Abstract
Background:
Besides the conventional clear-cell renal cell carcinoma (ccRCC), papillary RCC (pRCC) is the second most common renal malignancy. Papillary RCCs can further be subdivided into two distinct subtypes. Although a clinical relevance of pRCC subtyping has been shown, little is known about the molecular characteristics of both pRCC subtypes.
Methods:
We performed microarray-based microRNA (miRNA) expression profiling of primary ccRCC and pRCC cases. A subset of miRNAs was identified and used to establish a classification model for ccRCC, pRCC types 1 and 2 and normal tissue. Furthermore, we performed gene set enrichment analysis with the predicted miRNA target genes.
Results:
Only five miRNAs (miR-145, -200c, -210, -502-3p and let-7c) were sufficient to identify the samples with high accuracy. In a collection of 111 tissue samples, 73.9% were classified correctly. An enrichment of miRNA target genes in the family of multidrug-resistance proteins was noted in all tumours. Several components of the Jak-STAT signalling pathway might be targets for miRNAs that define pRCC tumour subtypes.
Conclusion:
MicroRNAs are able to accurately classify RCC samples. Deregulated miRNAs might contribute to the high chemotherapy resistance of RCC. Furthermore, our results indicate that pRCC type 2 tumours could be dependent on oncogenic MYC signalling.
Keywords: microRNA, markers, genetic, renal cell carcinoma, signal pathways
Renal cell carcinoma (RCC) accounts for nearly 4% of all malignancies. In Europe, the age-standardised incidence rates per 100 000 individuals are 15.8 for male individuals and 7.1 for female individuals, making RCC the tenth most common malignancy in this region (Ljungberg et al, 2011). According to the WHO classification of tumours of the urinary system (Eble et al, 2004), RCC is a collection of different subtypes rather than one entity. Besides conventional clear-cell RCC (ccRCC), papillary RCC (pRCC) is the second most common RCC type with an incidence of 10–15% (Nelson et al, 2007). On the basis of histomorphological characteristics, pRCCs can further be subdivided into two distinct subtypes (Delahunt and Eble, 1997). Survival analyses of pRCC patients indicated that type 2 pRCC (pRCC2) is associated with a worse clinical outcome than type 1 pRCC (pRCC1) (Pignot et al, 2007).
In cases of familiar pRCC syndromes, mutations of the c-MET proto-oncogene are correlated with a pRCC1 morphology (Lubensky et al, 1999), whereas in cases of Hereditary leiomyomatosis and renal cell cancer there is a high incidence for mutations in the fumarate hydratase (FH) gene, leading to the occurrence of pRCC2 (Gardie et al, 2011).
MicroRNAs (miRNAs) are short, non-coding RNA molecules that are ∼19–25 nucleotides in length. Associated with the argonaute (Ago) proteins within the RNA-induced silencing complex (RISC), miRNAs contribute to translational silencing (Meister, 2007). The miRNAs interact with the 3' UTR of their target genes (Lytle et al, 2007), ultimately leading to reduced protein expression. Extensive deregulation of miRNA expression has been observed in ccRCC cases (White et al, 2011a; Osanto et al, 2012). MicroRNA expression signatures are highly reproducible; therefore, the definition of a signature of malignancy (Jung et al, 2009) or a molecular classification of tumours according to their miRNA expression is feasible (Lu et al, 2005). For prostate cancer, we previously showed that a set of only three miRNAs was able to correctly discriminate between prostate cancer samples and the corresponding normal tissue from the same organ with an overall accuracy of 77% (Wach et al, 2012).
Here, we performed miRNA expression profiling in RCC subtypes. Global miRNA expression profiles were established using miRNA microarrays. A reduced set of five miRNAs was identified that was subsequently used to establish a sequential classification model. These miRNAs were able to discriminate between the tumour and the corresponding normal tissue samples with high accuracy. In addition, they were also able to distinguish between the histological tumour entities of ccRCC and pRCC subtypes 1 and 2.
Patients and methods
Patients and tissue samples
The use of the patient tumour samples for the molecular analyses was approved by the institutional ethical review board, and the study was conducted according to the standards set by the declaration of Helsinki. A first patient set was acquired from the local Institute of Pathology and was designated as the discovery set. It consisted of seven cases of ccRCC, eight cases of pRCC1 and eight cases of pRCC2. After a pathological review by an experienced uropathologist (AH), samples of the tumour and associated normal tissue were prepared. Briefly, formalin-fixed and paraffin-embedded (FFPE) tissue sections were prepared in 10-μm slices and microdissected to generate samples of tumour and the corresponding normal tissue. In addition, we incorporated one sample of normal tissue derived from a ccRCC case and one sample of pRCC1 tumour tissue.
A second patient cohort, the designated validation set, was provided by the tissue bio-repository at the University Cancer Center (UCC), University Hospital Erlangen. Haematoxylin and eosin-stained tissue sections were reviewed by an experienced uropathologist (AH). The RCC subtypes were classified according to the WHO guidelines for tumours of the urinary tract (Eble et al, 2004). This patient cohort consisted of 37 tumour and 32 normal tissue samples, derived from the corresponding normal tissue. Normal control tissue consisted of renal cortex, including parts of the normal tubular system, taken from the opposite pole of the kidney to avoid the cancer field effect. The tissue specimens were cryopreserved samples, and the tumour samples displayed a varying tumour cell content ranging from 40% to 100%. The pathological review revealed eight cases of pRCC1, nine cases of pRCC2 and one case of pRCC where no further subclassification was available. In addition, this patient cohort contained 19 cases of ccRCC.
RNA extraction
The RNA from frozen tissue samples was extracted using the Trizol reagent (Invitrogen, Karlsruhe, Germany) according to the manufacturer's instructions. The RNA from FFPE tissue preparation was extracted using the MasterPure complete DNA and RNA purification kit (Epicentre Biotechnologies, Madison, WI, USA), according to the manufacturer's instructions. All RNA preparations were treated with RNase-free DNaseI (Roche, Mannheim, Germany). The RNA yield and purity was determined using a microliter spectrophotometer (NanoDrop 1000, Thermo Scientific, Wilmington, DE, USA).
MiRNA microarrays
The miRNA microarray expression measurements were performed on GeneChip miRNA microarrays V1.0 (Affymetrix, Santa Clara, CA, USA) according to the manufacturer's instructions. The array contained sequence-specific probes for 678 human miRNAs, listed in miRBase v11.0 (http://www.mirbase.org). The signal intensity data were further analysed with Partek software v6.2 (Partek, St Louis, MO, USA). To identify miRNAs differentially expressed between the defined sample groups, ANOVA test statistics were applied. A principal component analysis (PCA) was applied to compress the multidimensional miRNA expression data to three dimensions while maintaining the variance (Jolliffe, 2002). The miRNA expression measurements are available at GEO (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE41282).
