Abstract
The effect of feeding C57BL/6 mice white button (WB) mushrooms or control (CTRL) diets for 6 wk was determined on the bacterial microflora, urinary metabolome, and resistance to a gastrointestinal (GI) pathogen. Feeding mice a diet containing 1 g WB mushrooms/100 g diet resulted in changes in the microflora that were evident at 2 wk and stabilized after 4 wk of WB feeding. Compared with CTRL-fed mice, WB feeding (1 g/100 g diet) increased the diversity of the microflora and reduced potentially pathogenic (e.g., Clostridia) bacteria in the GI tract. Bacteria from the Bacteroidetes phylum increased and the Firmicutes phylum decreased in mushroom-fed mice compared with CTRL. The changes in the microflora were also reflected in the urinary metabolome that showed a metabolic shift in the WB-fed compared with the CTRL-fed mice. The WB feeding and changes in the microbiome were associated with fewer inflammatory cells and decreased colitis severity in the GI mucosa following Citrobacter rodentium infection compared with CTRL. Paradoxically, the clearance of C. rodentium infection did not differ even though Ifn-γ and Il-17 were higher in the colons of the WB-fed mice compared with CTRL. Adding modest amounts of WB mushrooms (1 g/100 g diet) to the diet changed the composition of the normal flora and the urinary metabolome of mice and these changes resulted in better control of inflammation and resolution of infection with C. rodentium.
Introduction
Medicinal mushrooms have been used either whole or as extracts in traditional Oriental therapies. Numerous bioactive components have been identified and mushroom extracts are sold worldwide as dietary supplements and make up an over 5 million dollar industry in the US. Understanding the mechanisms by which mushrooms affect health is complicated by mushroom variety and the differing purities and compositions of extracts being tested. There is evidence that edible mushrooms and/or mushroom extracts regulate the immune system (1, 2). The changes in immune function of healthy mice are small when ingesting mushrooms (1–10 g/100 g of the diet) (2, 3). Chemical injury induced the production of TNFα and a protective effect of white button (WB)9 mushroom feeding on dextran sodium sulfate (DSS)-induced colitis was evident but only after the injury (3). Understanding how edible mushrooms affect health and disease is an active area of research.
A population of nearly 100 trillion microbiota (between 500 and 1000 different species) inhabits the human gut (4). Unlike the genome of a single organism, the metagenome (the combined genomic content of the intestinal flora) can rapidly vary as a function of diet, location, and a variety of other factors. The gut microbiota is essential for normal immune system development, displacement of pathogens, and extraction of additional energy (e.g., SCFAs) from otherwise nondigestible dietary substrates (5–7). Highlighting the importance of the gut microbiota, numerous human diseases and conditions have been attributed to a substantial change in the normal gut microbiota homeostasis (6). For example, inflammatory bowel disease (IBD) is associated with an overgrowth of bacteria that cause severe intestinal inflammation (8). Patients with active IBD have less diverse microbial communities than healthy controls (9).
The composition of the gastrointestinal (GI) microflora is a critical determinant of the severity of experimental colitis in many models, including DSS colitis (10). DSS treatment of mice induces inflammation in the GI tract and the inflammation is most notable in the colon (11). Colitis induced in the DSS model is characterized by the local overproduction of cytokines and then repair of the mucosa and recovery of the mice (12, 13). Feeding mice diets that contained 1–2 g WB mushrooms/100 g diet resulted in protection from DSS colitis and a decreased time to recovery (3). In addition to protecting the gut from chemical injury, the microflora protects the GI tract by preventing infection. Citrobacter rodentium is an extracellular enteric pathogen that causes a disease in mice that resembles human enteropathogenic Escherichia coli. In addition, C. rodentium infection in mice has been used as a model of IBD, where the infection causes inflammation in the gut and the inflammatory mediators, including inducible NO synthase, TNFα, and IL-12, are all important for regulating the extent of pathology in the gut (14). Infection and resolution of infection with C. rodentium in mice are affected by host immunity and the bacterial communities found in the gut (15).
We hypothesized that the beneficial effects of WB feeding might be due to alterations in the composition of the microbial flora. We tested the hypothesis using a model of colitis (C. rodentium infection), where changes in the composition of the microbial flora have been clearly shown to affect the development of colitis symptoms (15, 16). Compared with control feeding, WB feeding resulted in changes to the composition of the GI microflora, cathecholamine metabolism, and improved resolution of GI inflammation following an enteric infection.
Materials and Methods
Mice.
C57BL/6 male and female mice were bred and maintained at The Pennsylvania State University. The C57BL/6 breeders were originally from the Jackson Laboratories. Groups of 6- to 7-wk-old mice were used for experiments and all of the mice came from 1 of 2 breeding females. Experiments used both female and male mice or just male mice as indicated. Several factors are known to affect the composition of the microflora, including maternal factors and sex. Only male mice were used for some experiments to eliminate the sex effects on the microbiome. Groups of 6–8 mice were used in each experiment. All experimental procedures using mice were approved by the Office of Research Protection Institutional Animal Care and Use Committee at The Pennsylvania State University.
Diet.
At baseline, mice were fed nonpurified rodent diets from LabDiet 5001 (Animal Specialties and Provisions). For experiments, mice were started on purified diets complete for all nutrients as previously described (17, 18). Commercially available Agaricus bisporous (or common name WB) mushrooms were obtained from Modern Mushroom Farm. The whole mushrooms were freeze-dried and ground into a fine power. One-half of the mice were fed synthetic diets made in the laboratory [control (CTRL)], as previously described, and the other half of the mice were fed the CTRL diet with 1 g WB/100 g CTRL diet (3, 18, 19). Consistent with the literature (2, 3), there was no effect of WB feeding on the weight of the mice. Mice were placed in a clean cage without bedding for several minutes to collect feces left in the cages. Feces, spot urine samples, and blood were collected at the start of the diet treatment (time 0) and every 2 wk for a total of 6 wk.
