SUMMARY
Smad7 functions as an endogenous negative regulator of TGF-β/Smad signaling. TGF-β/Smad pathway is a major regulator of collagen production in connective tissue. Reduced expression of Smad7 has been reported in TGF-β-mediated fibrotic diseases, characterized by over production of collagen. Solar ultraviolet (UV) irradiation reduces collagen production by fibroblasts in human skin in vivo. We have investigated regulation of Smad7 gene expression by UV irradiation in human skin fibroblasts. UV irradiation transiently increased Smad7 mRNA and protein levels. Induction of Smad7 mRNA and protein was maximal within five hours, and returned to initial basal levels 24 hours post UV. UV irradiation induced Smad7 promoter reporter activity 3-fold. Smad7 promoter contains functional enhancer sequences that bind transcription factors Smad3 and activator protein-1 (AP-1). UV irradiation reduced protein binding to the Smad3 enhancer, and increased binding to the AP-1 enhancer. Deletion of AP-1 binding site in the Smad7 promoter completely abolished UV stimulation of Smad7 transcription. Deletion of Smad3 element had no effect on UV-induced promoter activity. UV irradiation increased mRNA and protein expression of AP-1 family members, c-Jun and c-Fos, which bound to the AP-1 element in the Smad7 promoter. Furthermore, over-expression of dominant negative c-Jun substantially reduced UV induction of Smad7 transcription. These data demonstrate that induction of Smad7 gene expression by UV irradiation is mediated via induction of transcription factor AP-1 in human skin fibroblasts.
Keywords: Smad7, Ultraviolet radiation, AP-1
INTRODUCTION
Transforming growth factor β (TGF-β)1 regulates a wide variety of cellular functions, including proliferation, differentiation, adhesion, migration, extracellular matrix production, and extracellular matrix degradation (1-3). TGF-β initiate their effects on cellular functions by binding to the cell surface TGF-β receptor complex. Receptor binding triggers phosphorylation and activation of Smad2 and Smad3, which regulates expression of TGF-β target genes (4-6). Activation of Smad2/3 is antagonized by the endogenous negative regulator, Smad7 (7). Expression of Smad7 is inducible by TGF-β itself, and this induction plays a critical role in negative feedback mechanisms that regulate TGF-β/Smad-mediated biological responses (7-10).
Aberrant expression of Smad7 disrupts the delicate balance inherent in TGF-β/Smad signaling, and thereby alters TGF-ß-dependent cellular responses (2,11-13). For example, the level of Smad7 expression has been reported to be reduced in the fibrotic disease scleroderma (14). This reduction could permit unrestrained activation of the TGF-β/Smad pathway, resulting in excessive production and deposition of collagen, which is observed in this disease (14). Conversely, Smad7 has been reported to be over-expressed in inflammatory bowel disease (15). This over-expression could down regulate TGF-β-mediated immunosuppression, resulting in hyper-secretion of proinflammatory cytokines, which is observed in this disease.
We have previously reported that ultraviolet (UV) irradiation induces Smad7 in human skin in vivo, and that this induction contributes to impairment of TGF-β/Smad signaling (16,17). We observed that UV irradiation induced Smad7 in both the outer compartment (epidermis) and inner compartment (dermis) of human skin (16). In the outer compartment of skin, TGF-β is a powerful negative regulator of cell (keratinocyte) proliferation (13,18-21). Therefore, induction of Smad7 contributes to keratinocyte hyperplasia, which occurs in response to UV irradiation (16,22,23). Increased keratinocyte growth is a characteristic of UV-induced skin cancer. In the inner compartment of skin, TGF-β is a major regulator of extracellular matrix production and deposition by fibroblasts (2,3,24,25). In dermal fibroblasts, impairment of the TGF-β/Smad pathway causes reduced production of type I procollagen, thereby leading to loss of collagen (26). Reduced collagen is a key factor in premature skin aging (photoaging), which is caused by UV irradiation (27). Therefore, induction of Smad7, with concomitant reduction of TGF-β responsiveness may be a key mediator of UV-induced skin cancer and photoaging. Accordingly, understanding mechanisms by which UV irradiation induces Smad7 gene expression is vital to our knowledge of the pathophysiology of these common diseases.
The 5′-flanking region of the Smad7 promoter contains binding sequences for transcription factors Smad3, AP-1, and Sp-1 (28). These regulatory elements and flanking sequences of the Smad7 gene promoter are highly conserved in human, rat and murine genes (29). Smad3, AP-1 and Sp-1 binding sequences are necessary for optimal induction of Smad7 transcription by TGF-β (28,30,31). Disruption of the Smad3 element abolishes TGF-β inducibility, but does not greatly impair basal expression. Disruption of AP-1 and Sp-1 binding elements substantially impairs basal and TGF-β-stimulated Smad7 expression.
Exposure of human skin in vivo to UV irradiation activates multiple cell surface cytokine and growth factor receptors, MAP kinase pathways, and rapidly induces transcription factor AP-1 activity (23,27,32,33). Based on these findings, we hypothesized that AP-1 plays a major role in UV induction of Smad7 expression. To test this hypothesis, we have investigated the mechanism of UV induction of Smad7. Our data demonstrate that AP-1 family members c-Jun and c-Fos, mediate UV activation of Smad7 transcription, in human skin dermal fibroblasts.
