Abstract
High-fat (HF) diets can produce obesity and have been linked to the development of nonalcoholic fatty liver disease and changes in the gut microbiome. To test the hypothesis that HF feeding increases certain predominant hind gut bacteria and development of steatohepatitis, C57BL/6 mice were fed an HF (45% energy) or low-fat (LF) (10% energy) diet for 10 wk. At the end of the feeding period, body weights in the HF group were 34% greater than those in the LF group (P < 0.05). These changes were associated with dramatic increases in lipid droplet number and size, inflammatory cell infiltration, and inducible nitric oxide (NO) synthase protein concentration in the livers of mice fed the HF diet. Consistent with the fatty liver phenotype, plasma leptin and tumor necrosis factor-α concentrations were also elevated in mice fed the HF diet, indicative of chronic inflammation. Eight of 12 pairs of polymerase chain reaction (PCR) primers for bacterial species that typically predominate hind gut microbial ecology generated specific PCR products from the fecal DNA samples. The amount of DNA from Lactobacillus gasseri and/or Lactobacillus taiwanensis in the HF group was 6900-fold greater than that in the LF group. Many of these bacteria are bile acid resistant and are capable of bile acid deconjugation. Because bile acids are regulators of hepatic lipid metabolism, the marked increase of gut L. gasseri and/or L. taiwanensis species bacteria with HF feeding may play a role in development of steatohepatitis in this model.
Introduction
Obesity, a condition of excess adipose storage in the body, is a major factor in the metabolic syndrome and for increased risk of type 2 diabetes (1). Because two-thirds of American adults are overweight or obese (2), there is an urgent need to understand the metabolic consequences of obesity. Chronic low-grade inflammation can accompany increased adiposity, which has been linked to the development of nonalcoholic fatty liver disease, an emerging clinical problem (3, 4).
The pathways that are active in promoting nonalcoholic fatty liver disease are poorly understood, but it is possible that the process may involve the hind gut microflora, which can affect extraction of energy from the diet (5–7). Germ-free mice have significantly less body fat than their normal counterparts (8–10). In addition, it has been found that lean and obese mice had different predominant bacteria in their respective hind gut microbiomes (11, 12). Oral treatment of lean mice with the feces of obese mice caused them to gain body fat (11, 12).
More than 400 species of bacteria are known to be present in the hind guts of most monogastric animals; however, a much smaller number of species, as few as a dozen (13–15), comprise most of the culturable biomass in the hind gut. These predominant species would appear to be most important in producing metabolites capable of affecting the host.
Studies have shown that the hind gut microflora can be affected by diet composition, particularly, with respect to insoluble, fermentable carbohydrate content and total fat level (6, 7, 11, 12). Recent studies demonstrated that the composition of the microbiota, along with its gene expression and functional metabolic pathways, rapidly changed when animals were switched from low-fat (LF)7 to high-fat (HF) diets (11, 12). However, studies have not addressed the possibility that the predominant hind gut bacterial species affected by HF feeding may be related to hepatic inflammation and steatosis. The present study addressed this issue and tested the hypothesis that HF feeding promotes both gut bacteria and the development of steatohepatitis in the C57BL/6 mouse model of obesity.
Materials and Methods
Animals, diets, and treatment.
This study was approved by the Animal Care and Use Committee of the Grand Forks Human Nutrition Research Center, and animals were maintained in accordance with NIH guidelines for the care and use of laboratory animals. Male C57BL/6 mice, 5 wk old, were obtained from Charles River Laboratories. Mice were individually housed in Plexiglas ventilated cages within a pathogen-free facility that maintained a 12-h light/dark cycle. Mice were given free access to food and deionized water and were allowed to acclimate in the facility for 2 d before being randomly assigned to 2 dietary treatment groups (n = 10 each). The feeding experiment was conducted for 10 wk, and treatments consisted of an LF purified diet (10% of calories from fat, D12450B; Research Diets) based on an AIN-76A or an HF diet (45% of calories from fat, D12451; Research Diets) (Supplemental Table 1) (16). At the termination of the experiment, mice were feed-deprived overnight (6 h) and then killed with a mixture of ketamine and xylazine. Liver and plasma were collected and stored at −80°C for analyses of leptin, interleukin 6 (IL-6), tumor necrosis factor α (TNF-α), and TGs and for Western blotting analysis.
Liver histology.