Denaturing gradient gel electrophoresis.
Total DNA was isolated from fecal samples using a QIAmp DNA stool minikit (Qiagen). Extracted fecal DNA was amplified with universal 16S rDNA primers that target the V3 region of the 16S rDNA, which is highly conserved across bacterial species (20). Denaturing gradient gel electrophoresis (DGGE) was performed with a DCode, Universal Mutation Detection System (Bio-Rad). PCR products were loaded onto the gel. The electrophoresis conditions were selected based on the results of perpendicular DGGE and set at 18 h at 70 V in a linear 30–60% denaturing gradient. Gels were analyzed using GelCompar II software (Applied Maths). The percent similarity (number of bands, location, and density) between different lanes was compared and used to generate clustering analysis that reports on similarity in DGGE banding patterns between samples. Standards (STDs) were Clostridium propionicum (ATCC strain 25522), Lactobacillus murinus (ATCC strain 35020), and Parabacteroides distasonis (ATCC strain 8503). The STDs were DGGE bands generated from DNA isolated from purified cultures of the 3 organisms and were used so that gels run on different days and with different samples could be compared with each other based on the migration of the STDs. The STDs ran from top to bottom: P. distasonis, L. murinus, and C. propionicum.
Metagenomic analysis.
Four DNA samples from 2 male CTRL-fed and 2 male WB-fed mice at 6 wk, when the DGGE profiles suggested that there were no longer changes in the microflora with diet, were sequenced on a 454 Titanium sequencer. The low numbers of samples sequenced was due in part to the high cost needed for sequencing. The other rationale for the low numbers of samples sequenced was that useful information could be determined using few samples. The goal of the metagenomic sequencing was to confirm the DGGE analysis that showed a change in the microflora with WB feeding and to give new information on the possible changes in phyla and class that occurred with WB feeding. The sequences obtained were analyzed using the MOTHUR software (21); all analysis scripts and datasets are available on the Penn State Bioinformatics Consulting Center Web site (22). The sequencing reads were filtered to remove reads that had an average read quality of <35. This initial filtering removed ∼30% of the reads, leaving 176,122 reads distributed over the 4 samples. Two additional read quality filtering steps were done. First, potential chimeric reads were removed (ChimeraSlayer developed by Broad Institute) and a preclustering step was applied that merged reads due to pyrosequencing errors. At the end of the quality filtering steps, 130,232 reads were retained and the read counts for each group were from 22,827 to 46,692 reads. To determine operational taxonomic units (OTUs), the filtered data were aligned via MOTHUR and against the SILVA 16S rRNA database containing 14,956 prealigned representative bacterial references. The resulting alignments were then clustered according to the furthest neighbor distance metric. For phylotyping analyses, the filtered data were classified using the SILVA taxonomy with the Bayesian classification method implemented in MOTHUR.
Rarefaction curves showed support for the use of the 99% level sequence similarity for further downstream analysis. At this similarity measure, 47,000 distinct OTUs were detected. The robustness of the samples corresponding to biological replicates was determined using the Jaccard and Bray Curtis similarity measures. The Berger-Parker index was measured to determine the abundance as a percent of the OTU with the highest counts.
Metabolomic analysis.
Spot urines were collected from mice fed CTRL or WB diets for 6 wk and profiled using ultra-performance liquid chromatography coupled with electrospray ionization quadrupole time-of-flight MS operating in positive ionization mode (Waters). Samples were prepared by diluting the urine samples with an equal volume of HPLC-grade water, vortexing, and centrifuging at 4°C at 14,000 × g to remove any particulates. The supernatant was transferred to an autosampler and samples were profiled as described (23) using a Waters ultra-performance liquid chromatography coupled to a Synapt G2S mass spectrometer (Waters). Peak alignment, deconvolution, and normalization were performed using MarkerLynx (Waters) and exported for analysis by SIMCA P13+ (Umetrics). Supervised orthogonal projection to latent structures was used to visualize the data and identify discriminating variables. Metabolite identities are based on searching against the Human Metabolome Database (24) or Metlin (25). Tandem MS was used to provide further structural information. In this case, collision energies were ramped from 15 to 45V while selecting for the ion of interest.
C. rodentium infection.
There were no sex differences [(26) and data not shown] in resistance to C. rodentium in C57BL/6 mice, but because of the male/female differences in the metabolome, only male mice were used for the C. rodentium infections. Male mice that were fed WB and CTRL diets for 6 wk were infected with C. rodentium and 3 mice each were killed by CO2 asphyxiation on d 10 and 14 postinfection. The C. rodentium strain ICC169 was a kind gift of Gad Frankel (London School of Medicine and Dentistry). For inoculations, bacteria were grown overnight in Luria Bertani broth containing 0.05 g/L nalidixic acid/mL (EMD Chemicals). Five × 109 CFU were gavaged orally to each mouse. Mice were individually housed for the C. rodentium infection to avoid shedding mice from infecting or reinfecting littermates. On d 10 and 14 postinfection, feces were collected, weighed, and suspended in PBS. Serial dilutions were plated onto nalidixic acid-containing plates. Bacterial colonies were counted after 24 h.
