Abstract
In addition to their well-known DNA-binding properties, homeodomains have the ability to efficiently translocate across biological membranes through still poorly-characterized mechanisms. To date, most biophysical studies addressing the mechanisms of internalization have focused on small synthetic peptides rather than full-length globular homeodomains. In this work, we characterized the conformational properties of chicken Engrailed 2 homeodomain (En2HD) in aqueous solution and in membrane mimetic environments using circular dichroism, Trp fluorescence, and NMR spectroscopy. En2HD adopts a well-defined three-helical bundle fold in aqueous solution. The Trp-48 residue, which is critical for internalization, is fully buried in the hydrophobic core. Circular dichroism and fluorescence reveal that a conformational transition occurs in anionic lipid vesicles and in micelles. En2HD loses its native three-dimensional structure in micellar environments but, remarkably, near-native helical secondary structures are maintained. Long-range interactions could be detected using site-directed spin labels, indicating that the three helices do not adopt extended orientations. Noncovalent paramagnetic probes yielded information about helix positioning and unveiled the burial of critical aromatic and basic residues within the micelles. Our results suggest that electrostatic interactions with membranes may be determinant in inducing a conformational change enabling Trp-48 to insert into membranes.
Introduction
Homeoproteins form a widespread class of transcription factors found in plants and animals (1). They were initially identified for their master role in the genetic control of embryonic development in insects and vertebrates (2). These proteins are characterized by the presence of an ∼60-residue DNA binding homeodomain that is highly conserved during evolution. More than 100 structures of wild-type or mutated homeodomains, in free forms or in complex with DNA, have been solved by x-ray crystallography and solution NMR spectroscopy. In these structures, homeodomains typically adopt a three-helix bundle fold, the third helix acting as the major DNA recognition element.
In addition to their well-known role as nuclear transcription factors, homeoproteins possess the intriguing property to be secreted and internalized using unconventional pathways. This membrane-translocation property was first reported for the Antennapedia homeodomain from Drosophila (3,4), but it has been observed since for other homeodomains in both animals and plants (5–8). The physiological significance of homeodomain translocation has been investigated, and it is now clear that this internalization process also occurs with full-length homeoproteins and is involved in original cell communication pathways (9). For instance, it was shown to regulate axonal guidance (10) and synaptic maturation in brain (11).
Structure/function relationship studies based on mutational analysis and use of synthetic peptides enabled researchers to ascribe this membrane-translocation property to the third helix of the Antennapedia homeodomain (12), which was referred to as penetratin. We and others have previously shown that peptides corresponding to the third helical segment of other homeodomains efficiently internalized into cells (13–15) and, taking into account the high sequence conservation, it is likely that this property is shared by most homeodomains. Penetratin was the first member of the still-growing class of cell-penetrating peptides (CPPs) that are able to translocate across biological membranes and deliver attached cargoes of various size and structure inside different cell lines. These peptides have received considerable attention for the vectorization of bioactive molecules into cells (16,17).
The internalization mechanism of these CPPs has been investigated by many authors but is still not fully elucidated. Although mechanisms involving vesicle formation by endocytosis have been proposed, it is clear that energy-independent, direct translocation of cell membranes is implicated (18). Several models have been suggested, including the formation of inverted micelles (19), ion pair complexes involving Arg residues, and an electroporation-like mechanism (20). Structure-function relationship studies of penetratin have proved that the chiral recognition by a receptor is not involved (19) and that the helical secondary structure is not required either (21). The electrostatic interactions with anionic lipids are critical because the removal of a single basic residue in penetratin sequence by Ala scanning decreases the membrane-binding affinity and the cell internalization efficiency (22). Hydrophobic residues in the third helix are also crucial; in particular, the deletion of the strictly conserved Trp-48 and Phe-49 residues abolishes the internalization (4).
To date, most biophysical studies have been carried out on penetratin (23–26) and few other peptides derived from the third helix of different homeodomains (13). Strikingly, only a few studies have examined the properties of the third helix in the context of the whole homeodomain being a more relevant biological object. It is likely that the interactions of the third helix with residues in the first and second helices in homeodomains may influence the membrane binding and translocation processes. Notably, the Trp-48 residue, which is critical for membrane translocation, is buried in the hydrophobic core of the homeodomain. The major aim of this study was to examine the properties of the third helix in the context of the full homeodomain and characterize the conformation of the homeodomain in membrane-mimetic environments.
We focused our work on Engrailed 2 homeodomain (En2HD). Engrailed 2 is involved in brain development and physiology in Vertebrates and the implication of its translocation properties has been well documented in axonal guidance (10,27). Engrailed 2 also plays an important role in the survival and physiology of dopaminergic neurons and has been proposed to be used as a therapeutic protein in Parkinson’s disease (28,29). We first determined the three-dimensional structure of chicken En2HD by NMR and then investigated the influence of membranes on the conformation of En2HD by circular dichroism (CD) and fluorescence. High-resolution NMR structural studies were then carried out using micelles of two different compositions as membrane-mimetic systems.
Materials and Methods
Circular dichroism
CD spectra were measured on a Jasco 815 spectropolarimeter (JASCO, Easton, MD) over the wavelength range 190–260 nm, by using a 0.1-cm path-length quartz cell (internal volume 200 μL) from Hellma (Mullheim, Germany). CD spectra were recorded at 25°C, at 0.2-nm intervals and 10 nm.min−1 scan speed. Measurements were carried out in 10-mM sodium phosphate buffer at pH 7.4. Protein concentration was 25 μM in the presence of detergents (20 mM sodium dodecyl-sulfate (SDS) or dodecylphosphatidylcholine (DPC)) or lipids (peptide/lipid ratio 1:50).
Fluorescence measurements
The interaction between membrane mimics and Engrailed homeodomain was monitored by Trp fluorescence. Fluorescence experiments were recorded on a MOS-200 spectrophotometer using a Xenon short arc lamp at 25°C and by using 0.3 × 0.3-cm path-length quartz cell (internal volume 130 μL) from Hellma. The fluorescence spectra were recorded between 290 and 440 nm (bandwidth 5 nm) with an excitation wavelength at 280 nm. Protein concentration was 25 μM in the presence of detergents (20 mM) or lipids (peptide/lipid ratio 1:50).
NMR data collection and analysis
NMR experiments were recorded on a Bruker DMX 500 MHz spectrometer equipped with a 5-mm TXI probe or on a Bruker Avance III 500 MHz spectrometer equipped with a 5-mm TCI cryoprobe (Bruker Daltonics, Fremont, CA). NMR experiments were processed with the software programs XWINNMR 2.6, TOPSPIN 2.1, or NMRPIPE (30). NMR spectra were assigned using the XEASY (31) or SPARKY (32) software programs.
NMR in aqueous solution
NMR samples contained 1 mM 15N or 15N, 13C uniformly labeled En2HD protein in H2O/D2O (90/10) or D2O, containing 50 mM sodium succinate buffer, pH 4.7, 0.02% (w/v) NaN3, and 0.1 mM sodium 3-(trimethylsilyl)propane-1-sulfonate as an internal chemical shift reference. The sample temperature was set to 303 K. Sequence-specific assignments of backbone 15N, 1HN, 13Cα, 1Hα, 13C′, and side-chain 13Cβ, 1Hβ resonances were obtained from three-dimensional triple resonance HNCA, HN(CO)CA, HNCO, HN(CA)CO, HNCACB, CBCA(CO)NH, and HBHA(CBCACO)NH spectra. Other aliphatic side-chain resonances were assigned using three-dimensional 15N-edited TOCSY-HSQC and three-dimensional 13C HCCH-TOCSY experiments. Aromatic resonances assignment was accomplished using a combination of two-dimensional (HB)CB(CGCD)HD, three-dimensional CT-HMQC-TOCSY, and three-dimensional NOESY-HMQC experiments. Assignments have been deposited in the Biological Magnetic Resonance Bank (http://www.bmrb.wisc.edu/) under BMRB accession No. 19049.
