Abstract
Single-molecule studies of enzymes open a window into their dynamics and kinetics. A single molecule of the catalytic domain of cAMP-dependent protein kinase (PKA) was attached to a single-walled carbon nanotube device for long duration monitoring. The electronic recording clearly resolves substrate binding, ATP binding, and cooperative formation of PKA’s catalytically functional, ternary complex. Using recordings of a single PKA molecule extending over 10 minutes and tens of thousands of binding events, we determine the full transition probability matrix and conversion rates governing formation of the apo, intermediate, and closed enzyme configurations. We also observe kinetic rates varying over two orders of magnitude from one second to another. Anti-correlation of the on- and off-rates for PKA binding to the peptide substrate, but not ATP, demonstrates that regulation of enzyme activity results from altering the stability of the PKA-substrate complex, not its binding to ATP. The results depict a highly dynamic enzyme offering dramatic possibilities for regulated activity, an attribute useful for an enzyme with crucial roles in cell signaling.
INTRODUCTION
The enzyme cAMP-dependent protein kinase A (PKA) regulates cell function by phosphorylating a wide-range of proteins involved in cell signaling, transcription, and metabolism.1–4 The phosphoryl transfer reaction catalyzed by PKA requires the enzyme to simultaneously bind adenosine-5’-triphosphate (ATP) and a phosphate acceptor peptide or protein. In the presence of Mg2+, this ternary complex transfers the gamma phosphate group of ATP to the hydroxyl group of a serine or threonine residue on the substrate.1,2,5,6 A key player in cell function and cell division, PKA also offers an archetypical protein for the study of kinase regulation and function. Furthermore, single molecule studies of PKA could address such outstanding issues as the basis for its regulated activity, the breadth of its dynamic range, and the full spectrum of kinetic parameters forming the basis of its activity.
Recently, we demonstrated a new electronic method for monitoring and deciphering enzymatic processing and kinetics with single molecule resolution.7–9 Single molecules of T4 lysozyme were non-covalently bioconjugated to single-walled carbon nanotube (SWNT) electronic devices. The motions by the lysozyme active site during chemical catalysis move charged functionalities close to the SWNT to convert conformational changes of the protein into an electronic signal.9 Analysis of long-duration records from single molecules established lysozyme as a processive enzyme, with multiple independent timescales governing its productive and non-productive motions.7
