Abstract
Memory T cells cross-reactive with epitopes encoded by related or even unrelated viruses may alter the immune response and pathogenesis of infection by a process known as heterologous immunity. Because a challenge virus epitope may react with only a subset of the T cell repertoire in a cross-reactive epitope-specific memory pool, the vigorous cross-reactive response may be narrowly focused, or oligoclonal. We show here, by examining human T cell cross-reactivity between the HLA-A2-restricted influenza A virus-encoded M158-66 epitope (GILGFVFTL) and the dissimilar Epstein-Barr virus-encoded BMLF1280-288 epitope (GLCTLVAML), that under some conditions heterologous immunity can lead to a significant broadening rather than a narrowing of the T cell receptor repertoire. We suggest that dissimilar cross-reactive epitopes might generate a broad rather than narrow T cell repertoire if there is a lack of dominant high affinity clones, and this hypothesis is supported by computer simulation.
Keywords: human, T cells, T cell receptors, repertoire development, Epstein Barr Virus, influenza
Introduction
The T cell response to viral infections consists of expanded clones of T cells arising from antigen-inexperienced naïve populations of small clone size and from antigen-experienced memory populations of much larger clone size (1-3). The relative influence of the memory cells on the T cell response depends on the host's previous exposure to pathogens, which has consequently shaped their memory T cell repertoire. Memory T cells cross-reactive with related or even unrelated viruses may change the immune response to a newly encountered virus and alter viral clearance and pathogenesis, which is referred to as heterologous immunity(4, 5). Cross-reactive memory cells have the capacity to dominate immune responses initiating from naïve cells because the memory cells are present at much higher frequencies and because they are easier to activate than naïve T cells(1-3). In the case where a cross-reactive challenge epitope is not completely homologous with the epitope that initially generated the memory cell population, it is likely that only a subset of that memory T cell repertoire will react with the new epitope sufficiently to induce proliferation. The proliferation of a subset of the memory cell population would likely result in a vigorous response with a narrow oligoclonal T cell receptor repertoire (6). This phenomenon has been demonstrated in mouse models of infection with different strains of influenza virus bearing a point mutation in a cross-reactive epitope and with LCMV and Pichinde virus, which have cross-reactive epitopes bearing 6 of 8 amino acids in common (6-8). Further, narrowly focused HCV T cell responses cross-reactive with influenza A virus (IAV) have correlated with fulminant hepatitis in humans (9). Narrow TCR repertoires have been observed with several human infections, including CMV, HCV, and HIV, and this property may allow for the generation of epitope escape variants (10-13). These narrow repertoires may or may not be the consequence of heterologous immunity, but they are generally considered undesirable.
Although it seems intuitively obvious that expanding a subset of a T cell memory pool should result in a narrow oligoclonal response, it may not always be the case. One needs to consider the relative numbers and affinities of the naïve T cell precursors in comparison to the cross-reactive memory T cell clones and consider what proportion of the memory cells specific to the immunizing epitope would be able to cross-react with the challenge epitope. Here we questioned whether narrowing of the T cell repertoire is always a consequence of heterologous immunity and show, by examining human T cell cross-reactivity between IAV and Epstein Barr virus (EBV), that it clearly is not.
We previously revealed the existence of cross-reactive T cells with specificity for two immunodominant, HLA-A2-presented, epitopes, IAV M158-66 and EBV BMLF1280-288, which have relatively little sequence similarity in comparison to most previously reported cross-reactive epitopes (14). This cross-reactive T cell population was functionally heterogeneous in regards to cytokine production, perhaps due to differences in the quality or strength of the signal emanating from each TCR. Structural diversity occurs in both M1- and BMLF1-specific TCR repertoires, but in unique ways. M1-specific TCR repertoires primarily involve one Vβ family, Vβ17, and often express the CDR3β motif xRSx (15-18). However, within the Vβ17 family there exist hundreds of T cell clones that differ in their nucleotide sequence (19). BMLF1-specific TCR repertoires, on the other hand, include multiple Vβ families that are often comprised of relatively few T cell clones (20-22). While there are at least four common BMLF1-specific Vβ families (Vβ2, 4, 16, and 22), their combination and hierarchy differs between individuals. The present study demonstrates that the cross-reactive T cell population specific for these two dissimilar epitopes, IAV-M1 and EBV-BMLF1, includes a wide array of TCR with little evidence for clonal dominance and repertoire narrowing. Furthermore, similar to the mouse model of cross-reactivity, each cross-reactive TCR repertoire examined was unique to the individual, revealing no common or predictable cross-reactive TCR structure (6). These results suggest that the breadth of the cross-reactive T cell repertoire may depend on various factors, including the level of structural similarity between epitopes, which we hypothesize impacts clonal dominance and which is supported by computer simulation of human immune responses.
Materials and Methods
Subjects
Five IAV-immune patients with acute EBV infection between the ages of 18-23 were volunteers from the University of Massachusetts (UMass) Student Health Services at Amherst, MA. HLA-typing and EBV serology were performed as previously described (14). Positive staining with HLA-A2-tetramers loaded with influenza-M1 epitope was used as an indication that these individuals had been exposed to influenza A virus in the past. For this study, a 50 ml blood sample was provided from patients within a year after presentation with symptoms of IM. Four healthy donors between the ages of 42 and 50 were volunteers from the research community at UMass Medical School (Worcester, MA). HLA status and immunity to EBV and influenza A virus were assessed using HLA-A2-specific monoclonal antibody (BB7.2, Becton Dickinson (BD), San Diego, CA) and tetramer stains, respectively. EBV serology was further confirmed through the detection of IgG specific for the viral capsid protein (14). This study was approved by the Human Studies Committee at UMass Medical School.
Blood preparation and bulk T cell culture
PBMC were isolated using Ficoll-paque plus (Amersham Bioscience, Uppsala, Sweden). CD8+ cells were isolated using the Miltenyi Biotech (Auburn, CA) MACS system and were cultured using our published protocol (14). Briefly, CD8+ lymphocytes were plated at a 5:1 ratio with 1μm peptide-pulsed and thereafter washed irradiated T2 cells (ATCC #CRL-1992), which were used to re-stimulate the T cell lines weekly.
HLA-A2-restricted peptides
EBV-BMLF1280-288 (GLCTLVAML), IAV-M158-66 (GILGFVFTL) peptides were synthesized to >90% purity by Biosource (Camarillo, CA).
MHC-Class I tetramers
A detailed description of the protocol used by the tetramer facility at UMass Medical School has been previously published(23). Tetramers were assembled using the above peptide sequences and were conjugated to either APC (Caltag, Burlingame, CA), or Quantum Red (Sigma).
Extracellular/Intracellular staining and cell sorting
CD8+ T cells were plated at 106 per well and washed with FACS buffer (PBS, 2% FCS, 1% sodium azide). Tetramers were incubated alone at room temperature for 20 min, followed by additional 20 min incubation with Vβ-specific monoclonal antibodies (Beckman Coulter, Fullerton, CA). The TCR Vβ nomenclature used in this study was originally described by Arden et al.(24). Cells were fixed with FACS Lysing Solution (BD) for 5 min at room temperature and then washed with FACS buffer. Cells stained for intracellular IFNγ incubated with peptide and brefeldin A for 5 hours before being permeabilized with cytofix/cytoperm reagent (BD). Cells then incubated 30 min at 4°C with anti-IFNγ antibody (BD, clone B27). Cells for sorting were incubated with tetramer for 40 min at room temperature in 2% FCS/PBS buffer and were immediately isolated unfixed using the Mo-Flo cell sorter (Beckman Coulter, Fullerton, CA).
CDR3 spectratyping and sequencing
RNA was isolated from sorted CD8+ T cell populations using TRIzol (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions. We synthesized cDNA using a poly-T primer as previously described (14). The cDNA was amplified using published primer sequences specific for TCR Vβ and Vα families with those specific to the Cβ and Cα regions, as previously described (14, 25, 26). The Vβ and Vα nomenclature used in this study was originally described by Arden et al.(24). For spectratyping analysis, formamide dye was added to the PCR products, which were boiled for 3 min and then put on ice before being loaded onto a 5% polyacrylamide gel and run at 2500V. Gels were read and analyzed using a FluorImager 595 (Molecular Dynamics, Sunnyvale, CA) and ImageQuant software (Molecular Dynamics), respectively. For sub-cloning and sequencing clonotypes within a given Vβ or Vα family, PCR products were ligated into the pCR4-TOPO vector (Invitrogen), as previously described, and were sequenced at the UMass Nucleic Acid Facility (Worcester, MA) using universal primers (14).
