Abstract
NAC [no apical meristem (NAM), Arabidopsis thaliana transcription activation factor [ATAF1/2] and cup-shaped cotyledon (CUC2)] proteins belong to one of the largest plant-specific transcription factor (TF) families and play important roles in plant development processes, response to biotic and abiotic cues and hormone signalling. Our genome-wide analysis identified 110 StNAC genes in potato encoding for 136 proteins, including 14 membrane-bound TFs. The physical map positions of StNAC genes on 12 potato chromosomes were non-random, and 40 genes were found to be distributed in 16 clusters. The StNAC proteins were phylogenetically clustered into 12 subgroups. Phylogenetic analysis of StNACs along with their Arabidopsis and rice counterparts divided these proteins into 18 subgroups. Our comparative analysis has also identified 36 putative TNAC proteins, which appear to be restricted to Solanaceae family. In silico expression analysis, using Illumina RNA-seq transcriptome data, revealed tissue-specific, biotic, abiotic stress and hormone-responsive expression profile of StNAC genes. Several StNAC genes, including StNAC072 and StNAC101that are orthologs of known stress-responsive Arabidopsis RESPONSIVE TO DEHYDRATION 26 (RD26) were identified as highly abiotic stress responsive. Quantitative real-time polymerase chain reaction analysis largely corroborated the expression profile of StNAC genes as revealed by the RNA-seq data. Taken together, this analysis indicates towards putative functions of several StNAC TFs, which will provide blue-print for their functional characterization and utilization in potato improvement.
Keywords: abiotic stress, genome-wide analysis, Illumina RNA-seq, NAC transcription factor, potato
1. Introduction
Potato (Solanum tuberosum L.) is the most important non-grain food crop and is central to global food security. Considering its importance, much research on potato has been carried out during last decades. However, the fact remains that the global average yield of potato (15 tons/ha) is far below its yield potential (120 tons/ha), primarily due to various biotic and abiotic stresses.1 High and low temperatures, salinity and drought are the major abiotic stress factors limiting growth and productivity of the potato crop.2,3 Among biotic stresses, oomycete Phytophthora infestans that cause late blight is the most devastating disease of the potato with potential of causing 40–50% yield loss.4 Thus, improved tolerance of potato to these stresses may significantly increase the potato production. Tolerance or susceptibility against these stresses is governed by plant's ability to express a set of genes whose expression is often regulated by specific transcription factors (TFs).
The NAC [no apical meristem (NAM), Arabidopsis thaliana transcription activation factor [ATAF1/2] and cup shaped cotyledon (CUC2)] TFs were originally identified from consensus sequences from petunia NAM, Arabidopsis thaliana ATAF1 and 2 and CUC2.5 The NAC family is one of the largest plant-specific TF families, represented by 117 genes in Arabidopsis and 151 in rice,6 163 in poplar,7 152 each in soybean8 and tobacco9 and 74 in grape.10 NAC proteins regulate a variety of plant developmental processes, such as the development of shoot apical meristem,11,12 lateral root development,13 embryonic and floral development,12,14 stress-induced flowering,15,16 leaf senescence,17 regulation of cell cycle,18,19 hormone signalling13,18,20,21 and grain nutrient remobilization.22
Some NAC proteins also regulate plant stress responses, including both biotic and abiotic.23,24 The Arabidopsis RESPONSIVE TO DEHYDRATION 26 (RD26) cDNA was first identified as dehydration responsive gene25 that was later shown to encode a NAC TF and functions in a novel abscisic acid (ABA)-dependent stress-signalling pathway.20 Using yeast one hybrid, three Arabidopsis NAC proteins (ANAC019, ANAC055 and ANAC072/RD26) were identified, and overexpression of either of these genes significantly improved drought tolerance of transgenic plants.26 Similarly, overexpression of various NAC genes in transgenic rice conferred improved tolerance against abiotic stresses.27–32 As far as crops are concerned, most of the studies reporting the overexpression of NAC genes are limited to rice except, by Xue et al.33 who have overexpressed a wheat NAC gene, TaNAC69 in transgenic wheat that resulted in improved dehydration tolerance. Thus, it is important to identify and functionally characterize NAC TF families from economically important crop plants and to use functional NAC genes for generating these crops with improved stress tolerance. The NAC proteins also regulate plant response against various biotic cues, including viral,34 bacterial and fungal pathogens.35
Typically, NAC proteins posses a conserved N-terminal DNA-binding NAC domain, which is divided into five subdomains (A–E), while C-terminal region is highly diversified and contains a transcriptional regulatory domain (TRD).36 Some NAC proteins, referred as NTL (NAC with Transmembrane Motif 1-like), also contain transmembrane motifs (TMs) at their C-terminal end.37,38 Crystal structure of the NAC domain of Arabidopsis ANAC01939 and rice SNAC140 revealed the presence of a novel TF fold consisting of a twisted anti-parallel β-sheet. Recently, a new subfamily of NAC family, called TNAC, was identified in tobacco, which seemed to be restricted to Solanaceae family.9 The NAC domain of TNACs lacks the LPPG and YPNG motifs that are conserved in NAC family members, whereas the conserved D/EEE motif found in other NACs is replaced by D/ExE in TNACs.9
The recent completion of genome sequencing of the potato by the potato genome sequencing consortium (PGSC)41 provides opportunities to identify protein families at genome-wide level, to analyse them and to utilize the potential genes for potato improvement. Recently, Jupe et al.42 have identified 438 NB-LRR genes containing nucleotide-binding (NB) and leucine rich repeat (LRR) domain in the potato genome. Similarly in a separate report, 435 NBS-encoding R genes were identified in the potato genome.43 NAC TFs have not been studied in the potato, except by Collinge and Boller,44 who found that a potato NAC gene, StNAC, was rapidly and strongly induced after wounding, while under P. infestans infection its transcript was detected only at 48 h. However, precise function of this StNAC remains to be elucidated.
Given the critical roles played by NAC TFs in plants, we have identified a NAC TF family in the potato genome, provided nomenclature, performed phylogenetic analysis, mapped genes onto the 12 potato chromosomes, identified membrane-bound proteins and carried out expression analysis under various developmental stages, biotic and abiotic stresses and hormone treatments. In future, this study will provide leads to functionally characterize potato NAC TFs, to utilize them for potato improvement and also to identify and characterize NAC TFs in other Solanum species.
2. Materials and methods
2.1. Identification of NAC gene family in potato
All the files related to potato genome sequence data used for the identification and annotation of NAC proteins were downloaded from the PGSC data sharing site (http://www.potatogenome.net/index.php/Main_Page). The Hidden Markov Model (HMM) profile of the NAM domain (PF02365) retrieved from Pfam 26.0 (http://Pfam.sanger.ac.uk/) was exploited to identify the putative NAC proteins in S. tuberosum group Phureja DM 1-3 516 R44 (DM) protein (v3.4) database using HMM search, with an expected value (e-value) cut-off of 1.0. The sequences of all identified DM protein (DMP) models were subjected to Pfam analysis to confirm the presence of NAM domain, with an e-value cut-off of 1e−3. Keyword searches in NCBI (http://www.ncbi.nlm.nih.gov/), UniProt (www.uniprot.org) and PlantTFDB v2.0 (http://planttfdb.cbi.edu.cn/) databases were also performed to identify potato NAC proteins. Arabidopsis thaliana orthologs for potato NAC proteins were identified using BLASTp search against Arabidopsis proteins TAIR10 release (http://www.arabidopsis.org). Prediction of membrane-bound StNAC proteins was performed using the TMHMM server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
2.2. Mapping NAC genes on chromosomes, their nomenclature and gene duplication
The position of each potato NAC gene on potato chromosomes was identified using the potato genome browser at the PGSC site. For nomenclature, prefix ‘St’ for S. tuberosum was added followed by NAC and numbered according to its position from top to bottom on the potato chromosome 1–12. Alternatively, spliced forms were represented by Arabic numbers after ‘.’ sign. To search for potential duplicated potato NAC genes, MCScanX software was used.45 All 56 218 potato genes were compared against themselves using BLASTp, with criterion of tabular format (-m 8) and e-value of <1e−5. The resulting blast hits were incorporated along with chromosome coordinates of all protein-coding genes as an input for MCScanX and classified into segmental, tandem, proximal and dispersed duplications under default criterion.
2.3. Phylogenetic analysis and identification of conserved motifs
Multiple sequence alignment of the full-length protein sequences along with three representative Arabidopsis NAC proteins, ANAC019 (AT1G52890), ANAC055 (AT3G15500) and ANAC072/RD26 (AT4G27410),26 was performed using CluatalW2 program with default parameters. Phylogenetic tree was plotted using MEGA5.05 software by the Neighbor-joining method with 1000 bootstrap replicates.46 To study the phylogenetic relationship of potato NAC proteins along with their counterparts in Arabidopsis and rice, full-length NAC protein sequences were retrieved from TAIR10 (http://www.arabidopsis.org) and RGAP7 (http://rice.plantbiology.msu.edu/), respectively, as described.6 Multiple sequence alignment was performed, and unrooted tree was plotted as described above. The conserved motifs in full-length NAC proteins were identified using Multiple Expectation Maximization for Motif Elicitation (MEME) program version 4.9.0, with default parameters except the maximum number of motifs to find was set to 10.47 To predict the secondary structure of potato NAC domain, full-length NAC sequences were aligned along with the known NAC domain structures using Promals3D web program.48 We considered three known structures of NAC domains obtained from PDB accession number, 1UT4 (A. thaliana), 3SWM (A. thaliana) and 3ULX (Oryza sativa), which have most of the hits of StNAC proteins by BLAST PDB (e-value of <1e−04 and maximum identity of >40%).
2.4. Potato RNA-seq data analysis
For expression profiling of potato NAC genes, we utilized the Illumina RNA-seq data that were previously generated by the PGSC41 and analysed by Massa et al.49 The RNA-seq data of 40 libraries representing a wide range of developmental stages, abiotic and biotic stress treatments and hormone treatments were generated using Illumina Genome Analyser II platform (Supplementary Table S1).49 Transcript abundance is expressed as fragments per kilobase of exon model per million mapped reads (FPKM) values (Supplementary Table S2). Heat maps for only those genes were generated, which have positive FPKM values in at least one or more of the samples. For the developmental stage dataset, FPKM values were log2 transformed, before generating heat maps. For abiotic, biotic stress and hormone treatments, relative expression ratios were calculated relative to their respective controls. Heat maps were generated and hierarchical clustering done using the Institute for Genomic Research (TIGR) MeV v4.4.1 software package.50
2.5. Plant material, in vitro culture and stress treatments
The shoot cultures of potato cv Kufri Sutlej, procured from Central Potato Research Institute, Shimla (India), were maintained under in vitro conditions. Potato shoots were inoculated into the Murashige and Skoog51 (MS) medium through nodal cuttings and incubated under a 16-h photoperiod (70 ± 5 µmol m−2 s−1 photosynthetic photon flux density) at 25 ± 2°C and 50–60% relative humidity. After three weeks, shoots were subjected to NaCl (100 mM), polyethylene glycol 6000 (PEG, 10%), cold (4°C), heat (42°C), ABA (100 µM) and salicylic acid (SA, 300 µM) treatments for 4 and 24 h. After stipulated time, the plantlets were harvested, frozen in liquid nitrogen and stored at −80°C until used. Shoots grown on MS basal medium at 25°C served as control. For the collection of root, stem, old leaf and young leaf samples, in vitro raised plantlets of potato cv. Kufri Sutlej were hardened and grown under contained conditions. Two-month-old plants were uprooted, and samples were harvested and frozen in liquid nitrogen and stored at −80°C until used.