Here, we apply this electronic monitoring technique to examine the catalytic subunit of PKA. Electronic transduction clearly resolves each step in the formation of the active, ternary complex. The results allow independent measurements of multiple kinetic parameters involved in PKA catalysis.
EXPERIMENTAL SECTION
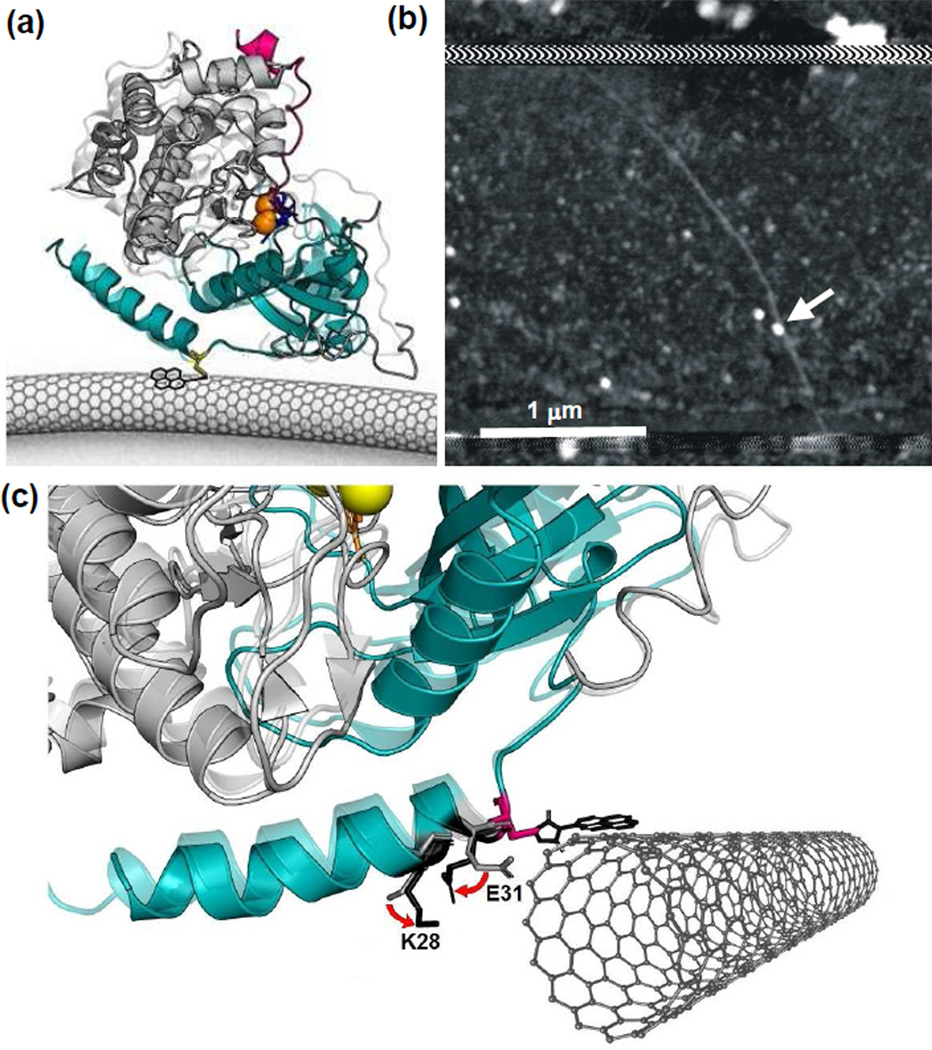
In order to examine PKA and its kinetics, individual molecules were attached to SWNT transistors using methods similar to past work.7–9 First, a surface-exposed, cysteine residue was introduced into a PKA variant by oligonucleotide-directed mutagenesis encoding a T32C substitution to provide a thiol functionality for site-specific attachment to SWNT devices; hereafter, this protein variant is termed PKA. After over-expression and purification, the enzyme was incubated with SWNT transistors that had been noncovalently functionalized with pyrene-maleimide linker molecules. The maleimide functionality of the linker formed stable thioether bonds with a PKA molecule. During functionalization, a solution of ATP and MgCl2 (2 mM each) was used to sterically block another, naturally-occurring cysteine sidechain in PKA.10 Figure 1a depicts the geometry of a SWNT device and its PKA attachment based on the position of the T32C site, and Figure 1b shows an atomic force microscopy image of an actual device after protein bioconjugation. Precise details of each step in the synthesis and bioconjugation processes are described more fully in the Supporting Information.
Figure 1.
PKA-labeled, SWNT-based nanocircuits for single molecule electronic monitoring (a) Schematic of a single core catalytic subunit of PKA (small lobe in cyan and large lobe in gray) attached to an SWNT-based circuit through a single cysteine (yellow). Two magnesium ions (orange) position ATP (blue) in the binding pocket. As PKA catalyzes the transfer of γ-phosphate from ATP to its substrate, Kemptide (magenta), the protein undergoes a conformational change from the open (opaque, PDB: 1J3H) to the closed (transparent, PDB: 1ATP) state. (b) Example atomic force micrograph of a SWNT device with a single protein attachment (arrow). Horizontal stripes at top and bottom show the extent of a passivating polymer that covers the distant source and drain electrode connections. (c) Schematic diagram of the PKA-SWNT interface, showing close-up X-ray crystal structures of PKA in its closed (opaque) and open (transparent) conformations (PDB: 1ATP and 1J3H, respectively). The C32 attachment to the pyrene-maleimide linker molecule provides a fixed reference point. In the vicinity of C32, residues 28 and 31 have charged side chains that move appreciably from the open configuration (light gray) to the closed conformation (black). The pyrene and SWNT are independently free to rotate around the C32 site, but this illustration features an energetically likely orientation.