EXPERIMENTAL PROCEDURES
Materials
Dulbecco’s Modified Eagle’s Media (DMEM), fetal bovine serum, trypsin solution, penicillin/streptomycin, and L-glutamine were purchased from Invitrogen-Life Technology (Rockville, MD). [γ-32P]ATP and [α-32P]dCTP were obtained from Perkin Elmer (Boston, MA). c-Jun and c-Fos antibodies were purchased from Transduction Laboratories (San Diego, CA) and Smad3 was obtained from Zymed Laboratories (South San Francisco, CA). Smad7 primary antibody was generously provided by Dr. Peter ten Dijke. Smad7 promoter reporter constructs were kindly provided by Rainer Heuchel. All other reagents were purchased from Sigma Chemical Company (St. Louis, MO).
Cell culture and UV irradiation
Human skin primary fibroblasts were prepared from keratome biopsy of healthy adult normal human skin as described previously (34). The cells used for this study were between passages 3 and 6. UV irradiation was performed as described previously (17,35). Briefly, sub-confluent cells were irradiated with UV (50 mJ/cm2) using an Ultralite Panelite lamp containing six FS24T12 UVB-HO bulbs (Daavlin, Byran, OH). A Kodacel filter was used in order to eliminate wavelengths below 290 nm (UVC). The irradiation intensity was monitored with an IL400A phototherapy radiometer and a SED240/UVB/W photodetector (International Light, Newbury, MA).
RNA isolation and Northern blot analysis
Total RNA from cultured human skin fibroblasts were prepared using a commercial kit (RNeasy Midi kit, Qiagen, Chatsworth, CA) according to the manufacturer’s protocol. Northern blot was performed as described previously (17). Briefly, total RNA (20 μg) was resolved by 1.2% agarose electrophoresis, transferred to nylon membranes, and hybridized with Smad7 cDNA probes (17) labeled with [32P]dCTP by random priming. Each blot was stripped and re-hybridized with 36B4 internal control gene transcript to monitor the sample load in each lane. The intensities of each band were quantified by STORM PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Quantitative real-time RT-PCR
Quantitative real-time RT-PCR was performed as previously described (35). PCR primers and probes were from Applied Biosystems custom oligonucleotide synthesis service. c-Jun and c-Fos primers and FAM labeled probes for real-time RT-PCR were as follows: c-Jun; sense primer, 5′-CCTCAACGCCTCGTTCCT-3′; anti-sense primer, 5′-TCAGGGTCATGCTCTGTTTCAG-3′; Probe, 5′-CCGAGAGCGGACCTTATGGCTACAGTAACCCC-3′; c-Fos sense primer, 5′-GCCGAGCGCAGAGCATT-3′; anti-sense primer, 5′-CCCTTCGGATTCTCCTTTTCTC-3′; Probe, 5′-TCTTCTGGAGATAACTGTTCCACCTTGCC-3′. Primers and VIC-labeled probe for 36B4 were described previously (35). Multiplex PCR reactions contained primers and probes for target gene and 36B4. Target gene and 36B4 mRNA levels were quantified based on standard curves, and gene levels were normalized to the housekeeping gene 36B4 (a ribosomal protein used as an internal control for quantitation).
Western analysis
Western blot was performed as described previously (17). Briefly, proteins from whole cell extract were resolved on 12% SDS-PAGE, transferred to PVDF membrane, and blocked with PBST (0.1% Tween 20 in PBS) containing 5% milk. Primary antibodies were diluted in the PBST solution (1:1000) and incubated with PVDF membrane for two hours at room temperature. Blots were washed three times with PBST solution and incubated with appropriate secondary antibody for one hour at room temperature. After washing three times with PBST, the blots were developed with ECF (Vistra ECF Western Blotting System, GE Healthcare, Piscataway, NJ) following the manufacturer’s protocol. The intensities of each band were quantified by STORM PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Immunohistology
Immunohistology was performed as described previously (17). Cells (1×104) were plated on eight-well chamber slides and exposed to UV as described above. Cells were washed once with PBS, and fixed in 2% paraformaldehyde for two hours at room temperature. The slides were pre-incubated with normal rabbit serum for one hour. Subsequently, the slides were incubated for one hour at room temperature with rabbit polyclonal antibody against Smad7. The slides were then incubated with a biotinylated anti-rabbit antibody for 30 minutes. The slides were then incubated with avidin biotin peroxidase complex for 30 minutes. 3-Amino-9-ethyl carbazole (Sigma) was used as chromogen. Between steps, the slides were rinsed for 10 minutes in Tris-buffered saline with 0.1% Triton-X-100. All sections were lightly counterstained with haematoxylin.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed as described previously (17), using nuclear extracts from the human skin fibroblasts. Oligonucleotides for EMSA probes, designed from Smad7 promoter, were as follows: Smad binding element (SBE) probe, 5′-TTTTAAAGCGACAGGGTGTCTAGACGGCCACG-3′; AP-1 binding element (AP-1E) probe, 5′-GGCCACGTGACGAGGCCGGAGCCGG-3′; SBE/AP-1E probe, 5′-TTTTAAAGCGACAGGGTGTCTAGACGGCCACGTGACGAGGCCG-GAGCCGG-3′ were synthesized by Operon Technologies, Inc. (Alameda, CA). AP-1 consensus (5′-CGCTTGATGACTCAGCCGGAA-3′) and mutant (5′-CGCTTGATGACTTGGCCGGAA-3′) probes were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). For competition experiments, a 100-fold molar excess of cold competitors was pre-incubated with nuclear extract for 30 minutes on ice before labeled probes were added. The gel was transferred to 3M Whatman paper, vacuum-dried and quantified by STORM PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Electrophoretic mobility shift assay coupled with Western blot
The major shifted bands were excised from EMSA gel and then proteins were extracted from the gel by incubation overnight at 37°C, in buffer (1× PBS, 10mM Tris-HCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.5% SDS, 2 μg/ml leupeptin, and 100 μg/ml pheylmethylsulfonyl fluoride). Gel debris was removed by centrifugation and the supernatant was concentrated by Centricon-10 (Amicon, Inc., Beverly, MA). The samples were then heated for five minutes at 95°C and analyzed by Western blot.