Colonic mRNA analyses.
The proximal colon was collected and weighed and mRNA was extracted using an RNeasy mini kit (Qiagen). cDNA was reverse transcribed and real-time PCR was conducted using a ABI 7500 Fast RT PCR machine (Applied Biosystems). The analyses included Ifn-γ, Il-12, Il-17, Il- 22, Il-23, and Gapdh expression.
Histological analysis of colon tissue.
The terminal colon was removed and fixed in 10% formalin. Paraffin-embedded sections were prepared and stained with hematoxylin and eosin at The Pennsylvania State University. For histological grading of colitis, 6 criteria were used: cryptitis, goblet cell hyperplasia, inflammation, erosion, ulcers, and edema. The lesions were scored from 0 to 4, where 0 = no colitis/epithelial thickening; 1 = increased number of leukocytes in the mucosa and/or slight epithelial cell hyperplasia; 2 = multiple loci of inflammation, leukocytic infiltration of mucosa and submucosa and/or pronounced epithelial cell hyperplasia (2- to 3-fold increase in crypts); 3 = extensive leukocytic infiltrate in mucosa, submucosa, ulceration, depletion of mucin-secreting goblet cells, and/or marked epithelial cell hyperplasia (3- to 10-fold increase in crypts); and 4 = extensive transmural leukocytic infiltrate, crypt abscesses, and/ or marked epithelial cell hyperplasia (crypts >10-fold or greater).
Statistics.
Two-tailed Student’s t tests were used to calculate differences between CTRL and WB groups for the metabolomics analyses. Males and females were tested separately for cinnamoylglycine, dopamine glucuronide, and hippuric acid variables only. Two-way ANOVA with Bonferroni post hoc tests were used to calculate diet × time effects for all of the other analyses using Prism software (GraphPad). Variables with unequal variance (several of the cytokine mRNA values) were log-transformed to normally distribute them prior to evaluating the data for significance by ANOVA. Pearson chi-square goodness-of-fit tests were done for metagenomic analyses. P ≤ 0.05 was considered significant. Values are presented as mean ± SE.
Results
DGGE banding patterns.
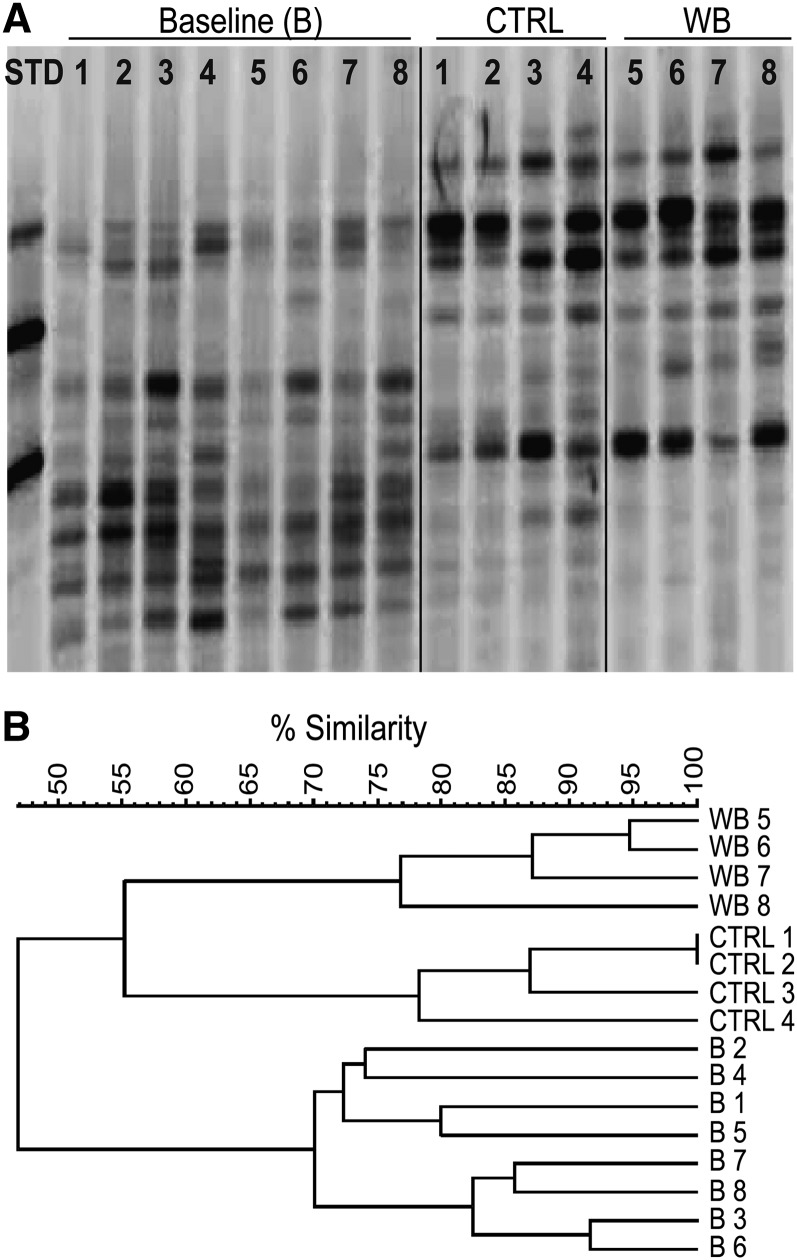
The DGGE banding patterns of individual mice fed a nonpurified diet at baseline had banding patterns that showed 70% similarity across the samples (Fig. 1A,B) (lanes 1–8). Mice fed the CTRL (lanes 1–4) or WB (lanes 5–8) diet for 2 wk experienced a shift in the bacterial species in their feces that is reflected in a change in the detectable DGGE bands (Fig. 1A). The CTRL DGGE banding patterns were 78–99% similar to each other (CTRL lanes 1–4) and the WB DGGE banding patterns were 77–95% similar to each other (WB lanes 5–8) (Fig. 1B). Male mice (95–99% similarity; WB lanes 5 and 6, CTRL lanes 1 and 2) had greater similarity to each other than female mice (77–78% similarity; WB lanes 7 and 8, CTRL lanes 3 and 4) at 2 wk fed either the WB or CTRL diet (Fig. 1B). Samples from mice fed the same diets or at baseline were more similar to each other than to samples from mice in the other groups (Fig. 1B). WB mushrooms altered the composition of the microflora at 2 wk (Fig. 1B). The diet effects persisted at 4 and 6 wk, with WB and CTRL DGGE banding patterns being more similar within groups than across groups (data not shown). Changes in diet resulted in alterations in the DGGE banding patterns of bacterial DNA in the feces.