2.6. RNA isolation and quantitative real-time PCR
Total RNA was isolated from 100 mg of frozen tissue using iRIS solution following the method as described.52 First-strand cDNA synthesis was done using RevertAid™ RNAse H minus cDNA synthesis kit as per manufacturer's instructions (Fermentas Life Sciences, USA). The primers for quantitative real-time PCR (qRT-PCR) analysis were designed using the Primer3 v.0.4.0 software (http://frodo.wi.mit.edu/; Supplementary Table S3). Reverse primers were designed preferentially from 3′-untranslated region wherever possible, because it is generally more unique than coding sequence and closer to the reverse transcriptase (RT) start site. To check the primer specificity, amplicons obtained after PCR were sequenced using the BigDye terminator sequencing kit on an automated DNA sequencer (3730 ×l DNA Analyser, Applied Biosystems, USA). The amplicon sequences are presented in Supplementary Table S3. The qRT-PCR assays were performed with three biological and three technical replicates. Each reaction was performed in 20 µl reaction mixture containing diluted cDNA sample as template and 2× Power SYBR Green PCR master mix (Applied Biosystems), and 200 nM each of forward and reverse gene specific primers. The reactions were performed using the MX 3000P Real-Time PCR system (Stratagene) with the following programme: 95°C (90 s) [94°C (30 s), corresponding annealing temperature (30 s), 72°C (30 s)] × 40 cycles. The specificity of the amplification was also determined by dissociation curve analysis in each case. To normalize the variance in cDNA input, elongation factor 1-α (ef1α) gene was used as the internal control as suggested earlier.53 The relative expression ratio of each gene was calculated using the comparative Ct value method.54
3. Results and discussion
3.1. Identification and nomenclature of the NAC family members in potato
To identify the putative NAC proteins in potato genome, HMM search was performed using the HMM profile of the NAM domain. This HMM search resulted in identification of 145 protein models (DMPs), which were encoded by 118 gene models (DMGs; Supplementary Table S4). Subsequently, all 145 protein sequences were subjected to Pfam analysis, with e-value cut-off of 1e−3, which resulted in identification of 136 NAC proteins encoded by 110 genes, because nine DMPs, either with no N-terminal NAM domain or with its e-value of >1e−3 were excluded. A keyword search against the NCBI, UniProt and PlantTFDB databases resulted in identification of 12, 7 and 40 previously annotated potato NAC proteins sequences, respectively (Supplementary Table S5). A careful analysis confirmed the presence of these proteins in the list of 136 NAC proteins identified through HMM search in potato genome. Hence, we show that potato NAC family is comprised of 136 NAC proteins, which are encoded by 110 genes (Table 1). Thus, NAC family in the potato is also comprised of >100 genes as reported for Arabidopsis, rice, poplar, soybean, tobacco, maize and grape.6–10 The annotations for potato NAC proteins reported in the NCBI and UniProt databases were highly disordered and uninformative (Supplementary Table S5). Thus, a uniform nomenclature has been assigned to 136 potato NAC proteins. Potato NAC proteins are designated as StNAC followed by Arabic number 1–110 based on the position of their corresponding genes on chromosomes 1–12 and from top to bottom (Table 1). Alternatively, spliced proteins are designated by same name by adding Arabic number 1, 2 and so on after ‘.’ sign. Similar criteria have also been adapted for the nomenclature of NAC proteins in soybean8 and WRKY proteins in maize.55 Of 110 StNAC genes, 19 (∼17%) undergo alternative splicing (Table 1). However, in rice, of 151 NAC genes, 15 (∼10%) were reported to produce alternative spliced transcripts.56 The higher frequency of splicing events in potato NAC family than that of rice is in agreement with the previous reports, where in potato genome 9875 genes (25.3%) have been shown to undergo alternative splicing,41 whereas in rice genome, 8772 (15.7%) genes undergo alternative splicing.57 The higher frequency of alternative splicing events in potato NAC family indicates more functional divergence of StNACs than that of rice. The length of StNAC proteins identified in this study ranges from 56 to 901 amino acids (aa) with an average of ∼312 aa. Whereas, in Populus, the size of NAC proteins ranges from 117 to 718 aa with an average of 342 aa.7 In potato, the StNAC054 (56 aa) is the smallest StNAC protein, wherein NAM domain appears to be truncated at C-terminal end (Supplementary Fig. S1). Whereas, StNAC036.1 is the largest StNAC protein (901 aa) and contains two NAM domains. However, the NAM domain at its C-terminal end (StNAC036.1C) appears to be truncated lacking subdomain A and B (Supplementary Fig. S2).
Table 1.
List of NAC transcription factor genes in potato (Solanum tuberosum L.) along with their corresponding proteins, CDS and protein length, duplications and Arabidopsis thaliana orthologs
| Gene | Protein | Protein identifier | Chromosome no. | CDS length (bp) | Protein length (aa) | Duplications | At ortholog locus | At locus description | Score (bits) | e-value |
|---|---|---|---|---|---|---|---|---|---|---|
| StNAC001 | StNAC001 | PGSC0003DMP400000341 | chr01 | 741 | 246 | Dispersed | AT5G62380.1 | ANAC101, VND6 | 47 | 1.00e−05 |
| StNAC002 | StNAC002 | PGSC0003DMP400058270 | chr01 | 423 | 140 | Dispersed | AT4G01520.1 | ANAC067 | 36 | 0.007 |
| StNAC003 | StNAC003 | PGSC0003DMP400069271 | chr01 | 825 | 274 | Dispersed | AT3G10480.1 | ANAC050 | 71 | 6.00e−13 |
| StNAC004 | StNAC004 | PGSC0003DMP400031815 | chr01 | 1212 | 403 | Dispersed | AT2G02450.1, AT2G02450.2 | ANAC034, ANAC035 | 312 | 2.00e−85 |
| StNAC005 | StNAC005 | PGSC0003DMP400051813 | chr01 | 588 | 195 | Dispersed | AT5G64530.1 | ANAC104, XND1 | 221 | 4.00e−58 |
| StNAC006 | StNAC006 | PGSC0003DMP400037231 | chr02 | 1689 | 562 | Dispersed | AT1G65910.1 | ANAC028 | 427 | e−119 |
| StNAC007 | StNAC007.1 | PGSC0003DMP400015241 | chr02 | 738 | 245 | Dispersed | AT2G17040.1 | ANAC036 | 231 | 5.00e−61 |
| StNAC007.2 | PGSC0003DMP400015242 | 504 | 167 | AT2G17040.1 | ANAC036 | 202 | 7.00e−53 | |||
| StNAC008 | StNAC008 | PGSC0003DMP400067304 | chr02 | 813 | 270 | Proximal | AT5G46590.1 | ANAC096 | 41 | 0.001 |
| StNAC009 | StNAC009 | PGSC0003DMP400041300 | chr02 | 1029 | 342 | Tandem | AT4G27410.2 | ANAC072, RD26 | 39 | 0.006 |
| StNAC010 | StNAC010 | PGSC0003DMP400057983 | chr02 | 1086 | 361 | Tandem | AT5G46590.1 | ANAC096 | 42 | 5.00e−04 |
| StNAC011 | StNAC011 | PGSC0003DMP400041296 | chr02 | 981 | 326 | Tandem | AT5G46590.1 | ANAC096 | 39 | 0.004 |
| StNAC012 | StNAC012 | PGSC0003DMP400041297 | chr02 | 978 | 325 | Tandem | AT2G46770.1 | ANAC043, NST1 | 45 | 6.00e−05 |
| StNAC013 | StNAC013 | PGSC0003DMP400058560 | chr02 | 558 | 185 | Tandem | AT3G10500.1 | ANAC055, ATNAC3 | 48 | 4.00e−06 |
| StNAC014 | StNAC014 | PGSC0003DMP400060071 | chr02 | 804 | 267 | Tandem | AT3G10500.1 | ANAC055, ATNAC3 | 37 | 0.02 |
| StNAC015 | StNAC015 | PGSC0003DMP400036603 | chr02 | 945 | 314 | Dispersed | AT2G43000.1 | ANAC042 | 235 | 4.00e−62 |
| StNAC016 | StNAC016.1 | PGSC0003DMP400054964 | chr02 | 882 | 293 | Segmental | AT4G28500.1 | ANAC073 | 332 | 2.00e−91 |
| StNAC016.2 | PGSC0003DMP400054965 | 561 | 186 | AT4G28500.1 | ANAC073 | 205 | 2.00e−53 | |||
| StNAC017 | StNAC017.1 | PGSC0003DMP400002396 | chr02 | 972 | 323 | Segmental | AT5G61430.1 | ANAC100, ATNAC5 | 365 | e−101 |
| StNAC017.2 | PGSC0003DMP400002397 | 774 | 257 | AT5G61430.1 | ANAC100, ATNAC5 | 245 | 2.00e−65 | |||
| StNAC018 | StNAC018.1 | PGSC0003DMP400002374 | chr02 | 1191 | 396 | Dispersed | AT5G39820.1 | ANAC094 | 285 | 3.00e−77 |
| StNAC018.2 | PGSC0003DMP400002375 | 1056 | 351 | AT1G26870.1 | ANAC009 | 213 | 2.00e−55 | |||
| StNAC019 | StNAC019.1 | PGSC0003DMP400022332 | chr02 | 843 | 280 | Dispersed | AT3G01600.1 | ANAC044 | 277 | 6.00e−75 |
| StNAC019.2 | PGSC0003DMP400022333 | 1182 | 393 | AT3G01600.1 | ANAC044 | 287 | 8.00e−78 | |||
| StNAC020 | StNAC020 | PGSC0003DMP400023688 | chr03 | 576 | 191 | Singleton | AT2G24430.2 | ANAC039 | 38 | 0.004 |
| StNAC021 | StNAC021 | PGSC0003DMP400060025 | chr03 | 699 | 232 | Tandem | AT2G02450.1, AT2G02450.2 | ANAC034, ANAC035 | 55 | 3.00e−08 |
| StNAC022 | StNAC022 | PGSC0003DMP400061582 | chr03 | 786 | 261 | Tandem | AT3G10490.1/AT3G10490.2 | ANAC051/ANAC052 | 74 | 7.00e−14 |
| StNAC023 | StNAC023.1 | PGSC0003DMP400001112 | chr03 | 1203 | 400 | Segmental | AT5G24590.2 | ANAC091, TIP | 270 | 1.00e−72 |
| StNAC023.2 | PGSC0003DMP400001113 | 813 | 270 | AT5G24590.2 | ANAC091, TIP | 240 | 9.00e−64 | |||
| StNAC023.3 | PGSC0003DMP400001114 | 1917 | 638 | AT5G24590.2 | ANAC091, TIP | 270 | 2.00e−72 | |||
