Abstract
Maternally expressed gene 3 (MEG3) is an imprinted gene belonging to the imprinted DLK1–MEG3 locus located at chromosome 14q32.3 in humans. Its mouse ortholog, Meg3, also known as gene trap locus 2 (Gtl2), is located at distal chromosome 12. The MEG3 gene encodes a long noncoding RNA (lncRNA) and is expressed in many normal tissues. MEG3 gene expression is lost in an expanding list of primary human tumors and tumor cell lines. Multiple mechanisms contribute to the loss of MEG3 expression in tumors, including gene deletion, promoter hypermethylation, and hypermethylation of the intergenic differentially methylated region. Re-expression of MEG3 inhibits tumor cell proliferation in culture and colony formation in soft agar. This growth inhibition is partly the result of apoptosis induced by MEG3. MEG3 induces accumulation of p53 (TP53) protein, stimulates transcription from a p53-dependent promoter, and selectively regulates p53 target gene expression. Maternal deletion of the Meg3 gene in mice results in skeletal muscle defects and perinatal death. Inactivation of Meg3 leads to a significant increase in expression of angiogenesis-promoting genes and microvessel formation in the brain. These lines of evidence strongly suggest that MEG3 functions as a novel lncRNA tumor suppressor.
Introduction
A tumor suppressor gene is broadly defined as a gene whose product normally inhibits tumor initiation and progression. Inactivation of tumor suppressors plays a major role in the molecular pathogenesis of human neoplasms. Therefore, the search for new tumor suppressors and investigation of their functions are challenging and exciting areas in cancer research. To date, the known tumor suppressors are predominantly protein-coding genes. Their tumor suppression functions are mediated by their gene products – proteins. However, recent studies reveal that another class of genes, whose products are long noncoding RNAs (lncRNAs) with sizes >200 nt, may also play an important role in tumor suppression (Gibb et al. 2011). One such lncRNA gene is maternally expressed gene 3 (MEG3), which is an imprinted gene, located on chromosome 14q32 (Miyoshi et al. 2000). MEG3 was first identified as the ortholog of gene trap locus 2 (Gtl2) in mice (Schuster-Gossler et al. 1998), officially known as Meg3 and located on mouse distal chromosome 12. The MEG3 gene belongs to the DLK1–MEG3 imprinting locus, containing multiple maternally and paternally imprinted genes (Fig. 1), including at least three paternally expressed protein-coding genes and numerous maternally expressed noncoding RNAs (see review da Rocha et al. (2008)). The gene expression in this locus is tightly controlled by at least two differentially methylated regions (DMRs): the intergenic DMR (IG-DMR) and the MEG3-DMR. The imprinted expression of these genes plays an important role in development and growth (see review da Rocha et al. (2008)). In humans, MEG3 is expressed in many normal tissues. The loss of MEG3 expression has been found in various types of human tumors and tumor cell lines. In addition, re-expression of MEG3 inhibits in vitro tumor cell proliferation (Zhang et al. 2003, 2010a, Zhou et al. 2007, Braconi et al. 2011, Wang et al. 2012). Therefore, it has been hypothesized that MEG3 is a tumor suppressor gene. In this review, we will discuss the evidence from studies by us and others to support this hypothesis.
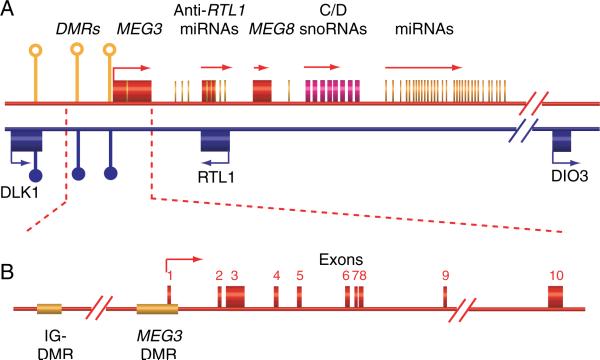
Figure 1.
(A) Schematic representation of the DLK1–MEG3 locus on human chromosome 14. The DLK1–MEG3 locus is w~837 kb long and consists of three known protein-coding genes including DLK1, RTL1, and DIO3, noncoding RNAs including at least three lncRNAs, and numerous small nucleolar RNAs (snoRNAs) and micro-RNAs (miRNAs). The maternal chromosome is in red and the paternal chromosome in blue. Differentially methylated regions are shown as circles. Filled circle, methylated; open circle, unmethylated. (B) The MEG3 gene is 35 kb long and consists of ten exons. The IG-DMR is ~13 kb upstream of the MEG3 gene. The MEG3-DMR overlaps with the MEG3 promoter.
Identification of MEG3 as a pituitary tumor-associated gene by representational difference analysis
A major focus of our research is the molecular pathogenesis of human pituitary adenomas. These tumors arise from the anterior pituitary, which contains somatotrophs (producing growth hormone), lactotrophs (prolactin), gonadotrophs (follicle-stimulating hormone (FSH) and luteinizing hormone (LH)), thyrotrophs (thyroid-stimulating hormone), and corticotrophs (adrenocorticotrophic hormone). Pituitary tumors are classified as clinically nonfunctioning adenomas (NFAs), which are mostly derived from gonadotroph cells (Chaidarun & Klibanski 2002), and clinically functioning adenomas, which are derived from the other four types of hormone-secreting cells. Patients with NFAs do not have symptoms associated with excess hormone secretion. All types of human pituitary adenomas are monoclonal in origin (Alexander et al. 1990, Herman et al. 1990, Biller et al. 1992), indicating that these tumors are derived from somatic cells with genetic or epigenetic mutations. To search for new genes linked to NFAs, we compared gene expression profiles between normal pituitaries and NFAs by representational difference analysis (RDA) techniques (Zhang et al. 2002). RDA identifies genes that are over- or under-expressed in tumor tissues compared with normal tissues. RNAs from four normal anterior pituitaries and four NFAs were used in the analysis (Zhang et al. 2002). Four genes identified as the most under-expressed genes in tumors are GADD45γ (GADD45G), histone deacetylase 6, MEG3, and delta-like 1 homolog (Drosophila; DLK1; Zhang et al. 2002). MEG3 and DLK1 were particularly noteworthy because they are known imprinted genes and belong to the same imprinted locus. Importantly, these genes are located on chromosome 14q32, a region proposed to contain putative tumor suppressors (Mutirangura et al. 1998, Bando et al. 1999). DLK1, also known as Pref-1, is a protein-coding gene and functions as an adipocyte differentiation inhibitor (Smas & Sul 1993). At that time, very little was known about the MEG3 gene and we chose to focus on this gene and its function.
