Abstract
Background:
Dysregulation of the Notch pathway has been identified to play an important role in the development and progression of colorectal cancer (CRC). In this study, we used a patient-derived CRC explant model to investigate the efficacy of the clinical γ-secretase inhibitor (GSI) PF-03084014.
Methods:
A total of 16 CRC explants were treated with PF-03084014. Knockdown of RBPjκ gene was used to determine the specificity of PF-03084014. Evaluation of the Notch and Wnt pathways in CRC explant tumours was performed by gene array and immunoblotting.
Results:
We identified a subset of CRC tumours that exhibited elevations of the Notch and Wnt pathways sensitive to PF-03084014. Treatment with the GSI resulted in a significant reduction in cleaved Notch, Axin2 (Wnt-dependent gene) and active β-catenin. In addition, knockdown of the RBPjκ gene showed that PF-03084014 has specificity for the Notch pathway in an HCT116 cell line xenograft model. Finally, an increase in apoptosis was observed in CRC001- and CRC021-sensitive tumours.
Conclusion:
This study provides evidence that inhibition of γ-secretase may be beneficial in a subset of patients with elevated levels of the Wnt and Notch pathways.
Keywords: colorectal cancer, Notch, γ-secretase inhibitor, Wnt
Colorectal cancer (CRC) is the second most common cancer in the United States and the third leading cause of cancer-related death (Siegel et al, 2012). Although earlier stages of CRC are highly curable, therapies in advanced and metastatic disease overall have been ineffective at reducing 5-year survival rates. Recent drug development has focused on targeting developmental pathways such as the Notch pathway as a potential therapy for CRC. The Notch pathway has a vital role in normal colon homeostasis by maintaining stem/progenitor cells and by regulating the differentiation of goblet cells (Fre et al, 2005; Radtke et al, 2006). Although mutations in the Notch receptors have not been described in CRC, aberrant activation of the Notch pathway has been shown to facilitate the development and progression of CRC (van Es et al, 2005; Sikandar et al, 2010; Sonoshita et al, 2011). For instance, in an intestinal adenomatous polyposis coli (APC) −/− mouse model, components of the Notch pathway were demonstrated to be upregulated in the crypts of the intestine; treatment with a γ-secretase inhibitor (GSI) resulted in the conversion of proliferative cells into more differentiated goblet cells resulting in a reduction in tumour burden. In addition, the Notch pathway has been described to be important in CRC progression. A decrease in AES (Grg5) resulted in enhanced Notch activity and tumour invasion that was prevented by inhibition of the Notch pathway (Sonoshita et al, 2011).
The Notch pathway is activated through the interaction of a Notch receptor (1, 2, 3, or 4) and ligand (DLL1, DLL3, DLL4, JAG-1, and JAG-2) between adjacent cells. This contact results in a series of proteolytic events that lead to the cleavage of the notch intracellular domain (NICD) by the γ-secretase complex and subsequent translocation of NICD into the nucleus and the transcription of Notch target genes (Pannuti et al, 2010). In CRC, activation of the Notch pathway appears to be oncogenic by regulating many signalling pathways involved in enhancing cellular survival and angiogenesis. In particular, Notch has been shown to augment cell-cycle progression and survival by activating the PI3K/AKT pathway (Nair et al, 2003; Sade et al, 2004) and transcriptionally inducing the expression of Hes-1, cyclin D1 and c-Myc (Ronchini and Capobianco, 2001; van Es et al, 2005; Zhang et al, 2012). The Notch pathway has also been shown to modulate tumour angiogenesis and is widely expressed in endothelial cells (Noguera-Troise et al, 2006). Specifically, VEGF from tumour cells induces DLL4 expression on endothelial cells and Notch signalling that results in an increase in vessel formation (Noguera-Troise et al, 2006).
Since the Notch pathway appears to be important in the development and progression of CRC as well as other cancers, drug development of GSIs is currently being evaluated in early phase clinical trials. PF-03084014 is a potent and selective inhibitor of γ-secretase activity and is currently in phase I clinical development. Unlike other agents in this class, PF-03084014 has favourable properties and daily dosing is feasible (Messersmith et al, 2011). In a preclinical model of T-cell acute lymphoblastic leukemia (T-ALL), this inhibitor has shown to induce cell growth arrest by hindering cell-cycle progression and enhancing apoptosis (Wei et al, 2010). In breast cancer cell line xenografts, PF-03084014 altered endothelial cell tube formation and had anti-tumour and anti-angiogenic activity (Zhang et al, 2012). However, the efficacy of PF-03084014 in CRC has not been examined. Therefore, we evaluated the effects of PF-03084014 in our patient-derived CRC explant model and show that a subset of tumours that exhibit increased levels of the Notch and Wnt pathways are sensitive to PF-03084014.
Materials and methods
CRC explant xenograft model
Patient-derived colorectal adenocarcinoma tumour specimens were obtained from consenting patients at the University of Colorado Hospital in accordance with protocols approved by the Colorado Multiple Institutional Review Board. Four- to six week-old female athymic (nu+/nu+) mice were obtained from Harlan Laboratories (Washington, DC, USA) under an approved research protocol by the Institutional Animal Care and Use Committee. The tumour pieces were implanted in mice and expansion of the F1–F3 generations was carried out as previously described (Rubio-Viqueira et al, 2006; Dangles-Marie et al, 2007). Tumours were expanded in the left and right flanks of 5–6 mice (10 evaluable tumours per group). Mice were randomised into vehicle or PF-03084014 (GSI) groups when tumour volumes reached ∼200 mm3. Mice were treated daily with PF-03084014 (125 mg kg−1 – BID) by oral gavage for 28 days. Mice were monitored daily for signs of toxicity, and the tumour size was evaluated twice per week by caliper measurements using the following formula: tumour volume=(length × width2) × 0.52.