NMR in micellar environments
Samples used for NMR assignment and structural data collection were prepared using 1 mM 15N-labeled En2HD protein in H2O/D2O (90:10) containing 50 mM sodium succinate buffer, pH 4.7, and 80 mM SDS or DPC detergents. The sample temperature was set to 303 K and 313 K for SDS and DPC samples, respectively. 1H, 15N resonances were assigned using a combination of three-dimensional 15N-edited TOCSY-HSQC and NOESY-HSQC experiments. Chemical shifts were indirectly referenced to external sodium 4,4-dimethyl-4-silapentane-1-sulfonate.
NMR positioning experiments in micelles
Previous NMR samples were diluted three times for titration with paramagnetic probes. SDS or DPC detergent was added to a final concentration of 40 mM. No chemical shift changes were observed on 1H-15N HSQC spectra upon sample dilution. A quantity of 5-doxylstearic acid (dissolved in methanol) was used at a concentration of 2.4–3.0 mM, corresponding to 4–5 molecules per micelle (assuming a detergent aggregation number of 60). Relaxations effects induced by 5-doxylstearic acid were estimated by measuring 1H-15N HSQC cross-peak intensity ratios in the absence and in the presence of the paramagnetic reagent. Gadodiamide (gadolinium-diethylenetriamine pentaacetic acid-bismethylamide) was used at a concentration of 1 mM. The paramagnetic relaxation enhancement (PRE) was determined by measuring the difference in R1 relaxation rates of amide protons upon gadodiamide addition (ΔR1) and expressed as relaxivity (ΔR1/[gadodiamide]). These relaxation rates were measured on 1H-15N HSQC experiments incorporating an inversion-recovery scheme at the beginning of the sequence. Amide proton longitudinal magnetization inversion was accomplished using a selective REBURP pulse of 2.7-ms duration. A series of 14 experiments was recorded with recovery delays of 10, 30, 50, 75, 100, 125, 150, 180, 200, 350, 500, 1000, 2000, and 4000 ms, and a recycle delay of 4 s.
Site-directed spin labeling experiments
Two-dimensional 1H-15N HSQC experiments were recorded on 0.2–0.3 mM 15N-labeled En2HD Cys-containing mutants tagged with the nitroxide probe, with a long relaxation delay (5 s) to ensure correct quantification. Relaxation enhancements were determined as the ratio of cross-peak intensities in the paramagnetic state and in the diamagnetic state. The diamagnetic state was obtained by reducing the spin label with 2 mM ascorbic acid (1 h incubation at 40°C). 1H, 15N backbone resonance assignments of tagged Cys mutants in the diamagnetic state were transferred from those of the wild-type protein and confirmed by analyzing three-dimensional 15N-edited TOCSY-HSQC and NOESY-HSQC experiments.
Results
Engrailed 2 homeodomain adopts a well-defined canonical fold in aqueous solution
Engrailed 2 homeodomain sequence is highly conserved in vertebrates. A unique substitution is observed between birds and mammals, residue Gly-39 in chicken Engrailed 2 being replaced by a serine in human counterpart (Fig. 1 A). The structure of Engrailed homeodomain from Drosophila has been determined both by x-ray crystallography (33) and solution NMR spectroscopy (34). The sequence alignments show 78% identity to Drosophila Engrailed with a very high conservation in the third helix segment.
Figure 1.
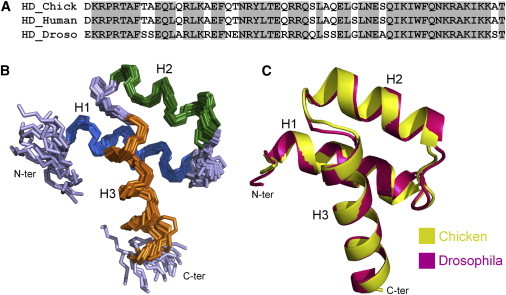
Comparison of chicken En2HD and Drosophila homeodomain structures. (A) Sequence alignment of chicken and human Engrailed 2, and Drosophila melanogaster Engrailed homeodomains. (B) NMR structure of chicken En2HD in aqueous solution (PDB:3ZOB). The backbone of the 20 lowest energy conformers is indicated. Structures were superimposed over residues 8–56. Disordered residues 1–3 and 57–60 at the extremities are not shown. (C) Superimposition of chicken En2HD solution NMR structure and Drosophila Engrailed homeodomain crystal structure (PDB:1ENH), represented as ribbon diagrams. Structures were superimposed on backbone atoms of residues 8–56.
Complete assignment of backbone and side-chain 1H, 13C, and 15N resonances of chicken En2HD was obtained from heteronuclear triple resonance experiments. The analysis of 1Hα, 13Cα, 13Cβ, 13C′, and 15N chemical shifts provided insight on the secondary structure of the protein. Fig. 2 A shows the chemical shift deviations of Hα proton resonances from random coil values (35). Three segments, encompassing residues 10–22, 28–37 and 42–56, have upfield-shifted Hα resonances (CSD < –0.1 ppm) that are characteristic of helical conformations. The secondary structure delineation was further confirmed by the observation of characteristic sequential and medium-range nuclear Overhauser effects (NOEs) (Fig. 2 B), associated with weak 3JHN-Hα coupling constants (<6 Hz) throughout the three segments.
Figure 2.
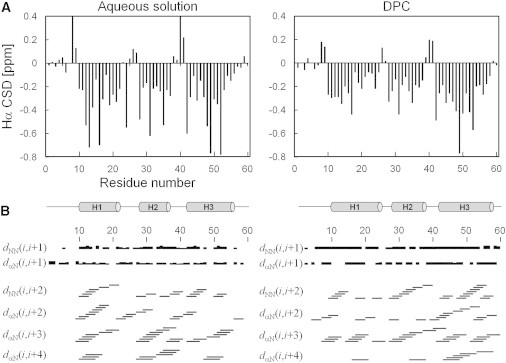
NMR parameters of En2HD in aqueous solution (left) and in DPC micelles (right). (A) Hα chemical shift deviations from random coil values. The location of helical secondary structure elements is shown at the bottom. (B) Summary of sequential and medium-range HN/HN and Hα/HN NOEs.
Structures of En2HD were calculated using a set of 868 distance restraints inferred from NOEs (see Table S1 in the Supporting Material). Backbone ϕ- and ψ-dihedral angle restraints were derived from chemical shift analysis and 3JHN-Hα coupling constants. Side-chain χ1 and χ2 torsional restraints were deduced from the measurement of heteronuclear coupling constants. The three-dimensional structure of En2HD is shown in Fig. 1 B. The NMR structures show no consistent restraint violations and have good stereochemical quality as estimated from the Ramachandran diagram analysis and low clash score (see Table S1). The root-mean square deviation (RMSD) from the mean structure for backbone atoms of ordered residues 8–55 is 0.6 Å.
En2HD adopts the characteristic homeodomain fold: helices H1 and H2 pack against each other in an antiparallel arrangement, whereas helix H3 is connected by a tight turn and makes a ∼60° angle with the other helices. The homeodomain is stabilized by a small hydrophobic core comprising four aromatic and six aliphatic residues. The van der Waals interaction involving residues Phe-8, Leu-16, and Phe-20 in helix H1, Leu-34 in helix H2, and Ile-45, Trp-48, and Phe-49 in helix H3 contribute to the packing of helices. Leu residues in the loop connecting helices H1-H2 (Leu-26) and in the tight turn between helices H2 and H3 (Leu-38 and Leu-40) also contribute to the hydrophobic core of En2HD. The side chains of all these residues but Leu-26, adopt unique conformations around their χ1 angle, in agreement with measured heteronuclear coupling constants.