MEG3 expression is lost in human clinically NFAs
To confirm the finding that MEG3 is under-expressed in NFAs, we first examined MEG3 expression in five normal anterior pituitaries and eight NFAs of gonadotroph origin by RT-PCR. MEG3s were detected in all five normal pituitaries. In contrast, MEG3 was not detected in any NFA (Zhang et al. 2003). Using in situ hybridization techniques, we detected MEG3 transcripts in gonadotroph cells in the normal human pituitary (Zhang et al. 2003). Therefore, the lack of MEG3 transcripts in NFAs is due to the loss of MEG3 expression. This observation was further validated in our follow-up studies including 16 more normal pituitaries and 50 additional NFAs of gonadotroph origin in which MEG3 expression was examined by conventional or quantitative RT-PCR (qRT-PCR) techniques (Zhao et al. 2005, Gejman et al. 2008, Cheunsuchon et al. 2011). In one study using qRT-PCR, we found that the average expression level of MEG3 in 19 NFAs was < 2% of that seen in normal pituitaries (Cheunsuchon et al. 2011). In an independent study, Mezzomo et al. (2012) reported that MEG3 expression was lost in all five NFAs, which stained positive for LH or FSH. These data demonstrated that MEG3 expression was lost in all human gonadotroph-derived NFAs. We also examined tumors that stained negative for LHβ and FSHβ, and MEG3 expression in these tumors was similar to that in gonadotroph NFAs (Cheunsuchon et al. 2011). In Mezzomo et al.'s (2012) study, four out of nine LHβ- and FSHβ-negative NFAs had lost MEG3 expression. Additional supportive data were obtained using PDFS, a folliculostellate cell line derived from a human NFA in our laboratory (Danila et al. 2000). Using RT-PCR, we did not detect any MEG3 expression in this cell line (Zhang et al. 2010a). These data indicate that loss of MEG3 expression is a common phenomenon in NFAs.
The anterior pituitary contains five types of hormone-secreting cells, all of which express MEG3, as demonstrated by in situ hybridization (Gejman et al. 2008). We examined MEG3 expression in functioning pituitary tumors derived from somatotrophs, lactotrophs, and corticotrophs. We found that MEG3 transcripts were readily detected in all those tumor types assessed by RT-PCR (Gejman et al. 2008). Although MEG3 expression levels appeared lower in functioning tumors than in normal pituitaries, this difference was not statistically significant (Cheunsuchon et al. 2011). Similar results were also reported by Mezzomo et al. (2012). These data indicate that MEG3 expression is exclusively lost in NFAs but not in functioning tumors and suggests that inactivation of the MEG3 gene may play an important role in the development of NFAs.
MEG3 expression is lost in other human cancers
In addition to the normal pituitary, MEG3 is highly expressed in the brain, adrenal gland, and placenta. Its transcripts are also detected in the testes, ovary, pancreas, spleen, mammary gland, and liver (Zhang et al. 2003). Multiple human cancer cell lines of various tissue origins have been examined for MEG3 expression. Loss of MEG3 expression has been found in all cancer cell lines examined including those derived from brain, bladder, bone marrow, breast, cervix, colon, liver, lung, meninges, and prostate (Table 1; Zhang et al. 2003, 2010a, Astuti et al. 2005, Zhou et al. 2007, Braconi et al. 2011, Wang et al. 2012). Loss of MEG3 expression is also found in primary tumors. MEG3 expression is lost or significantly reduced in 25% of neuroblastomas (Astuti et al. 2005), 81% of hepatocellular cancers (Braconi et al. 2011), and 82% of gliomas (Wang et al. 2012). Recently, we examined MEG3 expression in meningiomas and found that loss of MEG3 expression is strongly associated with tumor grade; MEG3 was detected in four of nine grade I, one of 11 grade II, and none of seven grade III meningiomas (Zhang et al. 2010a). These data suggest that loss of MEG3 expression contributes to tumor development in a wide range of tissues.
Table 1.
MEG3 expression is lost in human cancer cell lines
| Cancer |
Cell lines |
MEG3 status |
References |
|---|---|---|---|
| Brain | H4 | Not detected | Astuti et al. (2005) |
| Kelly | Zhang et al. (2003) | ||
| SK-N-AS | Wang et al. (2012) | ||
| SK-N-DZ | |||
| SK-N-F1 | |||
| U251 | |||
| U-87MG | |||
| Bladder | T24 | Not detected | Zhang et al. (2003) |
| 5637 | |||
| Bone marrow | K562 | Not detected | Zhang et al. (2003) |
| Breast | MCF7 | Not detected | Zhang et al. (2003) |
| T47D | |||
| Cervix | HeLa | Not detected | Zhang et al. (2003) |
| Colon | HCT116 | Not detected | Zhang et al. (2003) |
| H29 | Zhou et al. (2007) | ||
| Liver | HepG2 | Not detected | Braconi et al. (2011) |
| Huh-7 | |||
| PLC/PRF/5 | |||
| Lung | H1299 | Not detected | Zhang et al. (2003) |
| Meninges | CH157-MN | Not detected | Zhang et al. (2010a) |
| IOMM-Lee | |||
| Prostate | Du145 | Not detected | Zhang et al. (2003) |
DNA methylation plays a major role in silencing the MEG3 gene in tumors
One of the possible mechanisms whereby MEG3 expression is lost in NFAs is the allelic deletion of the gene. To investigate this possibility, two micro-satellite markers (D14S985 and WI-16835) flanking exons 4–9 of the MEG3 gene were quantified by qPCR in NFAs and normal pituitaries (Zhao et al. 2005). No significant differences in their respective levels were detected between NFAs and normal pituitaries (Zhao et al. 2005). For example, the relative levels for D14S985 are 1·15±0·17 in NFAs and 1·02±0·25 in normal pituitaries (Student's t-test, P=0·447). The relative levels for WI-16835 are 1·17±0·23 in NFAs and 1·04±0·34 in normal pituitaries (P=0·574). These data indicate that there are no genomic deletions in the MEG3 gene in NFAs. Individual exons of the MEG3 gene were also amplified by PCR and the sequences were confirmed. None of the ten exons carried deletions or point mutations in NFAs (Zhao et al. 2005). These findings suggest that genetic alterations do not play a role in MEG3 silencing in NFAs. In meningiomas, allelic deletion was notfoundinrelatively benign, grade I tumors (Zhang et al. 2010a). However, three of 11 grade II and four of seven grade III tumors had MEG3 gene allelic loss (Zhang et al. 2010a). These data indicate that genomic DNA deletion is associated with aggressive tumors.