| StNAC024 | StNAC024 | PGSC0003DMP400032120 | chr03 | 849 | 282 | AT1G69490.1 | ANAC029, ATNAP | 312 | 1.00e−85 | |
| StNAC025 | StNAC025 | PGSC0003DMP400054092 | chr03 | 753 | 250 | Dispersed | AT3G17730.1 | ANAC057 | 367 | e−102 |
| StNAC026 | StNAC026 | PGSC0003DMP400069047 | chr03 | 828 | 275 | Tandem | AT2G02450.1 | ANAC034, ANAC035 | 71 | 8.00e−13 |
| StNAC027 | StNAC027 | PGSC0003DMP400067675 | chr03 | 699 | 232 | Tandem | AT5G46590.1 | ANAC096 | 62 | 3.00e−10 |
| StNAC028 | StNAC028 | PGSC0003DMP400062654 | chr03 | 819 | 272 | Proximal | AT2G02450.1, AT2G02450.2 | ANAC034, ANAC035 | 68 | 6.00e−12 |
| StNAC029 | StNAC029 | PGSC0003DMP400067767 | chr03 | 792 | 263 | Proximal | AT2G02450.1, AT2G02450.2 | ANAC034, ANAC035 | 69 | 4.00e−12 |
| StNAC030 | StNAC030.1 | PGSC0003DMP400033928 | chr03 | 540 | 179 | Segmental | AT5G07680.1 | ANAC079, ANAC080,ATNAC4 | 270 | 3.00e−73 |
| StNAC030.2 | PGSC0003DMP400033929 | 999 | 332 | Segmental | AT5G61430.1 | ANAC100, ATNAC5 | 352 | 2.00e−97 | ||
| StNAC031 | StNAC031 | PGSC0003DMP400062169 | chr04 | 627 | 208 | Dispersed | AT5G46590.1 | ANAC096 | 50 | 1.00e−06 |
| StNAC032 | StNAC032 | PGSC0003DMP400005111 | chr04 | 852 | 283 | Dispersed | AT1G69490.1 | ANAC029, ATNAP | 316 | 1.00e−86 |
| StNAC033 | StNAC033 | PGSC0003DMP400055618 | chr04 | 912 | 303 | Tandem | AT1G01720.1 | ANAC002, ATAF1 | 324 | 4.00e−89 |
| StNAC034 | StNAC034 | PGSC0003DMP400009745 | chr04 | 945 | 314 | Dispersed | AT3G47570.1/AT5G53950.1 | LRR Protein Kinase/ANAC098, CUC2 | 92/43 | 3e−19/3e−04 |
| StNAC35 | StNAC35 | PGSC0003DMP400058145 | chr04 | 531 | 176 | Dispersed | AT5G17260.1 | ANAC086 | 43 | 1.00e−04 |
| StNAC036 | StNAC036.1 | PGSC0003DMP400054265 | chr04 | 2706 | 901 | Segmental | AT1G34190.1 | ANAC017 | 305 | 1.00e−82 |
| StNAC036.2 | PGSC0003DMP400054267 | 1797 | 598 | AT1G34190.1 | ANAC017 | 347 | 1.00e−95 | |||
| StNAC036.3 | PGSC0003DMP400054268 | 1485 | 494 | AT1G34190.1 | ANAC017 | 292 | 4.00e−79 | |||
| StNAC037 | StNAC037 | PGSC0003DMP400054262 | chr04 | 762 | 253 | Proximal | AT5G04410.1 | ANAC078, NAC2 | 65 | 4.00e−11 |
| StNAC038 | StNAC038 | PGSC0003DMP400054263 | chr04 | 546 | 181 | Proximal | AT5G04410.1/AT4G35580.1 | ANAC078, NAC2, NTL9 | 62 | 2.00e−10 |
| StNAC039 | StNAC039 | PGSC0003DMP400043482 | chr04 | 756 | 251 | Proximal | AT5G18270.2 | ANAC087 | 62 | 5.00e−10 |
| StNAC040 | StNAC040 | PGSC0003DMP400043483 | chr04 | 780 | 259 | Tandem | AT5G04410.1 | ANAC078, NAC2 | 64 | 1.00e−10 |
| StNAC041 | StNAC041 | PGSC0003DMP400043484 | chr04 | 774 | 257 | Tandem | AT5G04410.1 | ANAC078, NAC2 | 61 | 7.00e−10 |
| StNAC042 | StNAC042 | PGSC0003DMP400013984 | chr04 | 819 | 272 | Dispersed | AT1G52890.1 | ANAC019 | 91 | 8.00e−19 |
| StNAC043 | StNAC043.1 | PGSC0003DMP400048436 | chr04 | 1128 | 375 | Dispersed | AT2G24430.1/AT2G24430.2 | ANAC038/ANAC039 | 52 | 1.00e−06 |
| StNAC043.2 | PGSC0003DMP400048437 | 1128 | 375 | AT2G24430.2 | ANAC039 | 49 | 4.00e−06 | |||
| StNAC043.3 | PGSC0003DMP400048438 | 978 | 325 | AT5G07680.2 | ANAC080 | 48 | 1.00e−05 | |||
| StNAC044 | StNAC044 | PGSC0003DMP400017509 | chr04 | 1068 | 355 | Dispersed | AT1G76420.1 | ANAC031, CUC3 | 294 | 5.00e−80 |
| StNAC045 | StNAC045 | PGSC0003DMP400001544 | chr05 | 1308 | 435 | Tandem | AT1G25580.1 | ANAC008 | 480 | e−136 |
| StNAC046 | StNAC046 | PGSC0003DMP400054481 | chr05 | 1164 | 387 | Dispersed | AT1G26870.1 | ANAC009 | 315 | 4.00e−86 |
| StNAC047 | StNAC047 | PGSC0003DMP400029528 | chr05 | 1398 | 465 | Dispersed | AT4G29230.1 | ANAC075 | 424 | e−119 |
| StNAC048 | StNAC048 | PGSC0003DMP400002220 | chr05 | 849 | 282 | Dispersed | AT2G43000.1 | ANAC042 | 255 | 3.00e−68 |
| StNAC049 | StNAC049 | PGSC0003DMP400040416 | chr05 | 1176 | 391 | Proximal | AT3G10480.1 | ANAC050 | 286 | 2.00e−77 |
| StNAC050 | StNAC050.1 | PGSC0003DMP400040418 | chr05 | 495 | 164 | Tandem | AT5G04410.1 | ANAC078, NAC2 | 244 | 2.00e−65 |
| StNAC050.2 | PGSC0003DMP400040419 | 1608 | 535 | AT3G10500.1 | ANAC053 | 384 | e−106 | |||
| StNAC050.3 | PGSC0003DMP400040420 | 1464 | 487 | AT3G10500.1 | ANAC053 | 300 | 2.00e−81 | |||
| StNAC051 | StNAC051 | PGSC0003DMP400044233 | chr06 | 849 | 282 | Dispersed | AT4G28530.1 | ANAC074 | 266 | 2.00e−71 |
| StNAC052 | StNAC052 | PGSC0003DMP400037408 | chr06 | 447 | 148 | Dispersed | AT1G12260.1 | ANAC007, VND4 | 253 | 4.00e−68 |
| StNAC053 | StNAC053 | PGSC0003DMP400030689 | chr06 | 891 | 296 | AT1G01720.1 | ANAC002, ATAF1 | 386 | e−107 | |
| StNAC054 | StNAC054 | PGSC0003DMP400003753 | chr06 | 171 | 56 | Segmental | AT1G65910.1/AT3G03200.1 | ANAC028/ANAC045 | 67 | 2.00e−12 |
| StNAC055 | StNAC055 | PGSC0003DMP400045251 | chr06 | 990 | 329 | Dispersed | AT1G71930.1 | ANAC030, VND7 | 300 | 9.00e−82 |
| StNAC056 | StNAC056 | PGSC0003DMP400062271 | chr06 | 1566 | 521 | Dispersed | AT3G15500.1 | ANAC055, ATNAC3 | 49 | 6.00e−06 |
| StNAC057 | StNAC057 | PGSC0003DMP400050122 | chr06 | 918 | 305 | Dispersed | AT3G18400.1 | ANAC058 | 276 | 1.00e−74 |
| StNAC058 | StNAC058.1 | PGSC0003DMP400055799 | chr06 | 642 | 213 | Segmental | AT5G61430.1 | ANAC100, ATNAC5 | 286 | 6.00e−78 |
| StNAC058.2 | PGSC0003DMP400055800 | 813 | 270 | Segmental | AT5G61430.1 | ANAC100, ATNAC5 | 254 | 4.00e−68 | ||
| StNAC058.3 | PGSC0003DMP400055801 | 1011 | 336 | AT5G61430.1 | ANAC100, ATNAC5 | 362 | e−100 | |||
| StNAC059 | StNAC059 | PGSC0003DMP400046923 | chr06 | 1893 | 630 | Segmental | AT3G49530.1 | ANAC062 | 269 | 4.00e−72 |
| StNAC060 | StNAC060 | PGSC0003DMP400058755 | chr06 | 486 | 161 | Dispersed | AT1G77450.1 | ANAC032 | 84 | 6.00e−17 |
| StNAC061 | StNAC061 | PGSC0003DMP400010437 | chr06 | 1227 | 408 | Segmental | AT5G22290.1 | ANAC089 | 241 | 8.00e−64 |
| StNAC062 | StNAC062 | PGSC0003DMP400012636 | chr06 | 351 | 116 | Dispersed | AT1G65910.1 | ANAC028 | 187 | 2.00e−48 |
| StNAC063 | StNAC063 | PGSC0003DMP400034966 | chr06 | 855 | 284 | Dispersed | AT4G28530.1 | ANAC074 | 257 | 8.00e−69 |
| StNAC064 | StNAC064 | PGSC0003DMP400032661 | chr07 | 1470 | 489 | Dispersed | AT3G15500.1 | ANAC055, ATNAC3 | 174 | 2.00e−43 |
| StNAC065 | StNAC065 | PGSC0003DMP400068365 | chr07 | 549 | 182 | Tandem | AT1G79580.2/AT1G79580.3 | ANAC033 | 60 | 6.00e−10 |
| StNAC066 | StNAC066 | PGSC0003DMP400016573 | chr07 | 567 | 188 | Tandem | AT3G04060.1 | ANAC046 | 59 | 3.00e−09 |
| StNAC067 | StNAC067 | PGSC0003DMP400016578 | chr07 | 819 | 272 | Segmental | AT4G28500.1 | ANAC073 | 320 | 1.00e−87 |
| StNAC068 | StNAC068 | PGSC0003DMP400060971 | chr07 | 516 | 171 | Tandem | AT4G01540.1/AT4G01540.2 | ANAC068 | 60 | 7.00e−10 |
| StNAC069 | StNAC069 | PGSC0003DMP400021925 | chr07 | 864 | 287 | Dispersed | AT5G53950.1 | ANAC098, CUC2 | 211 | 3.00e−55 |
| StNAC070 | StNAC070 | PGSC0003DMP400012529 | chr07 | 615 | 204 | Dispersed | AT1G77450.1 | ANAC032 | 86 | 1.00e−17 |
| StNAC071 | StNAC071 | PGSC0003DMP400033522 | chr07 | 1020 | 339 | AT3G15510.1 | ANAC056, ATNAC2 | 306 | 2.00e−83 | |
| StNAC072 | StNAC072.1 | PGSC0003DMP400033523 | chr07 | 1071 | 356 | Tandem | AT4G27410.2 | ANAC072, RD26 | 363 | e−100 |
| StNAC072.2 | PGSC0003DMP400033524 | 486 | 161 | Tandem | AT3G15500.1 | ANAC055, ATNAC3 | 293 | 4.00e−80 | ||
| StNAC073 | StNAC073 | PGSC0003DMP400062002 | chr07 | 855 | 284 | Dispersed | AT2G43000.1 | ANAC042 | 261 | 4.00e−70 |
| StNAC074 | StNAC074 | PGSC0003DMP400038263 | chr07 | 1050 | 349 | Dispersed | AT1G56010.2 | ANAC022, NAC1 | 321 | 5.00e−88 |
| StNAC075 | StNAC075 | PGSC0003DMP400035655 | chr08 | 402 | 133 | Segmental | AT4G17980.1 | ANAC071 | 69 | 6.00e−13 |
| StNAC076 | StNAC076 | PGSC0003DMP400026135 | chr08 | 987 | 328 | Segmental | AT2G24430.2/AT2G24430.1 | ANAC038, ANAC039 | 288 | 5.00e−78 |
| StNAC077 | StNAC077.1 | PGSC0003DMP400010296 | chr08 | 1032 | 343 | Segmental | AT2G46770.1 | ANAC043, NST1 | 278 | 5.00e−75 |
| StNAC077.2 | PGSC0003DMP400010297 | 1047 | 348 | AT2G46770.1 | ANAC043, NST1 | 300 | 8.00e−82 | |||
| StNAC078 | StNAC078 | PGSC0003DMP400051536 | chr08 | 1047 | 348 | Segmental | AT2G24430.2/AT2G24430.1 | ANAC038, ANAC039 | 284 | 6.00e−77 |
| StNAC079 | StNAC079 | PGSC0003DMP400046613 | chr08 | 576 | 191 | Proximal | AT3G44350.2 | ANAC061 | 35 | 0.024 |
| StNAC080 | StNAC080 | PGSC0003DMP400046617 | chr08 | 585 | 194 | Proximal | AT5G64060.1 | ANAC103 | 39 | 0.003 |
| StNAC081 | StNAC081.1 | PGSC0003DMP400030569 | chr08 | 1002 | 333 | Dispersed | AT4G17980.1 | ANAC071 | 283 | 1.00e−76 |
| StNAC081.2 | PGSC0003DMP400030570 | 780 | 259 | AT4G17980.1 | ANAC071 | 275 | 2.00e−74 | |||
| StNAC082 | StNAC082 | PGSC0003DMP400008400 | chr08 | 897 | 298 | Dispersed | AT1G62700.1 | ANAC026, VND5 | 280 | 9.00e−76 |
| StNAC083 | StNAC083 | PGSC0003DMP400021401 | chr08 | 1113 | 370 | Segmental | AT2G46770.1 | ANAC043, NST1 | 304 | 6.00e−83 |
| StNAC084 | StNAC084 | PGSC0003DMP400006960 | chr09 | 633 | 210 | Dispersed | AT5G09330.1 | ANAC082 | 3.80E+01 | 0.005 |