The comparison of chicken En2HD structure with the crystal or solution structures of Engrailed homeodomain from Drosophila shows a remarkable conservation of the three-dimensional fold (Fig. 1 C). The RMSD calculated for backbone atoms of residues 8–55 is 0.6 Å. In particular, the 11 residues whose side chains are fully buried in the homeodomain structure (Phe-8, Leu-16, Phe-20, Leu-26, Leu-34, Leu-38, Leu-40, Gln-44, Ile-45, Trp-48, and Phe-49) are strictly conserved between Drosophila Engrailed homeodomain and chicken En2HD (Fig. 1 A) and adopt similar conformations.
CD and Trp fluorescence indicate alterations of the homeodomain structure in membrane environments
To investigate the influence of membrane environments on the structure of En2HD, we first recorded far-UV CD spectra in the absence and in the presence of large unilamellar vesicles (LUVs) made of DMPC or DMPG phospholipids. The CD spectra in aqueous solution and in the presence of zwitterionic DMPC LUVs are almost identical (Fig. 3 A), displaying two negative ellipticity minima near 208 and 222 nm that are characteristic of α-helical structure. Deconvolution of these spectra allowed us to estimate the proportion of helical secondary structure as 50% in both media, with a small proportion of β-sheet (5%) and turn (15%), and 30% of random coil. The quantification of the helical content tends to be slightly underestimated as compared with NMR data in aqueous solution. This discrepancy might be due to a significant contribution of side-chain aromatic chromophores to the CD signal (36,37), giving rise to an atypical [θ]222:[θ]208 ratio.
Figure 3.
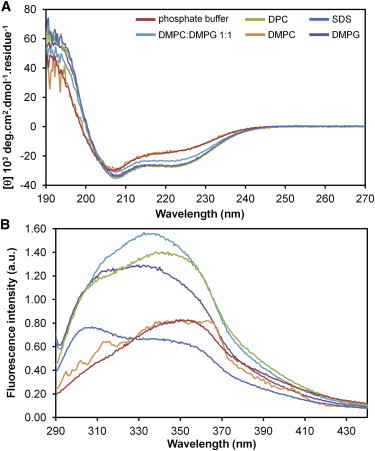
(A) CD spectra and (B) intrinsic tryptophan fluorescence spectra of En2HD in phosphate buffer, SDS micelles, DPC micelles, DMPC LUVs, DMPG LUVs, and DMPC/DMPG (1:1) LUVs. Samples contained 25 μM protein, and detergent and lipid concentrations, were 20 mM and 1.25 mM, respectively.
Interestingly, the addition of anionic lipids induces noticeable changes in the CD spectrum, with a lowering of the minima at 222 nm mainly, and 208 nm to a lesser extent. Larger effects are observed in the presence of pure DMPG LUVs in comparison with mixed DMPG/DMPC LUVs (1:1 ratio). The CD spectra clearly indicate helical structure but with modifications of the [θ]222:[θ]208 ratio. Such CD modifications induced by anionic lipids could be due to remodeling of the helical segments and/or changes in the environment of aromatic side chains. The deconvolution of CD spectrum in DMPG indicates an increase in the helical content (∼57%) and a decrease of random coil (25%), the proportion of β-sheet and turn remaining low (5 and 13%, respectively). Finally, CD spectra of En2HD were recorded in the presence of detergent micelles that are commonly used as membrane mimetic systems in solution NMR studies. The CD spectra of En2HD in the presence of zwitterionic DPC or anionic SDS micelles are very similar to those in the presence of LUVs made of DMPG. The deconvolution of CD spectra yields 63% of helical structure in both SDS and DPC micelles.
The interaction of En2HD in membrane environments was then followed by exploiting the intrinsic fluorescence of Trp-48, the only Trp residue in En2HD (Fig. 3 B). The emission spectra in aqueous solution and in the presence of zwitterionic DMPC vesicles are very similar, with a maximum at ∼350 nm. The addition of anionic lipids induces a shift toward shorter wavelengths with a concomitant increase in the fluorescence intensity. Such blue shift and enhanced quantum yield are typically observed when Trp residue inserts into lipid bilayers (38). Similar modifications of the Trp-48 fluorescence are also observed upon addition of zwitterionic DPC micelles. However, in the case of SDS micelles, the induced changes differ from those observed in the presence of phospholipids. It is likely that the more pronounced changes arise from the sulfate group environment in comparison with phospholipid headgroups.
Altogether, our results show that En2HD interacts with anionic lipids and undergoes conformational changes affecting its secondary structure and Trp-48 environment. Remarkably, the DPC micellar environment yields similar effects as assessed by CD and fluorescence data. Based on these results, we further investigated the structural changes affecting En2HD by liquid-state NMR in DPC micelles.
NMR spectroscopy shows that in DPC micelle environment the homeodomain secondary structure is maintained but its native three-dimensional fold is lost
1H, 15N NMR experiments were recorded on En2HD in the presence of DPC detergent below and above the CMC. The stepwise addition of DPC in the millimolar range leads to dramatic changes on the 1H-15N HSQC spectra of En2HD, with many resonances experiencing line-broadening or chemical shift changes (data not shown). This indicates a strong interaction of En2HD with detergent, occurring in intermediate or slow exchange regime. Titration with increasing concentrations of DPC gives rise to a unique set of chemical shifts with moderate line broadening at detergent concentrations >60 mM (Fig. 4 B), and no further evolution was observed beyond this concentration. Assuming a detergent aggregation number of 60 molecules per micelle, this indicates that no structural changes occur beyond a 1:1 ratio of micelle to protein. In all subsequent NMR studies, protein/detergent molar ratios of 1:60–1:100 were used, with protein concentration ranging between 0.3 and 1 mM. No modifications of HSQC spectra were observed under these conditions.
Figure 4.
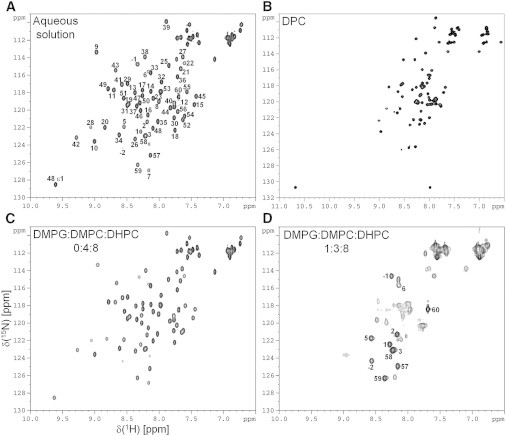
Two-dimensional 1H-15N HSQC spectra of En2HD in aqueous solution (A), in DPC micelles (B), in zwitterionic bicelles (C), and in anionic bicelles (D). Assignments of back-bone and side-chain Trp resonances are indicated in aqueous solution. All the spectra were recorded at 500 MHz and 30°C except the DPC spectrum, which was acquired at 40°C. DPC concentration was 80 mM. Zwitterionic bicelles contained 50 mM DHPC and 25 mM DMPC and anionic bicelles contained 50 mM DHPC, 18.75 mM DMPC, and 6.25 mM DMPG.
The comparison of 1H-15N HSQC spectra in DPC micelles and in aqueous solution shows large chemical shift changes (Fig. 4, A and B). In particular, the reduced dispersion in amide proton chemical shifts suggests an unfolding of En2HD in micelle environment. The destabilization of the native three-dimensional structure is further confirmed when examining the aliphatic region of one-dimensional 1H spectra (see Fig. S3 in the Supporting Material). Notably, the characteristic shieldings of Lys-52 methylenic γCH2 protons and Leu-16 methyl protons due to long-range contacts with aromatic rings in water are lost in micelles. Moreover, fewer NOE correlations are observed between aliphatic and aromatic protons on two-dimensional 1H-1H NOESY spectra, confirming that the native hydrophobic core of En2HD is not preserved in micelles.