MiRNA qRT–PCR
Quantification of miRNAs was conducted by applying a two-step reaction using miRCURY universal reverse transcription reagents and LNA-modified miRNA-specific primers (Exiqon, Vedbaek, Denmark) according to the manufacturer's instructions. Briefly, RNA (20 ng) was reverse transcribed using the miRCURY universal cDNA synthesis kit (Exiqon). The quantitative PCRs were performed in the StepOne plus real-time PCR system (Applied Biosystems, Foster City, CA, USA), using LNA-modified sequence-specific primer sets and SYBR green PCR mix (Exiqon). All reactions were measured in triplicate in a final volume of 10 μl. The thermal cycling conditions were chosen according to the manufacturer's recommendations. For the relative quantification, every sample was analysed in parallel for the expression of specific miRNAs and the endogenous reference RNA RNU44. The relative miRNA expression levels, normalised to RNU44, were calculated by applying the ΔCt method (Schmittgen and Livak, 2008).
Statistical analyses
The statistical analyses were performed using SPSS 19.0 (IBM, Armonk, NY, USA) and GraphPad Prism 4.0 (Graph Pad Software, La Jolla, CA, USA). All of the statistical tests were performed as two-sided tests, and P-values <0.05 were considered statistically significant. To determine the potential of miRNAs to discriminate between the tumour and normal samples or between the RCC subtypes, we applied binary logistic regression models. To select miRNAs essentially contributing to the discriminative model, we applied a backward elimination method with the likelihood ratio as the determinant (inclusion P<0.05; exclusion P>0.1). To calculate the receiver–operator characteristics, we used the probability function generated by the binary logistic regression algorithm.
The definition of potential miRNA target genes and the pathway analysis of the predicted target genes were conducted as previously described (Keller et al, 2011). Briefly, target genes of the best discriminative miRNAs were predicted by the miRanda algorithm using a significance threshold value of <0.01 and subjected to a gene set enrichment analysis (GSEA) using GeneTrail (Backes et al, 2007). The predicted miRNA target genes were mapped to pre-defined Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
Results
MiRNA microarray expression data
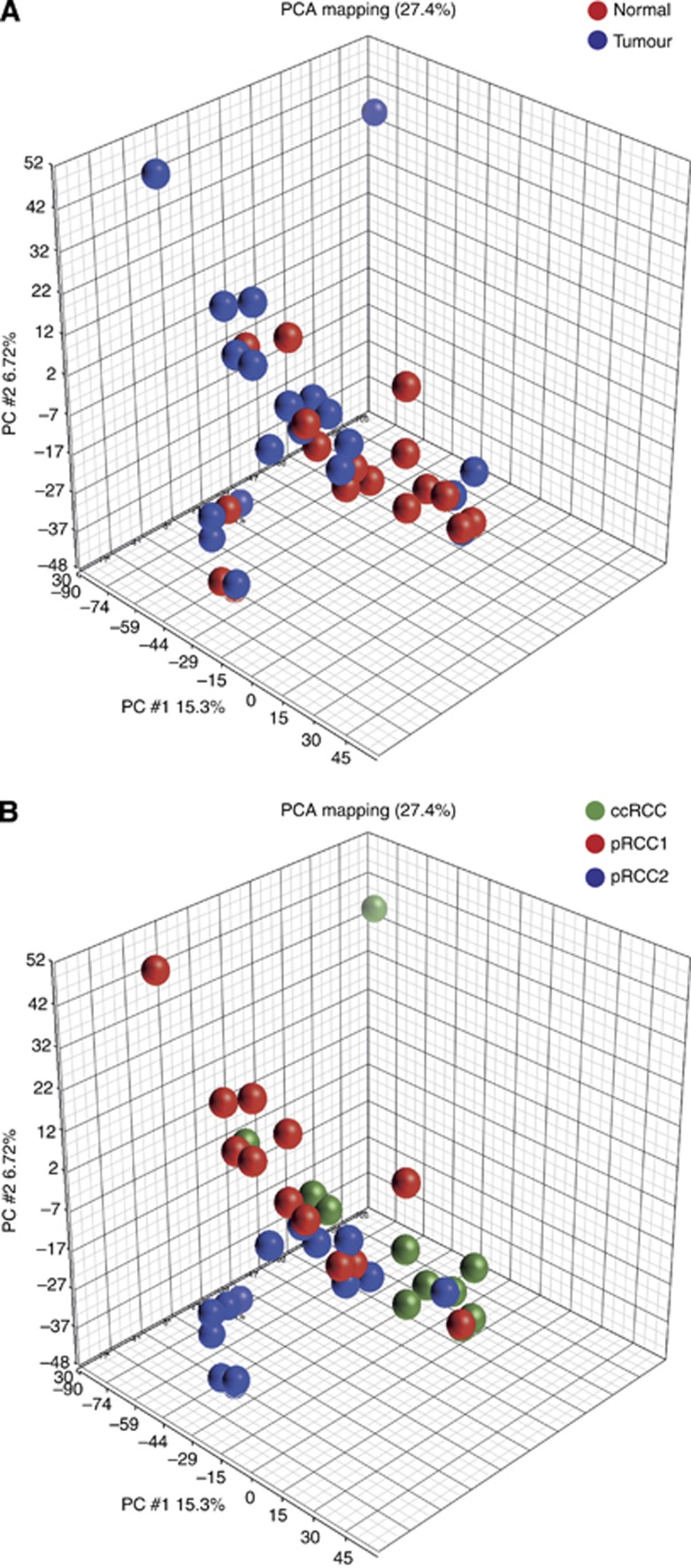
The miRNA expression profiling was performed on 38 out of 46 samples from the discovery patient set where the RNA extraction yielded sufficient amounts of RNA for hybridisation. These comprised six samples of ccRCC and five samples of corresponding normal tissue, seven samples of pRCC1 and five samples of corresponding normal tissue and seven samples of pRCC2 and eight samples of corresponding normal tissue. The PCA revealed that, depending on the entity the tissue was derived from, the samples displayed a tendency to form separate clusters (Figure 1A). Interestingly, the normal tissue samples preferentially associated with the corresponding tumour samples rather than with normal tissue samples from other RCC entities (Figure 1B). When comparing tumour and normal samples, 111 miRNAs (25 upregulated and 86 downregulated) displayed a significant differential expression. Tumour samples of ccRCC and pRCC (irrespective of the subtype) differed in the expression of 100 miRNAs (38 upregulated and 62 downregulated). Between the pRCC subtypes 1 and 2, 98 miRNAs (44 upregulated and 54 downregulated) were significantly differentially expressed. MicroRNAs differentially expressed in every comparison and the degrees of differential expression are given in Supplementary Table 1.
Figure 1.
Principal component analysis. The miRNA microarray expression data were used to construct a three-dimensional PCA. PCA stratified into normal (red) and tumour (blue) samples (A). PCA stratified into ccRCC (green), pRCC1 (red) and pRCC2 (blue) samples (B).