Transient transfection of Smad7 promoter reporter construct and luciferase assays
Cells were transfected with wild-type Smad7 promoter/luciferase construct (−613 to +112) or deletion constructs in the binding sites of Smad3 and AP-1 (28). Cells were co-transfected with β-galactosidase expression vector to provide an internal standard for transfection efficiency. In some studies, dominant-negative mutant c-Jun (TAM-67) (36) expression vector was transfected. All plasmids were transfected into dermal fibroblasts using Fugene 6 (Roche Molecular Biochemicals, Indianapolis, IN) or human dermal fibroblasts nucleofector kit (Amaxa Biosystems, Koln, Germany) according to the manufacturer’s protocols. After 24 hours of transfection, cells were irradiated with UV and harvested 24 hours post UV irradiation. Aliquots containing identical β-galactosidase activity were used for each luciferase assay. Luciferase activity was measured using an enhanced luciferase assay kit (BD Biosciences-PharMingen International, San Diego, CA) according to the manufacturer’s protocol.
Statistical analysis
Comparisons among treatment groups were made with the paired t-test (two groups) or the repeated measures of ANOVA (more than two groups). Multiple pair-wise comparisons were made with the Tukey Studentized Range test. All p values are two-tailed, and considered significant when <0.05.
RESULTS
UV irradiation increases Smad7 mRNA and protein in human skin fibroblasts
We first determined the kinetics of UV induction of Smad7 mRNA and protein in human skin fibroblasts. UV irradiation transiently increased the levels of Smad7 mRNA (Fig. 1A) and protein (Fig. 1B). Induction of Smad7 mRNA was maximal five hours post UV irradiation, and returned to initial basal level by 24 hours post UV irradiation. Induction of Smad7 protein was maximal between five and eight hours following UV irradiation, and returned to basal level 24 hours post irradiation. Immunohistology revealed that Smad7 protein was predominantly located in the cytoplasm of untreated cells, and Smad7 protein increased in the cytoplasm five to eight hours post UV (Fig.1C). These data indicate that UV irradiation increases the levels of Smad7 mRNA and protein in human skin fibroblasts.
Figure 1. UV irradiation increases Smad7 mRNA and protein in human skin fibroblasts.
A, UV irradiation induces Smad7 mRNA. Total RNA was prepared from cells at the indicated times post UV exposure (50 mJ/cm2). Smad7 and 36B4 (a ribosomal RNA used as an internal control for quantitation) mRNA was analyzed by Northern analysis. Inset shows representative Northern blot. Bars indicate means ± SEM for fold change in Smad7 mRNA levels following UV irradiation relative to levels in non-irradiated control cells (CTRL). *p<0.05 vs. non-irradiated control cells. N=3. B, UV irradiation induces Smad7 protein. Whole cell extracts were prepared from cells at the indicated times post UV exposure (50 mJ/cm2). Smad7 protein was quantified by Western analysis. Inset shows representative Western blot. Bars indicate means ± SEM for fold change in Smad7 protein following UV irradiation relative to levels in non-irradiated control cells (CTRL). *p<0.05 vs. non-irradiated control cells. N=3. C, Cellular localization of Smad7 protein following UV irradiation. Cellular localization of Smad7 protein was determined by immunohistochemical staining at the indicated times post UV (50 mJ/cm2) as described in Methods. Results are representative of three experiments.
UV irradiation induces AP-1-dependent Smad7 promoter activity in human skin fibroblasts
We next examined UV irradiation stimulation of Smad7 promoter activity. Smad7 promoter/luciferase construct (-613 to +112), containing Smad3, AP-1 and Sp-1 binding elements, was transfected into skin fibroblasts, and reporter activity was determined following sham or UV irradiation. UV irradiation stimulated promoter activity three-fold (Fig 2). Deletion of the Smad3 element reduced basal and UV-stimulated reporter activity proportionally, such that net UV induction was not altered. Deletion of the AP-1 binding element also reduced basal reporter activity. However, in contrast to deletion of the Smad3 element, deletion of the AP-1 element abolished UV induction of reporter activity. These data indicate that UV irradiation stimulates Smad7 promoter activity, and that this stimulation is dependent on the presence of the AP-1 binding site, in human skin fibroblasts.