FIGURE 1.
The DGGE banding pattern of bacterial DNA collected from the feces of mice at baseline and 2 wk after feeding 1 g/100 g WB or CTRL diets. (A) DGGE of the fecal bacterial DNA from 4 male (lanes 1, 2, 5, and 6) and 4 female (lanes 3, 4, 7, and 8) mice at B and 2 wk after they were switched to CTRL (lanes 1–4) or WB (lanes 5–8) diets. (B) Cluster analysis shows the relatedness of the banding profiles of the DGGE gels at B and after 2 wk of CTRL or WB feeding. The experiment was conducted using 8 mice and n = 4 samples/group and time point. One representative of 2 experiments is shown. B, baseline; CTRL, control; DGGE, denaturing gradient gel electrophoresis; STD, standard; WB, white button.
Metagenomic analyses.
To identify which phyla/classes of organisms were shifting in response to WB feeding, 2 of the DNA samples from each CTRL and WB group were sequenced. Because of the greater similarity of male samples to each other (Fig. 1), samples from male mice were sequenced. Jaccard and Bray Curtis similarity measures showed that microbial communities from the CTRL-fed samples were more similar to each other than the microbial communities found in the WB-fed samples. These analyses confirmed the less quantitative analyses using a larger number of mice and at different time points by DGGE (Fig. 1B). For further analyses, the 2 biological replicas were combined. The Berger-Parker index suggested that the CTRL group had substantially higher dominance than the WB group. Because the total numbers of sequences were close to being equal between the WB and CTRL groups, the results suggested that the CTRL group contained microbial communities that were less diverse than those present in the WB-fed group.
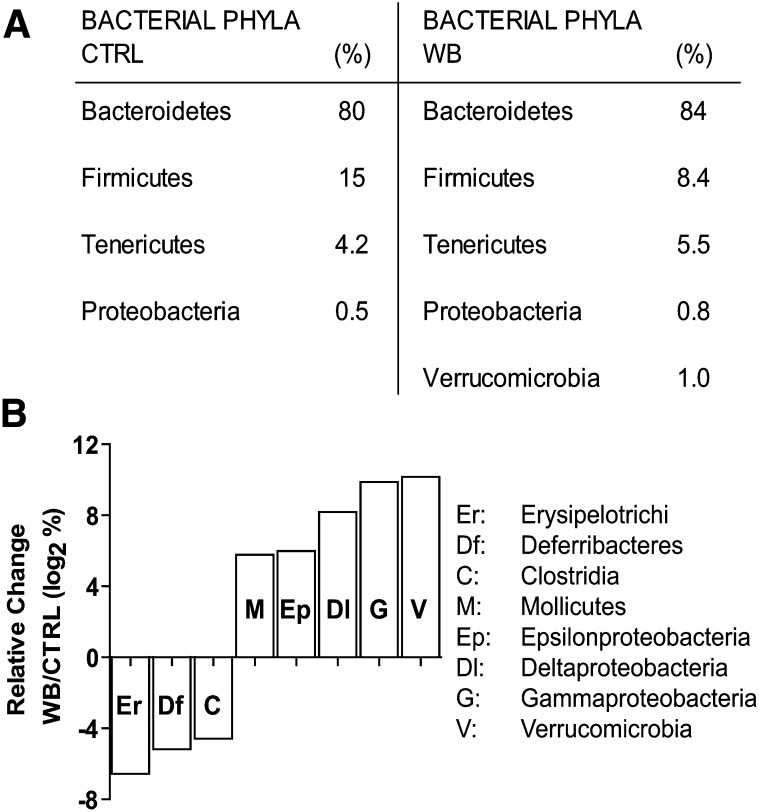
The frequencies of the bacteria at the phylum level present in the WB and CTRL samples after 6 wk showed that Firmicutes species were decreased in the WB-fed group (Fig. 2A). The decrease in Firmicutes phylum was associated with an increase in Bacteroidetes and the presence of Verrucomicrobia that was unique to the WB-fed mice. The data were plotted to show significant changes in the number of sequences identified in each phyla, order, and class as a result of the diet, and the class level plot is shown (Fig. 2B). The data were scaled logarithmically on the vertical axis to allow high and low abundance changes on the same plots. At the class level, bacteria belonging to the Erysipelotrichi, Deferribacteres, and Clostridia classes were decreased in the gut by the WB diets along with increased populations of Mollicutes, Epsilon-, Delta-, and Gamma Proteobacteria, and Verrucomicrobia.