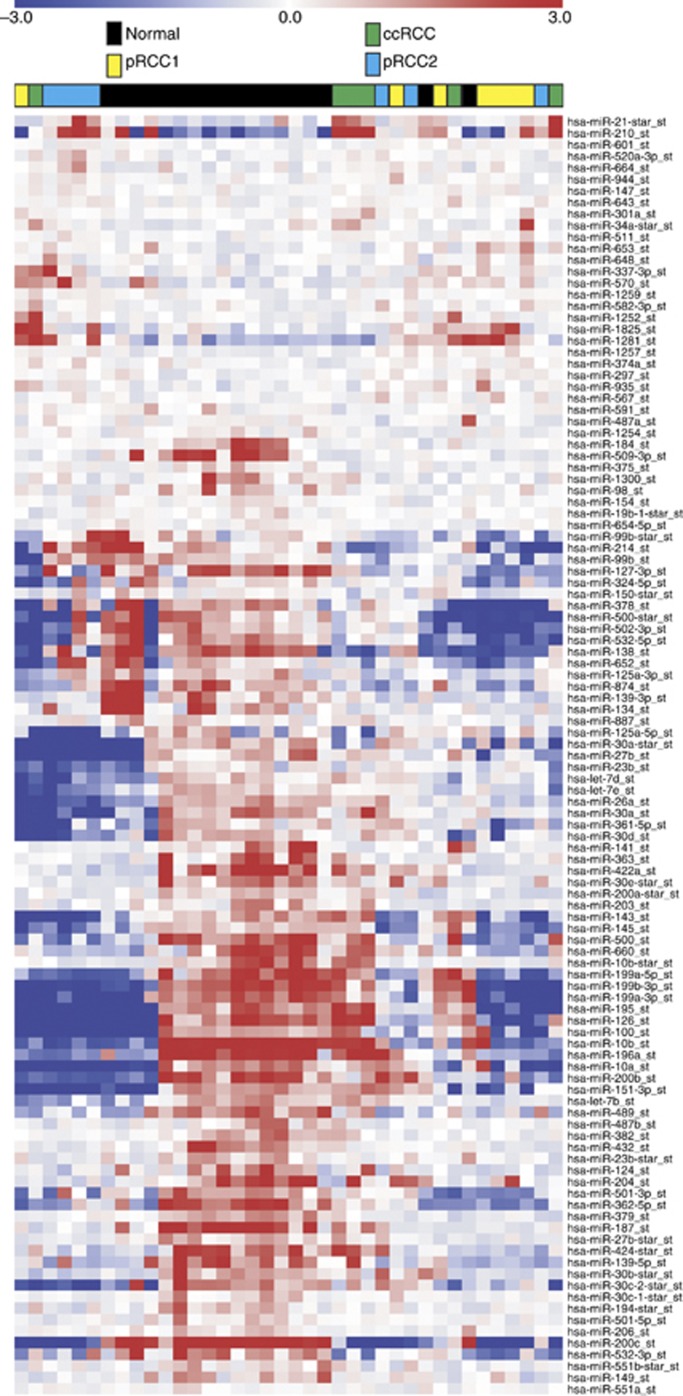
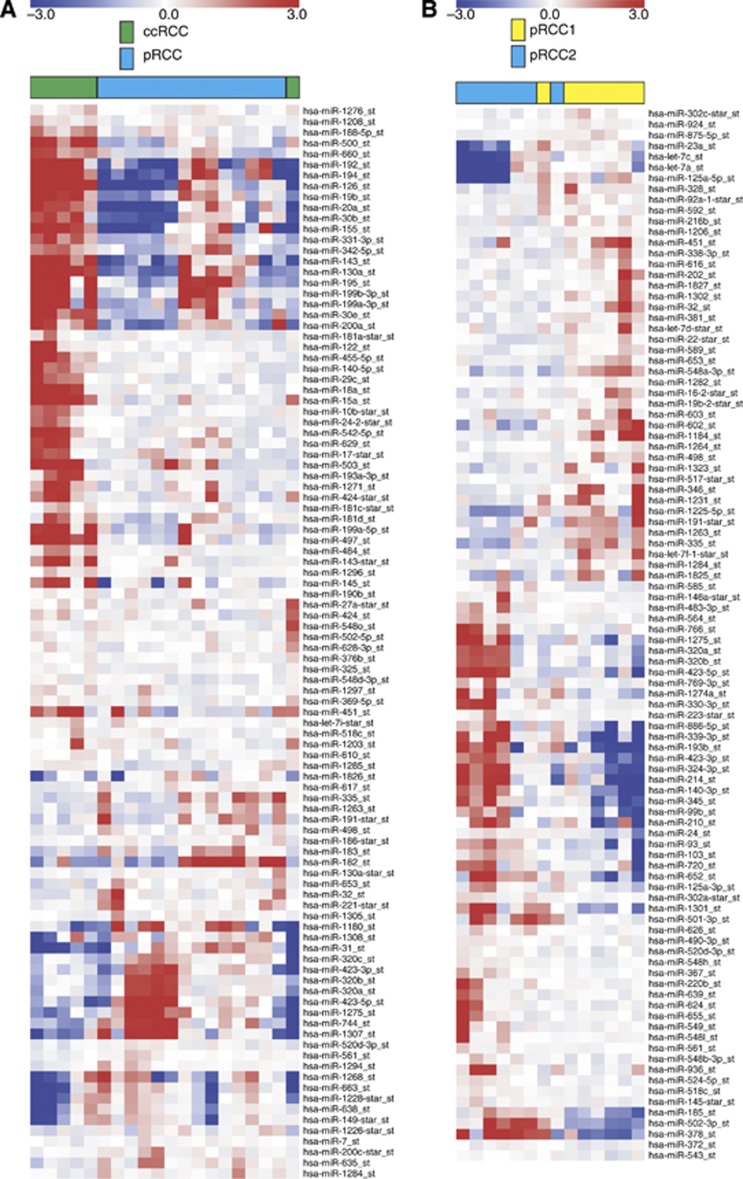
We then performed unsupervised hierarchical clustering using the Euclidean distance measure and average linkage. When analysing all of the samples (Figure 2), we reconfirmed that RCC is a very heterogeneous tumour entity. Tumour samples derived from pRCC1, pRCC2 and, to a lesser extent, those from ccRCC showed a tendency to form clusters. However, these clusters were not clearly separated from each other. Next, we examined the miRNAs differentially expressed between tumour and normal samples in each RCC entity. Within each entity, the tumour and the corresponding normal tissues formed distinct clusters (Supplementary Figure 1). Next, we concentrated on those miRNAs that were differentially expressed in ccRCC and pRCC samples, as well as those miRNAs differentially expressed in the pRCC subtypes 1 and 2. Tumour samples from ccRCC could be efficiently separated from pRCC. Only one sample of ccRCC was assigned to the pRCC group (Figure 3A). Furthermore, the subtypes of pRCC could also be separated with high accuracy. Here, only one case of pRCC1 was assigned to the pRCC2 group (Figure 3B).
Figure 2.
Hierarchical clustering. The miRNAs (111) identified as differentially expressed between the tumour and normal tissue samples by a one-way ANOVA were selected as markers for unsupervised hierarchical clustering. The tissue identifier (normal, ccRCC, pRCC1 and pRCC2) is shown on the top.
Figure 3.