Cell lines and culture
Human CRC cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in RPMI supplemented with 10% fetal bovine serum, 1% non-essential amino acids, and 1% penicillin/streptomycin, and were maintained at 37 °C under an atmosphere containing 5% CO2. The cells were routinely screened for the presence of Mycoplasma (MycoAlert; Cambrex BioScience, Charles City, IA, USA) and were exposed to PF-03084014 (5–0.08 μM) when they reached ∼70% confluence. All cell lines were tested and authenticated in the University of Colorado Cancer Center DNA Sequencing and Analysis Core. The CRC cell line DNA was tested using the Profiler Plus kit (Applied Biosystems, Foster City, CA, USA). The data obtained were compared with American Type Culture Collection data to ensure the cell lines have not changed.
Immunoblotting
Tumour tissues (50–75 mg per mouse) were minced on ice and homogenised using a Dounce homogenizer (Fisher Scientific, Pittsburgh, PA, USA) and centrifuged at 16 000 g at 4 °C for 10 min. The total protein in samples was determined using the Bio-Rad Dc Protein Assay kit, Bio-Rad, Hercules, CA, USA. Forty micrograms of sample were electrophoresed on 4–12% Bis-Tris precast gels (Life Technologies, Grand Island, NY, USA). After electrotransfer onto Immobilon-P membranes (Millipore, Billerica, MA, USA), membranes were blocked at room temperature with TBS (10 mmol l−1 Tris–HCl (pH 7.5), 0.5 mol l−1 NaCl, and 0.1% (v/v) Tween-20) containing 5% non-fat milk (Bio-Rad) for 1 h. Cleaved Notch, active β-catenin, β-catenin, Axin2, cleaved caspase 3, cleaved PARP, p-p65, BCLxL, and actin primary antibodies (Cell Signaling Technologies, Danvers, MA, USA) were diluted at 1 : 1000 in TBST containing 5% protease-free bovine serum albumin (Sigma-Aldrich, St Louis, MO, USA), and the membranes were incubated overnight at 4 °C with rocking. After washing three times with TBST, the membranes were incubated for 1 h at room temperature with anti-mouse IgG horseradish peroxidase-conjugated antibody at a final dilution of 1 : 50 000 in TBST. After washing three times with TBST, bound antibodies were detected by enhanced chemiluminescence (Millipore).
RBPjκ shRNA knockdown and xenograft
RBPjκ (TF302060) shRNA and pRS vector-negative control (scramble) plasmids (TR20003) were purchased from OriGene (Rockville, MD, USA). Stable clones were generated by transfecting HCT116 cell line in six-well plate with 1 μg of each of the shRNA plasmids using Fugene 6 (Roche, Nutley, NJ, USA) according to the manufacturer's recommendations. Seventy-two hours after transfection, the cells were placed under selection with 2.5 μg ml−1 puromycin, splitting 1 : 5 when the cells reached confluency. Multiple clones from the same transfection were pooled and grown under puromycin selection. Successful knockdown of specific genes and gene products was confirmed by semi-quantitative reverse transcription PCR. HCT116 parental, scramble, and RBPjκ knockdown were injected into the left and right flanks of 4- to 6-week-old female athymic (nu+/nu+) mice (Harlan Laboratories). Mice were randomised into the treatment group (PF-03084014) or vehicle group when tumour volumes reached ∼200 mm3. Mice were treated daily with PF-03084014 (125 mg kg−1 – BID) or vehicle by oral gavage. Mice were monitored daily for signs of toxicity, and the tumour size was evaluated twice per week by caliper measurements using the following formula: tumour volume=(length × width2) × 0.52.
RT–PCR
RT–PCR was used to evaluate the knockdown of RBPjκ for the shRNA experiments. Total RNA was extracted using the RNeasy Mini kit (Qiagen, Valencia, CA, USA). cDNA was synthesised using the Applied Biosystems high capacity cDNA reverse transcription kit, following the manufacturer's instructions. Validated and pre-designed primer/probes for RBPjκ and housekeeping gene(s) were purchased from Applied Biosystems. Samples were amplified using the ABI Step One Plus RT-PCR system (Applied Biosystem). Relative expression of the mRNA analysed was estimated using the formula: 2−ΔCT, where ΔCT=CT (mRNA)−CT (Housekeeper).
Notch and Wnt pathway analysis by gene array
Total RNA from the CRC explants was extracted using RNAeasy kit (Qiagen) and profiled by Affymetrix Human Gene 1.0 ST microarray (Affymetrix, Santa Clara, CA, USA). Microarray gene expression preparation and processing procedure were done as recommended by the manufacturer (Affymetrix, Inc.). Raw expression values were extracted and normalised by the Affymetrix Power Tools based on Robust Multiarray Average (RMA) approach. Multiple probe sets representing the same gene were collapsed by the maximum value. To analyse the pathway enriched in the sensitive vs resistant explants, we used the GSEA (gene set enrichment analysis) software version 2.0.6 obtained from the Broad Institute (http://www.broad.mit.edu/gsea) (Subramanian et al, 2005). We used the pathways defined by the Kyoto Encyclopedia of Genes and Genomes (KEGG) as the gene set in this study (Kanehisa et al, 2008). Gene set permutations were performed 1000 times for each analysis. We used the nominal P-value and Normalised Enrichment Score (NES) obtained from GSEA to sort the pathways enriched in sensitive explants.
Statistical analysis
An unpaired Student's t-test was used to determine whether the means between control and PF-03084014 were significant at end of treatment (∼28 days) and for comparison of differences between control and PF-03084014 in cleaved Notch, Axin2, and active β-catenin. The differences were considered as significant when the P-value was <0.05. All error bars are represented as the s.e.m.
