall polysaccharides.
In addition to the defects in recognition of the mutant, we have previously shown that the goa1Δ as well as the ndh51Δ are hypersusceptible to azoles, including fluconazole, but have wt sensitivity to micafungin and amphotericin B (Sun et al, 2013). Ndh51p is a subunit protein of the ETC complex 1 (CI). Thus, both a CI regulatory protein and a CI subunit protein have similar antifungal drug susceptibilities. The influence of another mitochondrial mutant on its antifungal susceptibility has been reported. The mitochondrial outer membrane protein Sam37p (Sorting and Assembly Machinery) is required for cell wall integrity and is essential for caspofungin tolerance (Dagley et al, 2011). The sam37Δ though, had no cell wall changes in the relative amounts of carbohydrate compared to control strains (Qu et al, 2012). However, CCR4-Pop2, an mRNA deadenylase and a regulator of mRNA stability and translation is required for cell integrity, mitochondrial functions, and phospholipid balance (Dagley et al., 2011). The corresponding mutant had reduced levels of cell wall β-glucans and was sensitive to caspofungin. Therefore, a relationship between trafficking of phospholipids and cell wall integrity has been suggested. In this regard, Singh et al, 2012, report that changes in specific phospholipid classes of the mitochondrial and plasma membranes of C. albicans were associated with a 200-fold increase in fluconazole resistance. An explanation for cell surface changes and antifungal susceptibility is the reduced membrane phospholipid content in certain mutants. The underlying changes in the goa1Δ cell wall may be different from that described in the sam37Δ and ccr4-pop2Δ mutants based upon their hypersusceptibility profiles mentioned above implying gene-specific affects on tolerance.
Nevertheless, the sensitivity to fluconazole may also suggest that a membrane alteration occurred in goa1Δ and the ndh51Δ. One of intermediate products of phospholipid homeostasis is phosphatidylinositol (PI) that facilitates ER, mitochondrial and plasma membrane homeostasis. PI is also a constituent of cell surface GPI-anchored glycoproteins such as the ALS family of GPI-anchored adhesins (Richard, et al., 2002). However, as mentioned above, the type of cell wall change may depend upon the specific mitochondrial defect.
The cell wall changes in goa1Δ caused a decrease in its recognition by cells such as macrophages. However, the mutant is killed to the same extent as control strains. How does this occur? The increased killing of the mutant can be at least partially explained by its changes in carbon metabolism previously published (Chen, et al., 2012; Sun, et al., 2013). Mitochondria usually provide cells with energy through oxidative phosphorylation when grown in nutrient-rich media. However, because of near starvation conditions in macrophages, especially in regard to glucose, during phagocytosis, growth slows, alternate carbon sources are utilized, and oxidative stress responses are induced (Miramón, et al., 2012). Subsequently, yeast-hyphal transition occurs allowing escape of C. albicans from macrophages. Accompanying or prior to morphogenesis is an activation of glycolysis/mitochondrial respiration and a down regulation of stress responses. The metabolic pathways for alternate carbon utilization during reduced glucose include those that are compromised in the mutant as outlined above. Thus, the necessary metabolic switch that allows wild type cells to adapt and escape macrophages is very likely absent in the mutant (Sun, et al., 2013).
It is quite likely that other deficiencies in the mutant result in killing by macrophages. We speculate that increased ROS in the mutant caused by dysfunctional mitochondria complex I (CI) (Li et al., 2011) may also contribute to its death. Thus, heightened mutant cell ROS as well as that produced by macrophages (both internal and external) may be another reason for cell death in goa1Δ. Consequently, the increase in killing of the mutant by macrophages (this study) and neutrophils (Bambach et al., 2009) may be partially explained by a down regulation of anti-oxidative enzymes and an inability to regulate a response to ROS (Chen, et al., 2012; Sun, et al., 2013).
Quite importantly, Goa1p or its transcriptional regulators could constitute targets that may be exploited in antifungal drug discovery. Preliminary data, in fact, indicate that a transcriptional regulator of GOA1 expression is fungal-specific. In fact, a new anti-C. albicans compound that targets mitochondria has been described (Nishikawa, et al., 2010).
Experimental procedures
Candida Strains and growth conditions
C. albicans strains goa1Δ mutant (GOA31) and the gene-reconstituted strain (GOA32) that were previously described were used for all experiments in this study. In addition, wild type SC5314 (WT) was used as a control (Bambach et al., 2009). The ndh51Δ and reconstituted strains were used in inhibitor studies. For most experiments, cells were initially grown at 30°C in YPD overnight.