The two-dimensional 1H-15N HSQC spectra could be reassigned using three-dimensional TOCSY-HSQC and NOESY-HSQC experiments. The analysis of Hα chemical shift deviations (Fig. 2 A) clearly shows the persistence of helical secondary structure in DPC, as already indicated by CD spectroscopy. Three continuous stretches of upfield-shifted Hα protons can be observed, which approximately correspond to the three native helices in aqueous solution. The delineation of the three α-helical segments was also confirmed by the observation of strong sequential HN-HN, medium-range HNi-HNi+2, Hαi-HNi+3, and Hαi-HNi+4 NOEs (Fig. 2 B) that are characteristic of α-helical conformations. Although the first helix showed a less negative Hα CSD for the central residue Ala-18, the persistence of medium-range NOEs suggests that this helix does not interrupt in its central portion.
Further analysis of Hα CSDs enables us to pinpoint the conformational propensities of individual residues in aqueous and micellar environments. Residues Phe-8, Thr-9, and Asn-41 invariantly adopt extended conformations, as indicated by positive Hα CSD values (>0.1 ppm) and helices H1 and H3 are initiated at the same residue position, Ala-10 and Glu-42, respectively, in both media. Helix H3 is slightly stabilized on its C-terminal side in DPC micelles and extends to residue Ile-56. The conformation of the H1-H2 connecting region appears to be more helical in micellar environment and differs from the native conformation. The short segment connecting helices H2 and H3 has turn propensity, as shown by the short stretch of positive Hα CSDs and the observation of medium-range NOEs. These NOE connectivities were also detected in aqueous solution but most of the native NOEs are missing in this region, in particular between Ala-35, Leu-40, and Glu-42, indicating that packing interactions between helices H2 and H3 are lost.
Overall, no long-range NOEs could be detected on two- or three-dimensional NOESY spectra in DPC micelles. The only observed NOEs between aromatic and aliphatic protons were assigned to sequential or medium-range connectivities involving side chains lying on the same face of helices. This indicates that the three helical segments may fold independently in the detergent micelles without tight packing between helices. The use of nondeuterated detergents enabled us to detect intermolecular NOEs between the detergent alkyl chains and aromatic residues, in particular involving Trp-48 (see Fig. S4). Thus it appears that the hydrophobic side chains of En2HD interact with the apolar detergent and are not involved in long-range intramolecular interactions.
Dynamics of the DPC-bound states by 15N relaxation
15N relaxation parameters were monitored to get information on the backbone dynamics and the size of the En2HD/micelle complexes (see Fig. S1). Heteronuclear 15N-{1H} NOEs in DPC micelles show that residues at the N- and C-termini are highly flexible whereas residues 10–55 of En2HD display lower conformational flexibility. The three helical segments exhibit heteronuclear NOE values at ∼0.6, indicating that they experience higher mobility than in aqueous solution but remain rigid on the picosecond-to-nanosecond timescale. Subtle motional differences are observed for the three helical segments. Helix H3 exhibits the highest NOE values in its central part, and shows fraying in its C-terminal part, as also evidenced by lower Hα CSDs. In contrast, helix H2 has the lowest NOE values. Both segments connecting helices H1-H2 and H2-H3 display decreased R2 and NOE values, indicating that they have greater flexibility than the helical parts but still have reduced mobility.
The correlation time of the protein could be estimated from the R1/R2 ratio measured for the most rigid residues. A value of 8.9 ns was obtained for En2HD at 40°C in DPC, corresponding to spherical particles of ∼25 kDa in aqueous solution. Under the assumption that En2HD forms with detergent tight complexes of roughly spherical shape, one could estimate that En2HD is associated with ∼50 DPC molecules. Because this value approximately corresponds to detergent aggregation numbers in pure DPC micelles, it can be concluded that En2HD forms a 1:1 protein/micelle complex.
Monitoring long-range contacts in DPC-bound homeodomain by intramolecular paramagnetic relaxation enhancements
Because our data unambiguously demonstrate that En2HD lacks a compact three-dimensional structure when bound to DPC micelles, we attempted to detect long-range interactions between α-helices using site-directed spin labeling. Indeed, while 1H-1H NOEs are observable for short interproton distances typically up to 5 Å, paramagnetic relaxation enhancements (PREs), which arise from strong dipolar interaction between unpaired electrons and nearby nuclei, allow the detection of long-range contacts within ∼25 Å from the spin label. Furthermore, while NOEs may be quenched by internal dynamics, PREs can be successfully used to detect transient long-range interactions in unstructured or partially disordered proteins (39).
The strategy was to covalently attach a nitroxide radical to En2HD using site-directed cysteine mutagenesis and chemical linkage. The spin label was introduced either at the N-terminus (Cys in position –3 with respect to homeodomain first residue), the C-terminus (C61 mutant), or in the turn connecting helices H2 and H3 (G39C mutant). The position 39 in the turn was selected because a Cys residue is observed at this position in several homeodomains of the HoxA family (1). Except for residues in close proximity with the mutated position, the insertion of a Cys residue in En2HD sequence did not induce significant shifts of the cross-peaks on two-dimensional 1H-15N HSQC spectra in aqueous solution, indicating that the mutations did not alter homeodomain fold. Similar observations were made in micelles. PREs were estimated by measuring the peak intensity ratios between two 1H-15N HSQC spectra recorded in the paramagnetic state and in the diamagnetic state, after reduction with ascorbic acid.
PREs were first measured for En2HD in aqueous solution (Fig. 5 A). The intensity profile of the C61 mutant shows that relaxation enhancements are observed locally up to 20 residues from the probe. Long-range effects are observed in two regions, in the loop connecting helices H1 and H2 (residues 21–26) and in the extended segment preceding helix 1 (residues 5–8). These effects are well accounted for by the NMR structure family, which shows that the disordered C-terminus can contact both affected segments in different conformers. The introduction of a spin label at the N-terminus affects residues located in the turn between helices H2 and H3, as expected from the three-dimensional structure. The PRE profile obtained with the G39C mutant has a bell shape extending to ∼15 residues on either side of the attached radical. A dramatic long-range effect is also observed in the N-terminal arm and the first helix, in agreement with the close proximity between Gln-12 and Gly-39 (distances between Cα atoms of 8.8 ± 0.4 Å). Overall, the PRE profiles obtained with the three mutants are in excellent agreement with the solution NMR structure and thus provide a characteristic signature of homeodomain native structure. The relaxation effects were also measured on side-chain HN groups in Asn, Gln, Arg, and Trp residues. Similar long-range contacts are observed and involve larger regions, presumably owing to increased conformational space explored by side chains.
Figure 5.
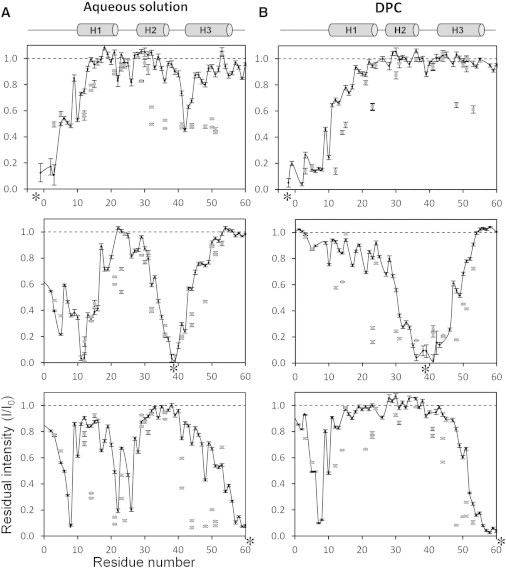
Paramagnetic relaxation enhancements induced by site-directed spin labeling in aqueous solution (A) and in DPC micelles (B). The paramagnetic tag was attached to a Cys residue (indicated by an asterisked (∗) label) in position –3 (upper), in position 39 (middle), or in position 61 (lower panel). The paramagnetic relaxation enhancements are shown as intensity ratios between the paramagnetic (I) and the diamagnetic state (I0). (Solid circles) Backbone amide groups; (shaded squares) side-chain groups.