Results
The effects of PF-03084014 on tumour growth in a CRC patient-derived explant model
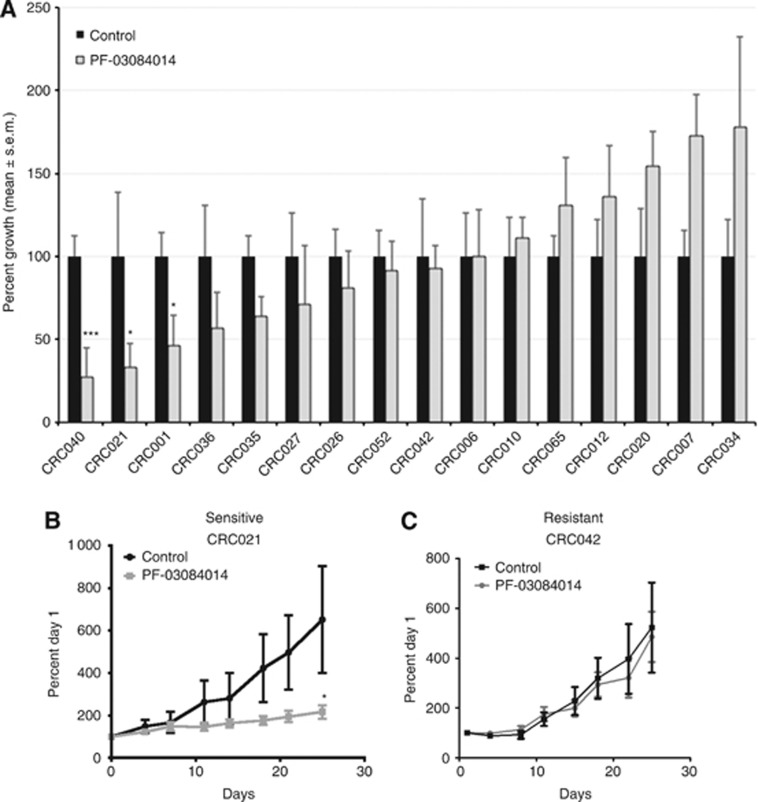
The efficacy of PF-03084014 (a GSI) on tumour growth was evaluated on 16 CRC patient-derived xenografts. Table 1 shows the type of tumour (colon or rectal), stage of disease, previous treatments, and common mutations of the CRC explant patients. As shown in Figure 1A, 3 out of 16 CRC explants (CRC040, CRC021, and CRC001) showed sensitivity (static effects, but no regression) to PF-03084014. A tumour growth index (TGI)⩽50% was considered as sensitive and TGI>50% as resistant. Figures 1B and C are representative graphs of a sensitive explant 021 (B) and resistant explant 042 (C). In addition to treating CRC explants in vivo, we also evaluated 23 CRC cell lines in vitro. As shown in Supplementary Figure 1, PF-03084014 had very little activity in vitro. The LS123 was the only cell line that showed some growth inhibitor effects to γ-secretase inhibition; however, the effects on proliferation never reached an IC50.
Table 1. Patient characteristics and mutational status of CRC explants.
| CRC explant | Primary | Stage | Age at consent | Previous treatment | KRAS | NRAS | PIK3CA | APC | CTNNB1 | BRAF | TP53 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CRC 001 |
Colon |
IV |
69 |
FOLFOX+bevacizumab |
G12D |
WT |
WT |
Mut |
WT |
WT |
WT |
| CRC 006 |
Colon |
IV |
42 |
Capecitabine+ Radiation, FOLFOX+bevacizumab, Irinotecan+cetuximab |
G12D |
WT |
WT |
Mut |
WT |
WT |
WT |
| CRC 007 |
Colon |
II |
47 |
None |
G13D |
WT |
WT |
WT |
Mut |
WT |
Mut |
| CRC 010 |
Rectal |
III |
52 |
None |
WT |
WT |
WT |
WT |
WT |
WT |
Mut |
| CRC 012 |
Rectal |
IV |
56 |
Capecitabine+ Oxaliplatin+bevacizumab |
G12V |
WT |
WT |
WT |
WT |
WT |
WT |
| CRC 020 |
Colon |
IV |
43 |
FOLFOX+bevacizumab |
WT |
G12S, Q61K |
Mut |
Mut |
WT |
WT |
WT |
| CRC 021 |
Rectal |
II |
72 |
None |
G12D |
WT |
WT |
WT |
WT |
WT |
Mut |
| CRC 026 |
Colon |
IV |
48 |
FOLFOX and bevacizumab |
WT |
Q61K |
WT |
Mut |
WT |
WT |
WT |
| CRC 027 |
Colon |
IV |
56 |
None |
G12A |
WT |
WT |
Mut |
WT |
WT |
Mut |
| CRC 034 |
Colon |
IV |
59 |
None |
WT |
WT |
WT |
WT |
WT |
WT |
WT |
| CRC 035 |
Colon |
IV |
44 |
FOLFOX |
G12S |
WT |
WT |
WT |
WT |
WT |
WT |
| CRC 036 |
Rectal |
IV |
42 |
Capecitabine+Radiation, FOLFOX+bevacizumab, Irinotecan+cetuximab |
G12A |
WT |
WT |
Mut |
WT |
WT |
WT |
| CRC 040 |
Rectal |
II |
63 |
None |
G12V |
WT |
Mut |
Mut |
WT |
WT |
WT |
| CRC 042 |
Rectal |
II |
73 |
FOLFIRI+bevacizumab |
G13D |
WT |
WT |
WT |
Mut |
WT |
Mut |
| CRC 052 |
Colon |
II |
51 |
None |
G12V |
WT |
WT |
Mut |
WT |
WT |
WT |
| CRC 065 | Colon | IV | 66 | FOLFIRI, FOLFOX | G12V | WT | WT | Mut | WT | WT | WT |
Figure 1.