After bioconjugation, devices were stored and then measured in a buffered aqueous solution (100 mM MOPs, 9 mM MgCl2, 100 µM TCEP, pH 7.2). Measurements were performed by applying a fixed, 100 mV source-drain bias along the SWNT conductor while continuously monitoring the source-drain current Isd(t). The attached PKA then interacted with its co-factor ATP or its peptide substrate Kemptide5 added singly or as mixtures to the electrolyte. Isd(t) recordings extended continuously for at least 300 s in each experiment; this Isd(t) measurement was digitized at 100 kHz and stored for subsequent analysis. To assist stability during long duration measurements, the electrolyte potential was controlled at −300 mV relative to the drain electrode using a Pt pseudo-reference electrode.
RESULTS and DISCUSSION
A number of different sites to attach PKA to the SWNT through cysteine thiol reaction with pyrene-maleimide were explored. The T32C variant of PKA emerged as the protein variant with the best overexpression levels. At the end of the A-helix, yet near the conserved start of the kinase core, the T32C bioconjugation site falls between regions of secondary structure; the position could thus limit the impact of the cysteine modification on protein stability. Charged residues in motion near to the attachment site during enzyme opening and closing allow the SWNT device to record conformational changes during enzymatic catalysis (Figure 1c).
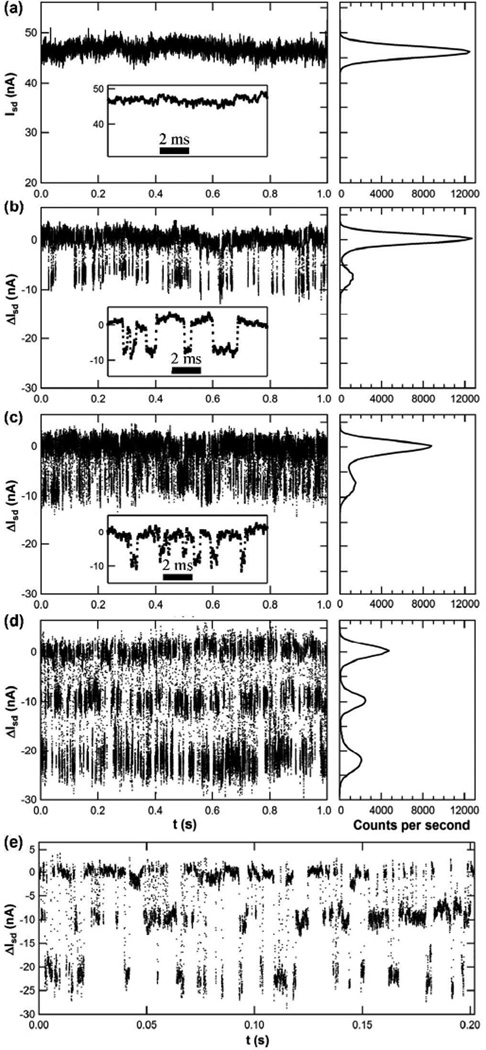
Figure 2 illustrates typical Isd(t) signals and signal distributions measured from a PKA-functionalized SWNT in various solutions. With PKA in buffer, Isd(t) merely fluctuated about a mean baseline value (Figure 2a) with a distribution determined by the 1/f noise of the SWNT device. Following the addition of ATP or Kemptide to the PKA-conjugated nanocircuit, new, quasi-stable levels of Isd(t) were observed below this baseline (Figures 2b–e). Figure 2b shows the results of measuring Isd(t) while the PKA was incubated in a 2 mM concentration of ATP, yielding a 2:9 molar ratio of ATP:Mg2+. In the presence of ATP, Isd(t) exhibited transient excursions ΔI of approximately −8 nA, or nearly −19%. Qualitatively similar excursions were observed when PKA was measured in an 100 µM solution of Kemptide substrate (Figure 2c). This concentration was slightly below the Kd for the PKA-Kemptide complex (230 ± 70 µM),11 which could decrease the apparent affinity for Kemptide measured here.
Figure 2.