Hierarchical clustering. The miRNAs identified as differentially expressed between RCC tumour entities were selected as markers for the unsupervised hierarchical clustering. The miRNAs (100) identified as differentially expressed between ccRCC and pRCC tumours by a one-way ANOVA (A). The miRNAs (98) identified as differentially expressed between pRCC1 and pRCC2 samples by a one-way ANOVA (B). The respective tissue identifiers are shown on the top.
Validation of miRNA expression data by qRT–PCR
First, we sought to validate the expression of selected miRNAs using an independent experimental method in the discovery set. We selected 10 miRNAs for validation by quantitative PCR. The selection criteria were the following: the miRNAs should have the potential to either discriminate between tumour and normal tissue, between ccRCC and pRCC, or between pRCC1 and pRCC2. The miRNAs miR-143, miR-145, miR-200c and miR-502-3p had the potential to differentiate between the tumour and the normal tissue samples. The miRNAs miR-143, miR-145, miR-15a and miR-10b had the potential to discriminate between ccRCC and pRCC tumours. The miRNAs let-7c, miR-93, miR-193b, miR-502-3p and miR-210 were potential discriminators between pRCC1 and pRCC2. Owing to a low RNA yield, four samples had to be omitted, resulting in a total of 42 samples. For normalisation, we used the reference gene RNU44. This reference gene has been successfully used in studying miRNA expression in RCC (Wotschofsky et al, 2011). We observed that with the exception of miR-193b all of the miRNAs were significantly deregulated as indicated by the microarray data. The following miRNAs were significantly downregulated in tumour tissues: MiR-200c (47.0-fold), miR-145 (11.0-fold), miR-10b (9.0-fold), miR-502-3p (6.9-fold), miR-143 (6.8-fold), let-7c (3.9-fold), miR-93 (2.7-fold) and miR-15a (2.1-fold). MiR-210 was significantly upregulated by 2.8-fold in the tumour tissue samples (Supplementary Figure 2).
Supervised establishment of predictive rules
Using the selected miRNAs, we sought to answer three questions. Is a given sample derived from the tumour or the normal tissue? Is a tumour sample derived from ccRCC or pRCC? Is a pRCC sample derived from pRCC subtype 1 or subtype 2? To establish predictive rules, we used a logistic regression analysis. By applying a stepwise backward elimination method, we selected those that contributed essentially to the predictive model. For the discrimination of tumour and normal tissue, a combination of miRNAs miR-145, miR-210, miR-200c and miR-502-3p was determined, to correctly discriminate between the tumour and the normal tissue samples with an accuracy of 92.9% (area under receiver–operator characteristic (ROC) curve (AUC)=0.975; P<0.001). Interestingly, miR-210 was an essential contributor, although this was not suggested by the microarray data. For discrimination between ccRCC and pRCC tumours, a combination of miR-145 and miR-502-3p was determined, which was able to correctly predict the RCC entity of tumour samples in all cases (AUC=1.0; P<0.001). The miRNA miR-502-3p was essential for distinguishing between ccRCC and pRCC. However, it was primarily selected for its potential to distinguish between the tumour and normal tissue samples. Finally, for discrimination of the two pRCC subtypes, a combination of miR-210 and let-7c was selected. This combination was able to correctly predict the pRCC subtype in 92.3% of the cases (AUC=0.857; P=0.03). The characteristics of the ROC analysis are given in Table 1 and Supplementary Figure 3.
Table 1. Receiver–operator characteristics and classification properties of miRNAs.
|
Discovery set (n=42) | |||||
|---|---|---|---|---|---|
| Discrimination | MiRNAs | AUC | 95% CI | P-value | Correct classification n (%) |
| Tumour/normal | miR-145, miR-200c, miR-210, miR-502-3p | 0.975 | 0.936–1.000 | <0.0001 | 39/42 (92.8) |
| ccRCC/pRCC | miR-145, miR-502-3p | 0.978 | 0.923–1.000 | 0.0006 | 19/20 (95.0) |
| pRCC type 1/2 |
miR-210, let-7c |
0.875 |
0.646–1.000 |
0.028 |
12/13 (92.3) |
|
All cases (n=111) | |||||
| Tumour/normal | miR-145, miR-200c, miR-210, miR-502-3p | 0.919 | 0.863–0.975 | <0.0001 | 96/111 (86.5) |
| ccRCC/pRCC | miR-145, miR-502-3p | 0.833 | 0.710–0.956 | <0.0001 | 38/49 (77.6) |
| pRCC type 1/2 | miR-210, let-7c | 0.900 | 0.741–1.000 | 0.0016 | 19/22 (86.4) |
Abbreviations: AUC=area under receiver–operator characteristic curve; ccRCC=clear-cell renal cell carcinoma; CI=confidence interval; miRNAs=microRNAs; pRCC=papillary renal cell carcinoma
Establishment and validation of a sequential classification model
Next, we tested the performance of the predictive rules to classify the samples of the discovery set in an unsupervised manner.
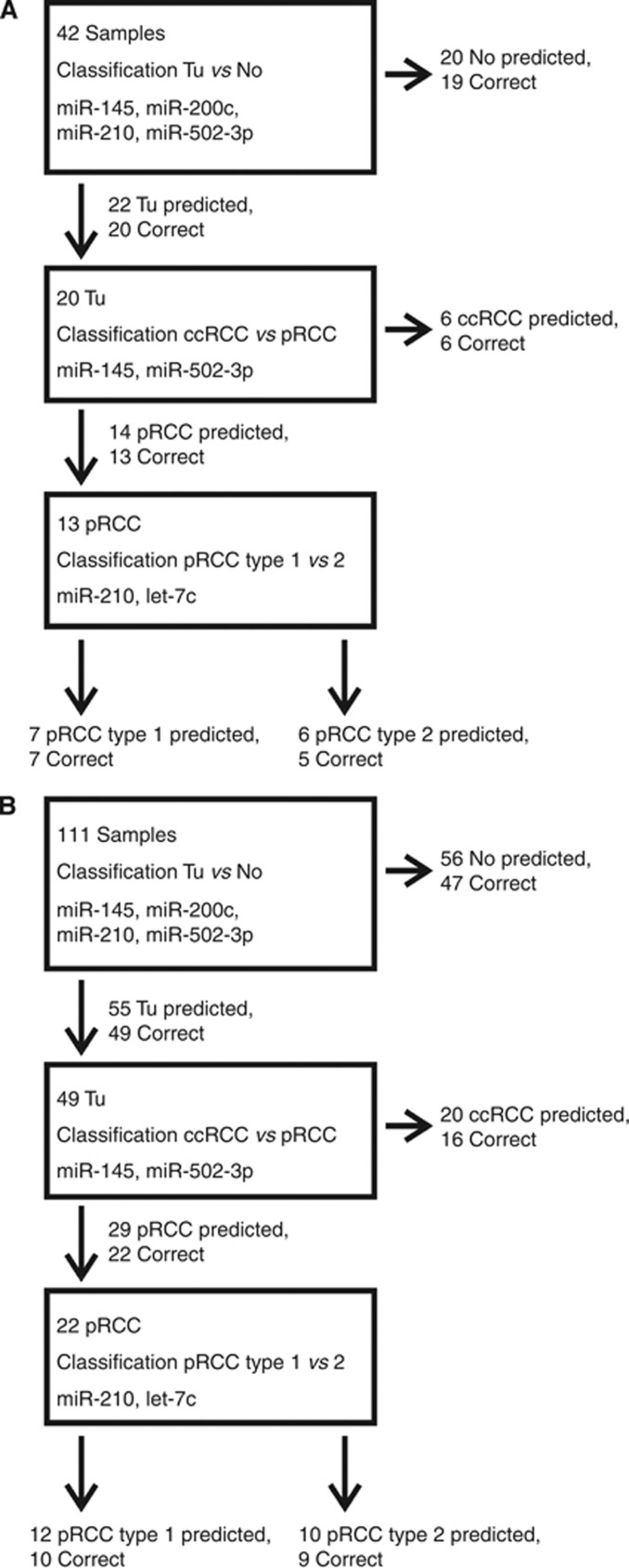
All 42 samples were classified as tumour or normal. Then, those samples that were predicted to be tumour samples were assigned to the ccRCC or the pRCC group. Subsequently, those tumour samples that were assigned to the pRCC group were further subclassified into pRCC1 or pRCC2. The dignity of any sample (tumour or normal) was correctly determined in 39 of 42 cases (92.8%). The RCC entity (ccRCC or pRCC) was correctly predicted in 19 of 20 cases (95.0%), and the subtype of pRCCs was correctly predicted in 12 of 13 cases (92.3%) (Figure 4A).