PF-03084014 effects on tumour growth in CRC explants. (A) Sixteen CRC explants were treated with PF-03084014 125 mg kg−1 per day BID by oral gavage for 28 days. Tumour size was evaluated twice per week by caliper measurements using the formula: tumour volume=(length × width2) × 0.52. Tumour growth index (TGI) was calculated by relative tumour growth of treated mice divided by relative tumour growth of control mice × 100. Cases with a TGI of <50% were considered as sensitive, and TGI of >50% were considered as resistant to PF-03084014. Three xenografts (CRC001, CRC021, and CRC040) were sensitive to PF-03084014 (TGI⩽50%) and thirteen xenografts were resistant to PF-03084014 (TGI>50%). Columns, mean (n=8–10 tumours per group); bars, s.e. *, significance (*P<0.05), (***P<0.001) compared with vehicle-treated tumours. (B) A representative figure of a sensitive explant CRC021 and (C) resistant explant CRC042.
PF-03084014 is a specific inhibitor of the Notch pathway
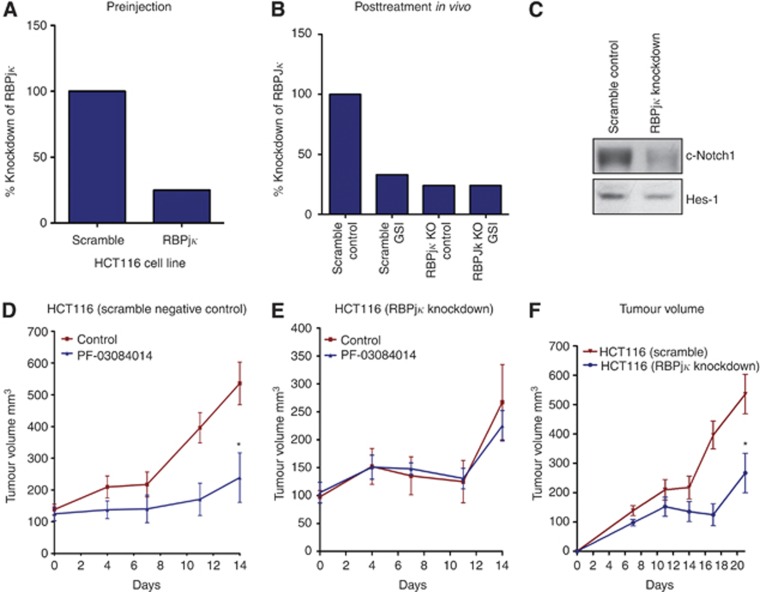
To assess the specificity of PF-03084014 on Notch pathway inhibition and tumour growth, we stably transfected the HCT116 cell line with shRNA to RBPjκ. RBPjκ is an essential transcription factor that binds intracellular cleaved Notch in the nucleus, ultimately leading to the transcription of Notch target genes. Before injecting in mice, we evaluated the gene expression of RBPjκ in the HCT116 cell line by RT–PCR. Gene expression of RBPjκ was decreased by 75% before injection (Figure 2A). In addition, examination of RBPjκ in xenograft tumours at the end of treatment showed a 76% reduction in RBPjκ in both the RBPjκ knockdown control and RBPjκ knockdown treated with GSI (Figure 2B). Furthermore, knockdown of RBPjκ resulted in a decrease in the notch pathway evidenced by a decrease in cleaved Notch1 and Hes-1 (Figure 2C). We next evaluated treatment effects of PF-03084014 on tumour growth in HCT116 scramble negative control and HCT116 RBPjκ knockdown cell lines in an in vivo xenograft model. PF-03084014 treatment significantly decreased the growth of HCT116 scramble group (Figure 2D). Of note, a significant reduction in tumour growth was also seen in the HCT116 parental cell line (data not shown). In contrast, PF-03084014 did not have any additional effects on growth of the RBPjκ knockdown group (Figure 2E). Furthermore, knockdown of RBPjκ in the HCT116 cell line resulted in a significant decrease (P<0.05) in growth when compared with the HCT116 scramble (Figure 2F). As expected, there was no difference in tumour growth between the scramble control and HCT116 control cell line (data not shown).
Figure 2.
Investigation of PF-03084014 as a specific inhibitor of the Notch pathway. (A) Evaluation of RBPjκ knockdown before injection and (B) after treatment of tumours in a xenograft model showed a 75% and 76% knockdown of RBPjκ, respectively. (C) RBPjκ knockdown reduced cleaved Notch1 activity and Hes-1 in xenograft tumours compared with scramble control. PF-03084014 treatment effects on HCT116 scramble control (D) and HCT116 RBPjκ knockdown (E) were evaluated in an in vivo xenograft model. Treatment significantly decreased the growth of HCT116 parental (data not shown) and scramble groups (D). In contrast, PF-03084014 did not have any additional anti-proliferative effects in the RBPjκ knockdown group (E). Knockdown of RBPjκ significantly decreased growth of the HCT116 cell line when compared with HCT116 scramble control (F). Columns, mean (n=8–10 tumours per group); bars, s.e. *, significance (*P<0.05).