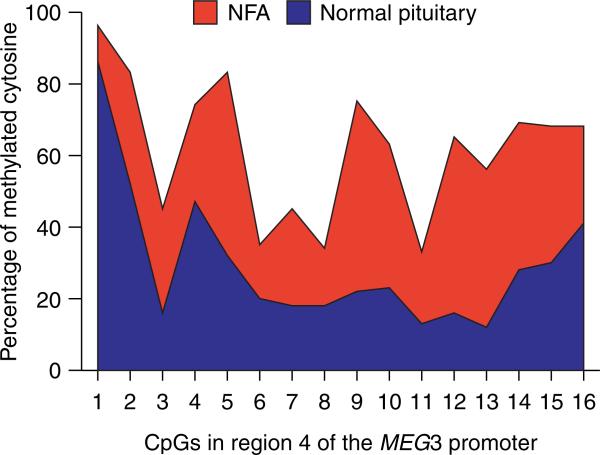
There are two DMRs located upstream of the MEG3 gene, IG-DMR and MEG3-DMR (Fig. 1B). A recent study by Kagami et al. (2010) analyzing patients carrying microdeletions at these DMRs indicates that the IG-DMR and MEG3-DMR are imprinting control centers for the DLK1–MEG3 locus in the placenta and the body respectively. In the body, however, the methylation status of the MEG3-DMR is governed by the IG-DMR, suggesting a hierarchical relationship between these two DMRs (Kagami et al. 2010). MEG3-DMR covers a DNA segment that starts ~1·5 kb upstream of the first MEG3 exon and extends to the first intron (Paulsen et al. 2001, Takada et al. 2002). Kagami et al. (2010) reported that MEG3 transcripts were not detected in cells obtained from a patient with a microdeletion at the MEG3-DMR, indicating that this region is required for expression of the MEG3 gene. The MEG3 promoter overlaps with the MEG3-DMR and is GC rich (Zhao et al. 2005). Bisulfite sequencing revealed that the methylation in regions 1 and 4 of the MEG3 promoter is significantly higher in NFAs than in normal pituitary (Zhao et al. 2005; Fig. 2). Interestingly, this promoter hypermethylation is also observed in meningiomas (Zhang et al. 2010a). Astuti et al. (2005) reported that the MEG3-DMR is completely methylated in neuroblastoma cell lines. These studies indicate that inactivation of the MEG3 gene in these tumors is in part attributed to promoter silencing by hypermethylation. We have identified a transcription factor-binding site within the MEG3 promoter, which is a cAMP response element (CRE). Deletion of the CRE significantly reduces promoter transcription activity (Zhao et al. 2006). In addition, the CRE sequences contain a CpG dinucleotide, suggesting that methylation of this CpG may prevent transcription factors from binding to the site. This likely plays a role in promoter silencing of the MEG3 gene in tumors.
Figure 2.
Hypermethylation of the MEG3 promoter in NFAs. Region 4 of the MEG3 promoter contains 33 CpG dinucleotides. The methylation status of the first 16 CpGs are shown (Zhao et al. 2005). The percentage of methylated CpGs in tumors is in red and that in normal pituitaries is in blue.
The IG-DMR is ~13 kb upstream of the MEG3 gene (Paulsen et al. 2001, Takada et al. 2002). Hypomethylation of this region in the maternal allele is required for activation of all MEGs, including MEG3, and correct imprinting of the DLK1–MEG3 locus. In mice, maternal deletion of the IG-DMR silences MEGs and activates paternally expressed genes, which are normally silenced on the maternal chromosome (Lin et al. 2007). In humans, MEG3 expression was lost in cells obtained from a patient carrying a small DNA deletion containing the IG-DMR (Kagami et al. 2010). Therefore, it is believed that the IG-DMR is required for activation of its downstream MEG (see review da Rocha et al. (2008)). An increase in IG-DMR methylation has been found in human NFAs (Gejman et al. 2008). Interestingly, the degree of the methylation increase correlates with meningioma tumor grade (Zhang et al. 2010a). For example, the percentages of methylated CpG in IG-DMR are 50·1, 56·4, 61·0 and 68·8% in normal meninges, grade I, grade II, and grade III meningiomas respectively. Hypermethylation of the IG-DMR is also reported in neuroblastoma cell lines, a Wilms’ tumor, two neuroblastomas, and a pheochromocytoma (Astuti et al. 2005). More importantly, treatment with a demethylating agent, 5-aza-2-deoxycytidine, resulted in re-expression of MEG3 in multiple human breast cancer tumor cell lines (Zhao et al. 2005), meningiomas (Zhang et al. 2010a), neuroblastomas (Astuti et al. 2005), and hepatocellular carcinomas (Braconi et al. 2011). Taken together, these findings strongly indicate that epigenetic modification of the IG-DMR and MEG3-DMR plays a significant role in inactivation of the MEG3 gene in human tumors.