Computer simulation
We used IMMSIM, an agent-based model of the immune system governed by probabilistic events. This program is available (http://www.immsim.org) and can be downloaded for the purpose of research and education. The IMMSIM model consists of epithelial cells in a grid of discrete “interaction sites,” where T cells, B cells, and APCs of the immune system encounter each other and antigens. Cellular and humoral responses mount whenever a virus infects and expresses antigens in the target epithelial cells. The interactions are governed by affinity and chance encounters (via computer generated random numbers). This model's past applications are found in several previous reports(6, 27-30). For the present modeling, we started by creating a group of 26 virtual individuals containing 7500 CD8 T lymphocytes from a theoretical repertoire of 65,536 different receptors. We injected the individuals with 70 virus particles sporting a single epitope (hex 0×F0) in order to obtain a stable representative primary response specific to this epitope that has a wide enough TCR memory repertoire (50 clones) to be used by the adoptive transfer feature (available on the recent versions of the simulator); this feature is then used to apply successive cross-reactive challenges. To study the response of structurally similar (“near”) versus dissimilar (“far”) cross-reactive epitopes in a comparable way and to limit the number of memory CD8 T cells needed for the simulations, we started by selecting a single medium cross-reactive epitope (hex 0×10) that we have already used in the model for cross-reactive (to 0×F0) challenge simulations (6). We increased or decreased the affinity (to the single cross-reactive epitope 0×10) in two or three of the most abundant T cell clones found in the primary repertoire. The change in affinity was accomplished according to the “more bits of mismatch - less affinity” rule of the simulator, where we modified the T cell receptor hexadecimal (hex) IDs increasing the “bits of mismatch” (to 0×10) to decrease the affinity, and vice versa to increase it. The ultimate result was that we obtained two slightly modified primary T cell repertoires, one acting as a similar (“near”) cross-reactive challenge (where some of the abundant T cells in the primary response are cross-reactive at high affinity), and the other acting as a dissimilar (“far”) cross-reactive challenge (where none of the abundant T cells in the primary response are cross-reactive at high affinity). These two T cell memory repertoires were then adoptively transferred into two groups of 26 virtual mice, and all mice were challenged with 1000 virus particles sporting the same cross-reactive epitope 0×10. A representative output for each group was selected for the plot, while a two-tailed t test was applied to all the results to determine the statistical significance.
Statistical analysis
Where indicated, the paired Student's t test was used to assess differences between groups.
Results
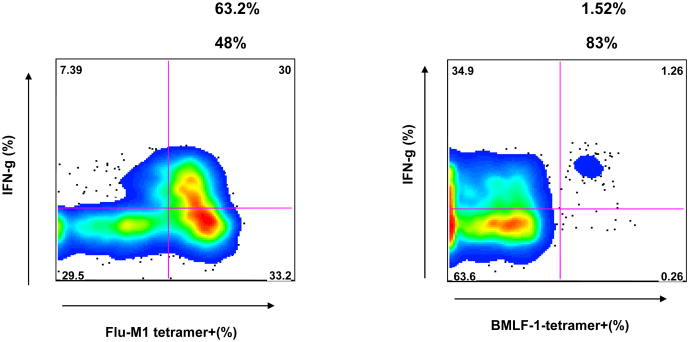
Cross-reactive CD8 T cells recognizing the two dissimilar HLA-A2-presented IAV-M1 and EBV-BMLF1 epitopes have been characterized in the peripheral blood of healthy immune donors and of patients with EBV-associated acute infectious mononucleosis (IM) by co-staining with M1- and BMLF1-loaded tetramers (14). These cross-reactive T cells could be expanded in vitro in the presence of M1, BMLF1, or both peptides simultaneously. These cultured T cells exist as a highly heterogeneous population in regards to their ability to functionally respond by proliferation, cytotoxicity or cytokine production to either peptide and in their ability to bind one or both peptide-specific tetramers. For instance, in the M1-specific lines, the cells were able to produce cytokines such as MIP-1β, IFNγ and TNFα in response to the cross-reactive ligand BMLF1. However, there was a hierarchy in the cytokine responsiveness of the cells in the order of MIP-1β>IFNγ> TNFα (14). MIP-1β is most readily secreted upon cross-reactive stimulation, for as high as 90% of an M1-stimulated line could produce MIP-1β in response to the BMLF1 peptide (14). Some subsets of functionally cross-reactive cells efficiently stained with only one tetramer type but produced cytokines or proliferated in response to the other peptide. Here we show such an example, where a subset (1.5%) of a CD8 T cell line grown in the presence of only M1 peptide was more efficient at staining with a BMLF1-loaded tetramer than with an M1-loaded tetramer. However, the BMLF1-tetramer positive population proliferated in response to M1 peptide stimulation and about 83% of the cells produced IFNγ upon M1 stimulation (Fig. 1). In this study, we sought to characterize the cross-reactive TCR repertoire and compare it to non-cross-reactive repertoires, so we chose to include two cross-reactive populations, defined either by M1/BMLF1 double-tetramer+ staining (cross-reactive population 1, CXR-1) or, as demonstrated in Fig. 1, BMLF1 single-tetramer+ T cells that responded functionally to M1 peptide stimulation (cross-reactive population 2, CXR-2).
Figure 1. BMLF1-tetramer positive T cells can produce IFNγ following M1 stimulation.
CD8 T cells isolated from patient D-002 were grown in the presence of M1 peptide for 3 weeks. After a 5 hour stimulation with M1 peptide, the M1-specific T cell line was stained extracellularly with either M1- or BMLF1-specific tetramer and intracellularly for IFNγ. The % of the M1-specific T cell line that stained with the indicated tetramer (top number) and the % of the tetramer-positive population that co-stained with anti-IFNγ (bottom number) are shown. Not shown are control peptide EBV-BRLF-1190 stimulation, whereby <0.06% of the BMLF1 tetramer+ population produced IFNγ, and control tetramer staining, CMVpp65 <.001%.
T cell lines accurately reflect the antigen-specific TCR repertoire ex vivo
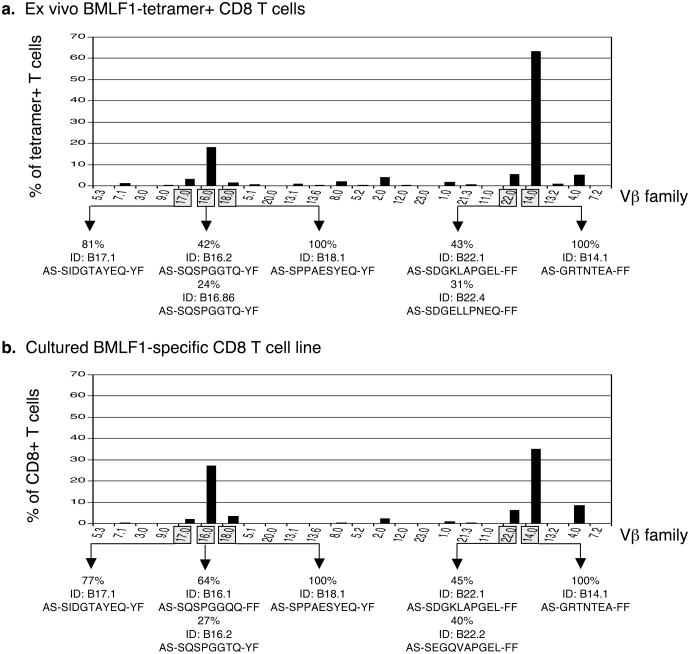
In order to fully investigate the organization and structural diversity of cross-reactive TCR repertoires, we first ensured that our culturing conditions supported the growth of most antigen-specific T cell clones. Since the population of BMLF1-specific memory CD8 T cells in healthy donor D-002 was large enough to sort ex vivo, we compared the TCR Vβ usage of freshly isolated BMLF1-tetramer+ cells with those from the same healthy donor grown in the presence of BMLF1 peptide for 4 weeks and found that the two repertoires were almost identical both in VB usage and individual clonotypes (Fig. 2). Directly ex vivo, Vβ14 and Vβ16 families predominated as compared to the ungated CD8 T cell population (Supplemental Figure 1) and were conserved during culture. Our culturing conditions also effectively supported the growth of low frequency Vβ families, including Vβ17, Vβ18, and Vβ22. We then sub-cloned and sequenced the TCR CDR3β expressed by BMLF1-specific T cells, whereby every unique nucleotide sequence was considered a unique Vβ clonotype. We found BMLF1-specific clonotypes using both high and low frequency Vβ families in both ex vivo and in vitro repertoires (Fig. 2). The same dominant clonotype within each of the Vβ14, 17, 18, and 22 families was present ex vivo and in vitro. The dominant Vβ16+ clonotype ex vivo (ID: B16.2) became co-dominant in vitro with another Vβ16+ clonotype (ID: B16.1) that differed by only 1 of 8 aa residues comprising the CDR3β. These results suggested that TCR repertoire analyses of cultured T cell lines accurately reflect the antigen-specific repertoire present in vivo.
Figure 2. The Vβ repertoire of a cultured T cell line accurately reflects the antigen-specific repertoire ex vivo.
(a) CD8 T cells were isolated from the peripheral blood of donor D-002 and immediately co-stained with BMLF1-specific tetramer and Vβ-specific antibodies. The bar graph illustrates the % of BMLF1 tetramer+ cells using each respective Vβ family. We then sorted BMLF1+ T cells and sub-cloned the CDR3β region using primers specific for 5 different Vβ families, which are indicated by the shaded boxes. The ID of the predominant clonotype(s), its frequency among all sequences analyzed within that family, and the amino acid sequence of its CDR3β loop is provided. (b) CD8 T cells derived from D-002 were cultured for 4 weeks in the presence of BMLF1-pulsed T2 cells before being stained with Vβ-specific antibodies. The bar graph illustrates the % of the T cell line (CD8 cells) using each respective Vβ family. The line was >90% BMLF1 tetramer+ (data not shown). The predominant clonotype(s) found within each of the 5 shaded Vβ families is displayed as described above.