The Notch pathway is upregulated in sensitive tumours and PF-03084014 treatment reduces Notch activity
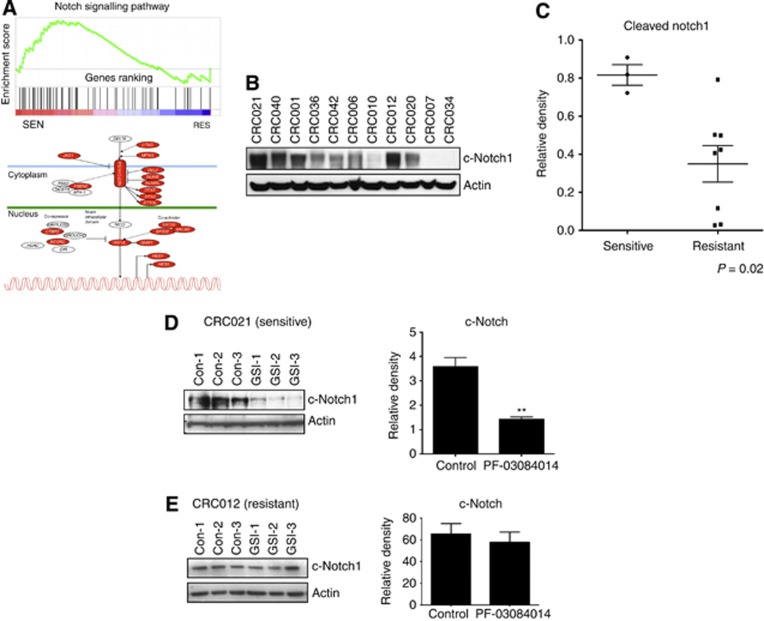
We next compared the Notch pathway between the three sensitive (CRC040, 021, and 001) and three most resistant explants (CRC020, 007, and 034) by gene array and KEGG pathway analysis to determine if tumours sensitive to PF-03084014 had elevated levels of the Notch pathway. As shown in Figure 3A, many components of the Notch pathway such as Notch receptors (Notch1, 3, and 4), the ligand JAG1 and co-activators were upregulated in the sensitive explants when compared with resistant explants. In addition, as shown in Figure 3B and C, cleaved Notch1 levels were significantly elevated in the GSI-sensitive tumours. Inhibition of γ-secretase with PF-03084014 reduced levels of cleaved Notch in the sensitive tumours CRC021 (D) CRC001 and CRC040 (Supplementary Figure 2), whereas no difference was seen in the CRC012-resistant explant (Figure 3E).
Figure 3.
Notch pathway analysis between sensitive (CRC040, 021, and 001) vs resistant tumours (CRC020, 007, and 034). (A) KEGG pathway analysis of the Notch pathway shows an increase in many components of the Notch pathway in sensitive tumours when compared with resistant tumours. Red indicates elevated gene expression. (B) Baseline levels of cleaved Notch1 are elevated in sensitive tumours compared with resistant tumours. (C) Densitometry of cleaved Notch1/Actin ratio showed a significant increase in sensitive tumours compared with resistant tumours (P<0.02). (D) PF-03084014 treatment decreased cleaved Notch activity in the sensitive tumour CRC021 (n=3 mice/tumour per group for control and GSI treated). Examination of cleaved Notch by densitometry showed a significant (*P<0.05) decrease with PF-03084014 treatment. (E) No treatment effects were observed in the resistant tumour CRC012.
PF-03084014-sensitive tumours exhibit elevations of the WNT pathway and treatment reduces Wnt signalling
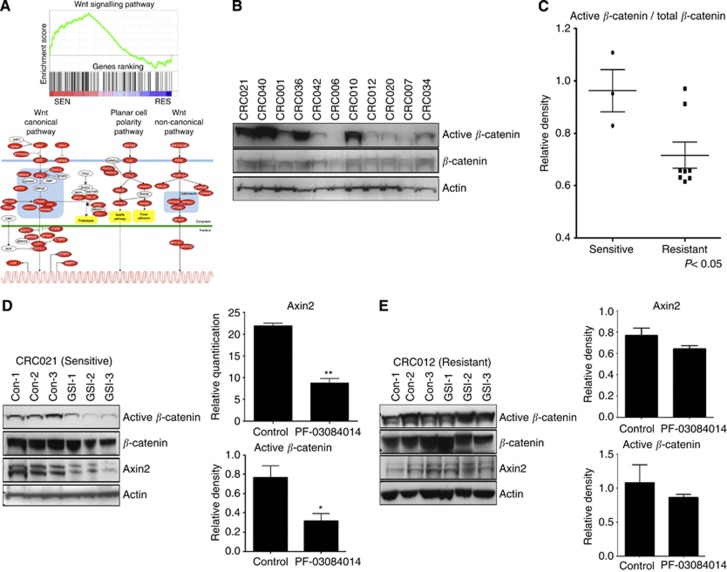
Interactions between the Notch and Wnt pathways have been described in CRC (Rodilla et al, 2009; Kwon et al, 2011). Therefore, we also examined gene expression levels of the Wnt pathway between sensitive and resistant tumours. KEGG analysis of the Wnt pathway revealed an increase in many components of the canonical Wnt pathways in sensitive tumours when compared with resistant tumours (Figure 4A). In addition, baseline levels of active β-catenin were significantly elevated in the sensitive tumours CRC001, CRC021, and CRC040 when compared with resistant tumours, confirming that the Wnt pathway is elevated in sensitive tumours (Figure 4B and C). We next were interested in determining whether PF-03084014 modulated the activation of the canonical Wnt pathway. As shown in Figure 4D, PF-03084014 resulted in a significant decrease in both active-β catenin and Axin2 (a Wnt-dependent gene) in the sensitive CRC021 explant. In contrast, no treatment effects on active β-catenin and Axin2 were seen in the resistant CRC012 explant (Figure 4E).
Figure 4.
Wnt pathway analysis between sensitive vs resistant tumours. (A) KEGG pathway analysis of the Wnt pathway shows an increase in many components of the canonical Wnt pathway in sensitive tumours when compared with resistant tumours. Red indicates elevated gene expression. (B) Baseline levels of active β-catenin are elevated in sensitive tumours compared with resistant tumours. (C) Densitometry of active β-catenin/β-catenin ratio showed a significant increase in sensitive tumours compared with resistant tumours (P<0.05). (D) PF-03084014 treatment decreased active β-catenin and Axin2 levels in the sensitive tumour CRC021. (E) No treatment effects were observed in the resistant tumour CRC012.