MEG3 inhibits tumor cell proliferation in vitro
One characteristic of a tumor suppressor is inhibition of tumor cell proliferation. Whether MEG3 negatively regulates cell proliferation was tested in a number of human cancer cell lines in several independent studies using four experimental assays: colony-formation, growth curves, BrdU incorporation, and anchorage-independent growth. In the colony formation assay experiments, MCF7, HeLa, H4, and CH157-MN cells were transfected with a plasmid expressing MEG3 or a control vector. The transfected cells were treated with the selection drug and the number of drug-resistant colonies was counted. The colony number in cells transfected with MEG3 was significantly lower than in cells transfected with controls (Zhang et al. 2003, 2010a). Secondly, in assessing cell proliferation using a growth curve assay, cells transfected with the MEG3 plasmid grew significantly slower than cells transfected with control vectors. Cell lines used for this assay were the cervical cancer HeLa (Zhang et al. 2003), hepatocellular cancer PRC/PRF/5 (Braconi et al. 2011), and gliomas U251 and U87 MG cell lines (Wang et al. 2012). Thirdly, using the BrdU incorporation assay, cells were transfected with a vector expressing both MEG3 and GFP. DNA synthesis in cells expressing MEG3 (GFP positive) was determined by BrdU incorporation. Re-expression of MEG3 significantly inhibited BrdU incorporation in cell lines HCT116, IOMM-Lee, and CH157-MN (Zhou et al. 2007, Zhang et al. 2010a). Finally, using the anchorage-independent growth assay, the cell line PRC/PRF/5 was transfected with a MEG3 expression construct and seeded into soft agar. MEG3 significantly suppressed colony-formation efficiency in the PRC/PRF/5 cell line (Braconi et al. 2011). These studies demonstrated that MEG3 inhibits proliferation of tumor cells. Braconi et al. (2011) reported that ectopic expression of MEG3 causes apoptosis in PRC/PRF/5 cells. Apoptosis induced by MEG3 has also been observed in U251 and U87 MG cells (Wang et al. 2012). These data suggest that MEG3 inhibition of tumor cell proliferation is due in part to induction of apoptosis. We are currently investigating whether MEG3 inhibits tumor growth in vivo in our laboratory. Our studies to date indicate that re-expression of MEG3 in PDFS cells suppresses tumor growth in nude mice (Zhou et al., manuscript in preparation).
MEG3 functionally interacts with tumor suppressor p53
The p53 (TP53) gene is mutated in most human cancers and has been subject to intensive investigation and review (see reviews Olivier et al. (2010) and Soussi (2011)). It functions as a transcription factor capable of regulating expression of many target genes leading to suppression of tumor development and growth (Vousden & Prives 2009). p53 has also been shown to mediate functions of other tumor suppressors, such as ARF (CDKN2A; Sherr & Weber 2000), BRCA1 (Ouchi et al. 1998), and PTEN (Mayo & Donner 2002). Experimental data from our studies demonstrated that p53 is a target of MEG3. Transfection of MEG3 expression constructs induces a significant increase in p53 protein levels in human cancer cell lines (Zhou et al. 2007). Normally, p53 protein levels are very low due to its rapid degradation via the ubiquitin-proteasome pathway (see review Brooks & Gu (2011)). The ubiquitination of p53 is mainly mediated by MDM2, an E3 ubiquitin ligase (Haupt et al. 1997, Kubbutat et al. 1997). Inhibition of MDM2 function plays a major role in p53 stabilization (see review Harris & Levine (2005)). We found that MEG3 downregulates MDM2 expression (Zhou et al. 2007), suggesting that MDM2 downregulation is one of the mechanisms whereby MEG3 activates p53. MEG3 stimulates p53-dependent transcription (Zhou et al. 2007, Zhang et al. 2010a, Wang et al. 2012). Interestingly, MEG3 does not stimulate expression of p21Cip1, a well-known p53 target gene (el-Deiry et al. 1993). Instead, MEG3 enhances p53-dependent expression of GDF15, an inhibitor of cell proliferation. Further investigation revealed that MEG3 enhances binding of p53 to the GDF15 promoter but not to the p21Cip1 promoter (Zhou et al. 2007). These findings indicate that MEG3 activates p53 and selectively activates expression of p53 target genes.
The product of the MEG3 gene is a noncoding RNA
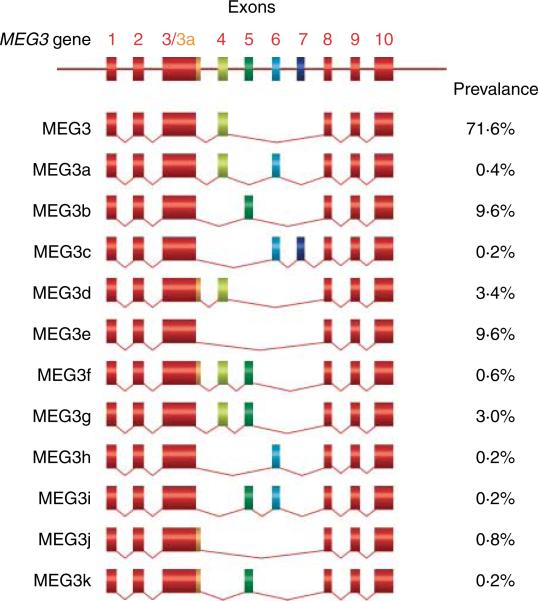
The MEG3 gene consists of ten exons (Fig. 1B; Miyoshi et al. 2000). Transcription from the MEG3 gene yields multiple MEG3 transcript isoforms due to alternative RNA splicing (Zhou et al. 2007, Zhang et al. 2010b). A total of 12 MEG3 RNA isoforms have been identified, named MEG3, MEG3a to MEG3k (Fig. 3). The predominantly expressed isoform is MEG3, containing exons 1–4 and 8–10 (GenBank accession number, NR_002766.2; Zhang et al. 2010b). The mature MEG3 RNA is ~1600 nt in length. No single large open reading frame (ORF) has been found in any of the MEG3 isoforms. There are three small major ORFs in MEG3 RNA with lengths ranging from 150 to 250 nt. No apparent consensus Kozak sequences are found around AUG in those ORFs, suggesting that these ORFs are not constructed for optimal translation. When transfected, all MEG3 iso-forms stimulate transcription from a p53-reporter construct and inhibit BrdU incorporation in HCT116 cells, although the strength of the activity varies among MEG3 isoforms (Zhou et al. 2007, Zhang et al. 2010b). The major isoform MEG3 was chosen for further investigation. A series of MEG3 expression constructs were generated to express individual ORFs, two ORFs in combination, or full-length molecules carrying point mutations that cause a frameshift in targeted ORFs leading to peptide premature termination if translated. These constructs were then tested for their ability to activate p53. We found that expression of individual ORFs failed to activate p53. In contrast, expression of full-length MEG3s in which the ORFs were destroyed by point mutations activated p53 comparable to wild-type MEG3 (Zhou et al. 2007). These results indicate that MEG3 function is not mediated by any individual ORFs. Instead, the full-length MEG3 transcript is essential for MEG3 function. These data strongly support the concept that MEG3 is an ncRNA.