Extensive TCR repertoire analyses performed on BMLF1-specific T cell populations derived from healthy donor D-002 confirmed many of the general characteristics of BMLF1-specific TCR structure and repertoire organization previously established (20-22, 31). All four of the common Vβ families (Vβ2, 4, 16, 22) were represented in the BMLF1-specific TCR repertoire of healthy donor D-002 (Fig. 2). Furthermore, we confirmed the observation that a given BMLF1-specific Vβ family is often comprised of only 1-2 clonotypes (31). For donor D-002, one or 2 clonotypes accounted for greater than 60% of each Vβ population investigated (Fig. 2). Where determined, the clonotypes expressed the conserved Jβ family and/or CDR3β amino acid (aa) motif specifically associated with their respective Vβ family. Vβ16+ clonotypes fit the CDR3β motif AS-SQSPGGTQ-YF and had fewer restrictions on their Jβ usage, while Vβ22+ clonotypes were more restricted by the usage of Jβ2.1 or Jβ2.2 than by the aa sequence of their CDR3β loop (generally fitting the published motif AS-S*G*V*PGEL-FF) (22) (Suppl. Table 1). These data also revealed novel features, not previously published, of the BMLF1-specific TCR repertoire, such as the usage of Vβ families 14, 17 and 18. All three Vβ families were comprised of one dominant clonotype with a CDR3β sequence that may be conserved in other donors with BMLF1-specific repertoires that include these particular Vβ families (Fig. 2). Overall, the 5 Vβ families analyzed here (Vβ14, 16, 17, 18, 22) represented over 80% of the BMLF1-specific repertoire of donor D-002. The total number of unique Vβ clonotypes identified out of the total number of sequences analyzed was as few as 18 out of 201 isolated ex vivo and 21 out of 160 isolated in vitro, suggesting that the BMLF1-specific TCR repertoire is relatively narrow or oligoclonal (Supplemental Table 1), consistent with previous observations (22).
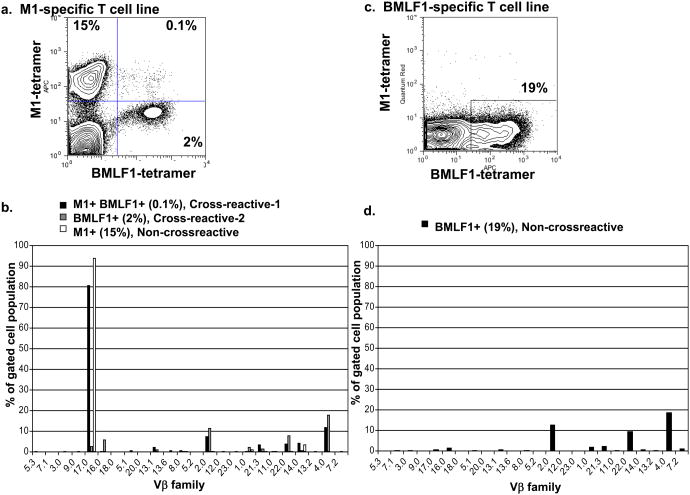
Multiple Vβ families can be used to co-recognize M1 and BMLF1
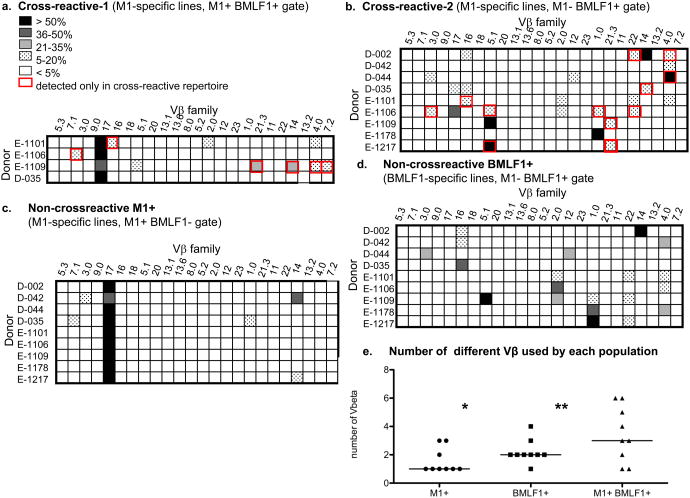
Subsets of T cells cultured in vitro with M1 peptide were considered cross-reactive with BMLF1 by their ability to bind BMLF1-loaded tetramer either simultaneously with M1-loaded tetramer (CXR-1) or exclusively while retaining the ability to proliferate and produce cytokines in response to M1 stimulation (CXR-2) (14) (Fig. 1). Both subsets of cross-reactive T cell populations were present within an M1-specific T cell line derived from IM patient E1101. CXR-1 cells co-stained with both tetramers (M1+ BMLF1+, 0.1% of the T cell line) and CXR-2 cells stained with only the BMLF1-loaded tetramer (M1- BMLF1+, 2% of the T cell line), but remained functionally responsive to M1 stimulation (Fig. 3). We gated on these distinct cross-reactive T cell subsets and used Vβ-specific monoclonal antibodies to determine their TCR usage (Fig. 3b). The M1+ BMLF1+ CXR-1 cells predominantly used Vβ17 (81%), but also Vβ families 2 (8%) and 4 (12%). The M1- BMLF1+ CXR-2 cells within an M1-specific T cell line were not dominated by one Vβ family, but instead used 4 Vβ families: 2 (12%), 4 (18%), 16 (6%), and 22 (8%). These cross-reactive Vβ repertoires were clearly different from those of the tetramer negative cell populations in these cultures (Supplemental Fig 2A,B). We performed the same Vβ analyses on the cross-reactive T cells derived from 4 additional IM patients and 4 healthy immune donors (Fig. 4a, b). Among these 9 individuals, we determined that each cross-reactive TCR repertoire contained 1 or more of 12 different Vβ families, including Vβ1, 2, 3, 4, 5.1, 7.2, 12, 14, 16, 17, 21.3 and 22. Thus, a wide range of TCR structures appeared capable of recognizing the two dissimilar M1 and BMLF1 peptides. Overall, the two most common Vβ families detected among the cross-reactive repertoires were Vβ4 and 17, used by 5 and 4 out of 9 individuals, respectively. While we were unable to screen for all of the known Vβ families, as monoclonal antibodies to all known Vβ families are not available, the cross-reactive repertoire of two individuals was comprised of as few as 1 Vβ family (IM patient E1178, Vβ1 and healthy donor D-042, Vβ4), while the cross-reactive repertoire of two other individuals was comprised of as many as 6 different Vβ families (IM patient E1109, Vβ4, 5.1, 7.2, 14, 17 and 21.3 and IM patient E1106, Vβ1, 2, 3, 5.1, 17, 22). We found no discernable difference in the breadth of the cross-reactive repertoires of IM patients (median=5, range=1-6, n=5) compared to healthy immune donors (median=3, range=1-4, n=4). Rather, the breadth of the cross-reactive Vβ repertoire was significantly (p<.02) more extensive in individuals that had two distinct, tetramer-defined (CXR-1, M1+BMLF1+ and CXR-2, M1-BMLF1+), populations of cross-reactive T cells, such as IM patients E1101, E1106, E1109, and healthy donor D-035 (median=5.5, range=3-6, n=4) when compared to those with only one cross-reactive population (CXR-2, M1-BMLF1+) (median=2, range=1-4, n=5). Most noteably, the breadth of the cross-reactive Vβ repertoire, including both cross-reactive populations, was significantly more extensive than that of either the M1+ or BMLF1+ non-cross-reactive populations (Fig. 4e, Supplemental Fig. 3).
Figure 3. Each tetramer-defined sub-population of a T cell line has a distinct Vβ repertoire.
(a) CD8 T cells isolated from patient E1101 were cultured for 3 weeks in the presence of M1-pulsed T2 cells before being co-stained with M1- and BMLF1-specific tetramers. (b) Following incubation with tetramers, cells (a) were stained with Vβ-specific antibodies. Each bar graph illustrates the % of cells within its respective tetramer-defined gate that use each Vβ family. (c) CD8 T cells isolated from patient E1101 were cultured for 3 weeks in the presence of BMLF1-pulsed T2 cells before being co-stained with M1- and BMLF1-specific tetramers. (d) Following incubation with tetramers, cells (c) were stained with Vβ-specific antibodies. The bar graph illustrates the % of BMLF1-tetramer+ cells that use each Vβ family. Control tetramer CMVpp65 in both M1 line and BMLF1 line was <.001%.
Figure 4. The Vβ repertoire of cross-reactive cells is often comprised of multiple Vβ families and is unique to each individual.
CD8 T cells were isolated from 4 healthy donors and 5 patients with IM. (a-c) CD8 T cells were cultured for 3-4 weeks in the presence of M1-pulsed T2 cells before being co-stained with M1- and BMLF1-specific tetramers followed by Vβ-specific antibodies. Three tetramer-defined gates were analyzed separately: (a) M1+ BMLF1+ (CXR-1), (b) M1- BMLF1+ (CXR-2), and (c) M1+ BMLF1- (non-cross-reactive M1). (d) CD8 T cells were cultured for 3-4 weeks in the presence of BMLF1-pulsed T2 cells before being stained with tetramers and Vβ-specific antibodies. The Vβ usage of M1- BMLF1+ (non-cross-reactive BMLF1) cells is shown. The degree of shading within each box represents the % of tetramer-gated cells that express that Vβ family, and a red outline of a box indicates that the Vβ family was detected in the cross-reactive repertoire but not the non-cross-reactive repertoires of that individual. e) Increased breadth of Vβ usage in the cross-reactive population when compared to the non-cross-reactive IAV-M1 population (* p=.05, paired t test) and the non-cross-reactive EBV-BMLF1 population (** p=.06, paired t test).