Livers were fixed in 10% neutral buffered formalin and embedded in paraffin. Five-micrometer sections were mounted on slides and stained with hematoxylin and eosin. Each section was then scored for the number and size of lipid droplets and the infiltration of inflammatory cells by using a standardized determination of morphology (17, 18). Image Pro Plus version 6.2 software (North Central Instruments) was used for quantification of digitized images.
Inducible NO synthase immunohistochemistry.
Liver tissues were sectioned at 5 μm, and sections were deparaffinized and rehydrated. The inducible NO synthase (iNOS) expression was assessed by using an immunohistochemistry detection kit (Abcam, Inc.). Rabbit polyclonal iNOS antibody (Abcam, Inc.) was diluted at 1:100. For histologic analysis of the iNOS, 10-mm2-section fields from each liver were captured by a Leica MZ6 stereomicroscope and a Leica DFC420 C digital camera (North Central Instruments). Image Pro Plus version 6.2 software was used for quantification of the immunostained area.
Plasma leptin, IL-6, TNF-α and TGs.
Leptin, IL-6, and TNF-α concentrations were measured in plasma by using ELISA kits (R&D Systems). Plasma TGs were assessed in duplicate by using a TG assay kit (Zen-Bio, Inc.).
Western blot analysis of phosphorylated AMP-activated protein kinase/total AMP-activated protein kinase.
Cell lysates were made by sonicating homogenized liver tissues at 4°C using a tissue lysis buffer (Cell Signaling Technologies) including 1 mmol/L phenylmethylsulfonyl fluoride, as previously described (19). The cell lysate was clarified by centrifugation at 14,000 × g for 30 min at 4°C; the supernatant was designated as whole-cell protein extract and kept at −80°C. The protein concentration was quantified by the Bradford dye-binding assay (Bio-Rad Laboratories). Equal amounts of protein extract, ∼40 μg, were resolved over 4–20% Tris-glycine gradient gels under denaturing and reducing conditions and electroblotted onto polyvinylidene fluoride membranes (Invitrogen). Membrane blots were blocked in PBS containing 0.05% Tween (v:v) supplemented with 1% (wt:v) nonfat dry milk (BioRad) at 4°C for overnight. Membranes were probed with antibodies against AMP-activated protein kinase (AMPK) or phospho-AMPKα (Thr172) and then incubated with an anti-rabbit (1:3000 dilution) HRP-conjugated secondary antibody (Santa Cruz Biotechnology) in blocking solution for 1 h at room temperature. Blots were washed as above, and proteins were detected by using an enzymatic chemiluminescence plus kit (Amersham Pharmacia Biotech) with the Molecular Dynamics Image-Quant system.
Detection and quantitation of predominant bacteria in fecal samples and 16S sequencing.
Fecal pellets were collected from each mouse for 5 d in wk 10 and stored at −20°C. The total fecal genomic DNA of each mouse was isolated from fecal pellets by using a genomic DNA isolation kit (Qiagen). The 12 pairs of PCR primers (13) for numerically predominant species of hind gut bacteria were previously identified (13–15). The specificity of these primers was updated by primer DNA sequence alignment against the National Center for Biotechnology Information’s database 16S reference data. Accordingly, the potential bacteria with which these 12 primer pairs reacted were as follows: 1) Bifidobacterium ruminantium; 2) Bifidobacterium longum; 3) Eubacterium biforme; 4) Eubacterium limosum, Eubacterium chllanderi; 5) Faecalibacterium prausnitzii; 6) Blautia luti; 7) Lactobacillus gasseri, Lactobacillus taiwanensis, Lactobacillus johnsonii; 8) Escherichia coli, Shigella sonnei; 9) Bacteroides xylanisolvens, Bacteroides finegoldii, Bacteroides ovatus, but only certain strains of Bacteroides thetaiotaomicron; 10) Bacteroides dotei, Bacteroides baku; 11) Parabacteroides distasonis; and 12) Clostridium clostridiiforme. Oligonucleotide primers for those species were synthesized by Integrated DNA Technologies with HPLC purification. The genomic DNA of each of these bacteria was assessed by using a QuantiTect SYBR Green PCR kit (Qiagen) with the comparative CT method (20) in an Applied Biosystems 7300 Real-Time PCR System, and a set of universal primers for the amplification of bacterial 16S ribosomal RNA (rRNA) was included to ensure the equal bacterial genomic DNA load template in above real-time PCR reactions (20, 21). The DNA samples were diluted so that the actual PCR was performed within a closer range of CT values, and specific PCR products were examined via 1.9% agarose gel electrophoresis. The PCR products whose amount was significantly changed because of HF feeding were gel-purified (Qiagen) and sequenced from both strands by using an ABI 377 automated sequencer.