Cell wall phenotypes
Drop plate and cell clumping assays were used to identify cell wall defects in goa1Δ. For drop plate assays, the overnight cultures of all strains were centrifuged, washed twice in saline, adjusted to 1×106 – 1×101 in 5μl, and spotted onto YPD agar medium with or without cell wall inhibitors. The cell wall inhibitor Congo red (CR) or calcofluor white (CFW) each was added to YPD at 25μg/ml. The sensitivity of strains to the cell wall inhibitors was determined after 48h on plates at 30°C. RPMI medium (Invine Scientific) was used to measure cell clumping (Li, et al., 2009). Overnight cultures were collected, washed, and transferred into RPMI media at an OD600 of 0.5, then incubated at 37°C for 6h with gentle shaking. Suspensions were vortexed for a few seconds prior to taking photographs.
Cell surface hydrophobicity (CSH)
Cyclohexane and xylene were applied to measure CSH for each strain (Gelis, et al., 2009). Overnight culture were grown to their exponential phase, washed three times with PBS, and adjusted at OD600 to 1.0 in 3 ml. The cell suspensions were mixed well with 150 μl of cyclohexane or xylene in acid-washed glass tubes. Cells were incubated at 30°C for 10 min, vortexed once for 1 min, and then transferred to room temperature for 20 min (A0). The absorbance of aqueous phase cells at OD600 were measured compared to cell suspensions prior to mixing with each hydrocarbon (A1). The cell surface hydrophobicity was determined by the percentage of (A1/AO)/100. Triplicate readings of samples were averaged for each strain.
Macrophage assays
The mouse macrophage cell line RAW264.7 (ATCC) was used for all assays described below. Cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 100U/ml penicillin and 100μg/ml streptomycin at 37°C with 5% CO2 in air atmosphere.
(i) Macrophage phagocytosis
RAW264.7 confluent cells were collected, washed, and seeded in 96-well plates at 5×105 in 100μl per well for 2 h. Following their attachment, cells from overnight cultures of Candida strains (1×108 ) were labeled with fluorescein isothiocyanate (FITC, Sigma, 1.25mM in 0.1mM sodium bicarbonate buffer with 0.5% DMSO, pH9.0) at 4°C overnight (Klippel, et al., 2010). After three PBS washes to remove unbound dye, labeled yeast cells suspended in DMEM + 10% FBS in 100μl were added at a ratio of macrophage:yeast=1:2, and plates were incubated for 1 h at 37°C in a 5% CO2 atmosphere. At each time point, 100μl of cell suspension was removed and 100 μl trypan blue (250μg/ml in PBS) was added to quench the fluorescence of yeasts which were bound but not internalized at room temperature for 1 min. The number of internalized and fluorescent yeasts was then estimated (Ex/Em=495/525) by a 96-plate reader (CytoFluor Series 4000, PerSeptive Biosystems), and verified by fluorescence microscopy as well.
(ii) Macrophage killing assay
As stated above, macrophage RAW264.7 cells (1 day old) were prepared and seeded in a 96-well plate at 5 ×104 in 150μl of DMEM overnight at 37°C as described above. The overnight cultures of strains were diluted into DMEM at 2 ×106/ml, and a 50μl aliquot of each strain was added into the first column wells, mixed with macrophages, and then serially diluted 1:4 a total of six times. Strains incubated without macrophages were diluted similarly. Plates were incubated for 24h at 37°C in a 5% CO2 atmosphere. Then, colonies were counted by microscopy in each well. The survival rate of each strain was calculated as the numbers of colonies with macrophages divided by the number of colonies in the absence of macrophages x100. Each experiment was done in duplicate, and p values were determined using the unpaired Student's t test.
(iii) Cytokine expression in macrophages infected with Candida strains
The expression of cytokines was measured by real-time PCR. RAW264.7 cells were prepared as previously described (Ghosh, et al., 2010). Briefly, 1×106 cells/well were seeded in 6-well plates in the appropriate medium and grown overnight at 37°C in a 5% CO2 atmosphere. The overnight cultures of all Candida strains were washed with PBS, suspended in DMEM, and then added to each macrophage culture at ratio of C. albicans: macrophages =4:1. Mixed cultures or controls were then incubated for 1 or 2 h. For all macrophage cytokine assays, cell pellets were washed three times with PBS. RNA extractions were performed by adding 1ml of Trizol (Invitrogen). The quality and concentration of RNAs were measured with a nanospectrophotometer, and approximately 0.8 μg of RNA was used to prepare cDNA. Quantitative real-time PCR (qPCR) was carried out in 20-μl reaction volumes that contained 1× iQ SyBR green Supermix (Bio-Rad), including a 0.2 μM concentration of each primer (Table 2), and 8 μl of a 1:8 dilution of each cDNA from each strain. The experiment was performed in triplicate using Bio-Rad iQ5, and the transcription level of each gene was normalized to GAPDH levels. Data are presented as the means ± standard deviations (SD). The 2–ΔΔCT (where CT is the threshold cycle) method of analysis was used to determine the fold change in gene transcription (Sun, et al., 2013).