| StNAC085 | StNAC085 | PGSC0003DMP400018183 | chr09 | 834 | 277 | Dispersed | AT4G28530.1 | ANAC074 | 275 | 3.00e−74 |
| StNAC086 | StNAC086.1 | PGSC0003DMP400006339 | chr09 | 468 | 155 | Dispersed | AT2G18060.1 | ANAC037, VND1 | 291 | 1.00e−79 |
| StNAC086.2 | PGSC0003DMP400006341 | 1044 | 347 | AT2G18060.1 | ANAC037, VND1 | 415 | e−116 | |||
| StNAC87 | StNAC87 | PGSC0003DMP400019955 | chr10 | 942 | 313 | Dispersed | AT2G46770.1 | ANAC043, NST1 | 310 | 9.00e−85 |
| StNAC88 | StNAC88 | PGSC0003DMP400043440 | chr10 | 1056 | 351 | Segmental | AT3G15510.1 | ANAC056, ATNAC2 | 310 | 8.00e−85 |
| StNAC089 | StNAC089 | PGSC0003DMP400009699 | chr10 | 591 | 196 | Dispersed | AT5G64530.1 | ANAC104, XND1 | 193 | 6.00e−50 |
| StNAC090 | StNAC090 | PGSC0003DMP400019203 | chr10 | 870 | 289 | AT2G17040.1 | ANAC036 | 300 | 6.00e−82 | |
| StNAC091 | StNAC091 | PGSC0003DMP400014381 | chr10 | 1077 | 358 | Tandem | AT1G01720.1 | ANAC002, ATAF1 | 62 | 4.00e−10 |
| StNAC092 | StNAC092 | PGSC0003DMP400014380 | chr10 | 702 | 233 | Tandem | AT5G04410.1 | ANAC078, NAC2 | 40 | 0.001 |
| StNAC093 | StNAC093 | PGSC0003DMP400014332 | chr10 | 906 | 301 | Proximal | AT5G04410.1 | ANAC078, NAC2 | 42 | 5.00e−04 |
| StNAC094 | StNAC094.1 | PGSC0003DMP400049938 | chr11 | 1578 | 525 | Tandem | AT5G64060.1 | ANAC103 | 242 | 5.00e−64 |
| StNAC094.2 | PGSC0003DMP400049939 | 1629 | 542 | AT5G64060.1 | ANAC103 | 244 | 1.00e−64 | |||
| StNAC094.3 | PGSC0003DMP400049940 | 1629 | 542 | AT5G09330.3/AT5G09330.4 | ANAC082 | 257 | 1.00e−68 | |||
| StNAC095 | StNAC095 | PGSC0003DMP400054120 | chr11 | 1203 | 400 | Tandem | AT3G10480.2 | ANAC050 | 290 | 8.00e−79 |
| StNAC096 | StNAC096 | PGSC0003DMP400054118 | chr11 | 1632 | 543 | Tandem | AT5G04410.1 | ANAC078, NAC2 | 394 | e−109 |
| StNAC097 | StNAC097.1 | PGSC0003DMP400016315 | chr11 | 573 | 190 | Dispersed | AT1G01720.1 | ANAC002, ATAF1 | 190 | 4.00e−49 |
| StNAC097.2 | PGSC0003DMP400016316 | 453 | 150 | AT1G01720.1 | ANAC002, ATAF1 | 271 | 1.00e−73 | |||
| StNAC097.3 | PGSC0003DMP400016317 | 876 | 291 | AT1G01720.1 | ANAC002, ATAF1 | 387 | e−108 | |||
| StNAC098 | StNAC098 | PGSC0003DMP400001684 | chr11 | 951 | 316 | Dispersed | AT1G71930.1 | ANAC030, VND7 | 298 | 3.00e−81 |
| StNAC099 | StNAC099.1 | PGSC0003DMP400034078 | chr11 | 1293 | 430 | Segmental | AT2G27300.1 | ANAC040, NTL8 | 239 | 2.00e−63 |
| StNAC099.2 | PGSC0003DMP400034080 | 1236 | 411 | AT2G27300.1 | ANAC040, NTL8 | 238 | 6.00e−63 | |||
| StNAC100 | StNAC100 | PGSC0003DMP400045708 | chr11 | 786 | 261 | Tandem | AT5G22380.1 | ANAC090 | 247 | 6.00e−66 |
| StNAC101 | StNAC101 | PGSC0003DMP400026903 | chr12 | 1056 | 351 | Tandem | AT4G27410.2 | ANAC072, RD26 | 355 | 2.00e−98 |
| StNAC102 | StNAC102 | PGSC0003DMP400017075 | chr12 | 975 | 324 | Dispersed | AT1G79580.3/AT1G79580.2 | ANAC033 | 307 | 5.00e−84 |
| StNAC103 | StNAC103 | PGSC0003DMP400009522 | chr12 | 1008 | 335 | Dispersed | AT3G18400.1 | ANAC058 | 281 | 6.00e−76 |
| StNAC104 | StNAC104 | PGSC0003DMP400027999 | chr12 | 474 | 157 | Dispersed | AT3G04060.1 | ANAC046 | 82 | 2.00e−16 |
| StNAC105 | StNAC105.1 | PGSC0003DMP400029635 | chr12 | 1761 | 586 | Segmental | AT1G34190.1 | ANAC017 | 358 | 5.00e−99 |
| StNAC105.2 | PGSC0003DMP400029636 | 1440 | 479 | AT1G34190.1 | ANAC017 | 300 | 2.00e−81 | |||
| StNAC106 | StNAC106 | PGSC0003DMP400021076 | 801 | 266 | AT5G13180.1 | ANAC083 | 255 | 3.00e−68 | ||
| StNAC107 | StNAC107 | PGSC0003DMP400064998 | 534 | 177 | AT3G15500.1 | ANAC055, ATNAC3 | 52 | 2.00e−07 | ||
| StNAC108 | StNAC108 | PGSC0003DMP400007702 | 783 | 260 | AT5G13180.1 | ANAC083 | 216 | 2.00e−56 | ||
| StNAC109 | StNAC109 | PGSC0003DMP400065497 | 894 | 297 | AT1G77450.1 | ANAC032 | 191 | 5.00e−49 | ||
| StNAC110 | StNAC110 | PGSC0003DMP400033187 | 753 | 250 | AT2G43000.1 | ANAC042 | 180 | 7.00e−46 |
In all StNAC proteins, only NAM domain is present, except in StNAC034, where an additional tyrosine kinase domain (PF07714) is also found. To check whether any NAC protein along with kinase domain is reported from any other organism, extensive BLAST searches of the NCBI database (All GenBank, EMBL, DDBJ and PDB) were performed. Interestingly, no protein was found to have NAM and protein kinase domains together, indicating that potato StNAC034 uniquely possess an additional tyrosine kinase domain. Tyrosine protein kinase catalyses ATP-dependent phosphorylation of the tyrosine residue on target proteins and plays a central role in many signalling pathways in plants.58 The NAC proteins have been shown to physically interact with protein kinase SnRK1 α-subunits AKIN10 and AKIN11.59 Thus, tyrosine protein kinase domain in StNAC034 may be responsible for regulating its activity by autophosphorylation. However, experimental evidences are required to establish the precise role of tyrosine kinase domain in the regulation of StNAC034 activity.
Since, Arabidopsis is considered a model plant system for plant biology research, and many of its NAC genes have been functionally characterized, its orthologous NAC proteins to StNACs have been assigned in this study (Table 1). Interestingly, this analysis has identified StNAC072 and StNAC101 as orthologs of Arabidopsis RD26 with strong e-value support. Previously, RD26 has been shown to be involved in the ABA-dependent stress-signalling pathway.20 Overexpression of rice OsNAC6, ortholog of Arabidopsis RD26, conferred dehydration and salinity stress tolerance in rice.28,29 Thus, functional characterization of these RD26 orthologs will be of immense interest.
3.2. Chromosomal distribution and duplication events among StNAC genes
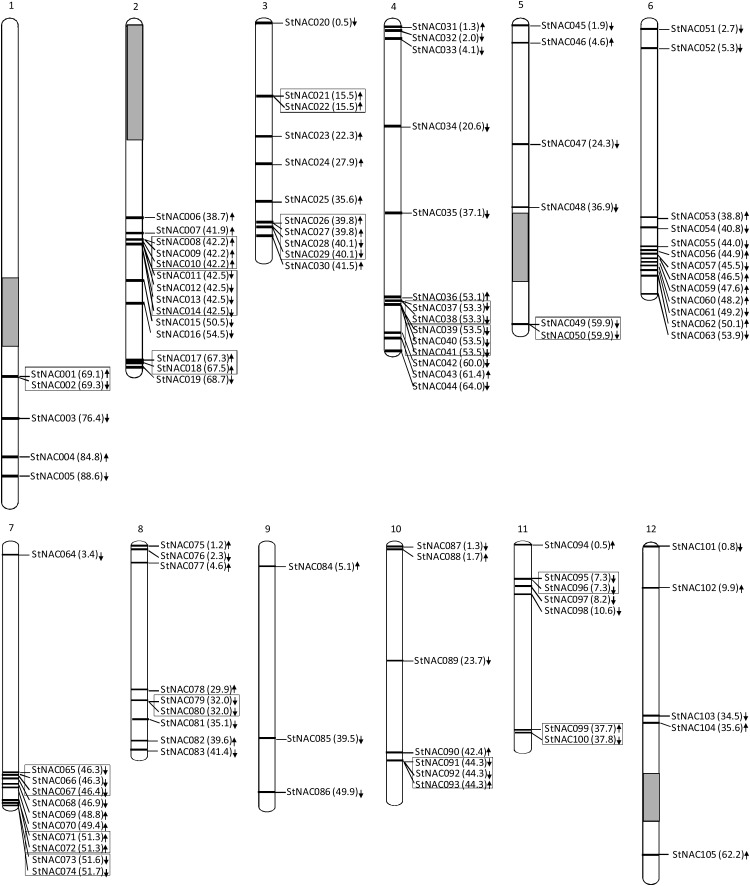
The physical map position of 105 StNAC genes on 12 potato chromosomes was identified. However, five StNAC genes could not be anchored on any of the potato chromosomes. Similarly, out of 438 NB-LRR genes, physical map position for 370 (84%) genes was predicted on potato chromosomes.42 The 105 members of the StNAC gene family are distributed non-randomly on 12 potato chromosomes (Fig. 1). Chromosomes 2 and 4 each contains the largest number of StNAC genes comprising 14 members (∼13%), whereas chromosome 9 contains only three members (∼3%; Supplementary Fig. S3). Based on the previously defined criteria,42 16 clusters comprising of 40 StNAC genes distributed on nine potato chromosomes were identified (Fig. 1). Chromosome 2 contains the maximum number of clusters (3) comprising of nine StNAC genes, whereas chromosomes 1, 5, 8 and 10 each contain single cluster. Genes belonging to a family are often distributed in clusters at certain chromosomal regions. NAC family genes in rice, poplar and soybean were also found to be distributed in clusters.6–8
Figure 1.
Chromosomal distribution of 105 potato NAC genes identified in this study. The chromosome number is indicated on the top of each chromosome. Values in parenthesis following each gene represent its position on the chromosome. Arrows pointing downward and upward represents forward and reverse orientation of the respective gene, respectively, on the chromosome. Sixteen clusters of StNAC genes are indicated in boxes. Grey bars on chromosome 1, 2, 5 and 12 represent known gaps in the chromosome assembly.
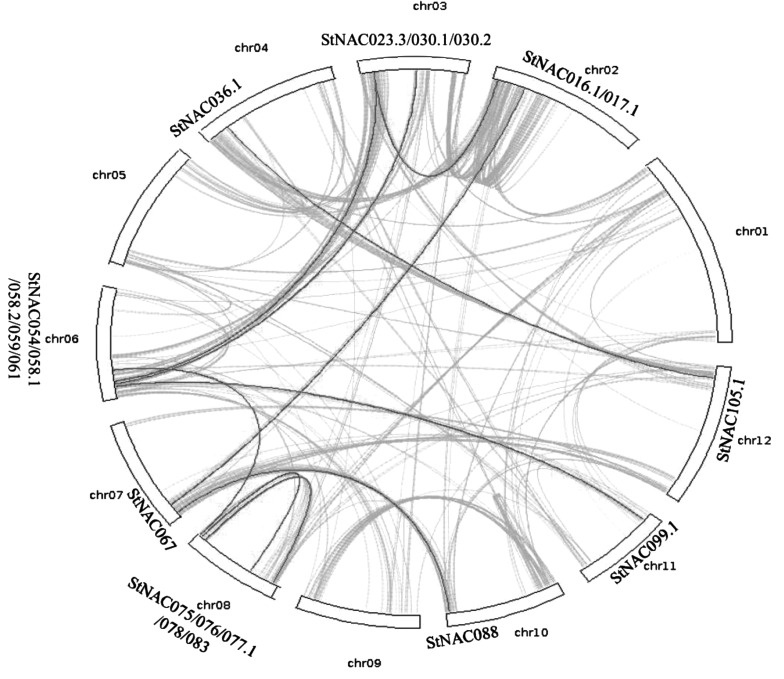
Sequencing and analysis of the potato genome revealed that it has undergone two rounds of whole-genome duplication.41 Moreover, the large size of StNAC gene family suggests that it has evolved through a large number of duplication events in potato. In whole potato genome, we have identified 12083 (23.47%) genes as tandem and 4253 (8.26%) genes as segmental duplicated (Supplementary Tables S6 and S7). Among StNAC genes, 20 were found to be segmentally duplicated, which are located on duplicated segments on chromosomes 2, 3, 4, 6, 7, 8, 10, 11 and 12 (Table 1 and Fig. 2). Maximum five StNACs are located in duplicated segments on each chromosomes 6 and 8, followed by three StNACs on chromosome 3, and two StNACs on chromosome 2. Duplicated segments on chromosome 4, 7, 10, 11 and 12 each contains one StNAC. Interestingly, all the StNAC gene containing chromosomal segments have a StNAC gene in its duplicated segment, suggesting that all the StNAC genes have been retained in potato after segmental duplications. Similarly, 9 NAC genes in rice6 and 21 NAC genes in grape10 were found to be segmentally duplicated. In addition, 27, 10 and 46 StNACs were also found to be tandem, proximal and dispersed duplicated, respectively (Table 1), which might have also contributed to the expansion of the StNAC family.
Figure 2.
Depiction of segmentally duplicated StNAC genes on 12 potato chromosomes. Grey lines indicate collinear blocks in whole potato genome, and black lines indicate duplicated StNAC gene pairs.