Figure 3.
Schematic representation of MEG3 RNA isoforms generated by alternative splicing. The common exons are in red. Exon 3a contains 40 extra nucleotides at the 3′-end of the exon 3, which is in gold color. The prevalence of each MEG3 RNA isoform is the average of values from five human tissues and cell lines (Zhang et al. 2010b).
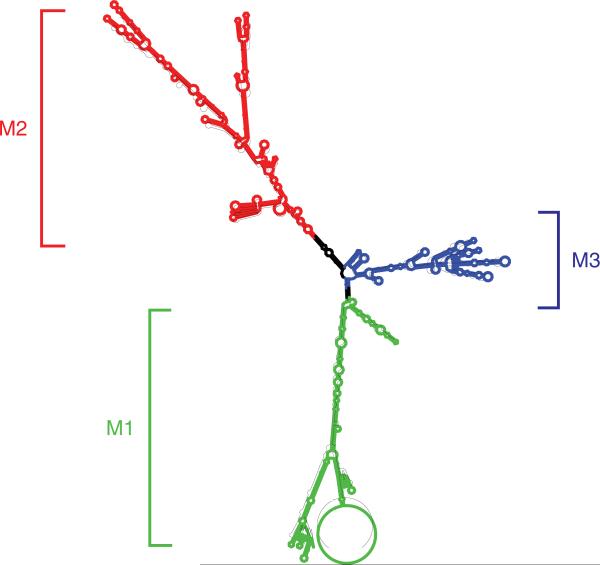
More direct evidence supporting the concept that MEG3 is an ncRNA came from structural studies. The computer program Mfold is widely used to predict the secondary structures of nucleic acids based on their thermostability (Mathews et al. 1999, Zuker 2003). MEG3 and its isoforms have similar secondary structures as predicted by Mfold (Fig. 4; Zhang et al. 2010b). For example, MEG3 RNA folds into three major motifs, M1, M2, and M3 (Fig. 4). A MEG3 RNA carrying a small deletion within M2 that does not significantly change the overall secondary folding activates p53. In contrast, a deletion of similar size within M2 that significantly alters its folding abolishes the p53-activating function of MEG3 (Zhou et al. 2007). Furthermore, when a half sequence of M2 was replaced with nucleotides, which differ dramatically from the original sequences but do not significantly alter the M2 folding, the resultant hybrid MEG3 functions as wild-type MEG3 in both p53-dependent transcription activation and inhibition of BrdU incorporation (Zhang et al. 2010b). These data demonstrate that MEG3 indeed functions as an ncRNA.
Figure 4.
MEG3 RNA folding is predicted by Mfold. The stems and loops of folding are categorized into three groups designated as M1, M2, and M3.
Meg3 gene deletion results in perinatal death in mice
Two Meg3 knockout (KO) mouse models have been reported. We created a Meg3 KO mouse model carrying a deletion of a 5 kb DNA, which contains the first five exons and a small portion of the Meg3 promoter (Zhou et al. 2010). As expected, mice carrying the paternal deletion are alive and normal. In these mice, there are no changes in expression of either Meg3 and its downstream MEGs or paternally expressed genes compared with wild-type controls. However, mice carrying the maternal deletion died perinatally and had major skeletal muscle defects. Expression of both Meg3 and its downstream MEGs was completely abolished. In contrast, the paternally expressed genes within the Dlk1–Meg3 locus were activated. These observations are very similar to those found in mice carrying the IG-DMR deletion (Lin et al. 2003). The other Meg3 KO mouse model was created by Takahashi et al. (2009) and carries a 10 kb deletion including the Meg3-DMR and the first five exons of the Meg3 gene. The phenotype of this mouse model is significantly different from our Meg3 KO mice as well as the IG-DMR KO mice. For example, Takahashi's mice with the maternal deletion died 4 weeks after birth, while those with the paternal deletion died perinatally. Surprisingly, mice with the homozygous deletion survived and grew to fertile adults (Takahashi et al. 2009). The expression pattern of the imprinted genes in Takahashi's mice is also different. All the MEGs except Meg3 were still moderately expressed in mice carrying the maternal deletion and their expression levels were significantly increased in mice carrying the paternal or homozygous deletion compared with those in wild-type mice (Takahashi et al. 2009). This dysregulated gene expression is likely due to the activity of the promoter controlling the neo expression cassette, which was used for selection of recombinant embryonic stem cells and is transcribed in the same direction as the Meg3 gene. Another phenotypic difference between Takahashi's Meg3 KO mice and our Meg3 KO mice is that in Takahashi's mice, the maternal Meg3 deletion did not affect the methylation status of the IG-DMR (Takahashi et al. 2009) but resulted in IG-DMR hypermethylation in our mice (Zhou et al. 2010). Taken together, data from these KO mice suggest that a function of MEG3 is to maintain an active (unmethylated) status at the IG-DMR by positively regulating expression of its downstream MEGs, which may act as inhibitors of IG-DMR methylation.
Deletion of the Meg3 gene promotes angiogenesis in vivo
To identify the target genes of Meg3 in vivo, we compared gene profiles in embryonic brains between Meg3 KO mice and wild-type mice using microarray techniques (Gordon et al. 2010). Among the genes that are up regulated in Meg3 KO mice are several genes in the angiogenesis pathway. Using qRT-PCR and immunohistochemical staining, we confirmed that expression of vascular endothelial growth factor alpha (VEGFA) and its type I receptor (VEFGR1) is significantly increased in Meg3 KO brain compared with their expression in the wild-type brain. Both VEGFA and VEFGR1 are primary regulators of angiogenesis (see review Veikkola & Alitalo (1999)). In agreement, the formation of cortical microvessels, identified by positive staining of Pecam1 and Vegfr2 (Kdr) markers, is significantly increased in the brains of Meg3 KO mice (Gordon et al. 2010). These data indicate that deletion of the Meg3 gene promotes angiogenesis. It is well known that novel blood vessel formation is required for tumor growth (see review Folkman (2002)). Therefore, enhanced angiogenesis may be one of the mechanisms by which inactivation of MEG3 contributes to tumor development.