Figure 2. Deletion of AP-1 binding site abolishes UV stimulation of Smad7 promoter activity.
Human skin fibroblasts were transiently transfected with wild-type Smad7 promoter/luciferase construct (-613 to +112) containing Smad3 element (SBE) and AP-1 element (AP-1E), or constructs with SBE or AP-1E deleted (depicted in the figure). Cells were co-transfected with β-galactosidase expression vector. Cells were irradiated with UV (50 mJ/cm2) 24 hours after transfection, and cell lysates were prepared 24 hours post UV. Smad7 promoter/luciferase activities were normalized to β-galactosidase activity. Bars indicate means ± SEM fold change in Smad7 promoter activity relative to non-irradiated, wild-type Smad7 control cells. *p<0.05 vs. non-irradiated, wild-type Smad7 control cells. N=3.
UV irradiation increases protein binding to the AP-1, but not Smad3 element in the Smad7 promoter
We next examined protein binding to the Smad3 and AP-1 elements in non-irradiated and UV irradiated skin fibroblasts. Electrophoretic mobility shift assays (EMSA) were performed using three probes with sequences identical to the Smad7 promoter. The SBE/AP-1E probe contained both Smad3 and AP-1 elements, the SBE probe contained only the Smad3 element, and the AP-1E probe contained only the AP-1 binding element (Fig. 3A). Nuclear proteins from non-irradiated fibroblasts formed two major specific retarded complexes with the SBE/AP-1E probe (Fig 3B). UV irradiation substantially increased the intensity of the upper retarded DNA/protein complex, and reduced the intensity of the lower complex (Fig. 3B). The upper retarded DNA/protein complex was abolished by excess unlabeled consensus AP-1, but not consensus SBE probe. In contrast, the lower complex was completely abolished by excess unlabeled consensus SBE, but not consensus AP-1 probe. These data indicate that the upper complex contains AP-1 family protein but not Smad3 protein, and the lower complex contains Smad3 protein, but not AP-1 family protein. Nuclear proteins from non-irradiated cells formed a single retarded complex with the SBE probe (Fig 3C). This single retarded DNA/protein complex migrated to the position of the lower band observed with the SEB/AP-1E probe (Fig. 3B and 3C). UV irradiation reduced the intensity of the SBE complex, similar to what was observed with the lower complex formed with the SBE/AP-1E probe (Fig. 3B and 3C). The AP-1E probe formed a single specific retarded complex with nuclear proteins from non-irradiated fibroblasts (Fig. 3D). The migration of this complex was similar to the upper retarded band formed with the SBE/AP-1E probe (Fig. 3B and 3D). UV irradiation increased the intensity of the AP-1 complex, as was observed with the upper retarded band formed with the SBE/AP-1E probe (Fig. 3B).
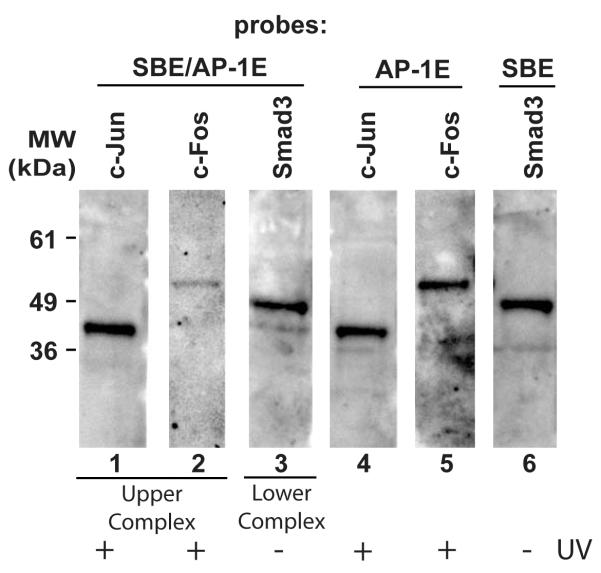
Figure 3. UV irradiation increases protein binding to the AP-1, but not Smad3 element of the Smad7 promoter.
A, Nucleotide sequence of the Smad7 proximal promoter. Smad3 (SBE) and AP-1 (AP-1E) binding sites are marked in capital letters. Heavy lines under the sequence denote EMSA probes containing SBE/AP-1E, SBE, and AP-1E. Arrow indicates transcription start site. B-E, Nuclear extracts were prepared at the indicated times post UV (50 mJ/cm2) and DNA/protein complex formation was analyzed by EMSA. Closed triangles indicate specific retarded complexes. Open triangle indicates non-specific bands. Results are representative of three experiments. B, UV irradiation alters DNA/protein complex formation with the SBE/AP-1E probe. C, UV irradiation reduces DNA/protein complex formation with the SBE probe. D, UV irradiation increases DNA/protein complex formation with the AP-1E probe. E, Competition of protein binding to the AP-1E probe by 100-fold excess unlabeled wild-type (AP-1) or mutant (Mut) AP-1 binding element. Nuclear extracts were prepared four hours post UV irradiation.