The cross-reactive TCR repertoire is unique to each individual
It has been previously documented that both M1-specific and BMLF1-specific TCR repertoires have features that are unique to each individual, such as the clonal composition of each individual's M1-specific Vβ17+ repertoire or the Vβ hierarchy of each individual's BMLF1-specific repertoire. This is consistent with the concept of a private specificity for each TCR repertoire (6, 18, 20, 22, 32-34). Thus, we predicted that this private specificity of each M1- and BMLF1-specific repertoire was responsible for the wide range of cross-reactive TCRs we observed and the individual variability between cross-reactive repertoires (Fig. 4).
We compared the Vβ repertoires of cross-reactive and non-cross-reactive T cell populations derived from each of the 9 individuals included in our study. T cells that grew in the presence of M1 peptide and only stained with M1-loaded tetramer were considered a non-cross-reactive M1-specific population, while T cells that grew in the presence of BMLF1 peptide and only stained with BMLF1-loaded tetramer were considered a non-cross-reactive BMLF1-specific population. It appeared that the specific combination of BMLF1-specific Vβ families used by each individual influenced the specific combination of cross-reactive Vβ families used by that same individual. For example, the non-cross-reactive BMLF1-specific repertoire of donor D-042 included Vβ4 and Vβ16 but not Vβ2 or Vβ22, and the cross-reactive repertoire of this donor reflected that by including one of the two Vβ families known to be used by the BMLF1-specific T cell population, Vβ4 (Fig. 4b, d). Thus, the composition of the non-cross-reactive Vβ repertoire appeared to influence the organization of the cross-reactive Vβ repertoire. The individual variability associated with both BMLF1-specific and, through association, cross-reactive Vβ repertoires likely stems from the clonal composition of each individual's precursor T cell population, which includes both naïve and memory T cell clones that can be activated by the BMLF1 epitope. A larger study cohort is necessary to reveal any common features of this cross-reactive repertoire that may be shared by multiple unrelated individuals.
Unique features of the cross-reactive TCR repertoire
Several features of the cross-reactive TCR repertoires made them unique. For instance, while both the CXR-1 (M1+ BMLF1+) and non-cross-reactive M1-specific populations derived from IM patient E1101 predominantly used Vβ17, the cross-reactive repertoire remained distinct by the inclusion of additional Vβ families (Vβ2 and 4) present at lower frequencies (Fig. 3b). Similarly, the CXR-2 (M1- BMLF1+) population within an M1-specific T cell line and the non-cross-reactive BMLF1-specific population within a BMLF1-specific T cell line were derived from the same IM patient, and both used Vβ families 2, 4 and 22. However, the cross-reactive repertoire remained distinct through the increased usage of Vβ16 (Fig. 3b, d).
Selective gating on cross-reactive T cell populations appeared to enhance the detection of Vβ families used more frequently by cross-reactive than non-cross-reactive T cells. Overall, the cross-reactive repertoire of 7 out of 9 individuals included 1 or more Vβ families not detected within that individual's non-cross-reactive repertoires (Fig. 4). IM patient E1217 represented the most extreme example of this, with the cross-reactive repertoire consisting entirely of two Vβ families (Vβ5.1 and Vβ21.3) that were not detected in this individual's non-cross-reactive repertoires. In some cases, the Vβ family strictly detected in one cross-reactive repertoire was an otherwise common component of the non-cross-reactive repertoires of other individuals, such as Vβ families 4, 16, 17 and 22. In other cases, the Vβ families detected only in a cross-reactive repertoire were rare even among the non-cross-reactive repertoires of other individuals, such as Vβ families 5.1, 7.2, and 21.3.
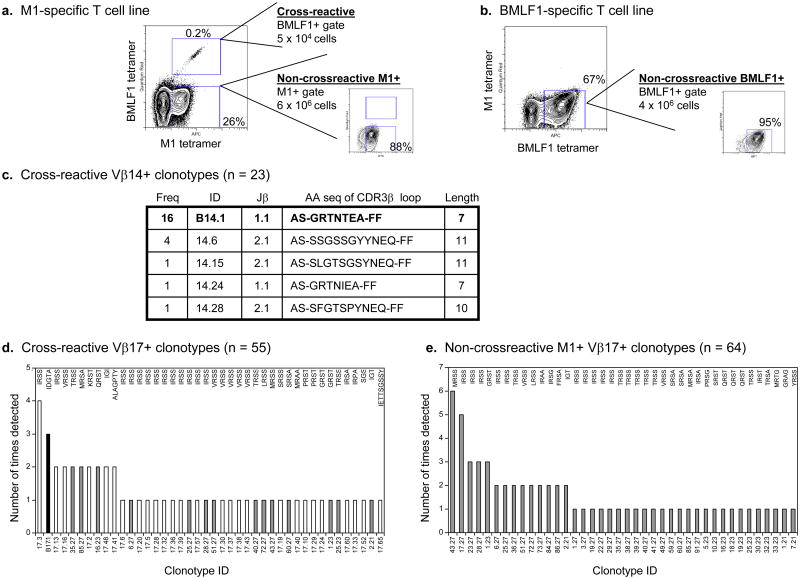
We also observed differences between cross-reactive and non-cross-reactive repertoires on a clonal level. We isolated the tetramer-defined cross-reactive and non-cross-reactive populations from T cell lines derived from one healthy donor, D-002 (Fig. 5a, b). Since both the cross-reactive and non-cross-reactive BMLF1-specific T cells predominantly used Vβ14 (Fig. 4b, d), we sub-cloned and sequenced the CDR3β loops of each Vβ14+ T cell population. We detected an increased number of Vβ14+ clonotypes in the cross-reactive repertoire (5 unique clonotypes out of 23 total sequences analyzed) compared to the highly restricted non-cross-reactive BMLF1-specific repertoire (1 unique clonotype out of 10 total sequences analyzed) (Figs. 5c and 2b). Three of the four novel cross-reactive Vβ14+ clonotypes used an alternative Jβ family (Jβ2.1) and a longer CDR3β loop (10-11aa length). Similarly, we sub-cloned and sequenced several novel cross-reactive Vβ17+ clonotypes, representing the dominant Vβ family of donor D-002's non-cross-reactive M1-specific repertoire. The majority of the cross-reactive Vβ17+ repertoire, 64% (27/42) of the unique clonotypes, was comprised of newly identified clonotypes that were not previously detected in either non-cross-reactive repertoire (Fig. 5d).
Figure 5. The Vβ repertoire of cross-reactive cells is comprised of multiple unique clonotypes using a combination of M1- and BMLF1-specific TCR elements.
(a) An M1-specific T cell line derived from donor D-002 was co-stained with M1- and BMLF1-specific tetramers. Two separate populations were sorted: M1- BMLF1+ (CXR-2) and M1+ BMLF1- (non-cross-reactive M1). (b) A BMLF1-specific T cell line derived from donor D-002 was co-stained with both tetramers and the BMLF1-tetramer+ (non-cross-reactive BMLF1) cells were collected (c) The CDR3β regions of cross-reactive Vβ14+ clonotypes were sequenced. Based on a unique nucleotide sequence, clonotypes were assigned an ID and their frequency is shown, i.e. the number of times that clonotype was detected among the total sequences analyzed. The length of the CDR3β region was determined according to Chothia et al., shown supported by two flanking framework regions (51). Bold indicates that the clonotype was previously detected within BMLF1-specific cell populations. (d) The CDR3β regions of cross-reactive Vβ17+ clonotypes were sequenced. Unique clonotypes are arranged based on their frequency among all sequences analyzed. Due to space limitations, the amino acid sequence of the CDR3β loop has been abbreviated where 4 residues represents a full length of 8. The bars are shaded to indicate simultaneous detection within an alternative cell population as follows: (x025A1) = unique to the cross-reactive population, = detection within the non-cross-reactive M1+ population, (x025A0) = detection within the non-cross-reactive BMLF1-specific cell populations. (e) The CDR3P regions of non-cross-reactive M1+ Vβ17+ clonotypes were sequenced and arranged as described above.
Similarities between the cross-reactive and non-cross-reactive TCR repertoires
Vβ repertoire comparisons revealed that the CXR-1 (M1+ BMLF1+) T cells often expressed TCRs similar in structure to those expressed by non-cross-reactive M1-specific T cells (Fig. 4a, c), while the CXR-2 (M1- BMLF1+) T cells found within M1-specific T cell lines more often expressed TCRs similar in structure to those expressed by non-cross-reactive BMLF1-specific T cells (Fig. 4b, d). Based on IM patients E1101, E1106 and E1109 and healthy donor D-035, cross-reactive T cells that efficiently bound M1-loaded tetramer predominantly used Vβ17, perhaps indicative of a relatively high avidity for the M1 peptide (15). On the other hand, cross-reactive T cells that preferentially bound BMLF1-loaded tetramer tended to express non-Vβ17 families that are commonly present in BMLF1-specific repertoires. Previous studies suggested that the BMLF1-specific TCR repertoires of most individuals include at least 1 of 4 common Vβ families (Vβ2, 4, 16, 22) (20-22, 31). Interestingly, each of these 4 Vβ families was represented at least once among the cross-reactive repertoires analyzed in this study (Fig. 4).