Conclusions
A bona fide tumor suppressor has to meet a minimum of three criteria: 1) it is functionally inactivated in tumors; 2) re-expression of the gene inhibits tumor growth in vitro and in vivo, and 3) knockout of the gene leads to tumor formation and/or developmental defects in an animal model (see reviews Weinberg (1991, 1993) and Hakem & Mak (2001)). Loss of MEG3 expression in various human tumors has been well documented. Re-expression of MEG3 inhibits proliferation, induces apoptosis, and inhibits anchorage-independent growth of human tumor cells. Its function, at least in part, is mediated by the tumor suppressor p53. Although Meg3 KO mice died prematurely, rendering it impossible to test whether inactivation of Meg3 leads to tumor formation, the requirement of Meg3 for embryonic development is consistent with the function of many known tumor suppressors. For example, Rb–/– embryos die in utero between E13·5 and E15·5 due to nervous system and hematopoietic defects (Clarke et al. 1992, Jacks et al. 1992). Pten–/– embryos die due to defective chorioallantoic development between E6·5 and E9·5 (Suzuki et al. 1998). In addition, in vivo angiogenesis activity is enhanced in the absence of MEG3, indicating that one of the functions of MEG3 is to inhibit angiogenesis. Taken together therefore, strong evidence exists to support the concept that MEG3 is a novel lncRNA tumor suppressor. Currently, we are generating a Meg3 conditional KO mouse model in which the Meg3 gene will be specifically deleted in targeted tissues. We will be able to determine whether inactivation of MEG3 leads to development of tumors in vivo and provides definite evidence demonstrating that MEG3 is an ncRNA tumor suppressor. These studies will reveal novel functions and mechanisms of large noncoding RNAs in the development and pathogenesis of human tumors, which is a new field of investigation for the future.
Acknowledgments
Funding
This work was supported by grants from the National Institutes of Health (A K, R01DK40947), the Guthart Family Foundation, and the Jarislowsky Foundation.
Footnotes
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- Alexander JM, Biller BM, Bikkal H, Zervas NT, Arnold A, Klibanski A. Clinically nonfunctioning pituitary tumors are monoclonal in origin. Journal of Clinical Investigation. 1990;86:336–340. doi: 10.1172/JCI114705. (doi:10.1172/JCI114705) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astuti D, Latif F, Wagner K, Gentle D, Cooper WN, Catchpoole D, Grundy R, Ferguson-Smith AC, Maher ER. Epigenetic alteration at the DLK1–GTL2 imprinted domain in human neoplasia: analysis of neuroblastoma, phaeochromocytoma and Wilms’ tumour. British Journal of Cancer. 2005;92:1574–1580. doi: 10.1038/sj.bjc.6602478. (doi:10.1038/sj.bjc.6602478) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando T, Kato Y, Ihara Y, Yamagishi F, Tsukada K, Isobe M. Loss of heterozygosity of 14q32 in colorectal carcinoma. Cancer Genetics and Cytogenetics. 1999;111:161–165. doi: 10.1016/s0165-4608(98)00242-8. (doi:10.1016/S0165-4608(98)00242-8) [DOI] [PubMed] [Google Scholar]
- Biller BM, Alexander JM, Zervas NT, Hedley-Whyte ET, Arnold A, Klibanski A. Clonal origins of adrenocorticotropin-secreting pituitary tissue in Cushing's disease. Journal of Clinical Endocrinology and Metabolism. 1992;75:1303–1309. doi: 10.1210/jcem.75.5.1358909. (doi:10.1210/jc.75.5.1303) [DOI] [PubMed] [Google Scholar]
- Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S, Negrini M, Miotto E, Croce CM, Patel T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750–4756. doi: 10.1038/onc.2011.193. (doi:10.1038/onc.2011.193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CL, Gu W. p53 regulation by ubiquitin. FEBS Letters. 2011;585:2803–2809. doi: 10.1016/j.febslet.2011.05.022. (doi:10.1016/j.febslet.2011.05.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaidarun SS, Klibanski A. Gonadotropinomas. Seminars in Reproductive Medicine. 2002;20:339–348. doi: 10.1055/s-2002-36708. (doi:10.1055/s-2002-36708) [DOI] [PubMed] [Google Scholar]
- Cheunsuchon P, Zhou Y, Zhang X, Lee H, Chen W, Nakayama Y, Rice KA, Tessa Hedley-Whyte E, Swearingen B, Klibanski A. Silencing of the imprinted DLK1–MEG3 locus in human clinically nonfunctioning pituitary adenomas. American Journal of Pathology. 2011;179:2120–2130. doi: 10.1016/j.ajpath.2011.07.002. (doi:10.1016/j.ajpath.2011.07.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AR, Maandag ER, van Roon M, van der Lugt NM, van der Valk M, Hooper ML, Berns A, te Riele H. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359:328–330. doi: 10.1038/359328a0. (doi:10.1038/359328a0) [DOI] [PubMed] [Google Scholar]
- Danila DC, Zhang X, Zhou Y, Dickersin GR, Fletcher JA, Hedley-Whyte ET, Selig MK, Johnson SR, Klibanski A. A human pituitary tumor-derived folliculostellate cell line. Journal of Clinical Endocrinology and Metabolism. 2000;85:1180–1187. doi: 10.1210/jcem.85.3.6424. (doi:10.1210/jc.85.3.1180) [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. (doi:10.1016/0092-8674(93)90500-P) [DOI] [PubMed] [Google Scholar]
- Folkman J. Role of angiogenesis in tumor growth and metastasis. Seminars in Oncology. 2002;29:15–18. doi: 10.1053/sonc.2002.37263. (doi:10.1053/sonc.2002.37263) [DOI] [PubMed] [Google Scholar]