FIGURE 2.
Bacterial metagenomic analysis of the phylum and class level differences evident in the DNA isolated from the feces of 2 male mice fed CTRL or 2 male mice fed WB diets for 6 wk. (A) The frequency of the different phyla sequenced from CTRL- and WB-fed mice. (B) Relative change in the class of bacteria found in WB-fed mice relative to CTRL. Values are the mean frequencies of the bacterial sequences from 2 mice in each treatment group. Values that are negative show class-level species that are inhibited by WB feeding. Values that are positive show class-level species that are more prevalent in WB-fed mice. Only significant changes are graphed (P < 0.001). CTRL, control; WB, white button.
Urinary metabolome.
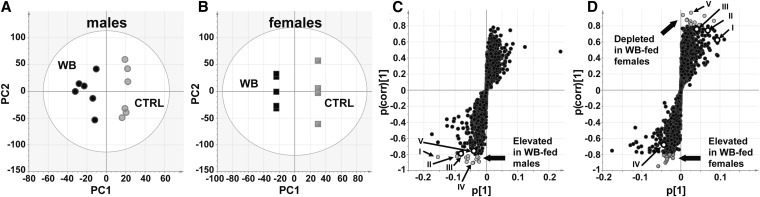
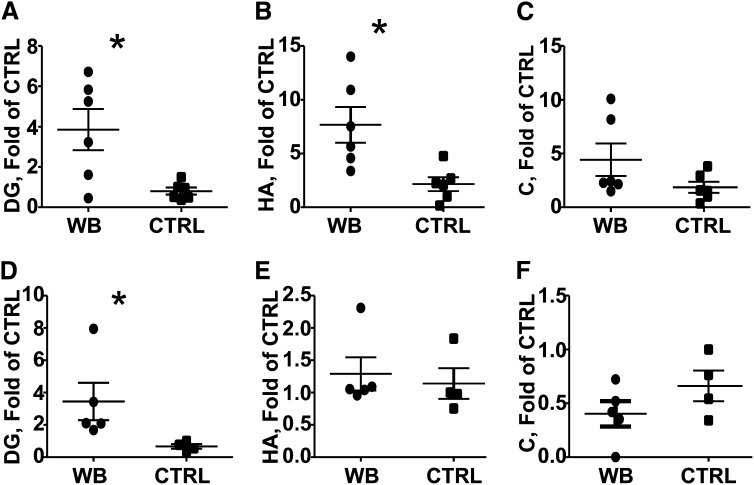
Principal component analysis showed that the WB diets altered the urinary metabolome in male (Fig. 3A) and female (Fig. 3B) mice. Interestingly, male samples showed more variability than females (Fig. 3A), as indicated by the increased scatter of the urine samples collected from males. Metabolites putatively identified by accurate mass and tandem MS fragmentography included hippuric acid, dopamine glucuronide, and cinnamoylglycine (Fig. 3; Table 1). Hippuric acid was identified as 105.034+ (hippuric acid fragment), 202.049+ (hippuric acid sodium adduct), and 180.066+ (hippuric acid) (Fig. 4;Table 1). Dopamine glucuronide was identified as the metabolite corresponding to 330.117+ and cinnamoylglycine was identified as the metabolite corresponding to 228.065+ (Fig. 4;Table 1). The concentrations of dopamine glucoranide were significantly higher in the WB-fed male and female urine samples, whereas the concentrations of hippuric acid were significantly higher only in males (Fig. 4).
FIGURE 3.
The urinary metabolites in male and female mice fed WB or CTRL diets for 6 wk. The scatter plot from the orthogonal projection to latent structure analysis is shown for urine samples from male WB-fed and CTRL-fed mice (A) and female WB-fed and CTRL-fed mice (B). Variables having a p(corr)[1] value >0.8 or <−0.8 are shaded gray in the loadings plots from males (C) and females (D). Variables depleted in WB-fed males did not meet the cutoff criteria and are therefore not shown. p[1] is a measure of covariance and p(corr)[1] a measure of model fit. White circles indicate variables that met the cutoff criteria in one gender but not the other. Variable labels I, II, III, IV, and V are described in Table 1. Values for A and B are individual data points for n = 4–6 mice/group and treatment. CTRL, control; PC, principal component; WB, white button.
TABLE 1.
Putative identities of urinary metabolites in mice fed 1 g WB mushrooms/100 g and CTRL diets for 6 wk by tandem MS fragmentography1
| Variable identification | Observed m/z [M+H]+ | Retention time | Empirical formula | Mass error | Putative identification |
| min | ppm | ||||
| I | 105.034 | 2.2 | C7H5O | 0 | Hippuric acid fragment |
| II | 202.049 | 2.2 | C9H9NO3Na | 7 | Hippuric acid [Na+] adduct |
| III | 180.066 | 2.2 | C9H10NO3 | 2 | Hippuric acid |
| IV | 330.117 | 0.3 | C14H20NO8 | 4 | Dopamine glucuronide |
| V | 228.065 | 3.9 | C11H11NO3Na | 8 | Cinnamoylglycine |
| [Na+] adduct |
CTRL, control; WB, white button.
FIGURE 4.
Urinary metabolites identified from Table 1 by accurate mass and tandem MS fragmentography in 6-wk-old male and female mice fed WB or CTRL diets, n = 4–6 mice/group. (A,D) DG, (B,E) HA, and (C,F) C. A, B, and C are males and D, E, and F are from females. Values are individual data points and mean ± SE of n = 4–6 mice/group and treatment. *WB-fed mice different from CTRL-fed mice, P < 0.05 (2-tailed Student’s t test). C, cinnamoylglycine; CTRL, control; DG, dopamine glucronide; HA, hippuric acid; WB, white button.