To distinguish any clonal differences between cross-reactive and non-cross-reactive T cells of the same Vβ family, we sub-cloned and sequenced the CDR3β regions of sorted populations from T cell lines derived from healthy donor D-002 (Fig. 5a, b). This donor's non-cross-reactive BMLF1-specific population predominantly used Vβ14 (66%), which was used by 57% of the cross-reactive population (Fig. 4b, d). Although there were a greater number of unique clonotypes in the cross-reactive repertoire, the most frequently detected Vβ14+ clonotype in the cross-reactive repertoire (ID: B14.1, Jβ1.1, 7aa length) was the same clonotype that dominated the non-cross-reactive BMLF1-specific repertoire of this donor. Similar results were obtained when we sub-cloned the Vβ17 family, which was predominantly used by the non-cross-reactive M1-specific population (81% Vβ17+) and used at a low frequency by the cross-reactive population (< 1% Vβ17+) (Fig. 4b, c). This small population of Vβ17+ cross-reactive cells was surprisingly diverse at the clonal level. It was comprised of 42 unique clonotypes out of 55 total sequences analyzed, and the organizational pattern mimicked that typical of a memory M1-specific TCR repertoire in a healthy donor (Fig. 5d) (19, 36). For instance, there was no apparent skewing of the repertoire by one dominant clonotype or any alteration of the Jβ usage, which was previously observed during an acute EBV infection when a select population of cross-reactive clones may have proliferated in vivo (14). In agreement with the literature, IRSS was the dominant CDR3β loop sequence found within the non-cross-reactive M1+ Vβ17+ repertoire (used by 25% (10/40) of the unique clonotypes) and remained dominant within the cross-reactive population (used by 31% (13/42) of the unique cross-reactive clonotypes), including the most frequently detected clonotype (ID: 17.3, frequency: 7% of total sequences) (Fig. 5d, e) (16-18). Interestingly, one of the two most frequently detected clonotypes within the cross-reactive population was previously detected within the BMLF1-tetramer+ population isolated ex vivo, suggesting that the dominant Vβ17+ BMLF1-specific memory T cell clones may be those that are cross-reactive with influenza-M1 (Figs. 5d, 2a, Supplementary Table 1). However, concurrent α-chain analyses are required to determine if the similarities in β-chain usage observed in this study truly indicate that the same T cell clones dominate the cross-reactive and BMLF1-specific populations.
The Vα repertoire of cross-reactive cells is comprised of multiple Vα families and is distinct from that of non-cross-reactive M1+ or BMLF1+ populations
The α-chain analyses of the same tetramer-defined cross-reactive and non-cross-reactive T cell populations derived from healthy donor D-002 (Fig. 5a, b) require using only the PCR-based technique known as CDR3α spectratyping, as very few monoclonal antibodies specific to Vα families are available. This technique allowed us to detect the presence of specific Vα families as well as the presence of clonal T cell expansions within specific Vα families that were distinguished based on CDR3α length; however, this technique did not allow us to quantify the frequency of Vα family usage (25, 37). In an initial screen using primers specific for 24 different Vα families, the cross-reactive repertoire included at least 9 different Vα families (Vα1b, 2, 6, 10, 11, 15, 20, 23 and 24) (Fig. 6a). Thus, just as was observed upon Vβ analysis, a wide range of TCR structures appeared capable of interacting with the two dissimilar peptides M1 and BMLF1.
Figure 6. The Vα repertoire of cross-reactive cells is comprised of multiple Vα families and is distinct from that of non-cross-reactive M1+or BMLF1+populations.
CDR3α spectratyping analysis was performed on (a) the cross-reactive population described in Fig. 5a where arrows indicate positive reactions, or on (b) the non-cross-reactive BMLF1-specific population described in Fig. 5b, or simultaneously on (c) all three tetramer-defined cross-reactive and non-cross-reactive populations described in Fig. 5a, b where B = non-cross-reactive BMLF1+ cells, X = cross-reactive cells, and M = non-cross-reactive M1+ cells. (d) The CDR3α regions of Vα15+ T cells found within each of the 3 separate T cell populations were sequenced. Clonotypes with unique nucleotide sequences were assigned an ID and their frequency among all sequences analyzed is shown. The length of the CDR3α region was determined according to Chothia et al., shown supported by two flanking framework regions (51). Bold indicates that the clonotype was simultaneously detected within the non-cross-reactive BMLF1+ population.
When comparing the Vα repertoires of the cross-reactive and non-cross-reactive populations, we noted several similarities as well as some distinguishing features between them. Non-cross-reactive BMLF1-specific T cells used Vα2, 10, 11, 15, 22 and 23, along with some additional Vα families that may have been at the limit of detection using this protocol (Fig. 6b). All but Vα22 were also detected in the cross-reactive repertoire (Fig.6 a). The Vα repertoire of the non-cross-reactive M1-specific population also included a wide range of Vα families, most consistently including Vα1b, 2, 10 and 24, but with many additional Vα families being detected such as Vα6, 11, 15, 20 and 23 (Fig. 6c). All of these families were also detected in the cross-reactive repertoire (Fig. 6a). In order to compare the clonal composition of the cross-reactive and non-cross-reactive populations that expressed the same Vα family, we ran the corresponding PCR reactions on the same poly-acrylamide gel and compared the relative lengths of the CDR3α loops (Fig. 6c). CDR3 length differences were an indication of clonal differences between the three T cell populations analyzed in this study. Such differences were evident within the Vα11 family, where cross-reactive T cells clearly expressed longer CDR3α loops than either non-cross-reactive BMLF1-specific or non-cross-reactive M1-specific T cells (Fig. 6c). Similarly, Vα20+ cross-reactive T cells appeared to express a unique CDR3α loop length compared to either non-cross-reactive T cell population. However, there were also cases where the cross-reactive and non-cross-reactive T cells expressed similar CDR3α loop lengths. Cross-reactive T cells that expressed Vα2 or Vα15 had CDR3α lengths similar to BMLF1-specific T cells, while cross-reactive T cells that expressed Vα10 had CDR3α lengths similar to M1-specific T cells (Fig. 6c).
As has been reported in the literature, BMLF1-specific T cells often co-express Vα15 with one of 3 common Vβ families (Vβ2, 4, or 16), perhaps indicating an important role for this particular α-chain in binding the BMLF1 peptide (20-22). Since CDR3 spectratyping analysis revealed similarities between cross-reactive and non-cross-reactive Vα15+ T cells, we sequenced and compared Vα15+ clonotypes isolated from both populations (Fig. 6d). The same Vα15+ clonotype dominated (ID: B1) the cross-reactive (frequency: 70% of the sequences analyzed) and non-cross-reactive (frequency: 76% of the sequences analyzed) BMLF1-specific repertoires. However, the cross-reactive repertoire also included Vα15+ clonotypes that expressed unique Jα families (Jα11 and Jα13) and CDR3α loops 6aa long, a length that was more frequent in the cross-reactive repertoire (18% of sequences analyzed) than in the non-cross-reactive BMLF1-specific repertoire (2% of sequences analyzed). These unique cross-reactive Vα15+ clonotypes also did not resemble any of the non-cross-reactive M1-specific Vα15+ clonotypes we identified (Fig. 6d).
Computer Simulation: Lack of high affinity cross-reactive clones leads to a broader repertoire distribution
Using a murine model of viral infection, we previously identified a cross-reactive T cell population that recognized two distinct but structurally similar epitopes, differing by only two amino acids, derived from the nucleoprotein (NP) of two viruses, LCMV and PV (7). Following heterologous virus infection, TCR analyses of this cross-reactive T cell population revealed the development of a more restricted TCR repertoire compared to that prior to the infection or that seen in a naive mouse infected with the second virus (6). Computer studies with virtual immune mice challenged with a cross-reactive epitope mimicked our experimental data, showing that T-cell cross-reactivity can modulate clonal dominance and narrowing of the TCR repertoire. This system was further useful in predicting that repertoire narrowing could be a function of the proportion of high avidity cross-reactive T cells (6).
However, in our current studies of cross-reactive T cells in human viral infections, the cross-reactive epitopes are structurally dissimilar, with only three amino acids in common and with two of them important for binding to the MHC molecule. In this case, we observed that the cross-reactive T cell repertoire was actually broader than the non-cross-reactive repertoires. We were interested in using the virtual immune system to determine whether the dichotomy in the cross-reactive TCR repertoires observed in the human and mouse systems could, in part, be explained by the level of structural similarity between the epitopes involved. Based on our earlier observations, we hypothesized that if there are a few high frequency memory clones specific to the primary infection pathogen that also have high affinity to the subsequent challenging pathogen, this type of clone will dominate the TCR repertoire to the challenge pathogen's cross-reactive epitope and lead to a narrower repertoire. This is more likely to occur when epitopes are structurally similar (termed “near” cross-reactivity in Fig. 7). When epitopes are structurally dissimilar (termed “far” cross-reactivity in Fig. 7), this type of high affinity clone may not exist. The resulting repertoire would be broader, lacking any dominant pre-existing cross-reactive memory clones. For this investigation, we used the IMMSIM model, a stochastic agent-based computer simulator by which we can do experiments on “virtual” immune systems of mice or humans (6, 28-30).