In order to confirm the specificity of the UV-induced retarded complexes formed with the AP-1E probe, we performed competition EMSA. DNA/protein complex formation with the labeled AP-1E probe was completely abolished by excess unlabeled consensus AP-1, but not by mutant AP-1 probe (Fig. 3E).
To identify proteins bound to the SBE/AP-1E, SBE, and AP-1E probes, we performed super-shifts using antibodies to Smad and AP-1 family members. However, these experiments did not yield reproducible results. Therefore, we performed EMSA coupled with Western blot for the identification proteins bound to the probes. The upper retarded complex formed by UV-irradiated cells with the SBE/AP-1E probe, the lower retarded complex formed by non-irradiated cells with the SBE/AP-1E probe, the complex formed by non-irradiated cells with the SBE probe, the complex formed by UV-irradiated cells with the AP-1E probe, and equivalent gel areas from lanes without probe (used for control), were excised and subjected to SDS-PAGE immunoblotting with antibodies to c-Jun, c-Fos and Smad3 (Fig. 4). C-Jun and c-Fos were readily detected in the upper retarded complex (Fig 4, lanes 1 and 2) and Smad3 was detected in the lower retarded complex (Fig 4, lane 3) formed with the SBE/AP-1E probe. The AP-1E probe bound c-Jun and c-Fos (Fig. 4, Lanes 4 and 5), but not Smad3 (data not shown), while the SBE probe bound Smad3 (Fig. 4, Lane 6), but not c-Jun or c-Fos (data not shown). No immunoreactivities were detected in extracts from gel control lanes (data not shown). These data demonstrate that UV irradiation increases binding of c-Jun and c-Fos, and reduces binding of Smad3 to the Smad7 promoter.
Figure 4. Identification of Smad7 proximal promoter DNA binding proteins by EMSA coupled with Western analysis.
Specific upper and lower shifted complexes formed by UV-irradiated or non-irradiated with SBE/AP-1E, SBE, and AP-1E probes from the proximal Smad7 promoter (as described in the legend to Figure 3) were excised and subjected to Western analysis with antibodies to c-Jun, c-Fos, and Smad3. Lanes 1 and 2, c-Jun and c-Fos proteins were detected in the upper complex formed by UV-irradiated (2 hours) with the SBE/AP-1E probe. Lane 3, Smad3 protein was detected in the lower complex formed by non-irradiated cells with the SBE/AP-1E probe. Lanes 4 and 5, c-Jun and c-Fos proteins were detected in the complex formed by UV-irradiated (4 hours) cells with the AP-1E probe. Lane 6, Smad3 protein was detected in the complex formed by non-irradiated cells with the SBE probe. Results are representative of three independent experiments.
Over-expression of dominant negative c-Jun abolished UV activation of Smad7 promoter
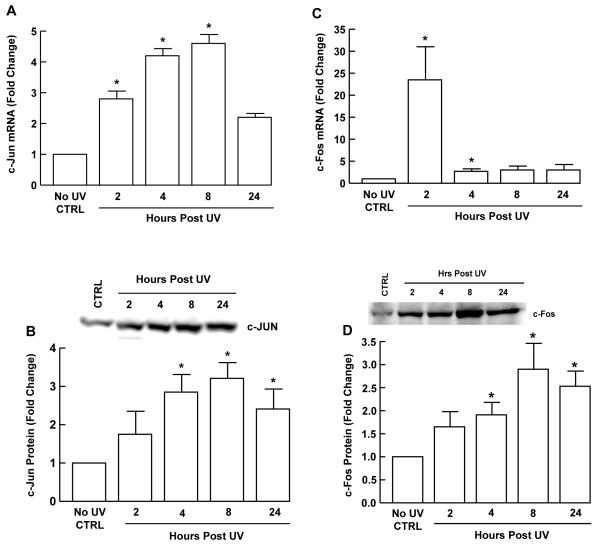
The above data indicate that AP-1 plays a major role in UV induction of Smad7 gene expression. To further substantiate this conclusion, UV induction of c-Jun and c-Fos mRNA and protein levels were determined by quantitative real-time RT-PCR and Western analysis, respectively. UV irradiation induced c-Jun and c-Fos mRNA (Fig. 5A, 5C) within two hours, and maximum induction of c-Jun and c-Fos proteins (Fig. 5B, 5D) was observed at eight hours post UV in human skin fibroblasts.
Figure 5. UV irradiation induces c-Jun and c-Fos in human skin fibroblasts.
Cells were UV-irradiated (50 mJ/cm2) and total RNA, or whole cell protein extracts were prepared at the indicated times post irradiation. A, c-Jun and C, c-Fos mRNA levels were quantified by real-time RT-PCR, normalized to mRNA for 36B4, which encodes a ribosomal protein, used as an internal control. B, c-Jun and D, c-Fos protein levels were determined by Western analysis. Insets show representative Western blots. Bar heights indicate means ± SEM fold change relative to levels in non-irradiated control cells (CTRL). *p<0.05 vs. CTRL. N=3-5.
Finally, we utilized dominant mutant c-Jun to examine AP-1 function. As shown in Figure 6, over-expression of dominant negative c-Jun reduced basal Smad7 promoter activity approximately 50%. Importantly, UV induction of Smad7 promoter activity was essentially abolished by over-expression of dominant negative c-Jun. Taken together, the above data demonstrate that UV induction of AP-1 functions to induce Smad7 gene expression in human skin fibroblasts.