C. rodentium infection.
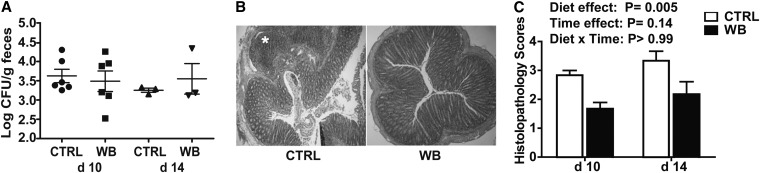
The weights of CTRL- and WB-fed mice did not differ following C. rodentium infection. There were no differences in the numbers of C. rodentium in the feces of WB- and CTRL-fed mice at either d 10 or 14 postinfection (Fig. 5A). CTRL mice had significant induction of inflammation and hyperplasia in the colon at d 10 postinfection (Fig. 5B). Severe inflammation and an ulcer were found in CTRL sections at d 10 postinfection ( ). There was significantly more inflammation in the d 14 than in the d 10 histopathology sections from both groups (Fig. 5C). In addition, significantly more severe colitis was noted in colons of the CTRL-fed mice compared with those of WB-fed mice (at both d 10 and 14 postinfection) (Fig. 5C). A DGGE analysis of the changes in the microbiota at d 14 postinfection showed that samples from the WB-fed mice clustered together (84% similarity) and separately (68% similarity between WB and CTRL) from the CTRL group (Supplemental Fig. 2).
FIGURE 5.
The shedding of C. rodentium in the feces and histopathology scores of male mice fed WB or CTRL diets for 6 wk. (A) The numbers of C. rodentium isolated from the feces of CTRL- and WB-fed mice at d 10 and 14 postinfection. (B) A representative histopathology section (provided in color as Supplemental Fig. 1) of the colons of CTRL- (scored 3) or WB- (scored 2) fed mice at d 10 postinfection. *An ulcer in the section from the CTRL-fed mouse. No ulcers were found in WB sections of the colon. (C) A summary of the histopathology scores given for CTRL- and WB-fed mice d 10 and 14 post-C. rodentium infection. One representative experiment of 2 experiments is shown. Values are individual data points and mean ± SE of n = 3–6 mice/group and treatment. CTRL, control; WB, white button.
Cytokine mRNA expression.
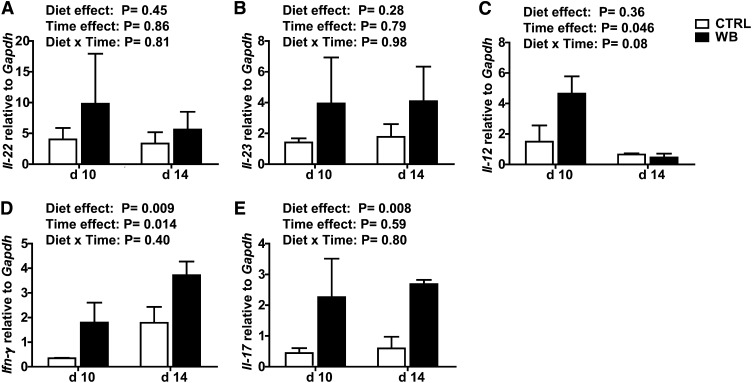
Il-22 and Il-23 did not differ in colons from the CTRL- and WB-fed mice, nor did they change with time (Fig. 6A,B). Il-12 mRNA was not different in colons from WB-fed mice compared with CTRL-fed mice at d 10 postinfection and the expression was higher at d 10 than 14 postinfection in the colons of the WB-fed mice (Fig. 6C). Ifn-γ mRNA was higher at d 14 postinfection compared with d 10 postinfection and higher in the WB samples compared with CTRL (Fig. 6D). Il-17 did not change between the d 10 and 14 time points but was significantly higher in the colon from WB-fed mice compared with CTRL (Fig. 6). At 10 d postinfection, Il-12 was produced that corresponded with the increased Ifn-γ response at d 14 (Fig. 6). Conversely, Il-22 and Il-23 were produced at constant levels between d 10 and 14 (Fig. 6). Colons from WB-fed mice had more Ifn-γ and Il-17 than those from CTRL-fed mice (Fig. 6).
FIGURE 6.
The colonic mRNA expression of inflammatory cytokines in C. rodentium-infected male WB- or CTRL-fed mice. Relative amount of mRNA for Il-22 (A), Il-23 (B), Il-12 (C), Ifn-γ (D), and Il-17 (E) to Gapdh expression in each sample. Values are in mean ± SE, n = 3 mice/group and time point. CTRL, control; WB, white button.
Discussion
Feeding mice modest amounts of WB mushrooms protected the gut from injury and hastened the resolution of inflammation following infection with C. rodentium. Previously, we reported that WB feeding resulted in protection from DSS-induced colitis (3). In this study, the beneficial effects of WB feeding were evident in a second model of GI injury (infection). The effects of the WB feeding are likely due to shared protective mechanisms present in both models. For both DSS colitis and C. rodentium infection, the composition of the GI microflora is critical for determining host susceptibility.