Statistical analysis.
Results are given as means ± SEMs. Data were analyzed by using Student’s t test (JMP, version 9.0; SAS Institute). For the real-time PCR data, ΔCT was analyzed; however, the fecal DNA amount (2−ΔCt) was reported in the text. Regression analysis (Excel 2007) was used to test whether there was a linear relationship between ΔCT and the area of lipid droplets. Differences with a P value <0.05 were considered significant.
Results
Body weight, hepatic lipid accumulation, and inflammatory cell infiltration.
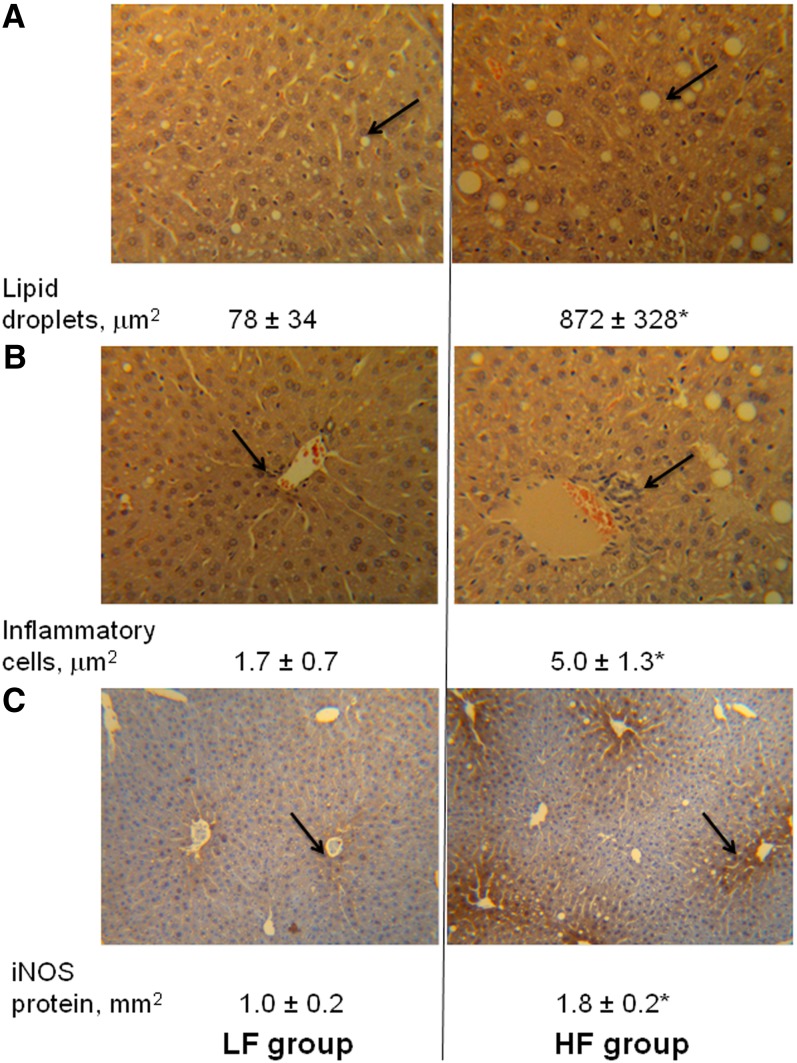
At the end of the 10-wk feeding period, the mean body weights in the HF group (36.7 ± 1.0 g) were greater than those in the LF group (27.3 ± 1.0 g) (P < 0.0001). Correspondingly, histologic examination with liver sections revealed that the areas (number and size) of lipid droplets, infiltration of inflammatory cells, and iNOS expression in the HF group were 10.1-, 1.9-, and 0.8-fold greater than that in the LF group, respectively (Fig. 1).
FIGURE 1.