Figure 4.
Classification of tissue samples using miRNA expression. Tissue samples were classified in an unsupervised manner. All of the tissue samples were classified into tumour or normal samples. Those samples that were predicted to be derived from tumour tissue were further classified into ccRCC or pRCC. Those samples that were predicted to be derived from pRCC tissue were further classified into pRCC1 or pRCC2. For every classification step, the utilised miRNAs, the number of predicted samples and the number of correct predictions are given. The samples (42) in the discovery set (A). The samples (111) in the combined patient sets (B).
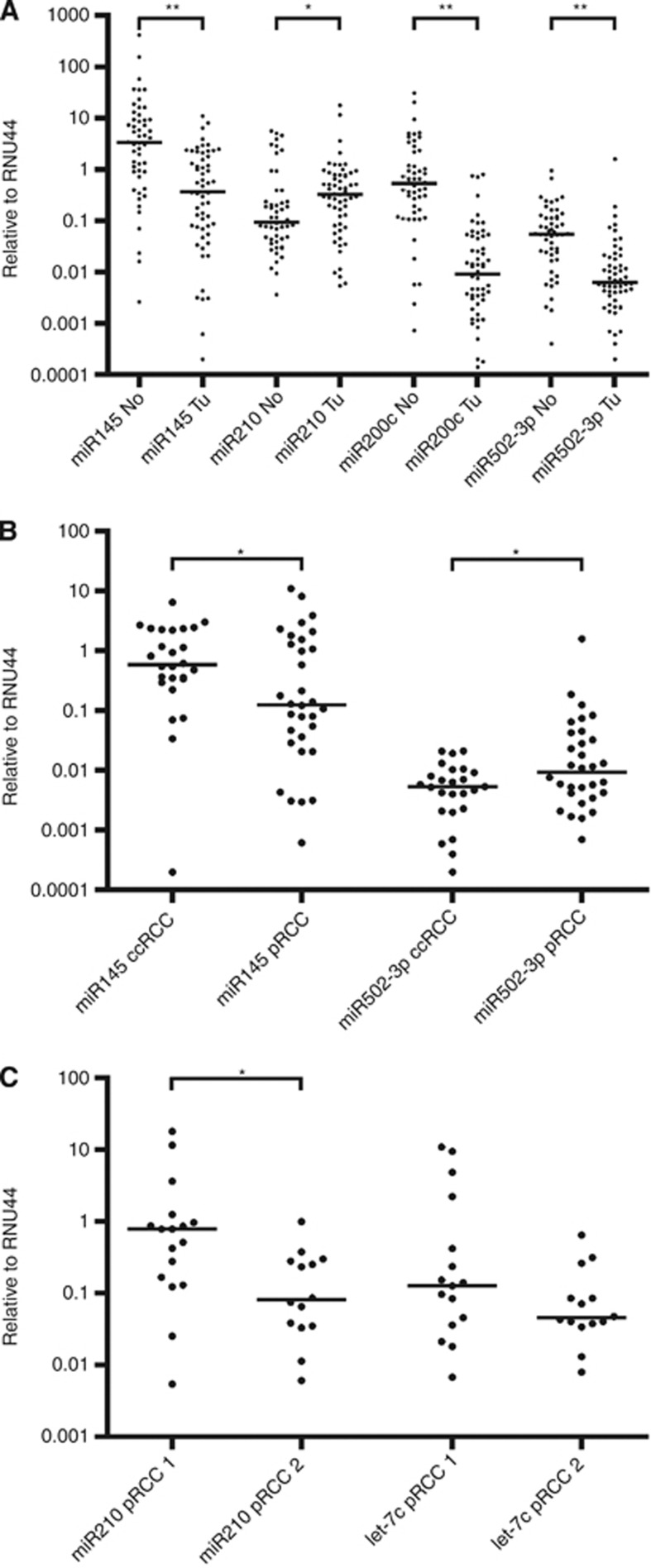
Finally, we applied the classification model to the designated validation cohort. The established set of five miRNAs was measured by quantitative PCR, and we applied the sequential classification model using the defined parameters. The dignity was correctly predicted in 57 of 69 cases (82.6%). Within the correctly classified tumour samples, the RCC entity was correctly determined in 19 of 29 cases (65.5%). The subtype of pRCCs was correctly predicted in seven of nine cases (77.7%). When combining both sets of samples, the resulting rates of correct classification were 96 of 111 (86.5%) for the discrimination between tumour and normal, 38 of 49 (77.6%) for the discrimination between ccRCC and pRCC and 19 of 22 (86.4%) for the identification of the pRCC subtype (Figure 4B). Quantitative PCR measurements of the five selected miRNAs are given in Figure 5, and the results of the classification model are given in Supplementary Table 2.
Figure 5.
Quantification of miRNAs. The miRNAs that were identified as discriminators were quantified in the combined cohort of 111 tissue samples. Quantification of miR-145, miR-210, miR-200c and miR-502-3p, which discriminates between tumour and normal tissue (A). Quantification of miR-145 and miR-502-3p, which discriminates between ccRCC and pRCC tumours (B). Quantification of miR-210 and let-7c, which discriminates between pRCC1 and pRCC2 (C); *P<0.05; **P<0.01.
To further characterise the discriminative properties, we performed ROC analyses using the predicted class probability from the binary logistic regression calculations as a measure (Table 1, lower part). Overall, for the determination of the dignity of the sample (tumour or normal), the resulting AUC was 0.919 (P<0.0001). For the prediction of the RCC entity, the AUC was 0.833 (P<0.0001), and for the determination of the histological subtype of pRCCs the AUC was 0.900 (P=0.0016). The characteristics of the discovery set of samples were not significantly better than those of the complete cohort. The highly similar characteristics of the discovery set and the complete cohort of samples demonstrated that the optimisation of the binary logistic regression algorithm did not lead to a model overfitted towards the discovery set.