PF-03084014 induces an increase in apoptosis by decreasing the phosphorylation of p65 and NF-κB-dependent transcription of BCLxL
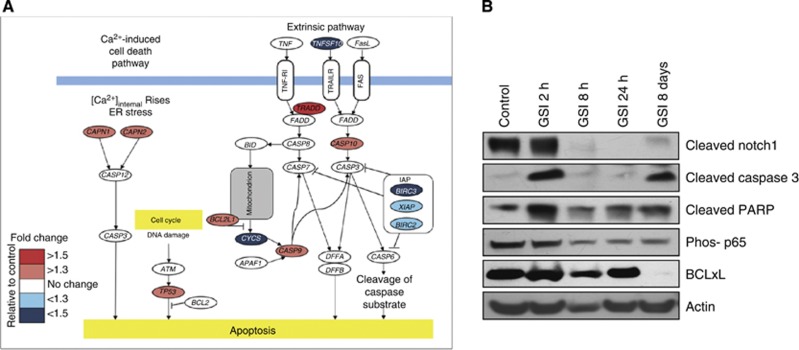
We investigated the effects of PF-03084014 on apoptosis in the sensitive explants CRC001 and CRC021 by gene array and western blot, respectively. As shown in Figure 5A, treatment with PF-03084014 increased the gene expression of several different apoptotic genes 3 days after treatment with PF-03084014 in CRC001. In addition, PF-03084014 treatment induced an increase in cleaved caspase 3 and PARP at 2 h and 8 days post treatment. A decrease in the phosphorylation p65 (a subunit of NF-κB) was observed at 8 h, 24 h, and 8 days post PF-03084014 treatment, while BCLxL (a NF-κB regulated gene) was significantly decreased 8 days post treatment.
Figure 5.
Assessment of PF-03084014 on apoptosis. (A) Post gene array 3 days after PF-03084014 treatment on gene expression of apoptotic genes in CRC001-sensitive explant. (B) Western blot analysis of cleaved Notch1, cleaved caspase 3, cleaved PARP, phos p65, and BCLxL in CRC021 2 h, 8 h, 24 h and 8 days after treatment with PF-03084014.
Discussion
In this study, we set out to determine the efficacy of PF-03084014, a clinical GSI, in a preclinical patient-derived CRC explant model. Herein, we show that inhibition of Notch signalling is effective only in a small subset of patient tumours. Of note, the effects of PF-03084014 on proliferation on 23 CRC cell lines in vitro were minimal, suggesting that the tumour microenvironment (stroma/angiogenesis) in vivo may be essential to see anti-proliferative effects. These results are consistent with a study by Zhang et al (2012) showing a lack of in vitro activity with this compound in 33 of 35 breast cancer cell lines. In our CRC explant model, three (CRC001, CRC021, and CRC040) out of sixteen tumours (∼19%) responded to γ-secretase inhibition. We examined whether a common mutation seen in CRC was associated with sensitivity to GSI; however, no association between mutational status (KRAS, PIK3CA, APC, CTNNB1, BRAF, or p53) and sensitivity to PF-03084014 was identified. A study by Fischer et al (2011) showed anti-tumour effects in a preclinical early passage CRC xenograft model using a DLL-4 antibody. In particular, they found a significant reduction in tumour growth in four out of six DLL-4-treated xenografts. Together, this shows that dysregulation of the Notch pathway has a role in tumorigenesis in a subset of CRC tumours.
RBPjκ is an essential transcription factor involved in the activation of the Notch pathway (Nam et al, 2006; Friedmann et al, 2008; Kopan and Ilagan, 2009). Similarly to treatment with a GSI, genetic manipulation of RBPjκ also resulted in the conversion of proliferative cells into post mitotic cells in an APC−/− mouse model (van Es et al, 2005). Since RBPjκ is directly involved in Notch-dependent activation, we stably transfected shRNA to knockdown RBPjκ in the HCT116 cell line to examine the specificity of PF-03084014 on the Notch pathway in a xenograft model. We demonstrated that treatment with PF-03084014 had similar anti-tumour activity on the HCT116 parental and scramble groups when compared with the RBPjκ untreated group. In addition, treatment of the HCT116 RBPjκ group did not provide any additional anti-tumour effects. These results imply that growth of the HCT116 cell line is partially dependent on the Notch pathway and PF-03084014 specifically targets the Notch pathway. We cannot rule out, however, that inhibition of other γ-secretase targets, such as CD44, EpCAM, and ADAM10 (Kopan and Ilagan, 2004; Maetzel et al, 2009; Tousseyn et al, 2009) may have contributed to anti-tumour activity independent of Notch in the CRC explants.
Since we identified differences in sensitivity to PF-03084014 in our CRC explant model, we investigated whether the Notch pathway is upregulated in tumours that responded to treatment compared with resistant tumours. Recently, in a breast cancer preclinical model, PF-03084014 had significant anti-tumour activity (>50% of tumours responded to treatment) in tumours that exhibited significant increases in Notch target genes (Zhang et al, 2012). Utilising baseline gene array, we profiled the Notch pathway by KEGG and observed that sensitive tumours had elevated levels of many components of the Notch pathway when compared with resistant tumours. In addition, we demonstrated that activated cleaved Notch1 levels were significantly increased in sensitive tumours, confirming the gene array analysis. We also showed that PF-03084014 decreases the Notch pathway in the sensitive tumour CRC021 as evidenced by a significant reduction in cleaved Notch activity. These results suggest that dysregulation of the Notch pathway may be important in driving tumour growth in PF-03084014-sensitive tumours. Since, there are many components of the Notch pathway and we used a GSI in this study, it remains unknown whether a particular Notch receptor (1, 2, 3, or 4) or ligand (DLL1, DLL3, DLL4, JAG-1, or JAG-2) or a combination of them are culpable in driving tumour growth in the sensitive explants. For instance, the Notch1 receptor has been identified to be associated with poorly differentiated tumours and worse survival in CRC (Chu et al, 2010; Zhang et al, 2010). In addition, DLL4 has been described to modulate endothelial sprout and angiogenesis in CRC (Noguera-Troise et al, 2006). It is likely that newer therapeutics targeting the individual receptors and/or ligands will provide more insight into the important Notch receptors or ligands involved in CRC tumorigenesis.