3.3. Structural and phylogenetic analysis of StNAC proteins
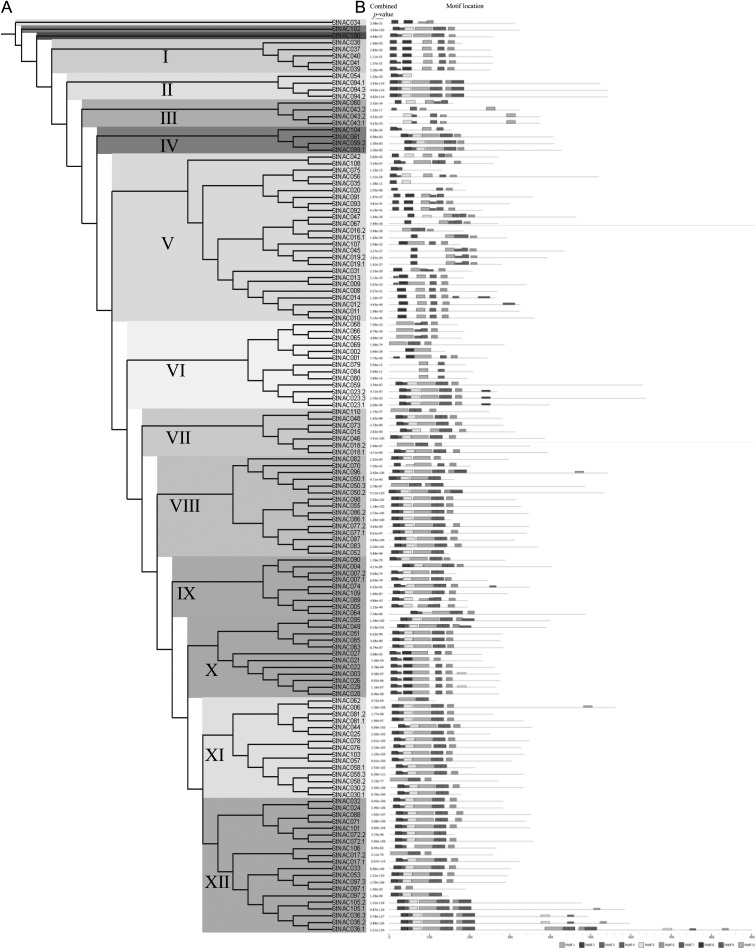
Multiple sequence alignment of full-length StNAC proteins along with three representative Arabidopsis NAC proteins,26 such as ANAC019, ANAC055 and ANAC072/RD26, revealed that most of the StNAC proteins contain highly conserved N-terminal NAC domain, divided into five subdomains (A– E) and a highly variable C-terminal transcriptional regulation domain as described previously (Supplementary Fig. S1).36 However, of 136, 13 StNACs lack conserve A and/or B subdomains, and four StNACs do not contain conserve C and/or D subdomains. Such NAC proteins may be described as NAC-like proteins similar to the description of these proteins in soybean and rice.8,60 All the StNAC proteins, except StNAC054 and StNAC075, contain a conserved nuclear localization signal sequence (NLS) lying within the D subdomain. Phylogenetic tree made from multiple sequence alignment of all 136 StNAC proteins divided them into 12 distinct subgroups (Fig. 3A). Subgroup V consists of the maximum (25) number of StNAC proteins, while subgroup II, III and IV each contain minimum four StNAC proteins. In similar studies, phylogenetic analysis divided poplar and soybean NACs into 10 and 6 subgroups, respectively.7,8 These observations indicate that NAC proteins in potato posses more diversity than poplar and soybean. To further examine the diversity in potato NAC genes, conserved motifs were predicted by using MEME program (Fig. 3B and Supplementary Fig. S4). In general, NAC proteins clustered in same subgroups, share similar motif composition, indicating functional similarities among members of the same subgroup (Fig. 3B). Interestingly, most of the conserved motifs were found lying within the N-terminal NAC domain, indicating that these motifs may be essential for the function of NAC proteins. While, none of the conserved motifs were found at the diversified C-terminal ends of the NAC proteins. Motifs 2, 5, 1, 3 and 6 representing the subdomains A, B, C, D and E, respectively, were present in most of the StNAC proteins. We have also predicted the secondary structure of conserved motifs corresponding to subdomains A–E covering the whole NAC domain (Supplementary Fig. S5). Previously, it was shown that NAC domain monomer consists of a twisted anti-parallel β-sheet, which packs against an N-terminal α-helix on one side and a short helix on the other side.39 Similarly in our analysis, a β-sheet in subdomain B was found to be flanked with a α-helix in subdomain A and another α-helix in subdomain B. In total, six β-sheets and two α-helices were predicted, which is in agreement with the previous report.39 However, in order to gain further insights into the structural features of StNAC domains, three-dimensional structure determination by X-ray crystallography would be required in future.
Figure 3.
Phylogenetic relationship and conserved motif compositions of StNAC proteins. (A) Multiple sequence alignment of 136 full-length StNAC proteins was done using ClustalW2, and the phylogenetic tree was constructed using MEGA5.05 by the Neighbor-joining method with 1000 bootstrap replicates. The tree was divided into 12 phylogenetic subgroups designated as I to XII marked with different colour backgrounds. (B) Schematic representation of the conserved motifs in the StNAC proteins as revealed by MEME analysis. Grey lines represent the non-conserved sequences, and each motif is represented by a box numbered at the bottom. The length of protein can be estimated using the scale at the bottom.
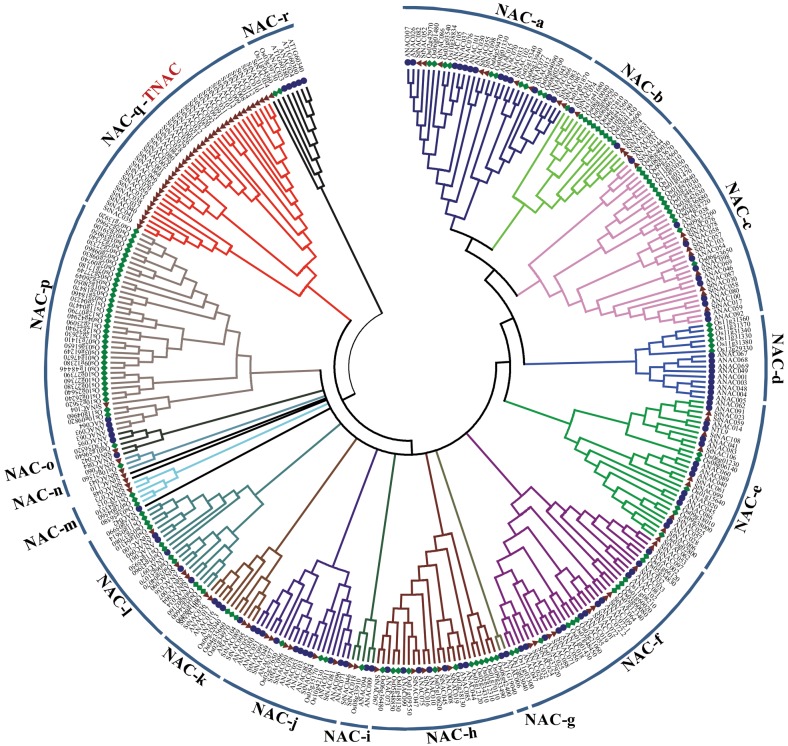
To examine the phylogenetic relationship of StNAC proteins with dicot (Arabidopsis) and monocot (rice) model plant systems, an unrooted tree was made from the alignments of full-length NAC protein sequences. The phylogenetic tree divided StNACs into 18 distinct subgroups (NAC-a to NAC-r) along with their Arabidopsis and rice orthologs (Fig. 4). In general, the Arabidopsis, rice and potato NAC proteins were distributed uniformly in all the subgroups. Exceptionally, NAC-d subgroup contains only Arabidopsis and rice NACs, but no potato NAC. Remarkably, NAC-q subgroup contains 36 potato NACs, but no Arabidopsis and rice NAC. This observation suggests that diversification and expansion of StNACs present in the NAC-q subgroup took place after the divergence of potato, Arabidopsis and rice. Previously, tobacco NAC family was shown to contain a Solanaceae-specific novel subfamily, TNAC, that contains approximately 50 TNAC genes.9 We sought to determine whether these 36 StNACs clustered in the NAC-q subgroup belong to the TNAC subfamily. Multiple sequence alignment of NAC domain sequences of all 136 StNACs along with three representative Arabidopsis NACs (ANAC019, ANAC055 and ANAC072), two tobacco NACs (NCBI accession numbers BAA78417and ADQ08688) and seven tobacco TNACS (NCBI accession numbers ACF19785, ACF19786, ACF19787, ACF19788, ACF19789, ACF19790 and ACF19791) was carried out, and an unrooted tree was made. Interestingly, StNACs classified in the NAC-q subgroup, clustered together with tobacco TNACS (Supplementary Fig. S6), while rest of the StNACs was clustered separately along with ANACs and tobacco NACs. Thus, we suggest that these 36 StNACs may be designated as TNACs, which were also subdivided into three clades represented by A, B and C as proposed earlier.9 Our analysis provides further evidence that TNAC subfamily is exclusive to Solanaceae family. However, their functional characterization would be required to ascertain if they play some unique role(s) in plant processes, in which NAC proteins have not been implicated, so far.
Figure 4.
Phylogenetic tree of NAC proteins of potato, Arabidopsis and rice. Multiple sequence alignment of full-length NAC proteins was done using ClustalW2, and the phylogenetic tree was constructed using MEGA5.05 by the Neighbor-joining method with 1000 bootstrap replicates. The tree was divided into 18 phylogenetic subgroups, designated as NAC-a to NAC-r. Members of potato, Arabidopsis and rice were denoted by triangle, circle and diamond respectively. Subgroup NAC-q represents the TNAC subgroup, which seems restricted to Solanaceae.
3.4. Membrane-bound StNAC subfamily
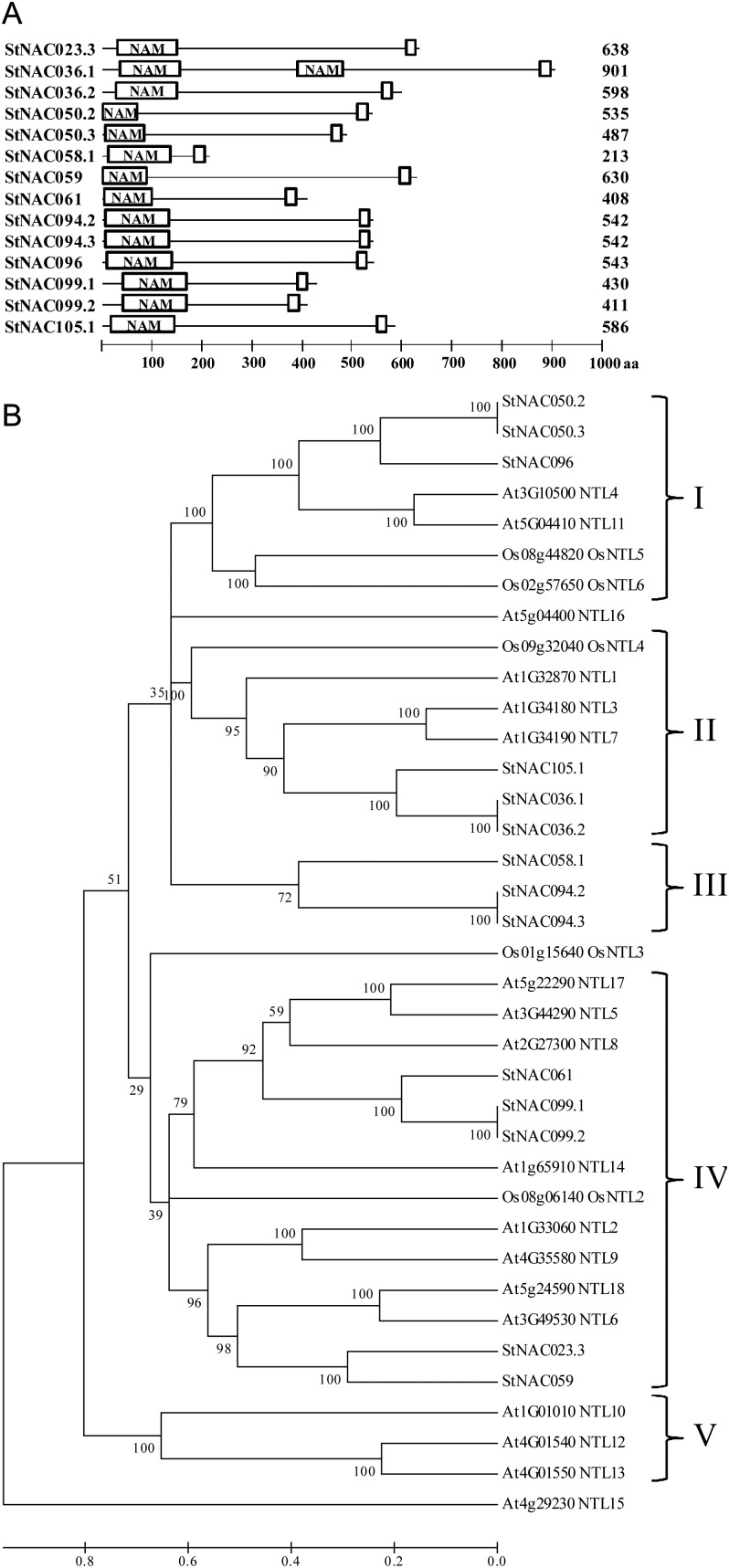
NAC membrane-bound TFs (MTFs) have been implicated in plant response to abiotic stress.15,17,37 Using TMHMM server v. 2.0, we identified 14 (∼10%) StNAC proteins containing α-helical TMs (Fig. 5A and Supplementary Table S8). Notably, primary transcripts of a large number of StNAC MTF genes (7 of 10) are alternatively spliced, which also code for proteins lacking the TM (Table 1 and Fig. 5A), suggesting that their activity may also be regulated at protein level through interaction between full-length and the alternatively spliced forms. Similar to Arabidopsis and rice NAC MTFs,38 all the identified StNAC MTFs also contain single TM at their C-terminal (Fig. 5A). Recently in soybean, of 152 GmNACs, 11 have been predicted to contain TMs. However, GmNAC013 and GmNAC136 were found to contain two TMs.8 Previously, 13 members of the Arabidopsis NAC family were predicted to be membrane-associated and named as NTL 1–13 (for NTM1 like).61 Later, a genome-wide analysis predicted 18 NTLs in Arabidopsis and 5 NTLs (OsNTLs) in rice.38 However, they have not assigned nomenclature for additional five Arabidopsis NTLs. Thus, to maintain uniformity, numbers from 14 to 18 are assigned to additional NTLs in this study. Phylogenetic analysis of the potato, Arabidopsis and rice NAC MTFs divided them into five clades (Fig. 5B). Maximum (14) NTLs were clustered together in Clade IV, followed by 7 each in Clades I and II, and 3 each in Clades III and V. In future, functional characterization of StNAC MTFs may identify candidate genes to engineering abiotic stress tolerance in potato and other Solanaceae plants, as well.