The effect of an HF diet on hepatic lipid accumulation and inflammatory status in C57BL/6 mice fed an HF or an LF diet for 10 wk. (A) The area of fatty droplets in a 3.8 × 106 μm2 liver tissue section stained with hematoxylin and eosin; (B) the area of inflammatory cells in a 1.5 × 107 μm2 liver tissue section stained with hematoxylin and eosin; and (C) the area of iNOS protein expression in a 10 mm2 liver tissue section stained with 3,3′-diaminobenzidine. Values are means ± SEMs, n = 9. *Different from LF, P < 0.05. HF, high-fat; iNOS, inducible NO synthase; LF, low-fat.
Plasma inflammatory cytokines and TGs and the phosphorylation state of hepatic AMPK.
HF feeding did not significantly affect plasma concentrations of TGs or IL-6 or hepatic phosphorylated:total AMPK ratio (Table 1). However, the concentrations of plasma leptin and TNF-α in the HF group were 5.2- and 0.3-fold greater than those in the LF group, respectively.
TABLE 1.
Plasma TGs, cytokines, and the hepatic phosphorylated:total AMPK ratio in C57Bl/6 mice fed an HF or an LF diet for 10 wk1
| HF group | LF group | |
| Plasma IL-6, ng/L | 0.68 ± 0.19 | 0.45 ± 0.18 |
| Plasma leptin, μg/L | 2.82 ± 0.37 | 0.45 ± 0.13* |
| Plasma TGs, mmol/L | 0.53 ± 0.04 | 0.51 ± 0.03 |
| Plasma TNF-α, ng/L | 2.47 ± 0.12 | 1.93 ± 0.17* |
| Hepatic phosphorylated: total AMPK | 0.20 ± 0.02 | 0.25 ± 0.03 |
Values are means ± SEMs, n = 10. *Different from HF, P < 0.05. AMPK, AMP-activated protein kinase; HF, high-fat; IL-6 interleukin 6; LF, low-fat; TNF-α, tumor necrosis factor α.
L. gasseri and/or L. taiwanensis bacteria correlated with lipid droplets in liver.
Eight out of 12 pairs of PCR primers (13) generated specific PCR products, and the 4 pairs of primers that did not yield specific PCR products were for the following bacteria: 1) B. ruminantium, 2) B. longum, 3) E. biforme, 4) and E. limosum and E. chllanderi. The DNA amounts of these 8 specific PCR products were assessed, and only 1 product was different because of HF feeding. On the basis of real-time PCR analysis, this suggested that the DNA amount of L. gasseri and/or L. taiwanensis and/ or L. johnsonii in the HF group (0.04 ± 0.02 arbitrary units) was 6910-fold greater than that in the LF group (6.3 × 10−6 ± 3.9 × 10−6 arbitrary units) (P < 0.01). To further confirm this bacterial determination, PCR products of 16S rRNA from 6 mice representing both LF and HF groups were sequenced from both strands and share the identical sequence (275 bp). When BLASTing the 275 bp against the National Center for Biotechnology Information’s 16S reference database, L. gasseri and L. taiwanensis species were the only 2 species with 100% sequence identity.
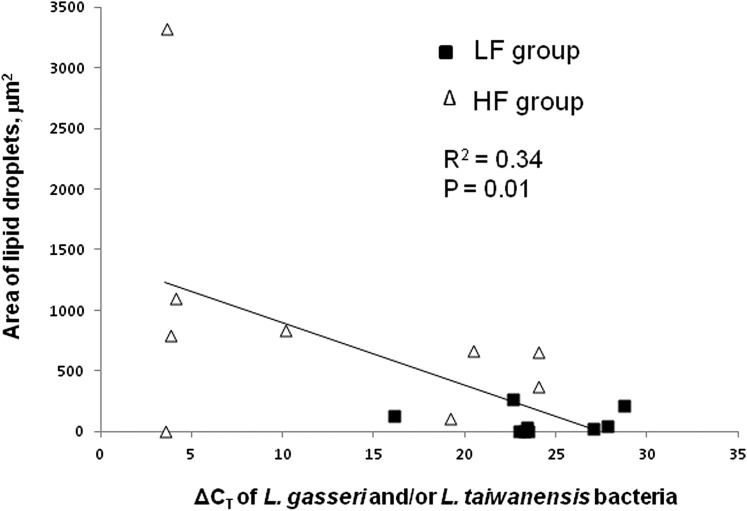
Further analyses revealed that there was a significant positive correlation of L. gasseri and/or L. taiwanensis DNA and the area of lipid droplets in liver (the smaller the ΔCT, the greater the DNA amount; Fig. 2).
FIGURE 2.