The observed PREs differ markedly in the presence of DPC micelles (Fig. 5 B). The probe attached in position 39 locally induces larger relaxation enhancements than in water, as can be seen for residues 31–46. This can be ascribed to an increased dynamics of both backbone and side chains for the attachment site in micelles, in comparison with aqueous solution. Weak PREs are detected in more distant regions but the magnitude of the effects indicates only transient contacts and the absence of preferential long-range interactions. When the probe is attached to the C-terminal residue, the characteristic long-range effects that were observed in segment 21–26 in water are lost, suggesting that En2HD explores nonnative conformations in micelles. However, long-range effects are still observed in the N-terminal 5–10 segment, with near-cancellation of the residual intensities of Ala-7 and Phe-8 residues. Consistently, when the nitroxide radical is attached to the N-terminal tail, Trp-48 and Arg-53 side chains are significantly affected, confirming that the N-terminus makes transient contact with the C-terminal extremity of En2HD in DPC micelles. Altogether, these data indicate that the three helices in En2HD do not adopt extended orientations in micelles but their relative orientation leads to a proximity of the C-terminus and the extended 5–10 segment that precedes helix H1.
Positioning of En2HD in DPC micelles using noncovalent paramagnetic probes
The localization of En2HD with respect to the micelle surface was examined using water-soluble (gadodiamide) and lipophilic (5-doxylstearic acid) paramagnetic probes. Gadodiamide is a neutral Gd(III) complex that partitions into the aqueous environment outside the micelle. It strongly enhances T1 relaxation of protons (40). We quantified the enhancement (PRE) of amide protons longitudinal relaxation by inserting before the HSQC sequence a T1 inversion-recovery block with selective inversion of the amide protons. In DPC detergent (Fig. 6 A), the water-soluble gadodiamide probe strongly enhances the longitudinal relaxation of amide protons in residues at the N- and C-termini, indicating that these residues are not buried inside the micelles. Residues located in the loop regions and at both extremities of the three helices are more affected than those lying in the central portion of the helices, suggesting that the core portions of the secondary structure elements are more buried within the micelles. The comparison of PREs indicates that helix H2 is slightly less buried than helices H1 and H3. A periodicity in the PRE values is apparent in the three helical segments and supports a parallel positioning to the micelle surface. In particular, hydrophobic residues in helices H1 and H2 display the lowest PREs (Leu-13, Leu-16, Phe-20, Leu-34, and Leu-38), indicating that these two helices orient their hydrophobic face toward the micelle interior.
Figure 6.
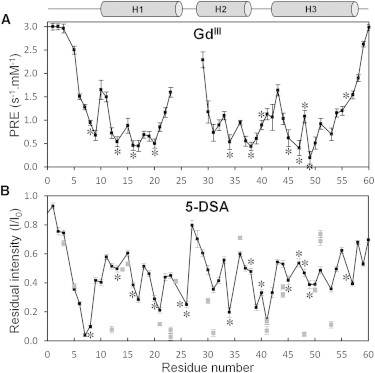
Use of paramagnetic reagents to probe the accessibility of En2HD residues in DPC micelles. (A) Paramagnetic relaxation enhancements of gadodiamide probe measured as amide proton relaxivities (differences in R1 longitudinal rates per mM concentration of gadodiamide). (B) Paramagnetic relaxation enhancements of 5-doxylstearic acid probe measured as residual intensities of HSQC cross-peaks. (Solid circles) Backbone amide groups; (shaded squares) side-chain groups. Hydrophobic side chains are indicated with asterisked (∗) labels.
Next, the use of the lipophilic 5-DSA (5-doxylstearic acid) 5-doxylstearic acid) enabled us to probe the immersion depth of En2HD within DPC micelles. 5-DSA is located in the micelle interior and enhances the relaxation of nuclei inserted in the micelle but located close to the micelle surface. The enhanced T2 relaxation was quantified by measuring the residual intensities of the HSQC correlations (Fig. 6 B). Periodic variations are observed in helical segments, confirming a parallel positioning of helices with respect to the micelle surface. The regions connecting helices (residues 26, 39, and 41) as well as the N-terminal segment 6–8 are the most affected by the 5-DSA probe, suggesting that they lie in close proximity with the spin label, around the fifth carbon position of the alkyl chain. Because the core helical segments are less affected by both lipophilic and water-soluble probes, it can be concluded that they should be buried below the position occupied by the C5-doxyl group within micelles. We also analyzed the relaxation enhancements on side-chain HN groups to get more insight on side-chain positioning in micelles. Trp-48 indolic HNε1 resonance is severely affected by the 5-DSA probe in DPC detergent, indicating that the aromatic group is located in the micelle interior but is not deeply buried. The analysis of side-chain HNε resonance of Arg residue shows different behaviors. Whereas most Arg residues are poorly affected by the 5-DSA probe, Arg-31 and Arg-53 are strongly attenuated in DPC micelle. Therefore, these two Arg residues are positioned inside the micelle but close to the headgroup.
Influence of anionic environments on homeodomain structure
To analyze the effect of anionic environments on En2HD structure, we next used SDS micelles as an anionic membrane mimetic (see Fig. S5). The titration with SDS detergent shows that En2HD forms a tight complex with the micelle and is associated with ∼60 SDS molecules, as inferred from the calculated correlation time based on relaxation data (8.6 ns at 40°C). As observed with DPC micelles, one- and two-dimensional NMR spectra exhibit reduced chemical shift dispersion (see Fig. S3 C and Fig. S5 A) and no long-range correlations could be detected on NOESY spectra, indicating that En2HD loses its native three-dimensional structure when bound to SDS micelle. Interestingly, the three-helix secondary structure is maintained, as evidenced by the analysis of Hα CSD and medium-range NOE (see Fig. S5, B and C). Subtle differences between SDS and DPC environments are nevertheless observed in the loop region connecting H1 and H2: helix H1 is slightly shorter in its C-terminal part whereas helix H2 is extended in its N-terminal part in SDS micelles. Overall, our data indicate that the micelle composition has little influence on En2HD structure. Indeed, site-directed spin labels (see Fig. S5 D) confirm the nonnative structure and the presence of long-range contacts between N- and C-terminal regions. Furthermore, the use of noncovalent paramagnetic probes enables us to position the three helices in parallel orientations close to the micelle surface. Notable is that the paramagnetic waves observed for helix H3 slightly differ in the two detergent systems (see Fig. S5 E) and suggest an influence of the detergent environment on the helix azimuthal angle.
Finally, we used small, isotropically tumbling bicelles as a better membrane mimetic to investigate En2HD interactions with a true membrane bilayer by liquid-state NMR. In the presence of small bicelles made of zwitterionic DMPC and DHPC (long-chain/short-chain lipid q ratio of 0.5), the 1H-15N HSQC spectrum of En2HD (Fig. 4 C) reveals only minor chemical shift changes, suggesting weak transient interactions with bicelles. Importantly, this demonstrates that En2HD native structure is maintained in a zwitterionic environment. In contrast, the addition of anionic lipids (25% DMPG, q = 0.5) yields to dramatic changes of the 1H-15N HSQC spectrum (Fig. 4 D), indicating a strong interaction with anionic bicelles. Residues in the unstructured N- and C-terminal parts exhibit weak chemical shift perturbation while resonances belonging to the core region of the homeodomain are no longer observed or show highly broadened signals, which can be ascribed to intermediate exchange on the NMR timescale. Interestingly, the upfield-shifted aliphatic resonances characteristic of homeodomain fold are no longer detected in anionic bicelles although they are observed in zwitterionic bicelles (see Fig. S3, D and E), suggesting that a large conformational change occurs upon interaction with anionic lipids.