The Wnt and Notch pathways have crucial roles in proliferation and maintenance of stem cells in the gut (van Es and Clevers, 2005). Inactivating mutations in APC and activating mutations in β-catenin occur in ∼80% and 5%, respectively, of CRC patients and have been identified to be key factors involved in the development of CRC (van Es and Clevers, 2005). Cross-talk between the Notch and Wnt signalling pathways has been described in CRC. In particular, binding of the Notch receptor to β-catenin has been shown to inhibit Wnt-dependent activity (Kwon et al, 2011). In addition, the Notch ligand JAG-1 is transcriptionally activated by β-catenin and a conditional knockout of JAG1 in an APC−/− mouse model resulted in a reduction in the development of intestinal tumours (Rodilla et al, 2009). Therefore, we first examined baseline differences between PF-03084014-sensitive tumours and resistant tumours in the Wnt pathway by gene array and KEGG pathway analysis. Similarly to the Notch pathway, we show that many components of the canonical Wnt pathway were increased in sensitive tumours compared with resistant tumours. No association between APC or β-catenin mutational status and sensitivity was observed. Analysis of active β-catenin by western blot confirmed that the Wnt pathway was significantly elevated in sensitive tumours. Furthermore, we demonstrated that PF-03084014 resulted in a significant reduction in levels of Axin2 and active β-catenin in the sensitive CRC021 explant. These findings suggest that PF-03084014 inhibits the Notch and Wnt pathways in sensitive tumours with elevated levels of Notch and Wnt, resulting in a reduction in tumour growth in our CRC explant model. However, more studies are needed to define the mechanism whereby Notch and Wnt interact.
Treatment with PF-03084014 reduced tumour growth in the sensitive explants by inducing apoptosis. We observed an increase in cleaved caspase 3 and PARP as early as 2 h post treatment as well as a decrease in the phosphorylation of p65, an NF-κB subunit that regulates genes that are important for cell survival. Since cleaved Notch levels were not decreased at 2 h post PF-03084014 treatment may suggest that off target effects were responsible for cleavage of caspase 3 and PARP. However, we demonstrate a significant decrease in cleaved Notch1 as well as in the phosphorylation of p65 and the NF-κB-dependent anti-apoptotic protein BCLxL at 8 h, 24 h and 8 days post treatment. This indicates that an induction of apoptosis by inhibiting the NF-κB pathway may be in part responsible for the anti-tumour effects observed in the PF-03084014-sensitive tumours.
In summary, our results provide insight into the role of the Notch pathway in CRC. PF-03084014 only had activity against a subset of tumours that exhibited elevated levels of the Notch and Wnt pathways and treatment reduced the activity of both pathways. These findings have potential for the treatment of patients with CRC. Patients with elevated levels of Notch and Wnt pathway may derive benefit from treatment with the Notch inhibitor PF-03084014. A phase I clinical trial of PF-03084014 is ongoing and the data readout should inform on patient selection and future development of the compound for CRC. Although this compound is distinct from others in the class by having linear pharmacokinetic (PK) properties and minimal gastrointestinal (GI) toxicity, further development will be facilitated by the identification of a robust biomarker.
Acknowledgments
This work was supported by a Pfizer IIR grant.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Chu D, Li Y, Wang W, Zhao Q, Li J, Lu Y, Li M, Dong G, Zhang H, Xie H, Ji G. High level of Notch1 protein is associated with poor overall survival in colorectal cancer. Ann Surg Oncol. 2010;17 (5:1337–1342. doi: 10.1245/s10434-009-0893-7. [DOI] [PubMed] [Google Scholar]
- Dangles-Marie V, Pocard M, Richon S, Weiswald LB, Assayag F, Saulnier P, Judde JG, Janneau JL, Auger N, Validire P, Dutrillaux B, Praz F, Bellet D, Poupon MF. Establishment of human colon cancer cell lines from fresh tumors versus xenografts: comparison of success rate and cell line features. Cancer Res. 2007;67 (1:398–407. doi: 10.1158/0008-5472.CAN-06-0594. [DOI] [PubMed] [Google Scholar]
- Fischer M, Yen WC, Kapoun AM, Wang M, O'Young G, Lewicki J, Gurney A, Hoey T. Anti-DLL4 inhibits growth and reduces tumor-initiating cell frequency in colorectal tumors with oncogenic KRAS mutations. Cancer Res. 2011;71 (5:1520–1525. doi: 10.1158/0008-5472.CAN-10-2817. [DOI] [PubMed] [Google Scholar]
- Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435 (7044:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- Friedmann DR, Wilson JJ, Kovall RA. RAM-induced allostery facilitates assembly of a notch pathway active transcription complex. J Biol Chem. 2008;283 (21:14781–14791. doi: 10.1074/jbc.M709501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36 (Database issue:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. Gamma-secretase: proteasome of the membrane. Nat Rev Mol Cell Biol. 2004;5 (6:499–504. doi: 10.1038/nrm1406. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137 (2:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C, Cheng P, King IN, Andersen P, Shenje L, Nigam V, Srivastava D. Notch post-translationally regulates beta-catenin protein in stem and progenitor cells. Nat Cell Biol. 2011;13 (10:1244–1251. doi: 10.1038/ncb2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maetzel D, Denzel S, Mack B, Canis M, Went P, Benk M, Kieu C, Papior P, Baeuerle PA, Munz M, Gires O. Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol. 2009;11 (2:162–171. doi: 10.1038/ncb1824. [DOI] [PubMed] [Google Scholar]