Figure 5.
Membrane-bound potato NAC proteins. (A) Protein structure of membrane-bound NAC TFs. The highly conserved NAM domain is shown at the N-terminal of the proteins. α-helical TMs located at the C-terminal are shown as open box. The number of total amino acid residues in each protein is shown at the right side of each protein structure. (B) Phylogenetic relationship of membrane-bound NAC proteins of potato with that of Arabidopsis and rice. Multiple sequence alignment of full-length NAC MTF proteins was done using ClustalW2, and the phylogenetic tree was constructed using MEGA5.05 by the Neighbor-joining method with 1000 bootstrap replicates. The tree was divided into five phylogenetic subgroups designated as I to V. The scale at the bottom represents relative divergence of the sequences examined, and bootstrap values are displayed next to the branch.
3.5. Differential expression of StNAC genes in various tissues/developmental stages
To identify overlapping and tissue-specific expression profile of StNAC genes, we utilized transcriptome data derived from Illumina RNA-Seq reads generated by PGSC41 and analysed by Massa et al.49 The potato RNA-seq data provide the expression of over 22 000 potato genes in 16 tissues representing major organs and developmental stages, grouped into five major classes; floral (carpels, petals, sepals, stamens and whole mature flower), fruit (immature, mature and inside of fruit), stolon/tubers (stolons, tuber1 and tuber2), leaf (leaves, petioles) and other tissues (shoots, roots and callus).
Transcript abundance of 69 StNACs in 16 different developmental stages and organs was obtained, while rest of the 41 StNACs either transcribe at too low level to be detected or have spatial and temporal expression pattern not covered in the RNA-seq libraries. Of these 69 StNACs, 20 (∼29%) are ubiquitously expressed in all 16 tissues, while 21 (∼30%) express in 1–5, 11 (10%) in 6–10 and 17 (∼15%) in 11–15 number of tissues (Fig. 6). Some of the StNACs also exhibit tissue-specific expression, for example, StNAC034 and StNAC075 express only in floral tissues, StNAC002, StNAC025, StNAC087 and StNAC091 in fruit tissues, StNAC073 in stolon/tuber tissues and StNAC082 specifically in root tissue (Fig. 6). These observations indicate that various StNACs may be associated with diversified functions similar to their Arabidopsis orthologs, for example, ANAC098 (CUC2; ortholog of StNAC034) regulates gynoecium development62 and Arabidopsis, vascular-related NAC domain 5 (VND5; ortholog of StNAC082), regulates the differentiation of root protoxylem vessels in co-operation with other VND proteins.63 The tissue-specific expression profiling of StNACs might enable the combinatorial usage of StNACs in transcriptional regulation of different tissues, whereas ubiquitously expressed StNACs might regulate the transcription of a broad set of genes. For example, a rice NAC gene, OsNAC10 predominantly expressed in roots and panicles and induced by drought, salinity and ABA, when overexpressed with root-specific promoter RCc3, improved root growth, enhanced drought tolerance and increased grain yield significantly under field drought conditions.31
Figure 6.
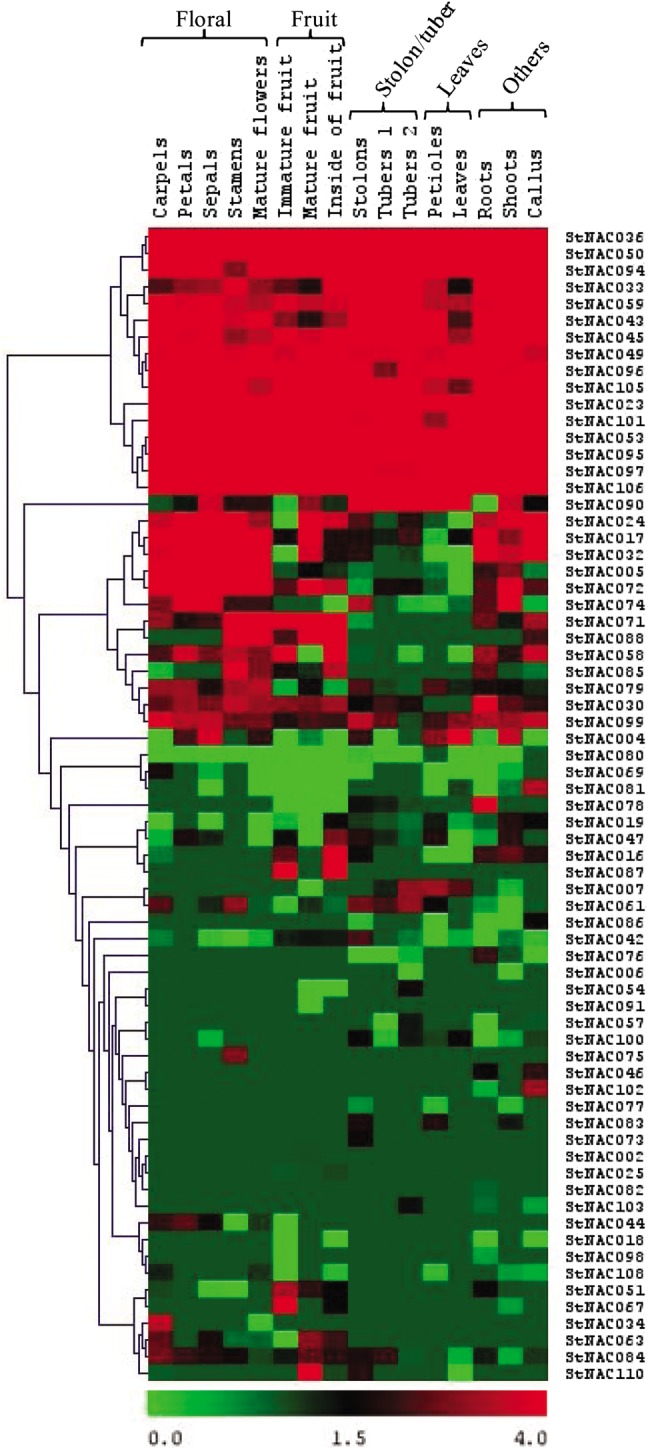
Heat map representation and hierarchical clustering of StNAC genes across different tissues and developmental stages. The Illumina RNA-seq data were reanalyzed, and the FPKM values were log2 transformed and heat map generated using TIGR MeV v4.1.1 software. Bar at the bottom represents log2 transformed values, thereby values 0, 1.5 and 4.0 represent low, intermediate and high expression, respectively.
3.6. Differential expression of StNAC genes during abiotic and biotic stresses
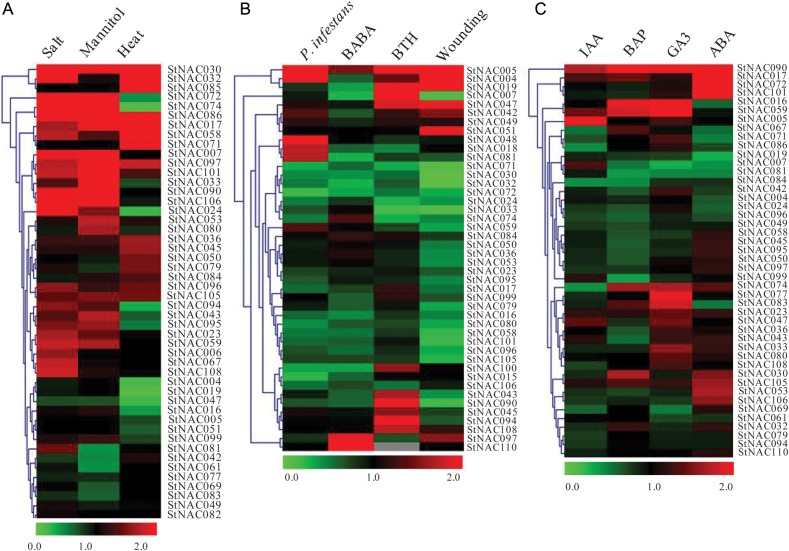
Several NAC proteins have been shown to play important roles in biotic and abiotic stress responses in plants.23,24 A microarray analysis in rice revealed induction of 46 NAC genes under abiotic and 26 by biotic stress.6 Thus, to identify the stress-responsive StNAC genes, we performed comprehensive expression profiling of StNAC genes using the Illumina RNA-Seq data. Abiotic stress treatments (24 h treatment of in vitro grown whole plants) include salt (150 mM NaCl), mannitol (260 µM) and heat (35°C). Relative transcript abundance for each treatment was calculated with respect to their respective controls.
Under abiotic stress treatments, 48 StNAC genes express in one or more of the conditions. Of these 48 StNACs, StNAC017, StNAC030, StNAC086 and StNAC097 were found to be induced under all the three stresses, namely salt, mannitol and heat treatments (Fig. 7A). Previously, overexpression of multiple stress-responsive NAC genes, such as OsNAC6, ONAC063, ONAC045 and SNAC2, conferred multiple abiotic stresses in transgenic plants.28–30 Some of the StNACs also exhibit induction under specific stress conditions, for example, StNAC024, StNAC067 and StNAC108 were induced specifically under salt stress, while StNAC053 and StNAC080 induced only under mannitol treatment and StNAC071 and StNAC085 induced under heat stress only (Fig. 7A). Interestingly, expression of Arabidopsis RD26 orthologs, StNAC072 and StNAC101, was highly induced by salt, mannitol and ABA treatments (Fig. 7A and C). Previously, expression of RD26 was found to be induced by dehydration and ABA and its overexpression conferred hypersensitivity to ABA in transgenic Arabidopsis, while RD26 repressed plants were insensitive.20 Overexpression of multiple stress-responsive rice NAC gene, OsNAC6 having high sequence similarity with Arabidopsis RD26, conferred dehydration and salinity stress tolerance in rice.28,29 Functional characterization of RD26 orthologs identified in this study may provide opportunities to develop abiotic stress tolerant transgenic potato and other Solanaceae crops.
Figure 7.
Heat map representation and hierarchical clustering of StNAC genes during (A) abiotic stress, (B) biotic stress and (C) hormone treatments. The Illumina RNA-seq data were reanalyzed, and the relative expression was calculated with respect to respective control (untreated) samples. Heat maps were generated using the TIGR MeV v4.1.1 software. Bar at the bottom of each heat map represents relative expression values, thereby values 0, 1.0 and 2.0 represent downregulated, unaltered and upregulated expression, respectively.