There was no effect of the WB feeding on the numbers of C. rodentium isolated from the feces. Therefore, the WB-mediated effect is not a result of more rapid clearance of the pathogen. Instead, protection corresponded to the increased diversity of the microflora in the gut of WB-fed mice. The Bacteroidetes phylum increased and Firmicutes phylum, including Clostridia, decreased following WB feeding. IBD is associated with an overgrowth of bacteria that cause severe intestinal inflammation (8). Patients with active IBD have less diverse microbial communities than healthy controls (9). Treatments that alter the composition of the gut microflora have been shown to be effective in some patients with IBD (27).
The intestinal microbiota also controls susceptibility to C. rodentium infection in mice (16, 28). Others have reported that lower numbers of the Bateroidetes phylum corresponded to increased susceptibility to C. rodentium (28) and the WB treatment increased the numbers of Bateroidetes. Also indicative of a shift in the bacterial microflora are the changes in hippuric acid and cinnamoylglycine in the urine of WB-fed mice. Germfree mice have been shown to have higher concentrations of tryptophan and lower concentrations of hippicuric acid and cinnamoylglycine than conventional mice, suggesting that changes in these metabolites are indicative of changes in the bacterial microflora (29). The increased frequency of Bateroidetes and greater diversity of the microflora in the WB-fed mice (compared with CTRL fed) was associated with the protection and healing of the GI tract.
Dopamine metabolites and tryptophan concentrations increased in the urine of WB- compared with CTRL-fed mice. Dopamine is produced from tryptophan and mushrooms have been shown to be a source of several indole compounds, including tryptophan (30). In addition, inflammatory cytokines induce production of catecholamines, which, like dopamine, increase the growth of E. coli in vitro and in vivo (31). The growth of C. rodentium, however, was not affected by WB feeding in vivo even though higher amounts of catecholamine metabolites were found in the urine. Instead, the increase in catecholamines may have been the result of increased inflammation (Fig. 6). Among the changes noted with WB feeding were several metabolites linked to catecholamine metabolism that are key hormones in the control of inflammation and bacterial growth (32).
Inflammation itself affects the microbial flora. Acute inflammation provides substrates that Salmonella and other pathogens can utilize to out-compete commensal organisms in the gut (33). WB-fed mice expressed significantly more of the inflammatory cytokines IL-12, IFNγ, and IL-17 in the colon compared with CTRL-fed mice. More inflammation might contribute to the shifts in the microbiome. However, this is a paradoxical finding, because there were fewer immune cells in the histopathology sections from WB-fed mice but higher expression of IFNγ and IL-17 mRNA. Germfree mice are unable to eradicate C. rodentium infection but do resolve the inflammation at the same rate as conventional mice (16). Therefore, the rate of C. rodentium clearance does not correspond to the generation or kinetics of inflammation but instead to the changes in the microflora (16). Previously, it was shown that WB feeding protected against colonic injury in DSS colitis and the protection was associated with an increase in TNFα production (3). Therefore, the WB diet increased the local expression of inflammatory cytokines in vivo and the increased production of these inflammatory cytokines is associated with a change in the bacterial flora that results in the more rapid repair of the injury. The mechanisms whereby this might occur warrant further investigation. WB feeding in 2 different models was associated with increased production of inflammatory cytokines and protection from injury (3).
The immune system and the GI microflora respond to changes in the diet. WB feeding increased the local inflammatory response, stimulated the production of catecholamines and their metabolites, and changed the composition of the gut flora. The mechanisms that result in the biological changes that occur with WB ingestion are likely to include direct stimulation of the innate immune systems that produce inflammation and affect the composition of the gut flora. It seems that WB feeding has a very specific and localized effect on the gut. Small increases in inflammatory cytokines induce changes in catecholamines and the complexity and composition of the microbial flora but do not alter the rate at which an enteric pathogen is cleared. The results raise a number of interesting areas for future investigation, including the kinetics and longevity of the WB-feeding effects. Consuming WB mushrooms might improve GI health by limiting the damage that occurs following injury or infection.
Supplementary Material
Acknowledgments
J.V., B.M.J., A.D.P., and M.T.C. designed research; J.V., R.L.S., I.A., J.F., and A.D.P. conducted research; J.V., J.H.O., I.A., J.F., A.D.P., and M.T.C. analyzed research; J.V., J.H.O., and M.T.C. wrote the paper; and M.T.C. had primary responsibility for final content. All authors read and approved the final manuscript.
Footnotes
Abbreviations used: CTRL, control; DGGE, denaturing gradient gel electrophoresis; DSS, dextran sodium sulfate; GI, gastrointestinal; IBD, inflammatory bowel disease; STD, standard; WB, white button.