Cytokine production of IL-6 was quantified by ELISA to confirm the expression profile after infection with strains (Ferwerda, et al., 2008). In a 96-well plate, adherent RAW264.7 macrophages (5×104 in 100 μl) were infected with 100 μl of yeast cells (1×104) in DMEM supplemented with 10% FBS, and co-incubated at 37°C in a 5% CO2 in air atmosphere for 3 h. As a negative control, macrophages were cultivated without yeasts. After centrifugation of the plates at 4°C for 5 min at 2,250 × g, the supernatants were collected, and stored at -80 °C until assays were performed. IL-6 levels were determined with an ELISA kit (ELISA Ready-SET-Gol, R&D Systems quantikine mouse IL-6 Kit) according to the protocol of the manufacturer. For analysis, background levels from non-infected macrophage cells were measured to obtain final determinations of IL-6 levels of co-cultivated cultures.
Epithelial cell assays
The FaDu epithelial cell line was chosen to measure differences among all Candida strains for the expression of epithelial receptors. FaDu cells (purchased from ATCC) were maintained at 37°C with 5% CO2 in MEM Earl's salts (Irvine Scientific) containing 10% FBS, 1 mM pyruvic acid, 2 mM L-glutamine, 0.1 mM nonessential amino acids, 100 IU/ml penicillin, and 100 IU/ml streptomycin. Cells were subjected to no more than 15 passages for our study (Phan et al., 2007; Zhu et al., 2012).
(i) Expression of epithelial cell receptors
The expression of epithelial cell EGFR and E-Cadherin around all strains of C. albicans hyphae was visualized using direct immunofluorescence. FaDu cells were prepared on fibronectin-coated coverslips in a 24-well tissue culture plate, and incubated with 2.5×105 C. albicans yeast cells for 90 min. The length of germ tubes and extent of hyphae formation among different strains was recorded. Cultures were fixed and permeabilized with 3% paraformaldehyde containing 0.5% triton X-100 in PBS, rinsed once with 1% BSA in PBS, and blocked with 5% goat serum in PBS. After extensive rinsing, the cells were incubated with either the anti-Ecadherin rabbit monoclonal antibody conjugated with Alexa 488 or an anti-EGFR rabbit monoclonal antibody conjugated with Alexa 555 (Cell Signaling Technology, catalog #3199 and #5108). Imaging was by confocal scanning laser microscopy (Zeiss LSM 510 META). Images of the organisms and host cells were acquired; and three to five optical sections were stacked along the z-axis to produce the final images (Zhu et al., 2012).
(ii) Endocytosis and binding of C. albicans by epithelial cells
The number of organisms that were endocytosed or associated with the host cells was quantified. After fixation, epithelial cells were stained with fluorescent conjugated EGFR and E-cadherin. Mixed images of organism and host cells were visualized in 10 high-power fields for each Candida strain with confocal scanning laser microscopy. At least 200 organisms were examined on each coverslip, and the results were expressed as the number of endocytosed or cell-associated organisms per high powered field.
(iii) ALS3 and SSA1 expression in C. albicans
The expression of ALS3 and SSA1 were measured by real-time PCR. All strains of C. albicans were prepared in YPD or in MEM Earl's salts (Irvine Scientific) containing 10% FBS for 4 hours. RNAs were extracted from each strain following the standard real-time PCR procedure as described above. The corresponding primer sets for each gene were listed in Table 1.
Table 1.