The biotic stress treatments (pooled samples at 24 h, 36 h, 72 h) include induction with P. infestans inoculum (Pi isolate US8:Pi02–007) and two chemical elicitors, acibenzolar-s-methyl (BTH, 100 µg/ml) and DL-β-amino-n-butyric acid (BABA, 2 mg/ml), using detached leaves and wounded leaves to mimic herbivory. A total of 44 StNACs were found to be expressed in one or more of the biotic stress conditions (Fig. 7B). Interestingly, StNAC005 was found to be induced under all the biotic stress conditions, except BABA treatment. Previously, its Arabidopsis ortholog, ANAC104 (AT5G64530.1; Table 1) was shown to be highly induced in Arabidopsis, challenged with plant pathogen Pseudomonas syringae pv. tomato DC3000 and human pathogen Escherichia coli O157:H7.64 StNAC004 was also induced under P. infestans infection and wounding, but downregulated under BABA treatment. Expression of StNAC018, StNAC048 and StNAC081 was induced only under P. infestans infection (Fig. 7B). Expression of StNAC097 and StNAC110 was induced only under BABA treatment, whereas expression of StNAC007, StNAC090 and StNAC094 was induced only under BTH treatment. StNAC051 was induced only under wounding stress. Previously, NAC proteins were shown to positively regulate defence response by activating pathogenesis-related genes, which in turn induce hypersensitive response and cell death at the site of infection.21 In contrast, NAC proteins have also been shown to negatively regulate defence response by suppressing defence-related gene expression.35 In future, it would be interesting to functionally characterize these biotic stress-responsive StNAC genes and to classify them as positive and negative regulators of pathogen defence response, especially against P. infestans infection.
3.7. Differential expression of StNAC genes during hormone treatments
NAC proteins have been shown to regulate a variety of plant processes by mediating hormone signalling. Thus, to identify hormone-responsive StNAC genes, we analysed the Illumina RNA-seq data, which include indole-3-acetic acid (IAA, 10 µM), 6-benzylaminopurine (BAP, 10 µM), gibberellic acid (GA3, 50 µM) and ABA (50 µM) treatment to in vitro grown whole plants for 24 h.49 Of 110 StNAC genes, 45 express under one or more of the hormone treatments (Fig. 7C). Interestingly, expression of StNAC090 was induced under all the phytohormone treatments that were analysed in this study. Expression of StNAC016 and StNAC059 was induced under both, BAP and GA3 treatments. In Fig. 5, we showed that StNAC059 is a membrane-bound NAC TF. A membrane-bound, cytokinin-inducible Arabidopsis NAC TF, NTM1 regulates cytokinin signalling during cell division.18 Similarly, Arabidopsis NTL8 regulates salt-responsive flowering via FLOWERING LOCUS T15 and mediates salt regulation of seed germination via the GA pathway.37 NTL8 expression was found to be induced by high salinity, but was unaffected by ABA. Similarly, StNAC059 expression was induced by salt stress (Fig. 7A), but remained unaffected by ABA treatment (Fig. 7C). Interestingly, StNAC059 and Arabidopsis NTL8 clustered together in Clade IV (Fig. 5B), indicating that they also share sequence similarity with each other. StNAC005 was induced only under IAA treatment. Overexpression of its Arabidopsis ortholog, ANAC104/XND1 (AT5G64530), resulted in extreme dwarfism associated with the absence of xylem vessels and little or no expression of tracheary element marker genes. Previously, differentiation of tracheary elements was shown to be enhanced by auxin.65 In addition, StNAC017, StNAC072, StNAC090 and StNAC101 were found to be highly responsive to ABA. These observations indicate that function of some of the NAC proteins might be conserved among species.
3.8. Validation of expression pattern of StNAC genes using qRT-PCR
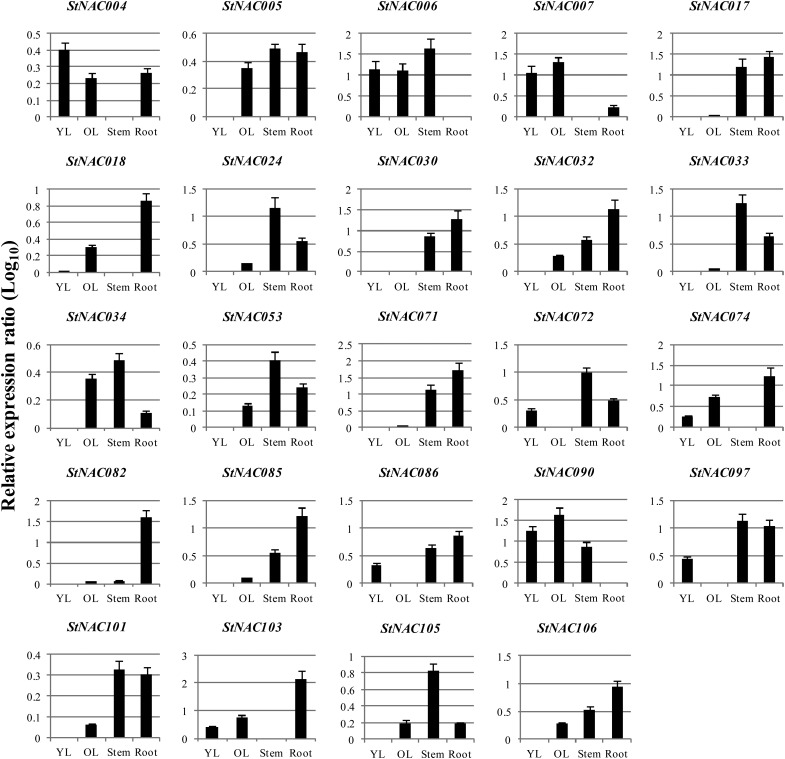
Expression profiling of members of large gene families using publicly available data (for, e.g. EST, microarray, MPSS and RNA-seq data), followed by validation of the expression pattern of selected genes using qRT-PCR, is a valuable approach, which provides preliminary indications about the function of newly identified genes and often been recently exploited.7,8 However, in some instances, data obtained from different methods may differ. Thus, in order to validate the expression pattern of StNAC genes, we have carefully selected few representative StNAC genes with diverse expression patterns and performed qRT-PCR analysis. As shown in Fig. 8, the qRT-PCR results of (22 of 24) representative StNAC genes in young leaf (YL), old leaf (OL), stem and root tissues of potato were found to be largely in good agreement with the RNA-seq data (Fig. 6). However, only in case of two genes (StNAC074 and StNAC034), qRT-PCR data differed from the RNA-seq data. These minor differences could be either due to difference in the stage of the plant at which the samples were collected or could be genotype dependent. For example, all the samples for RNA-seq analysis were collected from greenhouse grown plants, except root and shoot tissues, which were collected from in vitro grown plants,49 whereas, in the present study, all the samples were collected from in vitro raised hardened plantlets grown for 2 months in greenhouse.
Figure 8.
The relative expression ratio of 24 representative StNAC genes in young leaf (YL), old leaf (OL), stem and root tissues of potato determined using qRT-PCR. Relative expression ratios in different tissue samples have been calculated with reference to tissue sample in which the respective transcript exhibited the lowest expression. The relative expression values were log10 transformed. qRT-PCR data were normalized using potato elongation factor 1-α gene. The name of the gene is written on the top of each bar diagram. (Error bars indicate standard deviation.)
In another experiment, we have carried out qRT-PCR analysis of 16 representative StNAC genes under salt (100 mM NaCl), PEG 6000 (10%), heat (42°C) and ABA (100 µM) treatments to validate the expression pattern as revealed by RNA-Seq analysis. In addition, cold (4°C) and SA (300 µM) treatments were also included as one of the most prominent abiotic stresses and elicitor of the biotic stress response, respectively. The qRT-PCR results under these treatments also corroborate the expression profile as revealed by RNA-seq analysis. For example, expression of StNAC030 was induced after 4 h of salt stress imposition and maintained upto 24 h, whereas its expression was induced after 24 h of heat and ABA treatment (Fig. 9), corroborating the RNA-seq data (Fig. 7). Expression of Arabidopsis RD26 orthologs, StNAC072 and StNAC101, was also found to be highly induced by stress and ABA treatments, which is in agreement with the RNA-seq data (Fig. 7A and C) and previous reports.20 These results strongly suggest that preliminary expression profiling using publicly available expression data followed by its validation using qRT-PCR provide more reliable expression profile of members of large gene families in less time with reduced expenditure.
Figure 9.
The relative expression ratio of 16 representative StNAC genes analysed by qRT-PCR under stress treatments for 4 h (grey bars) and 24 h (black bars). The relative expression ratio of each gene was calculated relative to its expression in control sample. qRT-PCR data were normalized using potato elongation factor 1-α gene. The name of the gene is written on the top of each bar diagram. (Error bars indicate standard deviation.)
4. Conclusions
The present effort to identify and describe key attributes of uncharacterized NAC TFs in potato genome using high-throughput genome-wide survey, and utilization of available expression data coupled with molecular tools provides foundation of our understanding of their regulatory roles. Our comprehensive genome-wide analysis led to identification of 136 NAC TF proteins encoded by 110 genes in potato. A uniform nomenclature and annotation was provided to the identified genes and proteins, followed by their comparative phylogenetic analysis with Arabidopsis and rice NAC TFs. Phylogenetic analysis led to identification of TNAC subfamily comprising of 36 StNACs. Similar to tobacco, the presence of TNAC subfamily in potato provides further evidence of its existence in Solanaceae plants only. Considering the fact that most of the biological functions played by NAC TFs have been revealed using Arabidopsis NAC genes, we assigned Arabidopsis orthologs to each StNAC protein. The comparative analysis of StNACs with their respective Arabidopsis ortholog helped us to predict the potential functions of several StNAC proteins. The availability of potato transcriptome data generated by the Illumina RNA-seq approach has been exploited as a useful tool for preliminary analysis of gene expression and identified tissue-specific, stress- and hormone-responsive StNAC genes. Additional experiments through their over- and/or under-expression will help in determining the precise function of these genes. It will also be intriguing to identify and functionally to characterize their promoters, which may be utilized to engineer potato plants with improved performance under stressful conditions, in future. Thus, this analysis provides preliminary indications of putative function of several StNAC genes, which will help in channelizing directional efforts for their functional characterization.
Supplementary data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Acknowledgements
We sincerely acknowledge the Council of Scientific and Industrial Research, New Delhi, for financial support in the form of projects MLP0001 and BSC0107. This manuscript represents CSIR-IHBT communication number 3478.