Discussion
Our study, based on CD, Trp fluorescence, and NMR spectroscopy, reveals that the native conformation of Engrailed 2 homeodomain can be significantly altered in membrane environments. En2HD adopts the characteristic three-helical bundle fold in aqueous solution and the close similarity of CD, fluorescence, and NMR spectra suggests that the homeodomain fold is preserved in the presence of zwitterionic DMPC lipids. In contrast, the addition of anionic DMPG lipids induces changes in CD, NMR, and Trp fluorescence spectra. Notable is that the blue shift in the fluorescence emission spectrum indicates the environment of Trp-48 residue becomes more hydrophobic. This result provides functional insight on the membrane interactions of homeodomains because the conserved Trp-48 residue is known to play a critical role in the membrane translocation mechanism. Because lipid vesicles are usually not suitable for solution NMR, we investigated whether detergent micelles or small isotropically tumbling bicelles could mimic the effects observed with vesicles. The addition of anionic bicelles induced large changes in NMR spectra but severe broadening of En2HD amide resonances precluded high-resolution structural studies. We found that zwitterionic DPC induced comparable changes in CD and fluorescence signals to those observed in the presence of anionic lipids whereas anionic SDS micelles yielded more divergent fluorescence spectra. It might sound surprising that zwitterionic detergents could somehow mimic the effects of anionic lipids. However, similar results are often encountered for basic membranotropic peptides, where conformations in DPC micelles are closer to those found in DMPG vesicles than in DMPC vesicles (41). This can be explained by the higher detergent dynamics that favors peptide interaction with the hydrophobic chains, even in the absence of strong electrostatic interactions with polar headgroups.
Based on these results, we used solution NMR to gather information about En2HD conformation in DPC as membrane mimetics. En2HD loses its three-dimensional structure but, remarkably, the helical secondary structure is maintained with only slight adjustments of the length of the three helical segments. From titration data and use of noncovalent paramagnetic probes, we found that En2HD forms a tight 1:1 protein/micelle complex, each helical segment being embedded within the micelles in roughly parallel orientations. The core portions of the helices comprise the most deeply buried residues within micelles. Consistently, these segments tend to be the most rigid whereas connecting regions exhibit larger mobility on the picosecond to nanosecond timescale.
Site-directed spin labels enabled us to confirm the nonnative three-dimensional structure of En2HD in DPC micelles but also revealed long-range interactions between the C-terminus and the extended 5–10 segment. It is unlikely that these observed long-range PREs arise from a minor native conformer in fast exchange with the micelle-bound form because other characteristic native long-range contacts were not observed. Therefore En2HD does not adopt extended helix orientations when bound to micelles but the relative arrangement of helices leads to a proximity of the C-terminus with the segment immediately preceding helix H1. The analysis of NOE correlations supports the absence of packing interactions between helices, their hydrophobic face interacting with hydrocarbon chains of detergents. A model of En2HD conformation in DPC micelles was built using NOE- and PRE-based distance restraints (Fig. 7 A). Although this model may not reflect the ensemble of conformations explored by En2HD, it depicts one of the conformers that account for the transient long-range contacts inferred from PRE data, with the side chains emanating from the helices too far apart to make long-range contacts with each other. Notable is that such a helical arrangement, which fits well with the dimensions of a spherical micelle, could be induced by the curvature and size of this membrane mimetics. An alternative model might be drawn, with individual helices being in contact with different micelles, but this seems less plausible and is not supported by NMR titration and relaxation analysis.
Figure 7.
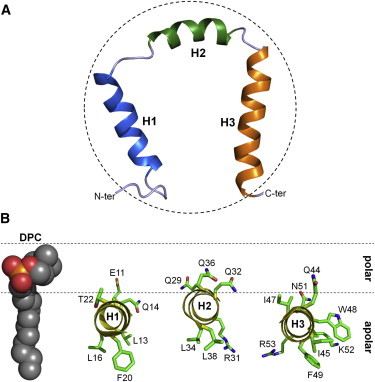
Model of DPC-bound En2HD structure. (A) Structural model based on chemical shifts, NOEs, and site-directed spin-labeled PREs data. The model was calculated with DYANA using 166 NOE-based distance constraints, 43 ϕ-, ψ-helical angular restraints, and two PRE-derived long-range distance constraints between the amide groups of Thr-60, Ala-7, and Phe-8, respectively (20 Å). (Dotted circle) The dimension of a spherical DPC micelle containing 60 monomers. (B) Positioning of En2HD three helices with respect to DPC micelle, inferred from paramagnetic effects induced by gadodiamide and 5-DSA probes.
The folding and unfolding of Drosophila Engrailed homeodomain have been extensively studied by Religa et al. (42). In particular, the structure of a denatured state and folding intermediate was characterized by NMR using an engineered mutant. This intermediate state has native helical secondary structure and the region encompassing helices H2 and H3 adopts the native helix-turn-helix motif. This motif was also shown to fold independently in a truncated 16–59 Engrailed homeodomain lacking the N-terminus and half of helix H1 (43), although this intrinsic folding property is not shared by all homeodomains (44). The nonnative state that we characterized in micelles also has nearly native secondary structure, showing that the three helical segments have high helical propensity. However, the comparison of NOEs in water and micelles indicates that the segment encompassing helices H2 and H3 does not adopt a stable helix-turn-helix motif in micelles.
The interaction of the third helix is of prime interest because this segment mediates the translocation properties. The helix positioning does not appear to be influenced by the environment of the full homeodomain because a similar parallel orientation has been observed by NMR for peptides corresponding to the third helix of different homeodomains (13,25). The parallel orientations of penetratin peptide in different membrane environments have also been established by a variety of biophysical techniques (45–48). Our NMR data show that the positioning of the third helix does not strongly differ from that of helices H1 and H2 but differences in the immersion depths were observed between the three helices. Because the time averaging of relaxation enhancement makes it sensitive to peptide conformational flexibility and motions within micelle, we did not attempt to derive distances from gadodiamide-based PREs. A qualitative model of helix positioning based on the results obtained with two distinct paramagnetic probes is presented in Fig. 7 B. Helix H2 tends to be slightly less buried than helices H1 and H3. The calculation of hydrophobic moments indicates that helices H2 and H3 are poorly amphipathic. The depth of immersion does not correlate with amphipathicity but seems to be related to helix hydrophobicity, helix H2 having fewer hydrophobic residues.
Accordingly, the central 47–51 portion of helix H3 that is composed of three consecutive hydrophobic side chains tends to be the most buried. We previously showed that the translocation efficiency does not imply a deep insertion of the third helix but that interactions near the membrane surface are a major determinant (13). The positions of the critical hydrophobic and cationic residues of the third helix with respect to the surface are therefore of prime importance. The side-chain indole HNε1 group of Trp-48 is strongly attenuated by the 5-doxylstearic acid probe in DPC, which indicates that this side chain is buried inside the micelle but remains close to the surface. The helical projection reveals that the central segment of the third helix is poorly amphipathic, basic residues Lys-52 and Arg-53 lying on the same face as hydrophobic Trp-48 and Phe-49 (Fig. 7 B). Interestingly, the HNε group of Arg-53 side chain is severely attenuated by the doxyl probe in DPC, indicating that the guanidinium group is also inserted within the micelle. The burial of Lys-52 and Arg-53 could be stabilized by π-cation interactions with Trp-48 and Phe-49 aromatic side chains, respectively (26,49). The observation of NOEs between Trp-48 HNε1 and Lys-52 Hε side-chain protons in micelles supports such π-cation interactions. In addition to these residues, Arg-31 in the second helix also appears to be buried inside the DPC micelle.
The electrostatic interactions between basic CPPs and anionic membranes are known to play a crucial role in membrane translocation. Trp fluorescence showed that an environment change of Trp-48 occurred in lipid vesicles only in the presence of anionic lipids. Based on our CD and NMR conformational study, we suggest that the membrane translocation of homeodomains involves unfolding of the native three-dimensional structure, with retention of helical secondary structure. The relative orientation of helices might be different between micelle mimetic systems and true lipid membranes, insofar as it may be dependent on parameters such as lipid composition, vesicle size, and curvature. For instance, NMR studies showed that α-synuclein forms two α-helices in an antiparallel arrangement in SDS micelles (50) while a long uninterrupted helix was deduced from electron paramagnetic resonance distance measurements when it is bound to small unilamellar vesicles (51). Recent studies suggested that α-synuclein actually populates both elongated and broken helical states upon small unilamellar vesicle binding (52). Thus, it is conceivable that homeodomain may explore different helical conformations depending on membrane environments. Our results suggest that electrostatic interactions with membranes may be determinant not only in providing membrane affinity but also in inducing a conformational change in homeodomain structure enabling Trp-48 residue to insert into membranes.