The positive relationship of the area of lipid droplets in liver and fecal DNA amounts of Lactobacillus gasseri and/or Lactobacillus taiwanensis species in C57BL/6 mice fed an HF or an LF diet for 10 wk; n = 18 (ΔCT is inversely related to DNA amount). HF, high-fat; LF, low-fat.
Discussion
This study undertook to test the hypothesis that HF feeding promotes certain predominant hind gut bacteria and the development of steatohepatitis in the C57BL/6 mouse model of obesity. This model uses a diet high in total fat, and particularly n6 fatty acids, to produce outcomes similar to those observed in obese humans (22, 23), namely, increased adiposity, production of proinflammatory cytokines, and fatty infiltration of the liver (Fig. 1).
Consistent with previous findings that metabolic syndrome caused by an HF diet results in activation of stress signaling pathways (24), the present results show that HF feeding increased plasma concentrations of proinflammatory cytokines, leptin, and TNF-α. However, because IL-6 concentrations were generally very low and not different between treatment groups, this suggested that IL-6 played little role in this model (Table 1). Leptin is an adipokine whose proinflammatory properties are known to contribute to the development and progression of inflammation in several autoimmune and nonautoimmune inflammatory conditions (25, 26). TNF-α is secreted in colonocytes and hepatic tissues in response to stimuli from the gut and circulation, respectively (27). HF feeding did not cause changes in plasma TG concentration, which is consistent with a previous finding (28) in the same mouse model, indicating that plasma TG concentration may not be a sensitive marker of early stages of steatohepatitis in C57BL/6 mice. The HF diet also did not affect hepatic concentrations of activated AMPK, which is known to stimulate ATP-producing catabolic pathways (e.g., fatty acid oxidation) and inhibit ATP-consuming processes (e.g., lipogenesis) (Table 1) (29, 30). That this endpoint was not significantly affected by diet indicates that it is not sensitive to early-stage hepatic lipid accumulation.
Emerging evidence suggests that hind gut microbiota may contribute to liver pathobiology (5). A close interplay exists between the gut and liver because metabolically active substances produced in the lumen of the colon are absorbed and delivered to the liver by enterohepatic recirculation (5). In fact, the mammalian liver derives nearly 70% of its blood supply from the portal vein, the direct venous outflow of the intestine (31). It is well known that HF feeding increases the secretion of bile and bile acid, which generally have strong antimicrobial activities (5–7), and bile acids are believed to be a determinant of the gut microbiota in response to an HF diet. Our results indicate that the HF feeding protocol used here greatly increased gut bacterial species L. gasseri and/or L. taiwanensis of the Lactobacillus acidophilus species group (32). This increase was from 0.00062% (LH feeding) to 4.3% (HF feeding) of total gut bacteria using the CT value of total bacterial 16S rRNA as the reference. Although much remains to be learned about the biological function of the newly established L. taiwanensis species (32), many L. acidophilus species bacteria including L. gasseri species are bile acid resistant, and also partially responsible for bile acid deconjugation, a process that efficiently reduces lipid absorption in the gut (33–36).
Bile acids not only act as metabolically active signaling molecules for hepatic lipid metabolism but they also play critical roles in inflammatory liver and colon diseases (37, 38). Thus, the dramatic increases in L. gasseri and/or L. taiwanensis and excessive adipose tissue/inflammatory fatty liver in mice fed the HF diet may reflect symbiotic relationships that are likely the results of coevolution of host and gut microbiota. This hypothesis is supported by the finding of a significant positive correlation of L. gasseri and/or L. taiwanensis bacterial DNA and hepatic lipid droplet areas, which was significant even after exclusion of the greatest (and most leveraging) Y-value (R2 = 0.3, P = 0.02; Fig. 2). L. gasseri and/or L. taiwanensis bacteria have been suggested to play a role in body weight control (39, 40). Further studies with germ-free mice are needed to determine the mechanistic role of these bacteria in the development of inflammatory fatty liver due to HF feeding.
These results demonstrate, for the first time to our knowledge, that in this model HF feeding causes obesity-related inflammatory fatty liver accompanied by a large increase in hind gut L. gasseri and/or L. taiwanensis, both of which are part of L. acidophilus species group of bacteria.