Primer sets used in this study
| Gene | Primers |
|---|---|
| GAPDH (Mouse, NM_008084.2) | Forward: CTTTGTCAAGCTCATTTCCTGG Reverse: TCTTGCTCAGTGTCCTTGC |
| Dectin-1 (Mouse, AF262985.1) | Forward: TGCAGTGGGTTTAGGAATCC Reverse: GGGCTTGTGGTTCTCTTTATTTC |
| TLR-2 (Mouse, AF165189.1) | Forward: ACTGTGTTCGTGCTTTCTGAG Reverse: ATGGCTTTCCTCTCAATGGG |
| TLR-4 (Mouse, AF185285.1) | Forward: GAGGACTGGGTGAGAAATGAG Reverse: GTAGTGAAGGCAGAGGTGAAAG |
| IL-6 (Mouse, NM_031168.1) | Forward: CAAAGCCAGAGTCCTTCAGAG Reverse: GTCCTTAGCCACTCCTTCTG |
| IL-10 (Mouse, NM_010548.2) | Forward: AGCCGGGAAGACAATAACTG Reverse: GGAGTCGGTTAGCAGTATGTTG |
| IFN-γ (Mouse, NM_008337.3) | Forward: CTTTGGACCCTCTGACTTGAG Reverse: TCAATGACTGTGCCGTGG |
| TNF-α (Mouse, NM_013693.2) | Forward: CTTCTGTCTACTGAACTTCGGG Reverse: CAGGCTTGTCACTCGAA TTTTG |
| MyD88 (Mouse, NM_010851.2) | Forward: AACAAAGGAACTGGGAGGC Reverse: GTCTGTTCTAGTTGCCGGATC |
| GAPDH (Human, NM_002046) | Forward: ACATCGCTCAGACACCATG Reverse: TGTAGTTGAGGTCAATGAAGGG |
| E-Cadherin (Human, NM_004360.3) | Forward: CCCAATACATCTCCCTTCACAG Reverse: CCACCTCTAAGGCCATCTTTG |
| EGFR (Human, NM_005228.3) | Forward: AAGCCATATGACGGAATCCC Reverse: GGAACTTTGGGCGACTATCTG |
| 18S RNA (C. albicans) | Forward: CGCAAGGCTGAAACTTAAAGG Reverse: AGCAGACAAATCACTCCACC |
| HSP70 (SSA1) (C. albicans, orf19. 4980) | Forward: ATTGCTGAAGGTTATTTGGGTTC Reverse: GGTGGCTTGTCTTTGAGAATC |
| ALS3 (C. albicans, orf19. 1816) | Forward: GGAATGCTGTTTTGGGTTGG Reverse: CACCATGAGCAGTCAAATCAAC |
Acknowledgements
Xiaodong She was funded by a postdoctoral fellowship from the Chinese Academy of Sciences. We wish to thank the Georgetown University Medical Center Biomedical Graduate Research Organization for funds to support this research project as well as an NIH-NIAID grant AI09029. We also wish to thank Dr. Brent Korba for help with tissue culturing.
References
- Bambach A, Fernandes MP, Ghosh A, Kruppa M, Alex D, Li D, et al. Goa1p of Candida albicans localizes to the mitochondria during stress and is required for mitochondrial function and virulence. Eukaryot Cell. 2009;8:1706–1720. doi: 10.1128/EC.00066-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. Hyphal Growth in Human Fungal Pathogens and Its Role in Virulence. International. J Microbiol. 2012;2012 doi: 10.1155/2012/517529. doi:10.1155/2012/517529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med. 2012;4:1–9. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- Calderone RA, Fonzi WA. Virulence factors of Candida albicans. Trends Microbiol. 2001;9:327–35. doi: 10.1016/s0966-842x(01)02094-7. [DOI] [PubMed] [Google Scholar]
- Chen H, Calderone R, Sun N, Wang Y, Li D. Caloric restriction restores the chronological life span of the goa1 null mutant of Candida albicans in spite of high cell levels of ROS. Fungal Genet Biol. 2012;49:1023–1032. doi: 10.1016/j.fgb.2012.09.007. [DOI] [PubMed] [Google Scholar]
- Cheng G, Wozniak K, Wallig MA, Fidel PL, Jr, Trupin SR, Hoyer LL. Comparison between Candida albicans agglutinin-like sequence gene expression patterns in human clinical specimens and models of vaginal candidiasis. Infect Immun. 2005;73:1656–1663. doi: 10.1128/IAI.73.3.1656-1663.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagley MJ, Gentle IE, Beilharz TH, Pettolino FA, Djordjevic JT, Lo TL, et al. Cell wall integrity is linked to mitochondrial and phospholipid homeostasis in Candida albicans through the activity of the post-transcriptional regulator Ccr4-Pop2. Mol Microbiol. 2011;63:968–989. doi: 10.1111/j.1365-2958.2010.07503.x. [DOI] [PubMed] [Google Scholar]
- Dranginis AM, Rauceo JM, Coronado JE, Lipke PN. A biochemical guide to yeast adhesions: glycoproteins for social and antisocial occasions. Microbiol Mol Biol Rev. 2007;71:282–294. doi: 10.1128/MMBR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferwerda G, Meyer-Wentrup F, Kullberg B-J, Netea MG, Adema GJ. Dectin-1 synergizes with TLR2 and TRL4 for cytokine production in human primary monocytes and macrophages. Cell Microbiol. 2008;10:2058–2066. doi: 10.1111/j.1462-5822.2008.01188.x. [DOI] [PubMed] [Google Scholar]