- Gejman R, Batista DL, Zhong Y, Zhou Y, Zhang X, Swearingen B, Stratakis CA, Hedley-Whyte ET, Klibanski A. Selective loss of MEG3 expression and intergenic differentially methylated region hypermethylation in the MEG3/DLK1 locus in human clinically nonfunctioning pituitary adenomas. Journal of Clinical Endocrinology and Metabolism. 2008;93:4119–4125. doi: 10.1210/jc.2007-2633. (doi:10.1210/jc.2007-2633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Molecular Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. (doi:10.1186/1476-4598-10-38) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon FE, Nutt CL, Cheunsuchon P, Nakayama Y, Provencher KA, Rice KA, Zhou Y, Zhang X, Klibanski A. Increased expression of angiogenic genes in the brains of mouse meg3-null embryos. Endocrinology. 2010;151:2443–2452. doi: 10.1210/en.2009-1151. (doi:10.1210/en.2009-1151) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakem R, Mak TW. Animal models of tumor-suppressor genes. Annual Review of Genetics. 2001;35:209–241. doi: 10.1146/annurev.genet.35.102401.090432. (doi:10.1146/annurev.genet.35.102401.090432) [DOI] [PubMed] [Google Scholar]
- Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. (doi:10.1038/sj.onc.1208615) [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. (doi:10.1038/387296a0) [DOI] [PubMed] [Google Scholar]
- Herman V, Fagin J, Gonsky R, Kovacs K, Melmed S. Clonal origin of pituitary adenomas. Journal of Clinical Endocrinology and Metabolism. 1990;71:1427–1433. doi: 10.1210/jcem-71-6-1427. (doi:10.1210/jcem-71-6-1427) [DOI] [PubMed] [Google Scholar]
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. (doi:10.1038/359295a0) [DOI] [PubMed] [Google Scholar]
- Kagami M, O'Sullivan MJ, Green AJ, Watabe Y, Arisaka O, Masawa N, Matsuoka K, Fukami M, Matsubara K, Kato F, et al. The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genetics. 2010;6:e1000992. doi: 10.1371/journal.pgen.1000992. (doi:10.1371/journal.pgen.1000992) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. (doi:10.1038/387299a0) [DOI] [PubMed] [Google Scholar]
- Lin SP, Youngson N, Takada S, Seitz H, Reik W, Paulsen M, Cavaille J, Ferguson-Smith AC. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1–Gtl2 imprinted cluster on mouse chromosome 12. Nature Genetics. 2003;35:97–102. doi: 10.1038/ng1233. (doi:10.1038/ng1233) [DOI] [PubMed] [Google Scholar]
- Lin SP, Coan P, da Rocha ST, Seitz H, Cavaille J, Teng PW, Takada S, Ferguson-Smith AC. Differential regulation of imprinting in the murine embryo and placenta by the Dlk1–Dio3 imprinting control region. Development. 2007;134:417–426. doi: 10.1242/dev.02726. (doi:10.1242/dev.02726) [DOI] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. Journal of Molecular Biology. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. (doi:10.1006/jmbi.1999.2700) [DOI] [PubMed] [Google Scholar]
- Mayo LD, Donner DB. The PTEN, Mdm2, p53 tumor suppressor–oncoprotein network. Trends in Biochemical Sciences. 2002;27:462–467. doi: 10.1016/s0968-0004(02)02166-7. (doi:10.1016/S0968-0004(02)02166-7) [DOI] [PubMed] [Google Scholar]
- Mezzomo LC, Gonzales PH, Pesce FG, Kretzmann Filho N, Ferreira NP, Oliveira MC, Kohek MB. Expression of cell growth negative regulators MEG3 and GADD45gamma is lost in most sporadic human pituitary adenomas. Pituitary. 2012 doi: 10.1007/s11102-011-0340-1. In press. (doi:10.1007/s11102-011-0340-1) [DOI] [PubMed] [Google Scholar]
- Miyoshi N, Wagatsuma H, Wakana S, Shiroishi T, Nomura M, Aisaka K, Kohda T, Surani MA, Kaneko-Ishino T, Ishino F. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes to Cells: Devoted to Molecular & Cellular Mechanisms. 2000;5:211–220. doi: 10.1046/j.1365-2443.2000.00320.x. (doi:10.1046/j.1365-2443.2000.00320.x) [DOI] [PubMed] [Google Scholar]
- Mutirangura A, Pornthanakasem W, Sriuranpong V, Supiyaphun P, Voravud N. Loss of heterozygosity on chromosome 14 in nasopharyngeal carcinoma. International Journal of Cancer. 1998;78:153–156. doi: 10.1002/(sici)1097-0215(19981005)78:2<153::aid-ijc5>3.0.co;2-y. (doi:10.1002/(SICI)1097-0215(19981005)78:2<153::AID-IJC5>3.0.CO;2-Y) [DOI] [PubMed] [Google Scholar]
- Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harbor Perspectives in Biology. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. (doi:10.1101/cshperspect.a001008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouchi T, Monteiro AN, August A, Aaronson SA, Hanafusa H. BRCA1 regulates p53-dependent gene expression. PNAS. 1998;95:2302–2306. doi: 10.1073/pnas.95.5.2302. (doi:10.1073/pnas.95.5.2302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen M, Takada S, Youngson NA, Benchaib M, Charlier C, Segers K, Georges M, Ferguson-Smith AC. Comparative sequence analysis of the imprinted Dlk1–Gtl2 locus in three mammalian species reveals highly conserved genomic elements and refines comparison with the Igf2-H19 region. Genome Research. 2001;11:2085–2094. doi: 10.1101/gr.206901. (doi:10.1101/gr.206901) [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. Genomic imprinting at the mammalian Dlk1–Dio3 domain. Trends in Genetics. 2008;24:306–316. doi: 10.1016/j.tig.2008.03.011. (doi:10.1016/j.tig.2008.03.011) [DOI] [PubMed] [Google Scholar]