Pathway analysis
We performed miRNA target prediction and GSEA using GeneTrail (Backes et al, 2007). For the miRNAs miR-145, miR-200c miR-210 and miR-502-3p, which discriminated between tumour and normal samples, a total of 1629 potential target mRNAs were identified. For miR-145 and miR-502-3p, which distinguished ccRCC from pRCC, there were 782 potential targets. For miR-210 and let-7c, which were used to discriminate between pRCC1 and pRCC2, 832 potential targets were identified. The predicted miRNA target genes were assigned to KEGG biological pathways, and those pathways enriched in miRNA target genes were identified (Supplementary Table 3). One prominent pathway influenced exclusively by those miRNAs discriminative for tumour and normal tissue is the family of ATP-binding cassette (ABC) transporter molecules. Here, a total of seven members are predicted to be regulated by the discriminative miRNAs, including three known multidrug-resistance proteins (MRP1, MRP2 and MRP8).
Those miRNAs that were essential in discriminating between ccRCC and pRCC may have an impact on several growth factor-related pathways, such as the vascular endothelial growth factor A (VEGF) signalling pathway or the Insulin signalling pathway. Several predicted target genes, such as mitogen-activated protein kinase kinase 2 (MAP2K2), AKT3 or BAD, are implicated in multiple pathways, demonstrating the potential for miRNAs to influence cellular processes such as proliferation, migration and apoptosis. The miRNAs miR-210 and let-7c showed the best discrimination between pRCC1 and pRCC2. They are predicted to target 10 genes involved in the Jak-STAT signalling pathway, including the v-myc myelocytomatosis viral oncogene homologue (MYC), signal transducer and activator of transcription 2 (STAT2), STAT6, and son of sevenless homologue 2 (SOS2). Furthermore, other predicted target genes imply a regulative impact on Hedgehog signalling via the wingless-type MMTV integration site family member 9B (WNT9B) and tumour protein p53 (TP53)-mediated signalling via BCL2-associated X protein (BAX). These results further support the hypothesis that pRCC2 is a distinct entity of RCC, which uses a unique set of signalling pathways.
Discussion
Several studies have recently demonstrated that miRNA expression profiling represents a useful tool for deciphering the genetic basis of malignant diseases. The miRNA expression profiles are highly reproducible among different patient cohorts, suggesting a possible application as diagnostic biomarkers (Schaefer et al, 2010; Faragalla et al, 2012; Wach et al, 2012). Similarly, miRNAs have been proposed as prognostic biomarkers in RCC (White et al, 2011b; Faragalla et al, 2012; Slaby et al, 2012). Here, we present the results of a comparative miRNA expression study with a special focus on the establishment of discriminative miRNA profiles between ccRCC and pRCC subtypes 1 and 2. One common limitation, when addressing pRCC subtypes, is that there are no validated immunohistochemical or molecular biomarkers available for distinguishing between the two subtypes. The method applied for routine pathology is the histomorphological characterisation according to the 2004 WHO classification system (Eble et al, 2004), which also was the basis for the development of our RCC classification method.
The results of our microarray analysis confirmed that RCC is a variety of different entities, each with its characteristic molecular pattern. Interestingly, matched tumour and normal tissue from the same RCC entity displayed a higher similarity in miRNA expression than normal tissue from different entities. It remains unclear whether this is due to pre-malignant changes in miRNA expression in normal tissue, or whether this reflects patient-specific individual miRNA expression profiles.
Recent studies made attempts at defining RCC tumours or tumour entities using miRNAs. One study used 35 miRNAs in a cluster analysis to correctly distinguish between the corresponding pairs of ccRCC and normal tissue (Juan et al, 2010). Another study used miRNA microarray expression data to define pairs of miRNAs that were used in a vote counting strategy (Youssef et al, 2011). Using six pairs of miRNAs, this strategy was able to distinguish (with a sensitivity of 97%) between RCC cases of different entities and normal tissue. In our analysis, a set of only four miRNAs could discriminate between tumour samples of different RCC entities and normal tissue with an accuracy of over 86%.
MicroRNAs that are differentially expressed between ccRCC and pRCC have been described. Petillo et al (2009) reported the differential expression of 27 miRNAs (9 primary transcripts and 18 mature miRNAs). Of the 18 mature miRNAs, 10 were identified by our microarray analysis as well. Youssef et al (2011) applied a vote counting strategy using 28 miRNAs to classify tumour samples into ccRCC, pRCC, chromophobe RCC and oncocytoma, with an overall accuracy of 87%. In addition, they developed a binary classification system using 11 miRNAs to distinguish between ccRCC and pRCC. Using the expression data of only two miRNAs, we were able to separate ccRCC from pRCC with an accuracy of >77%. Remarkably, of the 15 miRNAs that have been described as the most significantly deregulated among different RCC entities (Youssef et al, 2011), 9 were identified by our microarray analysis. To date, no attempt has been made to classify the pRCC subtypes using miRNAs. Using mRNA microarrays, it has been demonstrated that the top 100 deregulated mRNA transcripts were able to define patient subgroups, which were associated with the clinical outcome (Yang et al, 2005). In our analysis, only two miRNAs were able to discriminate the two pRCC subtypes with an accuracy of >86%. It should be noted that for the discovery set the normal tissue samples were derived from adjacent tissue in close proximity to the RCC lesion. Although the absence of any tumour cells was confirmed by pathological examination, we cannot formally exclude any type of cancer field effect. However, for the validation set, we sought to prevent this by preparing normal control tissue only from the opposite pole of the nephrectomy specimen.
We showed for the first time that expression values of only five miRNAs, measured by quantitative PCR, are able to classify the dignity or entity of the RCC samples with an accuracy comparable to microarray-based techniques. The most pronounced tumour-specific reduction was noted for miR-200c. This result is in line with several publications describing a pronounced downregulation of miR-200c in different RCC entities (Youssef et al, 2011). MiR-200c has been shown to target ZEB1 (Bracken et al, 2008), thereby influencing epithelial–mesenchymal transition (Bracken et al, 2009). An miRNA, which has been consistently reported to be downregulated in cancer, is miR-145. Overexpression of miR-145 causes a repression of c-MYC (Sachdeva et al, 2009), and an in vivo reduction in metastases (Peng et al, 2011). MiR-210 is commonly termed as the micromanager of the hypoxia pathway (Huang et al, 2010). MiR-210 expression is directly controlled by binding of Hif1α to a hypoxia-responsive element (HRE) in its proximal promoter (Huang et al, 2009). Tumour-specific upregulation of miR-210 has already been demonstrated in RCC cases (Juan et al, 2010); however, this study was primarily focused on ccRCC cases. For the first time, we have demonstrated that, in pRCC, miR-210 is expressed at significantly lower levels in pRCC2 than in pRCC1. Therefore, it is tempting to speculate that pRCC2, in contrast to ccRCC or pRCC1, might be less dependent on Hif1α/hypoxia-related signalling pathways. We also found that let-7c is essential for discriminating between pRCC1 and pRCC2. One prominent target gene of let-7c is MYC (Nadiminty et al, 2012). This result is in line with previous reports, which demonstrated overexpression of the MYC protein in high-grade pRCC2 (Furge et al, 2007), as well as an increased RNA expression of myc-induced nuclear antigen MINA (Yang et al, 2005).