Figure 6. Over-expression of dominant negative c-Jun abolished UV induction of Smad7 promoter activity in human skin fibroblasts.

Cells were co-transfected with Smad7 promoter/luciferase construct (-613 to +112) and β-galsctosidase expression vector, and empty or dominant negative c-Jun expression vector. Cells were UV irradiated (50mJ/cm2) 24 hours after transfection. Cell lysates were prepared 24 hours post UV irradiation, and promoter activity was determined by luciferase assay, and normalized to β-galactosidase activity. Bars indicate means ± SEM for fold change in Smad7 promoter activity relative to activity in non-irradiated control cells (CTRL). *p<0.05 vs. UV-irradiated cells. N=3.
DISCUSSION
Smad7 functions as an endogenous negative feedback regulator of TGF-β/Smad signaling to modulate the intensity and duration of TGF-β responses (7,8). The molecular mechanisms underlying the antagonistic effect of Smad7 on TGF-β/Smad pathway is relatively well characterized (1). Smad7 stably interacts with the activated TβRI to prevent association and phosphorylation of Smad2 and Smad3, and thus inhibits Smad2 and Smad3 nuclear translocation (7). Furthermore, Smad7 inhibits TGF-β responsiveness by promoting degradation of TβRI through recruitment of an E3 ubiquitin ligase to the TGF-β receptor (37-40). In addition to its inhibitory effects at the TGF-β receptor, Smad7 can function as transcription repressor in the nucleus, by recruiting histone deacetylases to TGF-β/Smad target genes (41).
We have previously reported that exposure of human skin in vivo to solar simulated UV irradiation induces Smad7 and impairs TGF-β/Smad signaling pathway (16,17). The data presented here demonstrate that UV irradiation stimulates Smad7 promoter activity, and that this activation is mediated by transcription factor AP-1, which is induced by UV irradiation, in human skin fibroblasts. The proximal promoter of the Smad7 gene contains binding elements for Smad3, AP-1 and Sp-1 (28). Smad3 binding element has been shown to be required for induction of Smad7 by TGF-β (30,31), while AP-1 and Sp-1 binding elements cooperate to enhance Smad3-dependent Smad7 gene expression (28).
Here we provide several lines of experimental evidence that support the conclusion that AP-1, but not Smad3, plays a critical role in UV activation of Smad7 promoter activity (Figures 2, 3, 6). Interestingly, we found that UV irradiation reduced Smad3 binding to its element in the Smad7 promoter. UV inhibition of Smad3 activation may be mediated by several mechanisms. Firstly, we have reported that UV irradiation down-regulates TβRII, which results in reduced phosphorylation and nuclear translocation of Smad3, in response to TGF-β (26). Secondly, as shown here, UV irradiation induces Smad7, which in turn interacts with TβRI to prevent association and activation of Smad3. Thirdly, it has been shown that c-Jun, which is induced by UV, is able to physically interact with Smad3 and thereby suppress Smad3-driven gene transactivation (42-44).
UV irradiation activates multiple cell surface growth factor and cytokine receptors, which initiates intracellular signal transduction cascades (22,23,27,33). These activated signaling cascades activate target transcription factors, including AP-1. We show here that UV irradiation induces c-Jun and c-Fos in human skin fibroblasts. A primary function of human skin fibroblasts is to produce type I collagen (and other extracellular matrix molecules), which is the major structural protein in skin connective tissue. This production of collagen by skin fibroblasts is strongly stimulated by TGF-β (26,45). We have previously reported that UV irradiation reduces type I procollagen production in human skin in vivo, and human skin fibroblasts (46). This inhibition is mediated in part by UV induction of AP-1 (46), which has been shown to interfere with TGF-β-activated Smad3 stimulation of type I procollagen promoter activity (45,47).
The data presented here strongly suggest that AP-1 represses type I procollagen production by a second, distinct mechanism, which involves stimulation of Smad7 expression, which impairs TGF-β responsiveness. In addition to driving Smad7 expression, AP-1 drives induction of matrix metalloproteinases by UV irradiation (22,23,27,33). UV-induced matrix metalloproteinases degrade skin collagen. This degradation of skin collagen is a major cause of premature skin aging and promotes a tissue environment conducive to skin cancer (33,48,49). The data presented herein support a new role for AP-1 in mediating UV-induced skin aging and cancer. By inducing Smad7 expression, AP-1 can interfere with type I procollagen production, and thereby exacerbate UV-induced degradation and loss of type I collagen.
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Rainer Heuchel and Dr. Peter ten Dijke who kindly provided us with reagents. We are indebted to Suzan Rehbine for the procurement of tissue specimens, Kathy Bucknell for technical assistance, Laura VanGoor for the preparation of graphic material, Ted Hamilton for statistical analysis, and Diane Fiolek for administrative assistance. This work was supported in part by the Babcock Foundation and NIH Grant AG19364-02 (GJF).