Supplementary Material
Acknowledgments
The authors thank LuAnn Johnson for conducting statistical analyses and Kay Keehr, James Lindlauf, and Karen LoneFight for their technical support. H.Z. designed the research; H.Z., J.L., F.-Q.Z., and M.I.J. conducted the research; H.Z., L.Y., M.I.J., and G.F.C. analyzed the data; and H.Z., G.F.C., and M.I.J. wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
Abbreviations used: AMPK, AMP-activated protein kinase; HF, high-fat; IL-6, interleukin 6; iNOS, inducible NO synthase; LF, low-fat; rRNA, ribosomal RNA; TNF-α, tumor necrosis factor α.
Literature Cited
- 1.Grundy SM, Cleeman JI, Daniels SR, Donato KA, Eckel RH, Franklin BA, Gordon DJ, Krauss RM, Savage PJ, et al. Diagnosis and management of the metabolic syndrome: an American Heart Association/National Heart, Lung, and Blood Institute Scientific Statement. Circulation. 2005;112:2735–52 [DOI] [PubMed] [Google Scholar]
- 2.Flegal KM, Carroll MD, Kit BK, Ogden CL. Prevalence of obesity and trends in the distribution of body mass index among US adults, 1999–2010. JAMA. 2012;307:491–7 [DOI] [PubMed] [Google Scholar]
- 3.Festi D, Colecchia A, Sacco T, Bondi M, Roda E, Marchesini G. Hepatic steatosis in obese patients: clinical aspects and prognostic significance. Obes Rev. 2004;5:27–42 [DOI] [PubMed] [Google Scholar]
- 4.Anstee QM, Goldin RD. Mouse models in non-alcoholic fatty liver disease and steatohepatitis research. Int J Exp Pathol. 2006;87:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Compare D, Coccoli P, Rocco A, Nardone OM, De Maria S, Cartenì M, Nardone G. Gut-liver axis: the impact of gut microbiota on non alcoholic fatty liver disease. Nutr Metab Cardiovasc Dis. 2012;22:471–6 [DOI] [PubMed] [Google Scholar]
- 6.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31 [DOI] [PubMed] [Google Scholar]
- 7.Yokota A, Fukiya S, Islam KB, Ooka T, Ogura Y, Hayashi T, Hagio M, Ishizuka S. Is bile acid a determinant of the gut microbiota on a high-fat diet? Gut Microbes. 2012;3:455–9 [DOI] [PubMed] [Google Scholar]
- 8.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mai V. Dietary modification of the intestinal microbiota. Nutr Rev. 2004;62:235–42 [DOI] [PubMed] [Google Scholar]
- 10.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. [DOI] [PMC free article] [PubMed]
- 13.Wang RF, Cao WW, Cerniglia CE. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl Environ Microbiol. 1996;62:1242–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drasar BS, Roberts AK. Control of the large bowel microflora. In: Hill MJ, Marsh PD, editors. Human microbial ecology. Boca Raton (FL): CRC Press, Inc.; 1990. p. 95–100.
- 15.Moore WE, Moore LH. Intestinal floras of populations that have a high risk of colon cancer. Appl Environ Microbiol. 1995;61:3202–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao JJ, Gregoire BR, Gao H. High-fat diet decreases cancellous bone mass but has no effect on cortical bone mass in the tibia in mice. Bone. 2009;44:1097–104 [DOI] [PubMed] [Google Scholar]
- 17.Panchal SK, Poudyal H, Waanders J, Brown L. Coffee extract attenuates changes in cardiovascular and hepatic structure and function without decreasing obesity in high-carbohydrate, high-fat diet-fed male rats. J Nutr. 2012;142:690–7 [DOI] [PubMed] [Google Scholar]
- 18.McCullough AJ. The clinical features, diagnosis and natural history of nonalcoholic fatty liver disease. Clin Liver Dis. 2004;8:521–33 [DOI] [PubMed] [Google Scholar]
- 19.Zeng H, Wu M, Botnen JH. Methylselenol, a selenium metabolite, induces cell cycle arrest in G1 phase and apoptosis via the extracellular-regulated kinase 1/2 pathway and other cancer signaling genes. J Nutr. 2009;139:1613–8 [DOI] [PubMed] [Google Scholar]
- 20.Giulietti A, Overbergh L, Valckx D, Decallonne B, Bouillon R, Mathieu C. An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods. 2001;25:386–401 [DOI] [PubMed] [Google Scholar]