Footnotes
Edited by Dr Satoshi Tabata
References
- 1.Papademetriou M.K. Proceedings of the workshop to commemorate the international year of the potato2008; Bangkok, Thailand. Food and Agricultural Organization of the United Nations, Regional Office for Asia and the Pacific; 2008. [Google Scholar]
- 2.Levy D., Veilleux R.E. Adaptation of potato to high temperature and salinity-a review. Am. J. Potato Res. 2007;84:487–506. doi:10.1007/BF02987885. [Google Scholar]
- 3.Arvin M.J., Donnelly D.J. Screening potato cultivars and wild species to abiotic stresses using a electrolyte leakage bioassay. J. Agric. Sci. Technol. 2008;10:33–42. [Google Scholar]
- 4.Fry W.E. Phytophthora infestans, the crop (and R gene) destroyer. Mol. Plant Pathol. 2008;9:385–402. doi: 10.1111/j.1364-3703.2007.00465.x. doi:10.1111/j.1364-3703.2007.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aida M., Ishida T., Fukaki H., Fujisawa H.,, Tasaka M. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997;9:841–57. doi: 10.1105/tpc.9.6.841. doi:10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nuruzzaman M., Manimekalai R., Sharoni A.M., et al. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465:30–44. doi: 10.1016/j.gene.2010.06.008. doi:10.1016/j.gene.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Hu R., Qi G., Kong Y., Kong D., Gao Q.,, Zhou G. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 2010;10:145. doi: 10.1186/1471-2229-10-145. doi:10.1186/1471-2229-10-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le D.T., Nishiyama R., Watanabe Y., et al. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 2011;18:263–76. doi: 10.1093/dnares/dsr015. doi:10.1093/dnares/dsr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rushton P.J., Bokowiec M.T., Han S., et al. Tobacco transcription factors: novel insights into transcriptional regulation in the Solanaceae. Plant Physiol. 2008;147:280–95. doi: 10.1104/pp.107.114041. doi:10.1104/pp.107.114041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang N., Zheng Y., Xin H., Fang L.,, Li S. Comprehensive analysis of NAC domain transcription factor gene family in Vitis vinifera. Plant Cell Rep. 2013;32:61–75. doi: 10.1007/s00299-012-1340-y. doi:10.1007/s00299-012-1340-y. [DOI] [PubMed] [Google Scholar]
- 11.Souer E., van Houwelingen A., Kloos D., Mol J.,, Koes R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordial boundaries. Cell. 1996;85:159–70. doi: 10.1016/s0092-8674(00)81093-4. doi:10.1016/S0092-8674(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 12.Aida M., Ishida T.,, Tasaka M. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development. 1999;126:1563–70. doi: 10.1242/dev.126.8.1563. [DOI] [PubMed] [Google Scholar]
- 13.Xie Q., Guo H.S., Dallman G., Fang S., Weissman A.M.,, Chua N.H. SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature. 2002;419:167–70. doi: 10.1038/nature00998. doi:10.1038/nature00998. [DOI] [PubMed] [Google Scholar]
- 14.Sablowski R.W.M.,, Meyerowitz E.M. A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell. 1998;92:93–103. doi: 10.1016/s0092-8674(00)80902-2. doi:10.1016/S0092-8674(00)80902-2. [DOI] [PubMed] [Google Scholar]
- 15.Kim S.G., Kim S.Y.,, Park C.M. A membrane-associated NAC transcription factor regulates salt-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Planta. 2007;226:647–54. doi: 10.1007/s00425-007-0513-3. doi:10.1007/s00425-007-0513-3. [DOI] [PubMed] [Google Scholar]
- 16.Yoo S.Y., Kim Y., Kim S.Y., Lee J.S.,, Ahn J.H. Control of flowering time and cold response by a NAC-domain protein in Arabidopsis. PLoS ONE. 2007;2:e642. doi: 10.1371/journal.pone.0000642. doi:10.1371/journal.pone.0000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S., Seo P.J., Lee H.J.,, Park C.M. A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 2012;70:831–44. doi: 10.1111/j.1365-313X.2012.04932.x. doi:10.1111/j.1365-313X.2012.04932.x. [DOI] [PubMed] [Google Scholar]
- 18.Kim Y.S., Kim S.G., Park J.E., et al. A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell. 2006;18:3132–44. doi: 10.1105/tpc.106.043018. doi:10.1105/tpc.106.043018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willemsen V., Bauch M., Bennett T., et al. The NAC domain transcription factors FEZ and SOMBRERO control the orientation of cell division plane in Arabidopsis root stem cells. Dev. Cell. 2008;15:913–22. doi: 10.1016/j.devcel.2008.09.019. doi:10.1016/j.devcel.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 20.Fujita M., Fujita Y., Maruyama K., et al. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004;39:863–76. doi: 10.1111/j.1365-313X.2004.02171.x. doi:10.1111/j.1365-313X.2004.02171.x. [DOI] [PubMed] [Google Scholar]
- 21.Jensen M.K., Hagedorn P.H., Torres-Zabala M., et al. Transcriptional regulation by an NAC (NAM-ATAF1,2-CUC2) transcription factor attenuates ABA signalling for efficient basal defence towards Blumeria graminis f. Sp. Hordei in Arabidopsis. Plant J. 2008;56:867–80. doi: 10.1111/j.1365-313X.2008.03646.x. doi:10.1111/j.1365-313X.2008.03646.x. [DOI] [PubMed] [Google Scholar]
- 22.Uauy C., Distelfeld A., Fahima T., Blechl A.,, Dubcovsky J. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science. 2006;314:1298–301. doi: 10.1126/science.1133649. doi:10.1126/science.1133649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakashima K., Takasaki H., Mizoi J., Shinozaki K.,, Yamaguchi-Shinozaki K. NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta. 2012;1819:97–103. doi: 10.1016/j.bbagrm.2011.10.005. doi:10.1016/j.bbagrm.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Puranik S., Sahu P.P., Srivastava P.S.,, Prasad M. NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 2012;17:6369–81. doi: 10.1016/j.tplants.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 25.Yamaguchi-Shinozaki K., Koizumi M., Urao S.,, Shinozaki K. Molecular cloning and characterization of 9 cDNAs for genes that are responsive to desiccation in Arabidopsis thaliana: sequence analysis of one cDNA clone that encodes a putative transmembrane channel protein. Plant Cell Physiol. 1992;33:217–24. [Google Scholar]
- 26.Tran L.S.P., Nakashima K., Sakuma Y., et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16:2481–98. doi: 10.1105/tpc.104.022699. doi:10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu H., Dai M., Yao J., et al. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA. 2006;103:12987–92. doi: 10.1073/pnas.0604882103. doi:10.1073/pnas.0604882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakashima K., Tran L.S., Van Nguyen D., et al. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007;51:617–30. doi: 10.1111/j.1365-313X.2007.03168.x. doi:10.1111/j.1365-313X.2007.03168.x. [DOI] [PubMed] [Google Scholar]
- 29.Hu H., You J., Fang Y., Zhu X., Qi Z.,, Xiong L. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol. Biol. 2008;67:169–81. doi: 10.1007/s11103-008-9309-5. doi:10.1007/s11103-008-9309-5. [DOI] [PubMed] [Google Scholar]
- 30.Zheng X., Chen B., Lu G.,, Han B. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem. Biophys. Res. Commun. 2009;379:985–9. doi: 10.1016/j.bbrc.2008.12.163. doi:10.1016/j.bbrc.2008.12.163. [DOI] [PubMed] [Google Scholar]
- 31.Jeong J.S., Kim Y.S., Baek K.H., et al. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010;153:185–97. doi: 10.1104/pp.110.154773. doi:10.1104/pp.110.154773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeong J.S., Kim Y.S., Redillas M.C., et al. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 2013;11:101–14. doi: 10.1111/pbi.12011. doi:10.1111/pbi.12011. [DOI] [PubMed] [Google Scholar]
- 33.Xue G.P., Way H.M., Richardson T., Drenth J., Joyce P.A.,, McIntyre C.L. Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol. Plant. 2011;4:697–712. doi: 10.1093/mp/ssr013. doi:10.1093/mp/ssr013. [DOI] [PubMed] [Google Scholar]
- 34.Selth L.A., Dogra S.C., Rasheed M.S., Healy H., Randles J.W.,, Rezaian M.A. A NAC domain protein interacts with Tomato leaf curl virus replication accessory protein and enhances viral replication. Plant Cell. 2005;17:311–25. doi: 10.1105/tpc.104.027235. doi:10.1105/tpc.104.027235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X., Basnayake B.M., Zhang H., et al. The Arabidopsis ATAF1, a NAC transcription factor, is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens. Mol. Plant Microbe Interact. 2009;22:1227–38. doi: 10.1094/MPMI-22-10-1227. doi:10.1094/MPMI-22-10-1227. [DOI] [PubMed] [Google Scholar]
- 36.Ooka H., Satoh K., Doi K., et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10:239–47. doi: 10.1093/dnares/10.6.239. doi:10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- 37.Kim S.G., Lee S., Seo P.J., Kim S.K., Kim J.K.,, Park C.M. Genome-scale screening and molecular characterization of membrane-bound transcription factors in Arabidopsis and rice. Genomics. 2010;95:56–65. doi: 10.1016/j.ygeno.2009.09.003. doi:10.1016/j.ygeno.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 38.Kim S.Y., Kim S.G., Kim Y.S., et al. Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucleic Acids Res. 2007;35:203–13. doi: 10.1093/nar/gkl1068. doi:10.1093/nar/gkl1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ernst H.A., Olsen A.N., Larsen S.,, Leggio L.L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 2004;5:297–303. doi: 10.1038/sj.embor.7400093. doi:10.1038/sj.embor.7400093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Q., Wang Q., Xiong L.,, Lou Z. A structural view of the conserved domain of rice stress-responsive NAC1. Protein Cell. 2011;2:55–63. doi: 10.1007/s13238-011-1010-9. doi:10.1007/s13238-011-1010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu X., Pan S., Cheng S., et al. Genome sequence and analysis of the tuber crop potato. Nature. 2011;475:189–95. doi: 10.1038/nature10158. doi:10.1038/nature10158. [DOI] [PubMed] [Google Scholar]
- 42.Jupe F., Pritchard L.,, Etherington G.J., et al. Identification and localization of the NB-LRR gene family within the potato genome. BMC Genomics. 2012;13:75. doi: 10.1186/1471-2164-13-75. doi:10.1186/1471-2164-13-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lozano R., Ponce O., Ramirez M., Mostajo N.,, Orjeda G. Genome-wide identification and mapping of NBS-encoding resistance genes in Solanum tuberosum group phureja. PLoS One. 2012;7:e34775. doi: 10.1371/journal.pone.0034775. doi:10.1371/journal.pone.0034775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collinge M., Boller T. Differential induction of two potato genes, Stprx2 and StNAC, in response to infection by Phytophthora infestans and to wounding. Plant Mol. Biol. 2001;46:521–9. doi: 10.1023/a:1010639225091. doi:10.1023/A:1010639225091. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y., Tang H., DeBarry J.D., et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. doi:10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamura K., Peterson D., Peterson N., Stecher G., Nei M.,, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. doi:10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bailey T.L., Boden M., Buske F.A., et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8. doi: 10.1093/nar/gkp335. doi:10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pei J., Kim B.H.,, Grishin N.V. PROMALS3D: a tool for multiple sequence and structure alignment. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. doi:10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Massa A.N., Childs K.L., Lin H., Bryan G.J., Giuliano G.,, Buell C.R. The transcriptome of the reference potato genome Solanum tuberosum Group Phureja clone DM1–3 516R44. PLoS One. 2011;6:e26801. doi: 10.1371/journal.pone.0026801. doi:10.1371/journal.pone.0026801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saeed A.I., Sharov V., White J., et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 51.Murashige T., Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962;15:473–97. doi:10.1111/j.1399-3054.1962.tb08052.x. [Google Scholar]
- 52.Ghawana S., Paul A., Kumar H., et al. An RNA isolation system for plant tissues rich in secondary metabolites. BMC Res. Notes. 2011;4:85–9. doi: 10.1186/1756-0500-4-85. doi:10.1186/1756-0500-4-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nicot N., Hausman J.F., Hoffmann L.,, Evers D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005;5:2907–14. doi: 10.1093/jxb/eri285. doi:10.1093/jxb/eri285. [DOI] [PubMed] [Google Scholar]
- 54.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. doi:10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 55.Wei K.F., Chen J., Chen Y.F., Wu L.J.,, Xie D.X. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 2012;19:153–64. doi: 10.1093/dnares/dsr048. doi:10.1093/dnares/dsr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manimekalai R., Kikuchi S.,, Satoh K. Alternative splicing events in NAC transcription factor genes in rice. Plant and Animal Genomes XIX Conference; January 15–19, 2011; San Diego, CA. 2011. ; (P259) [Google Scholar]
- 57.Campbell M.A., Haas B.J., Hamilton J.P., Mount S.M.,, Buell C.R. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics. 2006;7:327. doi: 10.1186/1471-2164-7-327. doi:10.1186/1471-2164-7-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kameyama K., Kishi Y., Yoshimura M., Kanzawa N., Sameshima M.,, Tsuchiya T. Tyrosine phosphorylation in plant bending. Nature. 2000;407:37. doi: 10.1038/35024149. doi:10.1038/35024149. [DOI] [PubMed] [Google Scholar]
- 59.Kleinow T., Himbert S., Krenz B., Jeske H.,, Koncz C. NAC domain transcription factor ATAF1 interacts with SNF1-related kinases and silencing of its subfamily causes severe developmental defects in Arabidopsis. Plant Sci. 2009;177:360–70. doi:10.1016/j.plantsci.2009.06.011. [Google Scholar]
- 60.Fang Y., You J., Xie K., Xie W.,, Xiong L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genomics. 2008;280:547–63. doi: 10.1007/s00438-008-0386-6. doi:10.1007/s00438-008-0386-6. [DOI] [PubMed] [Google Scholar]
- 61.Kim S.G., Lee A.K., Yoon H.K.,, Park C.M. A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. Plant J. 2008;55:77–88. doi: 10.1111/j.1365-313X.2008.03493.x. doi:10.1111/j.1365-313X.2008.03493.x. [DOI] [PubMed] [Google Scholar]
- 62.Nahar M.A., Ishida T., Smyth D.R., Tasaka M.,, Aida M. Interactions of CUP-SHAPED COTYLEDON and SPATULA genes control carpel margin development in Arabidopsis thaliana. Plant Cell Physiol. 2012;53:1134–43. doi: 10.1093/pcp/pcs057. doi:10.1093/pcp/pcs057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamaguchi M., Kubo M., Fukuda H.,, Demura T. Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 2008;55:652–64. doi: 10.1111/j.1365-313X.2008.03533.x. doi:10.1111/j.1365-313X.2008.03533.x. [DOI] [PubMed] [Google Scholar]
- 64.Thilmony R., Underwood W.,, He S.Y. Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J. 2006;46:34–53. doi: 10.1111/j.1365-313X.2006.02725.x. doi:10.1111/j.1365-313X.2006.02725.x. [DOI] [PubMed] [Google Scholar]
- 65.Shinohara N., Sugiyama M.,, Fukuda H. Higher extracellular pH suppresses tracheary element differentiation by affecting auxin uptake. Planta. 2006;224:394–404. doi: 10.1007/s00425-006-0224-1. doi:10.1007/s00425-006-0224-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.