Acknowledgments
We acknowledge funding from the Emergence-UPMC-2009 and Agence Nationale de la Recherche (grant No. ANR-BLAN2010-ParaHP) research programs.
Footnotes
Stéphane Balayssac’s present address is UMR 5068 CNRS, Université Paul Sabatier, Toulouse, France.
François-Xavier Cantrelle’s present address is UMR 8576 CNRS-Université Lille 1, Lille, France.
Supporting Material
References
- 1.Banerjee-Basu S., Baxevanis A.D. Molecular evolution of the homeodomain family of transcription factors. Nucleic Acids Res. 2001;29:3258–3269. doi: 10.1093/nar/29.15.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pearson J.C., Lemons D., McGinnis W. Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- 3.Joliot A., Pernelle C., Prochiantz A. Antennapedia homeobox peptide regulates neural morphogenesis. Proc. Natl. Acad. Sci. USA. 1991;88:1864–1868. doi: 10.1073/pnas.88.5.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Roux I., Joliot A.H., Volovitch M. Neurotrophic activity of the Antennapedia homeodomain depends on its specific DNA-binding properties. Proc. Natl. Acad. Sci. USA. 1993;90:9120–9124. doi: 10.1073/pnas.90.19.9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joliot A., Maizel A., Prochiantz A. Identification of a signal sequence necessary for the unconventional secretion of Engrailed homeoprotein. Curr. Biol. 1998;8:856–863. doi: 10.1016/s0960-9822(07)00346-6. [DOI] [PubMed] [Google Scholar]
- 6.Chatelin L., Volovitch M., Prochiantz A. Transcription factor hoxa-5 is taken up by cells in culture and conveyed to their nuclei. Mech. Dev. 1996;55:111–117. doi: 10.1016/0925-4773(95)00478-5. [DOI] [PubMed] [Google Scholar]
- 7.Noguchi H., Kaneto H., Bonner-Weir S. PDX-1 protein containing its own Antennapedia-like protein transduction domain can transduce pancreatic duct and islet cells. Diabetes. 2003;52:1732–1737. doi: 10.2337/diabetes.52.7.1732. [DOI] [PubMed] [Google Scholar]
- 8.Tassetto M., Maizel A., Joliot A. Plant and animal homeodomains use convergent mechanisms for intercellular transfer. EMBO Rep. 2005;6:885–890. doi: 10.1038/sj.embor.7400487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spatazza J., Di Lullo E., Prochiantz A. Homeoprotein signaling in development, health, and disease: a shaking of dogmas offers challenges and promises from bench to bed. Pharmacol. Rev. 2013;65:90–104. doi: 10.1124/pr.112.006577. [DOI] [PubMed] [Google Scholar]
- 10.Brunet I., Weinl C., Holt C. The transcription factor Engrailed-2 guides retinal axons. Nature. 2005;438:94–98. doi: 10.1038/nature04110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugiyama S., Di Nardo A.A., Hensch T.K. Experience-dependent transfer of Otx2 homeoprotein into the visual cortex activates postnatal plasticity. Cell. 2008;134:508–520. doi: 10.1016/j.cell.2008.05.054. [DOI] [PubMed] [Google Scholar]
- 12.Derossi D., Joliot A.H., Prochiantz A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J. Biol. Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- 13.Balayssac S., Burlina F., Lequin O. Comparison of penetratin and other homeodomain-derived cell-penetrating peptides: interaction in a membrane-mimicking environment and cellular uptake efficiency. Biochemistry. 2006;45:1408–1420. doi: 10.1021/bi0518390. [DOI] [PubMed] [Google Scholar]
- 14.Noguchi H., Matsushita M., Bonner-Weir S. Mechanism of PDX-1 protein transduction. Biochem. Biophys. Res. Commun. 2005;332:68–74. doi: 10.1016/j.bbrc.2005.04.092. [DOI] [PubMed] [Google Scholar]
- 15.Kilk K., Magzoub M., Gräslund A. Cellular internalization of a cargo complex with a novel peptide derived from the third helix of the islet-1 homeodomain. Comparison with the penetratin peptide. Bioconjug. Chem. 2001;12:911–916. doi: 10.1021/bc0100298. [DOI] [PubMed] [Google Scholar]
- 16.Dietz G.P.H., Bähr M. Peptide-enhanced cellular internalization of proteins in neuroscience. Brain Res. Bull. 2005;68:103–114. doi: 10.1016/j.brainresbull.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 17.Stewart K.M., Horton K.L., Kelley S.O. Cell-penetrating peptides as delivery vehicles for biology and medicine. Org. Biomol. Chem. 2008;6:2242–2255. doi: 10.1039/b719950c. [DOI] [PubMed] [Google Scholar]
- 18.Sagan S., Burlina F., Joliot A. Homeoproteins and homeoprotein-derived peptides: going in and out. Curr. Pharm. Des. 2013;19:2851–2862. doi: 10.2174/1381612811319160002. [DOI] [PubMed] [Google Scholar]
- 19.Derossi D., Calvet S., Prochiantz A. Cell internalization of the third helix of the Antennapedia homeodomain is receptor-independent. J. Biol. Chem. 1996;271:18188–18193. doi: 10.1074/jbc.271.30.18188. [DOI] [PubMed] [Google Scholar]
- 20.Binder H., Lindblom G. Charge-dependent translocation of the Trojan peptide penetratin across lipid membranes. Biophys. J. 2003;85:982–995. doi: 10.1016/S0006-3495(03)74537-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berlose J.P., Convert O., Chassaing G. Conformational and associative behaviors of the third helix of Antennapedia homeodomain in membrane-mimetic environments. Eur. J. Biochem. 1996;242:372–386. doi: 10.1111/j.1432-1033.1996.0372r.x. [DOI] [PubMed] [Google Scholar]
- 22.Drin G., Mazel M., Temsamani J. Physico-chemical requirements for cellular uptake of pAntp peptide. Role of lipid-binding affinity. Eur. J. Biochem. 2001;268:1304–1314. doi: 10.1046/j.1432-1327.2001.01997.x. [DOI] [PubMed] [Google Scholar]
- 23.Magzoub M., Eriksson L.E.G., Gräslund A. Conformational states of the cell-penetrating peptide penetratin when interacting with phospholipid vesicles: effects of surface charge and peptide concentration. Biochim. Biophys. Acta. 2002;1563:53–63. doi: 10.1016/s0005-2736(02)00373-5. [DOI] [PubMed] [Google Scholar]
- 24.Magzoub M., Eriksson L.E.G., Gräslund A. Comparison of the interaction, positioning, structure induction and membrane perturbation of cell-penetrating peptides and non-translocating variants with phospholipid vesicles. Biophys. Chem. 2003;103:271–288. doi: 10.1016/s0301-4622(02)00321-6. [DOI] [PubMed] [Google Scholar]
- 25.Lindberg M., Biverståhl H., Mäler L. Structure and positioning comparison of two variants of penetratin in two different membrane mimicking systems by NMR. Eur. J. Biochem. 2003;270:3055–3063. doi: 10.1046/j.1432-1033.2003.03685.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhang W., Smith S.O. Mechanism of penetration of Antp(43-58) into membrane bilayers. Biochemistry. 2005;44:10110–10118. doi: 10.1021/bi050341v. [DOI] [PubMed] [Google Scholar]
- 27.Wizenmann A., Brunet I., Prochiantz A. Extracellular Engrailed participates in the topographic guidance of retinal axons in vivo. Neuron. 2009;64:355–366. doi: 10.1016/j.neuron.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sonnier L., Le Pen G., Prochiantz A. Progressive loss of dopaminergic neurons in the ventral midbrain of adult mice heterozygote for Engrailed1. J. Neurosci. 2007;27:1063–1071. doi: 10.1523/JNEUROSCI.4583-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alvarez-Fischer D., Fuchs J., Prochiantz A. Engrailed protects mouse midbrain dopaminergic neurons against mitochondrial complex I insults. Nat. Neurosci. 2011;14:1260–1266. doi: 10.1038/nn.2916. [DOI] [PubMed] [Google Scholar]
- 30.Delaglio F., Grzesiek S., Bax A. NMRPIPE: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 31.Bartels C., Xia T.H., Wüthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 32.Goddard, T. D., and D. G.Kneller. SPARKY 3, University of California, San Francisco, CA.