- 21.Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148:257–66 [DOI] [PubMed] [Google Scholar]
- 22.Smith BK, Andrews PK, West DB. Macronutrient diet selection in thirteen mouse strains. Am J Physiol Regul Integr Comp Physiol. 2000;278:R797–805 [DOI] [PubMed] [Google Scholar]
- 23.Comuzzie AG, Allison DB. The search for human obesity genes. Science. 1998;280:1374–7 [DOI] [PubMed] [Google Scholar]
- 24.Sabio G, Das M, Mora A, Zhang Z, Jun JY, Ko HJ, Barrett T, Kim JK, Davis RJ. A stress signaling pathway in adipose tissue regulates hepatic insulin resistance. Science. 2008;322:1539–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.La Cava A. Proinflammatory activities of leptin in non-autoimmune conditions. Inflamm Allergy Drug Targets. 2012;11:298–302 [DOI] [PubMed] [Google Scholar]
- 26.Fantuzzi G, Faggioni R. Leptin in the regulation of immunity, inflammation, and hematopoiesis. J Leukoc Biol. 2000;68:437–46 [PubMed] [Google Scholar]
- 27.Prieto-Hontoria PL, Pérez-Matute P, Fernández-Galilea M, Bustos M, Martínez JA, Moreno-Aliaga MJ. Role of obesity-associated dysfunctional adipose tissue in cancer: a molecular nutrition approach. Biochim Biophys Acta. 2011;1807:664–78. [DOI] [PubMed]
- 28.Ha SK, Chae C. Inducible nitric oxide distribution in the fatty liver of a mouse with high fat diet-induced obesity. Exp Anim. 2010;59:595–604 [DOI] [PubMed] [Google Scholar]
- 29.Hardie DG. Minireview: the AMP-activated protein kinase cascade: the key sensor of cellular energy status. Endocrinology. 2003;144:5179–83 [DOI] [PubMed] [Google Scholar]
- 30.Long YC, Zierath JR. AMP-activated protein kinase signaling in metabolic regulation. J Clin Invest. 2006;116:1776–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Son G, Kremer M, Hines IN. Contribution of gut bacteria to liver pathobiology. Gastroenterol Res Pract. 2010;2010:453563. [DOI] [PMC free article] [PubMed]
- 32.Wang LT, Kuo HP, Wu YC, Tai CJ, Lee FL. Lactobacillus taiwanensis sp. nov., isolated from silage. Int J Syst Evol Microbiol. 2009;59:2064–8 [DOI] [PubMed] [Google Scholar]
- 33.Usman HA. Hypocholesterolemic effect of Lactobacillus gasseri SBT0270 in rats fed a cholesterol-enriched diet. J Dairy Res. 2001;68:617–24 [DOI] [PubMed] [Google Scholar]
- 34.Luchansky JB, Tennant MC, Klaenhammer TR. Molecular cloning and deoxyribonucleic acid polymorphisms in Lactobacillus acidophilus and Lactobacillus gasseri. J Dairy Sci. 1991;74:3293–302 [DOI] [PubMed] [Google Scholar]
- 35.Narushima S, Ito K, Kuruma K, Uchida K. Composition of cecal bile acids in ex-germfree mice inoculated with human intestinal bacteria. Lipids. 2000;35:639–44 [DOI] [PubMed] [Google Scholar]
- 36.Dumas ME, Barton RH, Toye A, Cloarec O, Blancher C, Rothwell A, Fearnside J, Tatoud R, Blanc V, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci USA. 2006;103:12511–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trauner M, Claudel T, Fickert P, Moustafa T, Wagner M. Bile acids as regulators of hepatic lipid and glucose metabolism. Dig Dis. 2010;28:220–4 [DOI] [PubMed] [Google Scholar]
- 38.Gadaleta RM, van Mil SW, Oldenburg B, Siersema PD, Klomp LW, van Erpecum KJ. Bile acids and their nuclear receptor FXR: relevance for hepatobiliary and gastrointestinal disease. Biochim Biophys Acta. 2010;1801:683–92. [DOI] [PubMed]
- 39.Million M, Angelakis E, Paul M, Armougom F, Leibovici L, Raoult D. Comparative meta-analysis of the effect of Lactobacillus species on weight gain in humans and animals. Microb Pathog. 2012;53:100–8 [DOI] [PubMed] [Google Scholar]
- 40.Million M, Maraninchi M, Henry M, Armougom F, Richet H, Carrieri P, Valero R, Raccah D, Vialettes B, et al. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int J Obes (Lond). 2012;36:817–25 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.