Footnotes
The abbreviations used are: TGF-β, transforming growth factor-β; Smad, Sma-and Mad-related protein; UV, ultraviolet; TβRI, TGF-β receptor type I; TβRII, TGF-β receptor type II; SBE, Smad binding element; PCR, polymerase chain reaction; EMSA, electrophoretic mobility shift assay; AP-1, activator protein-1; AP-1E, AP-1 binding element; MAP, mitogen activated protein kinase pathways.
REFERENCES
- 1.Massague J, Wotton D. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varga J. Arthritis Rheumatism. 2002;46:1703–1713. doi: 10.1002/art.10413. [DOI] [PubMed] [Google Scholar]
- 3.Verrecchia F, Mauviel A. J Invest Dermatol. 2002;118:211–215. doi: 10.1046/j.1523-1747.2002.01641.x. [DOI] [PubMed] [Google Scholar]
- 4.Heldin C-H, Miyazono K, ten Dijke P. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 5.Shi Y, Massague J. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 6.ten Dijke P, Hill C. Trends Biochem Sci. 2004;29:265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi H, Abdollah S, Qiu Y, Cai J, Xu Y-Y, Grinnell B, Richardson M, Topper J, Grimbrone M, Jr, Wrana J, Falb D. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 8.Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH, ten Dijke P. Nature. 1997;389:631–637. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 9.Afrakhte M, Moren A, Jossan S, Itoh S, Sampath K, Westermark B, Heldin CH, Heldin NE, ten Dijke P. Biochem Biophys Res Commun. 1998;249:505–511. doi: 10.1006/bbrc.1998.9170. [DOI] [PubMed] [Google Scholar]
- 10.Itoh S, Landstrom M, Hermansson A, Itoh F, Heldin C-H, Heldin N-E, ten Dijke P. J Biol Chem. 1998;273:29195–29201. doi: 10.1074/jbc.273.44.29195. [DOI] [PubMed] [Google Scholar]
- 11.Nakao A, Okumura K, Ogawa H. Trends Mol Med. 2002;8:361–363. doi: 10.1016/s1471-4914(02)02376-6. [DOI] [PubMed] [Google Scholar]
- 12.Fiocchi C. J Clin Invest. 2001;108:523–526. doi: 10.1172/JCI13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He W, Li A, Wang D, Han S, Zheng B, Goumans M-J, ten Dijke P, Wang X-J. EMBO J. 2002;21:2580–2590. doi: 10.1093/emboj/21.11.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong C, Zhu S, Wang T, Yoon W, Alvarez R, ten Dijke P, White B, Wigley F, Goldschmidt-Clermont P. Proc Nat Acad Sci USA. 2002;99:3908–3913. doi: 10.1073/pnas.062010399. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Monteleone G, Kumberova A, Croft N, McKenzie C, Steer H, MacDonald T. J Clin Invest. 2001;108:601–609. doi: 10.1172/JCI12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quan T, He T, Kang S, Voorhees J, Fisher G. J Invest Dermatol. 2002;119:499–506. doi: 10.1046/j.1523-1747.2002.01834.x. [DOI] [PubMed] [Google Scholar]
- 17.Quan T, He T, Voorhees J, Fisher G. J Biol Chem. 2001;276:26349–26356. doi: 10.1074/jbc.M010835200. [DOI] [PubMed] [Google Scholar]
- 18.Pittelkow M, Coffey R, Moses H. Ann NY Acad Sci USA. 1988;548:211–224. doi: 10.1111/j.1749-6632.1988.tb18809.x. [DOI] [PubMed] [Google Scholar]
- 19.Sellheyer K, Bickenbach J, Rothnagel J, Bundman D, Langley M, Kriieg T, Roche N, Roberts A, Roop D. Proc Nat Acad Sci USA. 1993;90:5237–5241. doi: 10.1073/pnas.90.11.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang DH, Sun LZ, Zborowski E, Willson JKV, Gong J, Verraraghavan J, Brattain MG. J Biol Chem. 1999;274:12840–12847. doi: 10.1074/jbc.274.18.12840. [DOI] [PubMed] [Google Scholar]
- 21.Blobe G, Schiemann W, Lodish H. New Eng J Med. 2000;342:1350–1358. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- 22.Fisher G, Kang S, Varani J, Bata-Csorgo Z, Wan Y, Datta S, Voorhees J. Arch Dermatol. 2002;138:1462–1470. doi: 10.1001/archderm.138.11.1462. [DOI] [PubMed] [Google Scholar]
- 23.Fisher GJ, Wang ZQ, Datta SC, Varani J, Kang S, Voorhees JJ. New Eng J Med. 1997;337:1419–1428. doi: 10.1056/NEJM199711133372003. [DOI] [PubMed] [Google Scholar]
- 24.Roberts A, Russo A, Felici A, Flanders K. Ann N Y Acad Sci. 2003;995:1–10. doi: 10.1111/j.1749-6632.2003.tb03205.x. [DOI] [PubMed] [Google Scholar]