- Messersmith WA, LoRusso P, Cleary JM, Dasari A, Zhang X, Shaik N, Courtney RD, Randolph S, Shapiro G.2011A phase I dose-escalation study of the novel gamma secretase inhibitor PF-03084014 in patients (pts) with advanced solid tumors J Clin Oncol 29(supplPlease provide page range for reference ‘Messersmith et al (2011).'abstr 3100. [Google Scholar]
- Nair P, Somasundaram K, Krishna S. Activated Notch1 inhibits p53-induced apoptosis and sustains transformation by human papillomavirus type 16 E6 and E7 oncogenes through a PI3K-PKB/Akt-dependent pathway. J Virol. 2003;77 (12:7106–7112. doi: 10.1128/JVI.77.12.7106-7112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124 (5:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- Noguera-Troise I, Daly C, Papadopoulos NJ, Coetzee S, Boland P, Gale NW, Lin HC, Yancopoulos GD, Thurston G. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature. 2006;444 (7122:1032–1037. doi: 10.1038/nature05355. [DOI] [PubMed] [Google Scholar]
- Osipo C, Golde TE, Osborne BA, Miele LA. Off the beaten pathway: the complex cross talk between Notch and NF-kappaB. Lab Investig. 2008;88 (1:11–17. doi: 10.1038/labinvest.3700700. [DOI] [PubMed] [Google Scholar]
- Pannuti A, Foreman K, Rizzo P, Osipo C, Golde T, Osborne B, Miele L. Targeting Notch to target cancer stem cells. Clin Cancer Res. 2010;16 (12:3141–3152. doi: 10.1158/1078-0432.CCR-09-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke F, Clevers H, Riccio O. From gut homeostasis to cancer. Curr Mol Med. 2006;6 (3:275–289. doi: 10.2174/156652406776894527. [DOI] [PubMed] [Google Scholar]
- Rodilla V, Villanueva A, Obrador-Hevia A, Robert-Moreno A, Fernandez-Majada V, Grilli A, Lopez-Bigas N, Bellora N, Alba MM, Torres F, Dunach M, Sanjuan X, Gonzalez S, Gridley T, Capella G, Bigas A, Espinosa L. Jagged1 is the pathological link between Wnt and Notch pathways in colorectal cancer. Proc Natl Acad Sci USA. 2009;106 (15:6315–6320. doi: 10.1073/pnas.0813221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronchini C, Capobianco AJ. Induction of cyclin D1 transcription and CDK2 activity by Notch(ic): implication for cell cycle disruption in transformation by Notch(ic) Mol Cell Biol. 2001;21 (17:5925–5934. doi: 10.1128/MCB.21.17.5925-5934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Viqueira B, Jimeno A, Cusatis G, Zhang X, Iacobuzio-Donahue C, Karikari C, Shi C, Danenberg K, Danenberg PV, Kuramochi H, Tanaka K, Singh S, Salimi-Moosavi H, Bouraoud N, Amador ML, Altiok S, Kulesza P, Yeo C, Messersmith W, Eshleman J, Hruban RH, Maitra A, Hidalgo M. An in vivo platform for translational drug development in pancreatic cancer. Clin Cancer Res. 2006;12 (15:4652–4661. doi: 10.1158/1078-0432.CCR-06-0113. [DOI] [PubMed] [Google Scholar]
- Sade H, Krishna S, Sarin A. The anti-apoptotic effect of Notch-1 requires p56lck-dependent, Akt/PKB-mediated signaling in T cells. J Biol Chem. 2004;279 (4:2937–2944. doi: 10.1074/jbc.M309924200. [DOI] [PubMed] [Google Scholar]
- Siegel R, Desantis C, Virgo K, Stein K, Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, Lin C, Leach C, Cannady RS, Cho H, Scoppa S, Hachey M, Kirch R, Jemal A, Ward E. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62 (4:220–241. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- Sikandar SS, Pate KT, Anderson S, Dizon D, Edwards RA, Waterman ML, Lipkin SM. NOTCH signaling is required for formation and self-renewal of tumor-initiating cells and for repression of secretory cell differentiation in colon cancer. Cancer Res. 2010;70 (4:1469–1478. doi: 10.1158/0008-5472.CAN-09-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoshita M, Aoki M, Fuwa H, Aoki K, Hosogi H, Sakai Y, Hashida H, Takabayashi A, Sasaki M, Robine S, Itoh K, Yoshioka K, Kakizaki F, Kitamura T, Oshima M, Taketo MM. Suppression of colon cancer metastasis by Aes through inhibition of Notch signaling. Cancer Cell. 2011;19 (1:125–137. doi: 10.1016/j.ccr.2010.11.008. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102 (43:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tousseyn T, Thathiah A, Jorissen E, Raemaekers T, Konietzko U, Reiss K, Maes E, Snellinx A, Serneels L, Nyabi O, Annaert W, Saftig P, Hartmann D, De Strooper B. ADAM10, the rate-limiting protease of regulated intramembrane proteolysis of Notch and other proteins, is processed by ADAMS-9, ADAMS-15, and the gamma-secretase. J Biol Chem. 2009;284 (17:11738–11747. doi: 10.1074/jbc.M805894200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es JH, Clevers H. Notch and Wnt inhibitors as potential new drugs for intestinal neoplastic disease. Trends Mol Med. 2005;11 (11:496–502. doi: 10.1016/j.molmed.2005.09.008. [DOI] [PubMed] [Google Scholar]
- van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, Clevers H. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435 (7044:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- Wei P, Walls M, Qiu M, Ding R, Denlinger RH, Wong A, Tsaparikos K, Jani JP, Hosea N, Sands M, Randolph S, Smeal T. Evaluation of selective gamma-secretase inhibitor PF-03084014 for its antitumor efficacy and gastrointestinal safety to guide optimal clinical trial design. Mol Cancer Ther. 2010;9 (6:1618–1628. doi: 10.1158/1535-7163.MCT-10-0034. [DOI] [PubMed] [Google Scholar]
- Zhang CC, Pavlicek A, Zhang Q, Lira ME, Painter CL, Yan Z, Zheng X, Lee N, Ozeck M, Qiu M, Zong Q, Lappin P, Wong A, Rejto PA, Smeal T, Christensen JG. Biomarker and pharmacological evaluation of the gamma-secretase inhibitor PF-03084014 in breast cancer models. Clin Cancer Res. 2012;18 (18:5008–5019. doi: 10.1158/1078-0432.CCR-12-1379. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li B, Ji ZZ, Zheng PS. Notch1 regulates the growth of human colon cancers. Cancer. 2010;116 (22:5207–5218. doi: 10.1002/cncr.25449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.