Literature Cited
- 1.Chang R. Functional properties of edible mushrooms. Nutr Rev. 1996;54:S91–3 [DOI] [PubMed] [Google Scholar]
- 2.Wu D, Pae M, Ren Z, Guo Z, Smith D, Meydani SN. Dietary supplementation with white button mushroom enhances natural killer cell activity in C57BL/6 mice. J Nutr. 2007;137:1472–7 [DOI] [PubMed] [Google Scholar]
- 3.Yu S, Weaver V, Martin K, Cantorna MT. The effects of whole mushrooms during inflammation. BMC Immunol. 2009;10:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20 [DOI] [PubMed] [Google Scholar]
- 5.Guarner F. Enteric flora in health and disease. Digestion. 2006;73 Suppl 1:5–12 [DOI] [PubMed] [Google Scholar]
- 6.Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003;361:512–9 [DOI] [PubMed] [Google Scholar]
- 7.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307 [DOI] [PubMed] [Google Scholar]
- 8.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–29 [DOI] [PubMed] [Google Scholar]
- 9.Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Folsch UR, Timmis KN, Schreiber S. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, Rennick DM, Sartor RB. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westbrook AM, Szakmary A, Schiestl RH. Mechanisms of intestinal inflammation and development of associated cancers: lessons learned from mouse models. Mutat Res. 2010;705:40–59. [DOI] [PMC free article] [PubMed]
- 12.Axelsson LG, Landstrom E, Goldschmidt TJ, Gronberg A, Bylund-Fellenius AC. Dextran sulfate sodium (DSS) induced experimental colitis in immunodeficient mice: effects in CD4(+) -cell depleted, athymic and NK-cell depleted SCID mice. Inflamm Res. 1996;45:181–91 [DOI] [PubMed] [Google Scholar]
- 13.Dieleman LA, Ridwan BU, Tennyson GS, Beagley KW, Bucy RP, Elson CO. Dextran sulfate sodium-induced colitis occurs in severe combined immunodeficient mice. Gastroenterology. 1994;107:1643–52 [DOI] [PubMed] [Google Scholar]
- 14.Mundy R, MacDonald TT, Dougan G, Frankel G, Wiles S. Citrobacter rodentium of mice and man. Cell Microbiol. 2005;7:1697–706 [DOI] [PubMed] [Google Scholar]
- 15.Hoffmann C, Hill DA, Minkah N, Kirn T, Troy A, Artis D, Bushman F. Community-wide response of the gut microbiota to enteropathogenic Citrobacter rodentium infection revealed by deep sequencing. Infect Immun. 2009;77:4668–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, Nunez G. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cantorna MT, Munsick C, Bemiss C, Mahon BD. 1,25-Dihydroxycholecalciferol prevents and ameliorates symptoms of experimental murine inflammatory bowel disease. J Nutr. 2000;130:2648–52 [DOI] [PubMed] [Google Scholar]
- 18.Cantorna MT, Humpal-Winter J, DeLuca HF. Dietary calcium is a major factor in 1,25-dihydroxycholecalciferol suppression of experimental autoimmune encephalomyelitis in mice. J Nutr. 1999;129:1966–71 [DOI] [PubMed] [Google Scholar]
- 19.Cantorna MT, Hayes CE, DeLuca HF. 1,25-Dihydroxycholecalciferol inhibits the progression of arthritis in murine models of human arthritis. J Nutr. 1998;128:68–72 [DOI] [PubMed] [Google Scholar]
- 20.Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bitiren M, Karakilcik AZ, Zerin M, Ozardali I, Selek S, Nazligul Y, Ozgonul A, Musa D, Uzunkoy A. Protective effects of selenium and vitamin E combination on experimental colitis in blood plasma and colon of rats. Biol Trace Elem Res. 2010;136:87–95. [DOI] [PubMed]
- 23.Patterson AD, Li H, Eichler GS, Krausz KW, Weinstein JN, Fornace AJ, Jr, Gonzalez FJ, Idle JR. UPLC-ESI-TOFMS-based metabolomics and gene expression dynamics inspector self-organizing metabolomic maps as tools for understanding the cellular response to ionizing radiation. Anal Chem. 2008;80:665–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Human Metabolome Database [cited 2012 Oct 11]. Available from: http://www.hmdb.ca.
- 25.Lyakh LA, Sanford M, Chekol S, Young HA, Roberts AB. TGF-beta and vitamin D3 utilize distinct pathways to suppress IL-12 production and modulate rapid differentiation of human monocytes into CD83+ dendritic cells. J Immunol. 2005;174:2061–70 [DOI] [PubMed] [Google Scholar]
- 26.Maaser C, Housley MP, Iimura M, Smith JR, Vallance BA, Finlay BB, Schreiber JR, Varki NM, Kagnoff MF, Eckmann L. Clearance of Citrobacter rodentium requires B cells but not secretory immunoglobulin A (IgA) or IgM antibodies. Infect Immun. 2004;72:3315–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandran P, Satthaporn S, Robins A, Eremin O. Inflammatory bowel disease: dysfunction of GALT and gut bacterial flora (II). Surgeon. 2003;1:125–36 [DOI] [PubMed] [Google Scholar]
- 28.Ghosh S, Dai C, Brown K, Rajendiran E, Makarenko S, Baker J, Ma C, Halder S, Montero M, Ionescu VA, et al. Colonic microbiota alters host susceptibility to infectious colitis by modulating inflammation, redox status, and ion transporter gene expression. Am J Physiol Gastrointest Liver Physiol. 2011;301:G39–49. [DOI] [PubMed]
- 29.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106:3698–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muszyńska B, Maslanka A, Ekiert H, Sulkowska-Ziaja K. Analysis of indole compounds in Armillaria mellea fruiting bodies. Acta Pol Pharm. 2011;68:93–7 [PubMed] [Google Scholar]
- 31.Verbrugghe E, Boyen F, Gaastra W, Bekhuis L, Leyman B, Van Parys A, Haesebrouck F, Pasmans F. The complex interplay between stress and bacterial infections in animals. Vet Microbiol. 2012;155:115–27 [DOI] [PubMed] [Google Scholar]
- 32.Green BT, Brown DR. Interactions between bacteria and the gut mucosa: do enteric neurotransmitters acting on the mucosal epithelium influence intestinal colonization or infection? In: Lyte M, Freestone PPE, editors. Interkingdom signaling in infectious disease and health. New York: Springer Science+Business Media LLC; 2010. p. 89–109. [DOI] [PubMed]
- 33.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Baumler AJ. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci USA. 2011;108:17480–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.