Using GSEA, we found that the ABC family of transmembrane transporters might be influenced in all RCC entities. Among the predicted miRNA targets are MRPs MRP1 (ABCC1), MRP2 (ABCC2) and MRP8 (ABCC11). MRP1 is overexpressed in primary RCC cases (Walsh et al, 2009). The MRPs are able to efficiently export anticancer drugs, thereby contributing to the low chemotherapy response rate of RCC (Gamelin et al, 1999). The resistance profile of MRP1, MRP2 and MRP8 includes the chemotherapeutics methotrexate and cisplatin (Chen and Tiwari, 2011). This points towards the possibility that deregulated miRNAs contribute to RCC chemoresistance.
In summary, we could confirm that miRNA expression profiles are capable not only of distinguishing between tumour and normal tissue samples but also of classifying the entity of RCC cases with high accuracy. Furthermore, we identified miRNAs capable of distinguishing between the two distinct histological variants of pRCC. The characteristic miRNAs and potential target genes that distinguish between pRCC1 and pRCC2 give rise to the hypothesis that type 2 pRCCs are characterised by an MYC expression signature, whereas type 1 pRCCs are predominantly characterised by an Hif1α/hypoxia-related gene expression signature.
Acknowledgments
This work was supported by a grant from the ELAN-Fonds of the University Hospital Erlangen (Grant No. 09.11.11.1) to SW. We thank American Journal Experts for providing language editing service.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Backes C, Keller A, Kuentzer J, Kneissl B, Comtesse N, Elnakady YA, Muller R, Meese E, Lenhof HP. GeneTrail--advanced gene set enrichment analysis. Nucleic Acids Res. 2007;35 (Web Server issue:W186–W192. doi: 10.1093/nar/gkm323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken CP, Gregory PA, Khew-Goodall Y, Goodall GJ. The role of microRNAs in metastasis and epithelial-mesenchymal transition. Cell Mol Life Sci. 2009;66 (10:1682–1699. doi: 10.1007/s00018-009-8750-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68 (19:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- Chen ZS, Tiwari AK. Multidrug resistance proteins (MRPs/ABCCs) in cancer chemotherapy and genetic diseases. FEBS J. 2011;278 (18:3226–3245. doi: 10.1111/j.1742-4658.2011.08235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delahunt B, Eble JN. Papillary renal cell carcinoma: a clinicopathologic and immunohistochemical study of 105 tumors. Mod Pathol. 1997;10 (6:537–544. [PubMed] [Google Scholar]
- Eble JN, Sauter G, Epstein JI, Sesterhenn IA. Pathology and Genetics of Tumours of the Urinary System and Male Genital Organs. IARC Press: Lyon; 2004. [Google Scholar]
- Faragalla H, Youssef YM, Scorilas A, Khalil B, White NM, Mejia-Guerrero S, Khella H, Jewett MA, Evans A, Lichner Z, Bjarnason G, Sugar L, Attalah MI, Yousef GM. The clinical utility of miR-21 as a diagnostic and prognostic marker for renal cell carcinoma. J Mol Diagn. 2012;14 (4:385–392. doi: 10.1016/j.jmoldx.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Furge KA, Chen J, Koeman J, Swiatek P, Dykema K, Lucin K, Kahnoski R, Yang XJ, Teh BT. Detection of DNA copy number changes and oncogenic signaling abnormalities from gene expression data reveals MYC activation in high-grade papillary renal cell carcinoma. Cancer Res. 2007;67 (7:3171–3176. doi: 10.1158/0008-5472.CAN-06-4571. [DOI] [PubMed] [Google Scholar]
- Gamelin E, Mertins SD, Regis JT, Mickley L, Abati A, Worrell RA, Linehan WM, Bates SE. Intrinsic drug resistance in primary and metastatic renal cell carcinoma. J Urol. 1999;162 (1:217–224. doi: 10.1097/00005392-199907000-00071. [DOI] [PubMed] [Google Scholar]
- Gardie B, Remenieras A, Kattygnarath D, Bombled J, Lefevre S, Perrier-Trudova V, Rustin P, Barrois M, Slama A, Avril MF, Bessis D, Caron O, Caux F, Collignon P, Coupier I, Cremin C, Dollfus H, Dugast C, Escudier B, Faivre L, Field M, Gilbert-Dussardier B, Janin N, Leport Y, Leroux D, Lipsker D, Malthieu F, McGilliwray B, Maugard C, Mejean A, Mortemousque I, Plessis G, Poppe B, Pruvost-Balland C, Rooker S, Roume J, Soufir N, Steinraths M, Tan MH, Theodore C, Thomas L, Vabres P, Van Glabeke E, Meric JB, Verkarre V, Lenoir G, Joulin V, Deveaux S, Cusin V, Feunteun J, Teh BT, Bressac-de Paillerets B, Richard S. Novel FH mutations in families with hereditary leiomyomatosis and renal cell cancer (HLRCC) and patients with isolated type 2 papillary renal cell carcinoma. J Med Genet. 2011;48 (4:226–234. doi: 10.1136/jmg.2010.085068. [DOI] [PubMed] [Google Scholar]
- Huang X, Ding L, Bennewith KL, Tong RT, Welford SM, Ang KK, Story M, Le QT, Giaccia AJ. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell. 2009;35 (6:856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Le QT, Giaccia AJ. MiR-210--micromanager of the hypoxia pathway. Trends Mol Med. 2010;16 (5:230–237. doi: 10.1016/j.molmed.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolliffe IT. Principal Component Analysis. Springer Verlag: New York; 2002. [Google Scholar]
- Juan D, Alexe G, Antes T, Liu H, Madabhushi A, Delisi C, Ganesan S, Bhanot G, Liou LS. Identification of a microRNA panel for clear-cell kidney cancer. Urology. 2010;75 (4:835–841. doi: 10.1016/j.urology.2009.10.033. [DOI] [PubMed] [Google Scholar]
- Jung M, Mollenkopf HJ, Grimm C, Wagner I, Albrecht M, Waller T, Pilarsky C, Johannsen M, Stephan C, Lehrach H, Nietfeld W, Rudel T, Jung K, Kristiansen G. MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. J Cell Mol Med. 2009;13 (9B:3918–3928. doi: 10.1111/j.1582-4934.2009.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Leidinger P, Bauer A, Elsharawy A, Haas J, Backes C, Wendschlag A, Giese N, Tjaden C, Ott K, Werner J, Hackert T, Ruprecht K, Huwer H, Huebers J, Jacobs G, Rosenstiel P, Dommisch H, Schaefer A, Muller-Quernheim J, Wullich B, Keck B, Graf N, Reichrath J, Vogel B, Nebel A, Jager SU, Staehler P, Amarantos I, Boisguerin V, Staehler C, Beier M, Scheffler M, Buchler MW, Wischhusen J, Haeusler SF, Dietl J, Hofmann S, Lenhof HP, Schreiber S, Katus HA, Rottbauer W, Meder B, Hoheisel JD, Franke A, Meese E. Toward the blood-borne miRNome of human diseases. Nat Methods. 2011;8 (10:841–843. doi: 10.1038/nmeth.1682. [DOI] [PubMed] [Google Scholar]