- 25.Lakos G, Takagawa S, Chen S-J, Ferreira A, Han G, KMasuda K, Wang X-J, DiPetro L, Varga J. Amer J Pathol. 2004;165:203–217. doi: 10.1016/s0002-9440(10)63289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quan T, He T, Kang S, Voorhees J, Fisher G. Amer J Pathol. 2004;165:741–751. doi: 10.1016/s0002-9440(10)63337-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisher GJ, Voorhees JJ. J Invest Dermatol. 1998;3:61–68. doi: 10.1046/j.1523-1747.1998.00112.x. [DOI] [PubMed] [Google Scholar]
- 28.Brodin G, Ahgren A, ten Dyke P, Heldin C-H, Heuchel R. J Biol Chem. 2000;275:29023–29030. doi: 10.1074/jbc.M002815200. [DOI] [PubMed] [Google Scholar]
- 29.Stopa M, Anhuf D, Terstegen L, CGatsios P, Gressner A, Dooley S. J Biol Chem. 2000;275:29308–29317. doi: 10.1074/jbc.M003282200. [DOI] [PubMed] [Google Scholar]
- 30.Nagarajan R, Zhang J, Li M, Chen Y. J Biol Chem. 1999;274:33412–33418. doi: 10.1074/jbc.274.47.33412. [DOI] [PubMed] [Google Scholar]
- 31.von Gersdorff G, Susztak K, Rezvani F, Bitzer M, Liang D, Bottinger E. J Biol Chem. 2000;275:11320–11326. doi: 10.1074/jbc.275.15.11320. [DOI] [PubMed] [Google Scholar]
- 32.Fisher G, Quan T, He T, Shao Y, Voorhees J. J Invest Dermatol. 2002;119:723. doi: 10.1046/j.1523-1747.2002.01834.x. [DOI] [PubMed] [Google Scholar]
- 33.Fisher GJ, Datta SC, Talwar HS, Wang ZQ, Varani J, Kang S, Voorhees JJ. Nature. 1996;379:335–339. doi: 10.1038/379335a0. [DOI] [PubMed] [Google Scholar]
- 34.Fisher GJ, Esmann J, Griffiths CEM, Voorhees JJ. J Invest Dermatol. 1991;96:699–707. doi: 10.1111/1523-1747.ep12470632. [DOI] [PubMed] [Google Scholar]
- 35.Quan T, He T, Kang S, Voorhees J, Fisher G. J Invest Dermatol. 2002;118:402–408. doi: 10.1046/j.0022-202x.2001.01678.x. [DOI] [PubMed] [Google Scholar]
- 36.Brown PH, Chen TK, Birrer MJ. Oncogene. 1994;9:791–799. [PubMed] [Google Scholar]
- 37.Kavsak P, Rasmussen R, Causing C, Booni S, Zhu H, Thomsen G, Wrana J. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 38.Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, Miyazono K. J Biol Chem. 2001;276:12477–12480. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki C, Murakami G, Fukuchi M, Shimanuki T, Shikaichi Y, Imamura T, Miyazono K. J Biol Chem. 2002;277:39919–39925. doi: 10.1074/jbc.M201901200. [DOI] [PubMed] [Google Scholar]
- 40.Tajima Y, Goto K, Yoshida M, Shinomiya K, Sekimoto T, Yoneda Y, Miyazono K, Imamura T. J Biol Chem. 2003;278:10716–10721. doi: 10.1074/jbc.M212663200. [DOI] [PubMed] [Google Scholar]
- 41.Bai S, Cao X. J Biol Chem. 2002;277:4176–4182. doi: 10.1074/jbc.M105105200. [DOI] [PubMed] [Google Scholar]
- 42.Verrecchia F, Pessah M, Atfi A, Mauviel A. J Biol Chem. 2000;275:30226–30231. doi: 10.1074/jbc.M005310200. [DOI] [PubMed] [Google Scholar]
- 43.Verrecchia F, Tacheau C, Schorpp-Kistnber M, Angel P, Mauviel A. Oncogene. 2001;20:2205–2211. doi: 10.1038/sj.onc.1204347. [DOI] [PubMed] [Google Scholar]
- 44.Verrecchia F, Vindevoghel L, Lechleider R, Uitto J, Roberts A, Mauviel A. Oncogene. 2001;20:3332–3340. doi: 10.1038/sj.onc.1204448. [DOI] [PubMed] [Google Scholar]
- 45.Chen SJ, Yuan WH, Mori Y, Levenson A, Trojanowska M, Varga J. J Invest Dermatol. 1999;112:49–57. doi: 10.1046/j.1523-1747.1999.00477.x. [DOI] [PubMed] [Google Scholar]
- 46.Fisher G, Datta S, Wang Z, Li X, Quan T, Chung J, Kang S, Voorhees J. J Clin Invest. 2000;106:661–668. doi: 10.1172/JCI9362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen S-J, Yuan W, Lo S, Trojanowska M, Varga J. J Cell Physiology. 2000;183:381–392. doi: 10.1002/(SICI)1097-4652(200006)183:3<381::AID-JCP11>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 48.Talwar HS, Griffiths CEM, Fisher GJ, Hamilton TA, Voorhees JJ. J Invest Dermatol. 1995;105:285–290. doi: 10.1111/1523-1747.ep12318471. [DOI] [PubMed] [Google Scholar]
- 49.Griffiths CEM, Russman AN, Majmudar G, Singer RS, Hamilton TA, Voorhees JJ. NEJM. 1993;329:530–535. doi: 10.1056/NEJM199308193290803. [DOI] [PubMed] [Google Scholar]