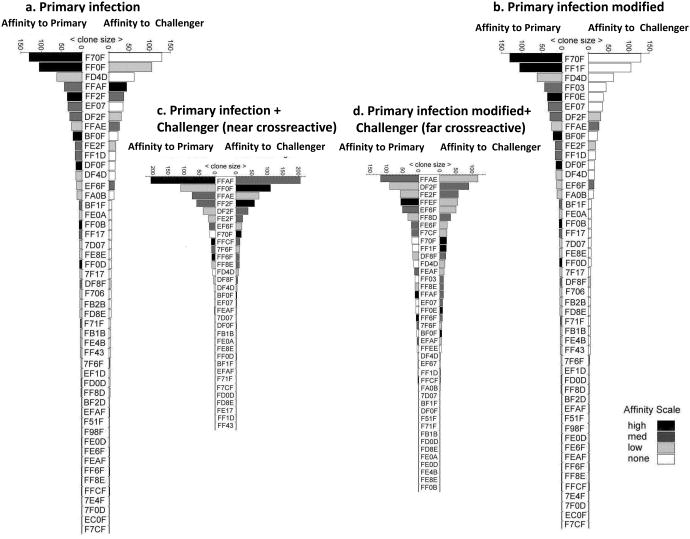
Figure 7. Computer simulation of cross-reactive response: Lack of high affinity cross-reactive clones leads to a broader repertoire distribution.
a.) Frequency hierarchy of clones responding to the primary infection. The clonal affinity to the primary immunogen is represented on the left and the affinity to the cross-reactive challenging pathogen is represented on the right. The bars are in shades of black to white, with black being clones having the highest affinity and white being clones having no affinity to the particular antigen. The individual clones are identified by a 4 digit code. b.) Frequency hierarchy of clones responding to the primary infection that has been modified to represent the distribution of clonotypes after the removal of the three highest frequency clonotypes with high affinity to the cross-reacting pathogen, thus simulating responses to a cross-reactive infection that is structurally dissimilar from the original antigen. In Fig. 7b, no high frequency high affinity clones to the primary epitope were cross-reactive with the challenging infection; clones FFOF, FFAF and FF2F have been replaced. c) The resulting repertoire upon secondary challenge of an individual having a memory TCR repertoire as depicted in (a) with a “near” or similar cross-reactive epitope. d) The resulting repertoire upon secondary challenge of an individual having a modified memory TCR repertoire as depicted in (b) with a “far” or dissimilar cross-reactive epitope. These results are representative of 3 sets of 26 virtual individuals each.
Figure 7a shows a computer generated repertoire after a primary infection and shows higher affinity clones to the primary pathogen epitope than to the cross-reactive epitope of the challenge pathogen. Fig. 7b shows what the repertoire would be to a cross-reactive epitope if we removed the three highest frequency clones having high affinity for the cross-reactive epitope from the memory pool of the primary infection. Both cross-reactive TCR repertoire distributions were evaluated after challenging with the cross-reactive epitope-containing pathogen in a simulation of the classic “adoptive transfer” technique, a feature introduced into IMMSIM's code to allow these kinds of studies. This feature allows us to analyse the diversity of secondary responses obtainable from the same memory repertoire, but in different individual environments, with remarkable accuracy and with similarity to in vivo adoptive transfer experiments. Upon challenging an individual with the unmodified memory TCR repertoire depicted in Fig. 7a with a structurally similar (“near”) cross-reactive epitope, the breadth of the cross-reactive repertoire shrank with notable skewing and domination of clone FFAF (Fig. 7c). In contrast, upon challenging an individual with the modified memory TCR repertoire depicted in Fig 7b with a structurally dissimilar (“far”) cross-reactive epitope, the breadth of the cross-reactive repertoire expanded (Fig. 7d). Note that the highest frequency clone FFAE now represents a lower proportion of the response than its counterpart in Fig. 7c. Interestingly, new unique clonotypes appeared in the repertoire during the latter experiment, which further increased diversity (FFEF, FFAF, FFEE, EF67). In Fig. 7c&d, after the secondary challenge, all of the higher frequency clones had high affinity for the second epitope, as they were driven to expand by their cross-reactivity to the second epitope. Following secondary challenge with the “near” cross-reactive epitope, the domination of a few clones having higher affinity for both the primary pathogen epitope and the cross-reactive epitope masked the detection of low frequency clones and resulted in a more polarized repertoire (Fig. 7c). The dominance of these high affinity memory clones is assured by their ability to out-compete the expansion of naïve clones. On the other hand, clones having lower affinity for the epitopes were present at lower frequencies upon challenge with the “far” cross-reactive epitope, allowing ample space for increased repertoire diversity (Fig. 7d). The data presented in Figure 7 represents one experiment out of 26 conducted. The TCR distributions presented here have been confirmed with statistical analyses that show during “near” cross-reactive epitope challenge experiments, 75% of the responding repertoire was composed of a mean of 5.3 different clones (95% confidence; range 4.9-5.6 clones), while during “far” cross-reactive epitope challenge experiments, 75% of the repertoire had a mean of 8.5 different clones (95% confidence; range 8.1-8.9 clones) (p<0.0001 n=26). Thus these computer simulation experiments conclude that the TCR repertoire responding to a structurally similar cross-reactive epitope is polarized by dominant high affinity clones, while that responding to a structurally dissimilar epitope has a broad distribution of lower affinity clones.
Discussion
Under conditions of heterologous immunity, reports have suggested that a cross-reactive population in response to a relatively similar epitope can lead to a narrower T cell repertoire and strong clonal dominance (6). Here, when examining cross-reactive repertoires in vitro that specifically recognize two dissimilar epitopes, IAV-M1 and EBV-BMLF1, we made a number of novel observations. First, upon validating the use of cultured cells in analysis of TCR repertoires, we showed that the cross-reactive repertoires were broader, using as many as 12 different Vβ families, and flatter, without selection of highly dominant clonotypes, when compared to the non-cross-reactive repertoire for each epitope. Second, spectratype analysis of the more difficult to study TCR Vα repertoire revealed an equally broad distribution utilizing 9 different Vα families. Third, the cross-reactive repertoires differed among 9 individuals tested, consistent with private specificity. Finally, the cross-reactive repertoires were enriched in otherwise low frequency T cell clones that expressed a TCR with a longer CDR3 loop length, often containing uncharged, non-bulky amino acid residues, such as glycines and serines. These features give TCRs added flexibility and, therefore, the ability to accommodate interactions with more than one epitope.
The mechanisms that shape T cell memory through α-TCR selection have been difficult to delineate due to the technical restraints associated with the lack of VA-family specific mAbs and ability of T cells to co-express two α-chains, with one usually being nonfunctional. We have recently examined the α-TCR repertoires of memory CD8 T cells reactive against the influenza A viral epitope, M158-66, restricted by HLA-A2.1 (38). The M158-66-specific, clonally diverse VB17 T cells expressed α-chains encoded by multiple AV-genes with different CDR3 sizes. A unique feature of these α-TCRs is the presence of poly-Gly/Ala runs in the CDR3, fitting to an AGA(Gn)GG-like amino acid motif much like those observed for the cross-reactive clones here. These non-template-encoded poly-Gly/Ala runs in the CDR3 of influenza A M1-specific memory pool are significantly enriched over that in naïve thymocytes, suggesting that these clones are preferentially selected during peripheral antigen exposure. The presence of these poly-Gly/Ala runs in the CDR3 of α- and β-chains might provide enhanced TCR flexibility and the ability to accommodate interaction with multiple epitopes (39, 40).
In this study, we hypothesized that the breadth of the cross-reactive T cell repertoire may depend on various factors such as the level of similarity between the epitopes. We tested this by computer simulation using IMMSIM analysis, which is capable of recapitulating a virtual immune response to viruses in individuals. We made the assumption that if epitopes are very dissimilar, it was less likely that high affinity cross-reactive clones exist that would rapidly dominate the response. In fact, the resulting repertoires did become broader and flatter upon exposure to the cross-reactive ligand compared to the starting memory repertoire responding to either cognate ligand alone. Such a diverse array of possible cross-reactive TCRs not only enhances the probability that a cross-reactive T cell response will occur during EBV infection but also expands or maintains the pool of cross-reactive memory T cell clones with the potential to effectively control EBV replication or contribute to protection from other new infections.
A difference between our murine and human models of heterologous immunity is the structure of the epitopes involved. The sequences of LCMV-NP205 and PV-NP205 are very similar, having 6 out of 8 aa residues in common, all of which are available to interact with the TCR. The sequences of IAV-M158 and EBV-BMLF1280 are quite dissimilar, having only 3 out of 9 aa residues in common with two of the amino acids important for binding to MHC. The cross-reactive repertoire of these two dissimilar epitopes appeared to include features of both antigen-specific TCR repertoires. M1-specific repertoires are polyclonal, comprised of many unique Vβ17+ clonotypes, while BMLF1-specific repertoires are often comprised of multiple different Vβ families. Thus, both antigen-specific TCR repertoires offered some level of structural diversity that, together, resulted in a cross-reactive TCR repertoire with the potential to include multiple Vβ families and multiple Vβ-specific clonotypes. This is in contrast to the LCMV or PV NP205-specific repertoire of a naïve host that is highly focused on only Vβ16 and that of an LCMV- or PV-immune host that is focused on two co-dominant Vβ families, Vβ16 and Vβ5 (6). Therefore, the cross-reactive memory repertoire in this murine system may have started with a more limited array of potential TCR structures and with the structural similarity the chances of having a dominant high affinity clone that recognizes both ligands equally is increased. There is some evidence that suggests in the mouse model that cross-reactive responses between LCMV GP34-41 and vaccinia virus a11r198-205, where sequence similarity is less than that between the highly homologous LCMV and PV NP epitopes, that the cross-reactive TcR repertoires can be broader and be associated with lower affinity responses that are less protective in vivo(41). Overall, the breadth of a responding T cell repertoire is likely dependent on the specific virus and epitope(s) involved.