- Schuster-Gossler K, Bilinski P, Sado T, Ferguson-Smith A, Gossler A. The mouse Gtl2 gene is differentially expressed during embryonic development, encodes multiple alternatively spliced transcripts, and may act as an RNA. Developmental Dynamics. 1998;212:214–228. doi: 10.1002/(SICI)1097-0177(199806)212:2<214::AID-AJA6>3.0.CO;2-K. (doi:10.1002/(SICI)1097-0177(199806)212:2<214::AIDAJA6>3.0.CO;2-K) [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Weber JD. The ARF/p53 pathway. Current Opinion in Genetics & Development. 2000;10:94–99. doi: 10.1016/s0959-437x(99)00038-6. (doi:10.1016/S0959-437X(99)00038-6) [DOI] [PubMed] [Google Scholar]
- Smas CM, Sul HS. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell. 1993;73:725–734. doi: 10.1016/0092-8674(93)90252-l. (doi:10.1016/0092-8674(93)90252-L) [DOI] [PubMed] [Google Scholar]
- Soussi T. TP53 mutations in human cancer: database reassessment and prospects for the next decade. Advances in Cancer Research. 2011;110:107–139. doi: 10.1016/B978-0-12-386469-7.00005-0. (doi:10.1016/B978-0-12-386469-7.00005-0) [DOI] [PubMed] [Google Scholar]
- Suzuki A, de la Pompa JL, Stambolic V, Elia AJ, Sasaki T, del Barco Barrantes I, Ho A, Wakeham A, Itie A, Khoo W, et al. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Current Biology. 1998;8:1169–1178. doi: 10.1016/s0960-9822(07)00488-5. (doi:10.1016/S0960-9822(07)00488-5) [DOI] [PubMed] [Google Scholar]
- Takada S, Paulsen M, Tevendale M, Tsai CE, Kelsey G, Cattanach BM, Ferguson-Smith AC. Epigenetic analysis of the Dlk1–Gtl2 imprinted domain on mouse chromosome 12: implications for imprinting control from comparison with Igf2-H19. Human Molecular Genetics. 2002;11:77–86. doi: 10.1093/hmg/11.1.77. (doi:10.1093/hmg/11.1.77) [DOI] [PubMed] [Google Scholar]
- Takahashi N, Okamoto A, Kobayashi R, Shirai M, Obata Y, Ogawa H, Sotomaru Y, Kono T. Deletion of Gtl2, imprinted non-coding RNA, with its differentially methylated region induces lethal parent-origin-dependent defects in mice. Human Molecular Genetics. 2009;18:1879–1888. doi: 10.1093/hmg/ddp108. (doi:10.1093/hmg/ddp108) [DOI] [PubMed] [Google Scholar]
- Veikkola T, Alitalo K. VEGFs, receptors and angiogenesis. Seminars in Cancer Biology. 1999;9:211–220. doi: 10.1006/scbi.1998.0091. (doi:10.1006/scbi.1998.0091) [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. (doi:10.1016/j.cell.2009.04.037) [DOI] [PubMed] [Google Scholar]
- Wang PJ, Ren ZQ, Sun PY. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. Journal of Cellular Biochemistry. 2012 doi: 10.1002/jcb.24055. In press. (doi:10.1002/jcb.24055) [DOI] [PubMed] [Google Scholar]
- Weinberg RA. Tumor suppressor genes. Science. 1991;254:1138–1146. doi: 10.1126/science.1659741. (doi:10.1126/science.1659741) [DOI] [PubMed] [Google Scholar]
- Weinberg R. Tumor suppressor genes. Neuron. 1993;11:191–196. doi: 10.1016/0896-6273(93)90177-s. (doi:10.1016/0896-6273(93)90177-S) [DOI] [PubMed] [Google Scholar]
- Zhang X, Sun H, Danila DC, Johnson SR, Zhou Y, Swearingen B, Klibanski A. Loss of expression of GADD45 gamma, a growth inhibitory gene, in human pituitary adenomas: implications for tumorigenesis. Journal of Clinical Endocrinology and Metabolism. 2002;87:1262–1267. doi: 10.1210/jcem.87.3.8315. (doi:10.1210/jc.87.3.1262) [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhou Y, Mehta KR, Danila DC, Scolavino S, Johnson SR, Klibanski A. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. Journal of Clinical Endocrinology and Metabolism. 2003;88:5119–5126. doi: 10.1210/jc.2003-030222. (doi:10.1210/jc.2003-030222) [DOI] [PubMed] [Google Scholar]
- Zhang X, Gejman R, Mahta A, Zhong Y, Rice KA, Zhou Y, Cheunsuchon P, Louis DN, Klibanski A. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Research. 2010a;70:2350–2358. doi: 10.1158/0008-5472.CAN-09-3885. (doi:10.1158/0008-5472.CAN-09-3885) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Rice K, Wang Y, Chen W, Zhong Y, Nakayama Y, Zhou Y, Klibanski A. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology. 2010b;151:939–947. doi: 10.1210/en.2009-0657. (doi:10.1210/en.2009-0657) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Dahle D, Zhou Y, Zhang X, Klibanski A. Hypermethylation of the promoter region is associated with the loss of MEG3 gene expression in human pituitary tumors. Journal of Clinical Endocrinology and Metabolism. 2005;90:2179–2186. doi: 10.1210/jc.2004-1848. (doi:10.1210/jc.2004-1848) [DOI] [PubMed] [Google Scholar]
- Zhao J, Zhang X, Zhou Y, Ansell PJ, Klibanski A. Cyclic AMP stimulates MEG3 gene expression in cells through a cAMP-response element (CRE) in the MEG3 proximal promoter region. International Journal of Biochemistry & Cell Biology. 2006;38:1808–1820. doi: 10.1016/j.biocel.2006.05.004. (doi:10.1016/j.biocel.2006.05.004) [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A. Activation of p53 by MEG3 non-coding RNA. Journal of Biological Chemistry. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. (doi:10.1074/jbc.M702029200) [DOI] [PubMed] [Google Scholar]
- Zhou Y, Cheunsuchon P, Nakayama Y, Lawlor MW, Zhong Y, Rice KA, Zhang L, Zhang X, Gordon FE, Lidov HG, et al. Activation of paternally expressed genes and perinatal death caused by deletion of the Gtl2 gene. Development. 2010;137:2643–2652. doi: 10.1242/dev.045724. (doi:10.1242/dev.045724) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. (doi:10.1093/nar/gkg595) [DOI] [PMC free article] [PubMed] [Google Scholar]