- Filler SG, Swerdloff JN, Hobbs C, Luckett PM. Penetration and damage of endothelial cells by Candida albicans. Infect Immun. 1995;63:976–983. doi: 10.1128/iai.63.3.976-983.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelis M, de Groot PW, Castillo L, Moragues MD, Sentandreu R, Gómez MM, et al. Pga13 in Candida albicans is localized in the cell wall and influences cell surface properties, morphogenesis and virulence. Fungal Genet Biol. 2009;49:322–331. doi: 10.1016/j.fgb.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Green CB, Cheng G, Chandra J, Mukherjee P, Ghannoum MA, Hoyer LL. RT-PCR detection of Candida albicans ALS gene expression in the reconstituted human epithelium (RHE) model of oral candidiasis and in model biofilms. Microbiology. 2004;150:267–75. doi: 10.1099/mic.0.26699-0. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Howe N, Volk K, Tati S, Nickerson KW, Petro TM. Candida albicans cell wall components and farnesol stimulate the expression of both inflammatory and regulatory cytokines in the murineRAW264.7macrophage cell line. FEMS Immunol Med Microbiol. 2010;60:63–73. doi: 10.1111/j.1574-695X.2010.00717.x. [DOI] [PubMed] [Google Scholar]
- Gow NA, Netea MG, Munro CA, Ferwerda G, Bates S, Mora-Montes HM, et al. Immune recognition of Candida albicans beta-glucan by dectin-1. J Infect Dis. 2007;196:1565–157. doi: 10.1086/523110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer LL, Green CB, Oh SH, Zhao X. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family-a sticky pursuit. Med Mycol. 2007;20:1–15. doi: 10.1080/13693780701435317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klippel N, Cui S, Groebe L, Bilitewski U. Deletion of the Candida albicans histidine kinase gene CHK1 improves recognition by phagocytes through an increased exposure of cell wall b-1,3-glucans. Microbiology. 2010;156:3432–3444. doi: 10.1099/mic.0.040006-0. [DOI] [PubMed] [Google Scholar]
- Kim HS, Choi EH, Khan J, Roilides E, Francesconi A, Kasai M, et al. Expression of genes encoding innate host defense molecules in normal human monocytes in response to Candida albicans. Infect Immun. 2005;73:3714–3724. doi: 10.1128/IAI.73.6.3714-3724.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriengkauykiat J, Ito JI, Dadwal SS. Epidemiology and treatment approaches in management of invasive fungal infections. Clin Epidemiol. 2011;3:175–191. doi: 10.2147/CLEP.S12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventakos K, Lewis RE, Kontoyiannis DP. Fungal Infections in Leukemia Patients: How Do We Prevent and Treat Them? Clin Infect Dis. 2010;50:405–415. doi: 10.1086/649879. [DOI] [PubMed] [Google Scholar]
- Lewis LE, Bain JM, Lowes C, Gillespie C, Rudkin FM, Gow NA, et al. Stage specific assessment of Candida albicans phagocytosis by macrophages identifies cell wall composition and morphogenesis as key determinants. PLoS Pathog. 2012;8:e1002578. doi: 10.1371/journal.ppat.1002578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Williams D, Lowman D, Monteiro MA, Tan X, Kruppa M, et al. The Candida albicans histidine kinase Chk1p: signaling and cell wall mannan. Fungal Genet Biol. 2009;46:731–41. doi: 10.1016/j.fgb.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Chen H, Florentino A, Alex D, Sikorski P, Fonzi WA, et al. Enzymatic dysfunction of mitochondrial complex I of the Candida albicans goa1 mutant is associated with increased reactive oxidants and cell death. Eukaryotic Cell. 2011;10:672–682. doi: 10.1128/EC.00303-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionakis MS, Lim JK, Lee CC, Murphy PM. Organ-specific innate immune responses in a mouse model of invasive candidiasis. J Innate Immun. 2010;3:180–199. doi: 10.1159/000321157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionakis MS, Netea MG. Candida and Host Determinants of susceptibility to Invasive Candidiasis. PLoS Pathog. 