Using computer simulation we were able to generate results that were similar to the in vivo results observed during murine LCMV-PVNP205 (similar “near” cross-reactive epitopes) and human IAV-MI-EBV-BMLFI (dissimilar “far” cross-reactive epitopes) leading to a partial explanation for the dichotomy in the effect cross-reactivity can have on the diversity of TCR repertoire. This is an example of how the IMMSIM model is useful to address biologically important situations that we cannot easily manipulate in vivo but could easily be occurring during viral infections in vivo. The computer simulation is very consistent with the concept that the abundance of high to moderate affinity memory clones govern dominance, bringing about a polarized repertoire and effectively limiting the expression of less abundant clones, thus causing repertoire reduction. This is a circumstance that is likely when there is a great deal of structural similarity between the two epitopes. In contrast, if there are small numbers of memory clones with moderate to high affinity to the cross-reactive epitope as might occur with more dissimilar cross-reactive epitopes this allows for some limited expansion of most of them as well as expansion of some new (unique) cross-reactive clones, leading to greater repertoire diversity.
Recent work using mutations in the H2Kb-restricted SIINFEKL epitope of ovalbumin and ovalbumin-specific transgenic T cells indicates that low affinity T cells initially expand with kinetics similar to that of high affinity T cells but leave the lymph node earlier and do not have the sustained expansion of higher affinity T cell clones, which eventually out compete the low affinity clones and dominate the response (42). The same may also be true for low affinity cross-reactive memory T cell clones that would appear early during infection due to their higher starting frequency but eventually be diluted by higher affinity less cross-reactive clones. In fact, we find the highest proportions of cross-reactive T cells during acute EBV infection and much lower proportions in the resting memory state (14). This editing of the lower affinity clones as the infection progresses may tend to eliminate the dominance of these clones and lead to a broader, less skewed repertoire when analyzed. This might contrast with the mouse model of LCMV and PV, where higher affinity T cells responding to more similar cross-reactive NP205 epitopes dominate the immune response, both during the acute and memory phase, upon PV infection of LCMV-immune mice (6). In conclusion, this study makes the point that a cross-reactive T cell response can be comprised of a diverse array of T cell clones. With structural and functional diversity, a responding T cell repertoire may be in better position to combat a viral infection. With age the naïve T cell population decreases (43-46) and memory T cells to previous infections are also deleted with each new infection (47-50) and each individual becomes more dependent on the diversity and potential cross-reactivity of memory T cells for any new response. Thus, the selection of a broad array of potentially cross-reactive memory T cells at low frequencies may ultimately become beneficial, maintaining a more diverse repertoire as the immune system ages. While there may be no current way to control the breadth of a T cell repertoire responding to a natural infection, one can aim to activate a broad T cell response during vaccination procedures.
Supplementary Material
Each unique clonotype is distinguished by its unique nucleotide sequence. * TCR variable (V) region nomenclature based on Arden et al., ** CDR3 loop length according to Chothia et al. shown flanked by two framework regions.
Acknowledgments
We thank Laurie Kenney, Robin Brody and Joyce Pepe for their helpful contributions to this study.
This research was supported by a Worcester Foundation grant and by US National Institute of Health (NIH) grants AI-49320, AI-42845, AI-054455, DR-32520 and an immunology training grant 5 T32 AI-07349-16. The contents of this publication are solely the responsibility of the authors and do not represent the official view of the NIH.
References
- 1.Dutton RW, Bradley LM, Swain SL. T cell memory. Annu Rev Immunol. 1998;16:201–223. doi: 10.1146/annurev.immunol.16.1.201. [DOI] [PubMed] [Google Scholar]
- 2.Sprent J, Surh CD. T cell memory. Annu Rev Immunol. 2002;20:551–579. doi: 10.1146/annurev.immunol.20.100101.151926. [DOI] [PubMed] [Google Scholar]
- 3.Welsh RM, Selin LK, Szomolanyi-Tsuda E. Immunological memory to viral infections. Annu Rev Immunol. 2004;22:711–743. doi: 10.1146/annurev.immunol.22.012703.104527. [DOI] [PubMed] [Google Scholar]
- 4.Chen HD, Fraire AE, Joris I, Brehm MA, Welsh RM, Selin LK. Memory CD8+ T cells in heterologous antiviral immunity and immunopathology in the lung. Nat Immunol. 2001;2:1067–1076. doi: 10.1038/ni727. [DOI] [PubMed] [Google Scholar]
- 5.Selin LK, Welsh RM. Plasticity of T cell memory responses to viruses. Immunity. 2004;20:5–16. doi: 10.1016/S1074-7613(03)00356-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornberg M, Chen AT, Wilkinson LA, Brehm MA, Kim SK, Calcagno C, Ghersi D, Puzone R, Celada F, Welsh RM, Selin LK. Narrowed TCR repertoire and viral escape as a consequence of heterologous immunity. J Clin Invest. 2006;116:1443–1456. doi: 10.1172/JCI27804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brehm MA, Pinto AK, Daniels KA, Schneck JP, Welsh RM, Selin LK. T cell immunodominance and maintenance of memory regulated by unexpectedly cross-reactive pathogens. Nat Immunol. 2002;3:627–634. doi: 10.1038/ni806. [DOI] [PubMed] [Google Scholar]
- 8.Haanen JB, Wolkers MC, Kruisbeek AM, Schumacher TN. Selective expansion of cross-reactive CD8(+) memory T cells by viral variants. J Exp Med. 1999;190:1319–1328. doi: 10.1084/jem.190.9.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Urbani S, Amadei B, Fisicaro P, Pilli M, Missale G, Bertoletti A, Ferrari C. Heterologous T cell immunity in severe hepatitis C virus infection. J Exp Med. 2005;201:675–680. doi: 10.1084/jem.20041058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borrow P, Lewicki H, Wei X, Horwitz MS, Peffer N, Meyers H, Nelson JA, Gairin JE, Hahn BH, Oldstone MB, Shaw GM. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- 11.Khan N, Cobbold M, Keenan R, Moss PA. Comparative analysis of CD8+ T cell responses against human cytomegalovirus proteins pp65 and immediate early 1 shows similarities in precursor frequency, oligoclonality, and phenotype. J Infect Dis. 2002;185:1025–1034. doi: 10.1086/339963. [DOI] [PubMed] [Google Scholar]
- 12.Meyer-Olson D, Shoukry NH, Brady KW, Kim H, Olson DP, Hartman K, Shintani AK, Walker CM, Kalams SA. Limited T cell receptor diversity of HCV-specific T cell responses is associated with CTL escape. J Exp Med. 2004;200:307–319. doi: 10.1084/jem.20040638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peggs K, Verfuerth S, Pizzey A, Ainsworth J, Moss P, Mackinnon S. Characterization of human cytomegalovirus peptide-specific CD8(+) T-cell repertoire diversity following in vitro restimulation by antigen-pulsed dendritic cells. Blood. 2002;99:213–223. doi: 10.1182/blood.v99.1.213. [DOI] [PubMed] [Google Scholar]
- 14.Clute SC, Watkin LB, Cornberg M, Naumov YN, Sullivan JL, Luzuriaga K, Welsh RM, Selin LK. Cross-reactive influenza virus-specific CD8+ T cells contribute to lymphoproliferation in Epstein-Barr virus-associated infectious mononucleosis. J Clin Invest. 2005;115:3602–3612. doi: 10.1172/JCI25078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawson TM, Man S, Williams S, Boon AC, Zambon M, Borysiewicz LK. Influenza A antigen exposure selects dominant Vbeta17+ TCR in human CD8+ cytotoxic T cell responses. Int Immunol. 2001;13:1373–1381. doi: 10.1093/intimm/13.11.1373. [DOI] [PubMed] [Google Scholar]
- 16.Lehner PJ, Wang EC, Moss PA, Williams S, Platt K, Friedman SM, Bell JI, Borysiewicz LK. Human HLA-A0201-restricted cytotoxic T lymphocyte recognition of influenza A is dominated by T cells bearing the V beta 17 gene segment. J Exp Med. 1995;181:79–91. doi: 10.1084/jem.181.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moss PA, Moots RJ, Rosenberg WM, Rowland-Jones SJ, Bodmer HC, McMichael AJ, Bell JI. Extensive conservation of alpha and beta chains of the human T-cell antigen receptor recognizing HLA-A2 and influenza A matrix peptide. Proc Natl Acad Sci U S A. 1991;88:8987–8990. doi: 10.1073/pnas.88.20.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naumov YN, Hogan KT, Naumova EN, Pagel JT, Gorski J. A class I MHC-restricted recall response to a viral peptide is highly polyclonal despite stringent CDR3 selection: implications for establishing memory T cell repertoires in “real-world” conditions. J Immunol. 1998;160:2842–2852. [PubMed] [Google Scholar]