- 33.Clarke N.D., Kissinger C.R., Pabo C.O. Structural studies of the engrailed homeodomain. Protein Sci. 1994;3:1779–1787. doi: 10.1002/pro.5560031018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Religa T.L. Comparison of multiple crystal structures with NMR data for engrailed homeodomain. J. Biomol. NMR. 2008;40:189–202. doi: 10.1007/s10858-008-9223-9. [DOI] [PubMed] [Google Scholar]
- 35.Wishart D.S., Bigam C.G., Sykes B.D. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 36.Chakrabartty A., Kortemme T., Baldwin R.L. Aromatic side-chain contribution to far-ultraviolet circular dichroism of helical peptides and its effect on measurement of helix propensities. Biochemistry. 1993;32:5560–5565. doi: 10.1021/bi00072a010. [DOI] [PubMed] [Google Scholar]
- 37.Banerjee R., Basu G. Direct evidence for alteration of unfolding profile of a helical peptide by far-ultraviolet circular dichroism aromatic side-chain contribution. FEBS Lett. 2002;523:152–156. doi: 10.1016/s0014-5793(02)02974-5. [DOI] [PubMed] [Google Scholar]
- 38.Christiaens B., Symoens S., Vanloo B. Tryptophan fluorescence study of the interaction of penetratin peptides with model membranes. Eur. J. Biochem. 2002;269:2918–2926. doi: 10.1046/j.1432-1033.2002.02963.x. [DOI] [PubMed] [Google Scholar]
- 39.Bertoncini C.W., Jung Y.-S., Zweckstetter M. Release of long-range tertiary interactions potentiates aggregation of natively unstructured α-synuclein. Proc. Natl. Acad. Sci. USA. 2005;102:1430–1435. doi: 10.1073/pnas.0407146102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Respondek M., Madl T., Zangger K. Mapping the orientation of helices in micelle-bound peptides by paramagnetic relaxation waves. J. Am. Chem. Soc. 2007;129:5228–5234. doi: 10.1021/ja069004f. [DOI] [PubMed] [Google Scholar]
- 41.Walrant A., Correia I., Alves I.D. Different membrane behavior and cellular uptake of three basic arginine-rich peptides. Biochim. Biophys. Acta. 2011;1808:382–393. doi: 10.1016/j.bbamem.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 42.Religa T.L., Markson J.S., Fersht A.R. Solution structure of a protein denatured state and folding intermediate. Nature. 2005;437:1053–1056. doi: 10.1038/nature04054. [DOI] [PubMed] [Google Scholar]
- 43.Religa T.L., Johnson C.M., Fersht A.R. The helix-turn-helix motif as an ultrafast independently folding domain: the pathway of folding of Engrailed homeodomain. Proc. Natl. Acad. Sci. USA. 2007;104:9272–9277. doi: 10.1073/pnas.0703434104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Banachewicz W., Religa T.L., Fersht A.R. Malleability of folding intermediates in the homeodomain superfamily. Proc. Natl. Acad. Sci. USA. 2011;108:5596–5601. doi: 10.1073/pnas.1101752108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bellet-Amalric E., Blaudez D., Renault A. Interaction of the third helix of Antennapedia homeodomain and a phospholipid monolayer, studied by ellipsometry and PM-IRRAS at the air-water interface. Biochim. Biophys. Acta. 2000;1467:131–143. doi: 10.1016/s0005-2736(00)00218-2. [DOI] [PubMed] [Google Scholar]
- 46.Brattwall C.E.B., Lincoln P., Nordén B. Orientation and conformation of cell-penetrating peptide penetratin in phospholipid vesicle membranes determined by polarized-light spectroscopy. J. Am. Chem. Soc. 2003;125:14214–14215. doi: 10.1021/ja0366989. [DOI] [PubMed] [Google Scholar]
- 47.Salamon Z., Lindblom G., Tollin G. Plasmon-waveguide resonance and impedance spectroscopy studies of the interaction between penetratin and supported lipid bilayer membranes. Biophys. J. 2003;84:1796–1807. doi: 10.1016/S0006-3495(03)74987-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caesar C.E.B., Esbjörner E.K., Nordén B. Membrane interactions of cell-penetrating peptides probed by tryptophan fluorescence and dichroism techniques: correlations of structure to cellular uptake. Biochemistry. 2006;45:7682–7692. doi: 10.1021/bi052095t. [DOI] [PubMed] [Google Scholar]
- 49.Lensink M.F., Christiaens B., Rosseneu M. Penetratin-membrane association: W48/R52/W56 shield the peptide from the aqueous phase. Biophys. J. 2005;88:939–952. doi: 10.1529/biophysj.104.052787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ulmer T.S., Bax A., Nussbaum R.L. Structure and dynamics of micelle-bound human α-synuclein. J. Biol. Chem. 2005;280:9595–9603. doi: 10.1074/jbc.M411805200. [DOI] [PubMed] [Google Scholar]
- 51.Jao C.C., Hegde B.G., Langen R. Structure of membrane-bound α-synuclein from site-directed spin labeling and computational refinement. Proc. Natl. Acad. Sci. USA. 2008;105:19666–19671. doi: 10.1073/pnas.0807826105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lokappa S.B., Ulmer T.S. Alpha-synuclein populates both elongated and broken helix states on small unilamellar vesicles. J. Biol. Chem. 2011;286:21450–21457. doi: 10.1074/jbc.M111.224055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marley J., Lu M., Bracken C. A method for efficient isotopic labeling of recombinant proteins. J. Biomol. NMR. 2001;20:71–75. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- 54.Augustyniak R., Balayssac S., Lequin O. 1H, 13C and 15N resonance assignment of a 114-residue fragment of Engrailed 2 homeoprotein, a partially disordered protein. Biomol. NMR Assign. 2011;5:229–231. doi: 10.1007/s12104-011-9306-5. [DOI] [PubMed] [Google Scholar]
- 55.Rouser G., Fkeischer S., Yamamoto A. Two dimensional then layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots. Lipids. 1970;5:494–496. doi: 10.1007/BF02531316. [DOI] [PubMed] [Google Scholar]
- 56.Raffard G., Steinbruckner S., Dufourc E.J. Temperature-composition diagram of dimyristoylphosphatidylcholine-dicaproyl phosphatidylcholine “bicelles” self-orienting in the magnetic field. A solid state 2H and 31P NMR study. Langmuir. 2000;16:7655–7662. [Google Scholar]
- 57.Shen Y., Delaglio F., Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Güntert P., Mumenthaler C., Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 59.Schwieters C.D., Kuszewski J.J., Clore G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 60.Bhattacharya A., Tejero R., Montelione G.T. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- 61.Kissinger C.R., Liu B.S., Pabo C.O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 Å resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990;63:579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- 62.Stollar E.J., Mayor U., Luisi B.F. Crystal structures of engrailed homeodomain mutants: implications for stability and dynamics. J. Biol. Chem. 2003;278:43699–43708. doi: 10.1074/jbc.M308029200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