- Ljungberg B, Campbell SC, Choi HY, Jacqmin D, Lee JE, Weikert S, Kiemeney LA. The epidemiology of renal cell carcinoma. Eur Urol. 2011;60 (4:615–621. doi: 10.1016/j.eururo.2011.06.049. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435 (7043:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Lubensky IA, Schmidt L, Zhuang Z, Weirich G, Pack S, Zambrano N, Walther MM, Choyke P, Linehan WM, Zbar B. Hereditary and sporadic papillary renal carcinomas with c-met mutations share a distinct morphological phenotype. Am J Pathol. 1999;155 (2:517–526. doi: 10.1016/S0002-9440(10)65147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3' UTR. Proc Natl Acad Sci USA. 2007;104 (23:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G. miRNAs get an early start on translational silencing. Cell. 2007;131 (1:25–28. doi: 10.1016/j.cell.2007.09.021. [DOI] [PubMed] [Google Scholar]
- Nadiminty N, Tummala R, Lou W, Zhu Y, Zhang J, Chen X, eVere White RW, Kung HJ, Evans CP, Gao AC. MicroRNA let-7c suppresses androgen receptor expression and activity via regulation of Myc expression in prostate cancer cells. J Biol Chem. 2012;287 (2:1527–1537. doi: 10.1074/jbc.M111.278705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson EC, Evans CP, Lara PN., Jr Renal cell carcinoma: current status and emerging therapies. Cancer Treat Rev. 2007;33 (3:299–313. doi: 10.1016/j.ctrv.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Osanto S, Qin Y, Buermans HP, Berkers J, Lerut E, Goeman JJ, van Poppel H. Genome-wide microRNA expression analysis of clear cell renal cell carcinoma by next generation deep sequencing. PLoS ONE. 2012;7 (6:e38298. doi: 10.1371/journal.pone.0038298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Guo W, Liu T, Wang X, Tu X, Xiong D, Chen S, Lai Y, Du H, Chen G, Liu G, Tang Y, Huang S, Zou X. Identification of miRs-143 and -145 that is associated with bone metastasis of prostate cancer and involved in the regulation of EMT. PLoS ONE. 2011;6 (5:e20341. doi: 10.1371/journal.pone.0020341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petillo D, Kort EJ, Anema J, Furge KA, Yang XJ, Teh BT. MicroRNA profiling of human kidney cancer subtypes. Int J Oncol. 2009;35 (1:109–114. doi: 10.3892/ijo_00000318. [DOI] [PubMed] [Google Scholar]
- Pignot G, Elie C, Conquy S, Vieillefond A, Flam T, Zerbib M, Debre B, Amsellem-Ouazana D. Survival analysis of 130 patients with papillary renal cell carcinoma: prognostic utility of type 1 and type 2 subclassification. Urology. 2007;69 (2:230–235. doi: 10.1016/j.urology.2006.09.052. [DOI] [PubMed] [Google Scholar]
- Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA. 2009;106 (9:3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, Jentzmik F, Miller K, Lein M, Kristiansen G, Jung K. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126 (5:1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3 (6:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Slaby O, Redova M, Poprach A, Nekvindova J, Iliev R, Radova L, Lakomy R, Svoboda M, Vyzula R. Identification of MicroRNAs associated with early relapse after nephrectomy in renal cell carcinoma patients. Genes Chromosomes Cancer. 2012;51 (7:707–716. doi: 10.1002/gcc.21957. [DOI] [PubMed] [Google Scholar]
- Wach S, Nolte E, Szczyrba J, Stohr R, Hartmann A, Orntoft T, Dyrskjot L, Eltze E, Wieland W, Keck B, Ekici AB, Grasser F, Wullich B. MicroRNA profiles of prostate carcinoma detected by multiplatform microRNA screening. Int J Cancer. 2012;130 (3:611–621. doi: 10.1002/ijc.26064. [DOI] [PubMed] [Google Scholar]
- Walsh N, Larkin A, Kennedy S, Connolly L. Expression of multidrug resistance markers ABCB1 (MDR-1/P-gp) and ABCC1 (MRP-1) in renal cell carcinoma. BMC Urol. 2009;9:6. doi: 10.1186/1471-2490-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White NM, Bao TT, Grigull J, Youssef YM, Girgis A, Diamandis M, Fatoohi E, Metias M, Honey RJ, Stewart R, Pace KT, Bjarnason GA, Yousef GM, Ballot J, Ooi W, Gullo G, Crown J, Clynes M, O'Driscoll L. miRNA profiling for clear cell renal cell carcinoma: biomarker discovery and identification of potential controls and consequences of miRNA dysregulation. J Urol. 2011a;186 (3:1077–1083. doi: 10.1016/j.juro.2011.04.110. [DOI] [PubMed] [Google Scholar]
- White NM, Khella HW, Grigull J, Adzovic S, Youssef YM, Honey RJ, Stewart R, Pace KT, Bjarnason GA, Jewett MA, Evans AJ, Gabril M, Yousef GM. miRNA profiling in metastatic renal cell carcinoma reveals a tumour-suppressor effect for miR-215. Br J Cancer. 2011b;105 (11:1741–1749. doi: 10.1038/bjc.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotschofsky Z, Meyer HA, Jung M, Fendler A, Wagner I, Stephan C, Busch J, Erbersdobler A, Disch AC, Mollenkopf HJ, Jung K. Reference genes for the relative quantification of microRNAs in renal cell carcinomas and their metastases. Anal Biochem. 2011;417 (2:233–241. doi: 10.1016/j.ab.2011.06.009. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Tan MH, Kim HL, Ditlev JA, Betten MW, Png CE, Kort EJ, Futami K, Furge KA, Takahashi M, Kanayama HO, Tan PH, Teh BS, Luan C, Wang K, Pins M, Tretiakova M, Anema J, Kahnoski R, Nicol T, Stadler W, Vogelzang NG, Amato R, Seligson D, Figlin R, Belldegrun A, Rogers CG, Teh BT. A molecular classification of papillary renal cell carcinoma. Cancer Res. 2005;65 (13:5628–5637. doi: 10.1158/0008-5472.CAN-05-0533. [DOI] [PubMed] [Google Scholar]
- Youssef YM, White NM, Grigull J, Krizova A, Samy C, Mejia-Guerrero S, Evans A, Yousef GM. Accurate molecular classification of kidney cancer subtypes using microRNA signature. Eur Urol. 2011;59 (5:721–730. doi: 10.1016/j.eururo.2011.01.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.