- 19.Naumov YN, Naumova EN, Hogan KT, Selin LK, Gorski J. A fractal clonotype distribution in the CD8+ memory T cell repertoire could optimize potential for immune responses. J Immunol. 2003;170:3994–4001. doi: 10.4049/jimmunol.170.8.3994. [DOI] [PubMed] [Google Scholar]
- 20.Annels NE, Callan MF, Tan L, Rickinson AB. Changing patterns of dominant TCR usage with maturation of an EBV-specific cytotoxic T cell response. J Immunol. 2000;165:4831–4841. doi: 10.4049/jimmunol.165.9.4831. [DOI] [PubMed] [Google Scholar]
- 21.Cohen GB, Islam SA, Noble MS, Lau C, Brander C, Altfeld MA, Rosenberg ES, Schmitz JE, Cameron TO, Kalams SA. Clonotype tracking of TCR repertoires during chronic virus infections. Virology. 2002;304:474–484. doi: 10.1006/viro.2002.1743. [DOI] [PubMed] [Google Scholar]
- 22.Lim A, Trautmann L, Peyrat MA, Couedel C, Davodeau F, Romagne F, Kourilsky P, Bonneville M. Frequent contribution of T cell clonotypes with public TCR features to the chronic response against a dominant EBV-derived epitope: application to direct detection of their molecular imprint on the human peripheral T cell repertoire. J Immunol. 2000;165:2001–2011. doi: 10.4049/jimmunol.165.4.2001. [DOI] [PubMed] [Google Scholar]
- 23.Catalina MD, Sullivan JL, Bak KR, Luzuriaga K. Differential evolution and stability of epitope-specific CD8(+) T cell responses in EBV infection. J Immunol. 2001;167:4450–4457. doi: 10.4049/jimmunol.167.8.4450. [DOI] [PubMed] [Google Scholar]
- 24.Arden B, Clark SP, Kabelitz D, Mak TW. Human T-cell receptor variable gene segment families. Immunogenetics. 1995;42:455–500. doi: 10.1007/BF00172176. [DOI] [PubMed] [Google Scholar]
- 25.Han M, Harrison L, Kehn P, Stevenson K, Currier J, Robinson MA. Invariant or highly conserved TCR alpha are expressed on double-negative (CD3+CD4-CD8-) and CD8+ T cells. J Immunol. 1999;163:301–311. [PubMed] [Google Scholar]
- 26.Maslanka K, Piatek T, Gorski J, Yassai M, Gorski J. Molecular analysis of T cell repertoires. Spectratypes generated by multiplex polymerase chain reaction and evaluated by radioactivity or fluorescence. Hum Immunol. 1995;44:28–34. doi: 10.1016/0198-8859(95)00056-a. [DOI] [PubMed] [Google Scholar]
- 27.Bahl K, Kim SK, Calcagno C, Ghersi D, Puzone R, Celada F, Selin LK, Welsh RM. IFN-induced attrition of CD8 T cells in the presence or absence of cognate antigen during the early stages of viral infections. J Immunol. 2006;176:4284–4295. doi: 10.4049/jimmunol.176.7.4284. [DOI] [PubMed] [Google Scholar]
- 28.Cheng Y, Ghersi D, Calcagno C, Selin LK, Puzone R, Celada F. A discrete computer model of the immune system reveals competitive interactions between the humoral and cellular branch and between cross-reacting memory and naive responses. Vaccine. 2009;27:833–845. doi: 10.1016/j.vaccine.2008.11.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohler B, Puzone R, Seiden PE, Celada F. A systematic approach to vaccine complexity using an automaton model of the cellular and humoral immune system. I. Viral characteristics and polarized responses. Vaccine. 2000;19:862–876. doi: 10.1016/s0264-410x(00)00225-5. [DOI] [PubMed] [Google Scholar]
- 30.Selin LK, Cornberg M, Brehm MA, Kim SK, Calcagno C, Ghersi D, Puzone R, Celada F, Welsh RM. CD8 memory T cells: cross-reactivity and heterologous immunity. Semin Immunol. 2004;16:335–347. doi: 10.1016/j.smim.2004.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Callan MF, Fazou C, Yang H, Rostron T, Poon K, Hatton C, McMichael AJ. CD8(+) T-cell selection, function, and death in the primary immune response in vivo. J Clin Invest. 2000;106:1251–1261. doi: 10.1172/JCI10590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bousso P, Casrouge A, Altman JD, Haury M, Kanellopoulos J, Abastado JP, Kourilsky P. Individual variations in the murine T cell response to a specific peptide reflect variability in naive repertoires. Immunity. 1998;9:169–178. doi: 10.1016/s1074-7613(00)80599-3. [DOI] [PubMed] [Google Scholar]
- 33.Cibotti R, Cabaniols JP, Pannetier C, Delarbre C, Vergnon I, Kanellopoulos JM, Kourilsky P. Public and private V beta T cell receptor repertoires against hen egg white lysozyme (HEL) in nontransgenic versus HEL transgenic mice. J Exp Med. 1994;180:861–872. doi: 10.1084/jem.180.3.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SK, Cornberg M, Wang XZ, Chen HD, Selin LK, Welsh RM. Private specificities of CD8 T cell responses control patterns of heterologous immunity. J Exp Med. 2005;201:523–533. doi: 10.1084/jem.20041337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naumov YN, Naumova EN, Clute SC, Watkin LB, Kota K, Gorski J, Selin LK. Complex T cell memory repertoires participate in recall responses at extremes of antigenic load. J Immunol. 2006;177:2006–2014. doi: 10.4049/jimmunol.177.3.2006. [DOI] [PubMed] [Google Scholar]
- 37.Yassai M, Naumova EN, Gorski J. Generation of TCR spectratypes by multiplex PCR for T cell repertoire analysis. In: Oksenberg JR, editor. The antigen T cell receptor: selected protocols and applications. Landes Company and Chapman & Hall; Austin, TX: 1997. pp. 327–372. [Google Scholar]
- 38.Naumov YN, Naumova EN, Yassai MB, Kota K, Welsh RM, Selin LK. Multiple glycines in TCR alpha-chains determine clonally diverse nature of human T cell memory to influenza A virus. J Immunol. 2008;181:7407–7419. doi: 10.4049/jimmunol.181.10.7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abergel C, Claverie JM. A strong propensity toward loop formation characterizes the expressed reading frames of the D segments at the Ig H and T cell receptor loci. Eur J Immunol. 1991;21:3021–3025. doi: 10.1002/eji.1830211218. [DOI] [PubMed] [Google Scholar]
- 40.Bentley GA, Mariuzza RA. The structure of the T cell antigen receptor. Annu Rev Immunol. 1996;14:563–590. doi: 10.1146/annurev.immunol.14.1.563. [DOI] [PubMed] [Google Scholar]
- 41.Cornberg M, Clute SC, Watkin LB, Saccoccio FM, Kim SK, Naumov YN, Brehm MA, Aslan N, Welsh RM, Selin LK. CD8 T cell cross-reactivity networks mediate heterologous immunity in human EBV and murine vaccinia virus infections. J Immunol. 2010;184:2825–2838. doi: 10.4049/jimmunol.0902168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zehn D, Lee SY, Bevan MJ. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 2009;458:211–214. doi: 10.1038/nature07657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu X, Beckman I, Ahern M, Bradley J. A comprehensive analysis of peripheral blood lymphocytes in healthy aged humans by flow cytometry. Immunol Cell Biol. 1993;71(Pt 6):549–557. doi: 10.1038/icb.1993.61. [DOI] [PubMed] [Google Scholar]
- 44.Provinciali M, Moresi R, Donnini A, Lisa RM. Reference values for CD4+ and CD8+ T lymphocytes with naive or memory phenotype and their association with mortality in the elderly. Gerontology. 2009;55:314–321. doi: 10.1159/000199451. [DOI] [PubMed] [Google Scholar]
- 45.Stulnig T, Maczek C, Bock G, Majdic O, Wick G. Reference intervals for human peripheral blood lymphocyte subpopulations from ‘healthy’ young and aged subjects. Int Arch Allergy Immunol. 1995;108:205–210. doi: 10.1159/000237155. [DOI] [PubMed] [Google Scholar]
- 46.Utsuyama M, Hirokawa K, Kurashima C, Fukayama M, Inamatsu T, Suzuki K, Hashimoto W, Sato K. Differential age-change in the numbers of CD4+CD45RA+ and CD4+CD29+ T cell subsets in human peripheral blood. Mech Ageing Dev. 1992;63:57–68. doi: 10.1016/0047-6374(92)90016-7. [DOI] [PubMed] [Google Scholar]
- 47.Selin LK, Lin MY, Kraemer KA, Pardoll DM, Schneck JP, Varga SM, Santolucito PA, Pinto AK, Welsh RM. Attrition of T cell memory: selective loss of LCMV epitope-specific memory CD8 T cells following infections with heterologous viruses. Immunity. 1999;11:733–742. doi: 10.1016/s1074-7613(00)80147-8. [DOI] [PubMed] [Google Scholar]
- 48.Selin LK, Vergilis K, Welsh RM, Nahill SR. Reduction of otherwise remarkably stable virus-specific cytotoxic T lymphocyte memory by heterologous viral infections. J Exp Med. 1996;183:2489–2499. doi: 10.1084/jem.183.6.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Welsh RM, Selin LK. Attrition of memory CD8 T cells. Nature. 2009;459:E3–4. doi: 10.1038/nature08091. discussion E4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hammarlund E, Lewis MW, Hansen SG, Strelow LI, Nelson JA, Sexton GJ, Hanifin JM, Slifka MK. Duration of antiviral immunity after smallpox vaccination. Nat Med. 2003;9:1131–1137. doi: 10.1038/nm917. [DOI] [PubMed] [Google Scholar]
- 51.Chothia C, Boswell DR, Lesk AM. The outline structure of the T-cell alpha beta receptor. Embo J. 1988;7:3745–3755. doi: 10.1002/j.1460-2075.1988.tb03258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Each unique clonotype is distinguished by its unique nucleotide sequence. * TCR variable (V) region nomenclature based on Arden et al., ** CDR3 loop length according to Chothia et al. shown flanked by two framework regions.