2013;9:e1003079. doi: 10.1371/journal.ppat.1003079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Bender JA, Fink GR. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryotic Cell. 2004;3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr KA, Balajee SA, Hawn TR, Ozinsky A, Pham U, Akira S, Aderem A, W. Conrad Liles WC. Differential role of MyD88 in macrophage-mediated Responses to opportunistic fungal pathogens. Infect. Immun. 2003;77:5280–5286. doi: 10.1128/IAI.71.9.5280-5286.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miramón P, Dunker C, Windecker H, Bohovych IM, Brown AJ, Kurzai O, et al. Cellular responses of Candida albicans to phagocytosis and the extracellular activities of neutrophils are critical to counteract carbohydrate starvation, oxidative and nitrosative Stress. PLoS ONE. 2012;7:e52850. doi: 10.1371/journal.pone.0052850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Gow NA, Munro CA, Bates S, Collins C, Ferwerda G, et al. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J Clin Invest. 2006;116:1642–1650. doi: 10.1172/JCI27114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa H, Yamada T, Shibata T, Uchihashi S, Fan H, Hayakawa H, et al. Uptake of T-2307, a novel arylamidine, in Candida albicans. J Antimicrob Chemother. 2010;65:1681–1687. doi: 10.1093/jac/dkq177. [DOI] [PubMed] [Google Scholar]
- Phan QT, Myers CL, Fu Y, Sheppard DC, Yeaman MR, Welch WH, et al. Als3 is a Candida albicans invasion that binds to cadherins and induces endocytosis by host cells. PLos Biol. 2007;5:e64. doi: 10.1371/journal.pbio.0050064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcaro I, Vidal M, Jouvert S, Stahl PD, Giaimis J. Mannose receptor contribution to Candida albicans phagocytosis by murine E-clone J774 macrophages. J Leukoc Biol. 2003;74:206–215. doi: 10.1189/jlb.1202608. [DOI] [PubMed] [Google Scholar]
- Qu Y, Jelicic B, Pettolino F, Perry A, Lo T, Hewitt V, Bantun F, et al. Mitochondrial sorting and assembly machinery subunit Sam37 in Candida albicans: insight into roles of mitochondria in fitness, cell wall integrity, and virulence. Eukaryot Cell. 2012;11:532–544. doi: 10.1128/EC.05292-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M, Ibata-Ombetta S, Dromer F, Bordon-Pallier F, Jouault T, Gaillardin C. Complete glycosylphosphatidylinositol anchors are required in Candida albicans for full morphogenesis, virulence and resistance to macrophages. Mol Microbiol. 2002;44:841–853. doi: 10.1046/j.1365-2958.2002.02926.x. [DOI] [PubMed] [Google Scholar]
- Romani L. Innate and adaptive immunity in Candida albicans infections and saprophytism. J Leukoc Biol. 2000;68:175–179. [PubMed] [Google Scholar]
- Rüping MJ, Vehreschild JJ, Cornely OA. Patients at high risk of invasive fungal infections: when and how to treat. Drugs. 2008;68:1941–62. doi: 10.2165/00003495-200868140-00002. [DOI] [PubMed] [Google Scholar]
- Singh A, Yadav V, Prasad R. Comparative lipidomics in clinical isolates of Candida albicans reveal crosstalk between mitochondria, cell wall integrity, and azole resistance. PLoS One. 2012;7:e39812. doi: 10.1371/journal.pone.0039812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Sun N, Fonzi1 W, Chen H, She X, Zhang L, Calderone R. Azole susceptibility and transcriptome profiling in the Candida albicans mitochondrial mutants. Antimicrob Agents Chemothe. 2013;57:532–542. doi: 10.1128/AAC.01520-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D, Lewis M. Pathogenesis and treatment of oral candidiasis. J Oral Microbiol. 2011;3:5771. doi: 10.3402/jom.v3i0.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Phan QT, Boontheung P, Solis NV, Loo JA, Filler SG. EGFR and HER2 receptor kinase signaling mediate epithelial cell invasion by Candida albicans during oropharyngeal infection. PNAS. 2012;109:14194–14199. doi: 10.1073/pnas.1117676109. [DOI] [PMC free article] [PubMed] [Google Scholar]