Representative ΔIsd (t) signals and signal distribution histograms for a PKA-conjugated SWNT device. (a) Unfiltered, raw data as measured in buffer, showing the absolute value of Isd(t). (b–e) ΔIsd (t) relative to the baseline current, as measured in the presence of (b) 2 mM ATP, (c) 100 µM Kemptide, and (d,e) both ATP and Kemptide. To the right of (a) through (d), the histograms indicate the signal distribution observed for each condition. The magnified view in the insets and (e) illustrates the high level of detail discernible in individual transitions. In the discussion, the high, mid and low current states are assigned to the open (apo), the substrate- or ATP-bound (intermediate), and the catalytically functioning (ternary or fully closed) enzyme conformations, respectively.
Differences in either amplitude or duration were investigated as possible metrics for distinguishing PKA-ATP from PKA-Kemptide ΔI excursions in mixtures of the two reagents. However, neither approach proved practical. The amplitude distributions of binding to either ATP or Kemptide almost completely overlapped, and the mean durations of each binding event concealed a wide, nonstationary distribution, which is discussed further below. Nevertheless, the mean duration of many thousands of excursions was distinguishably different in measurements made individually with either ATP or Kemptide, measuring 3.1 ms for ATP and only 1.8 ms for Kemptide. By also analyzing the waiting times between each excursion, we extracted rates for one cycle of binding and unbinding by each ligand of 125 s−1 and 286 s−1 for ATP and Kemptide, respectively. Thus, the enzyme cycles through binding and unbinding to Kemptide faster than to ATP. The results reported here confirm conventional ensemble measurements showing PKA can bind independently to both Kemptide and ATP, but the exact order is largely random.12,13
We interpret each transient excursion ΔIsd(t) to result from one PKA binding event. Table 1 shows that this interpretation results in average waiting times and rates for a complete cycle of binding and unbinding that agree well with ensemble values. Negative control measurements further support this analysis by showing that in the absence of an attached PKA, pyrene-maleimide functionalized SWNTs exhibit none of the low current excursions shown in Figure 2, which again are associated with ATP or Kemptide binding to PKA (see Supporting Information). Furthermore, ΔIsd(t) fluctuations were reliably eliminated by rinsing ATP and/or Kemptide off the device, and were restarted by reintroducing a binding partner. As an example of this, the data in Figure 2 is chronologically reversed: the baseline Isd(t) in Figure 2a was acquired after probing the device with ATP and Kemptide solutions; the results confirmed recovery of the baseline after testing.
Table 1.
PKA binding rates.a
| Single-Molecule | Ensemble | ||||||
|---|---|---|---|---|---|---|---|
| ATP | τ | ± | σ | <t> | ± | σ | <t> |
| bound (ms) | 1.99 | ± | 0.03 | 3.1 | ± | 4.3 | 5.329 |
| unbound (ms) | 0.77 | ± | 0.01 | 5.0 | ± | 19 | > 0.25b |
| binding / unbind- ing cycle (s−1) |
362c | ± | 5 | 125d | < 190b,e | ||
| Kemptide | τ | ± | σ | <t> | ± | σ | <t> |
| bound (ms) | 0.77 | ± | 0.01 | 1.8 | ± | 2.9 | <230 |
| Unbound (ms) | 0.67 | ± | 0.01 | 1.7 | ± | 3.7 | <2017 |
| binding / unbind- ing cycle (s−1) |
696c | ± | 8 | 286d | >46 | ||
Where τ is the characteristic time from an exponential fit to the data reported in Figure 3, and <t> is the arithmetic mean from the distribution shown in Figure 3.
Estimated by extrapolation to saturation conditions from the experimental data of Ni et al.29
1/(τbound + τunbound)
1/(<tbound> + <tunbound>
Estimated from ligand exchange with AMP-PNP.
The electrical signals can be well-explained by an electrostatic transduction mechanism previously demonstrated using SWNT devices and single T4 lysoyzme molecules.9 Like the previous experiments with lysozyme, PKA has two charged residues in close proximity to the SWNT conjugation site (Figure 1d). In the lysozyme example, the two proximal residues have positively charged sidechains that swing away from the SWNT during enzyme closing; for the hole-doped SWNTs of our experiments, such movement results in an increase in I(t).9 The proximal sidechain functionalities of PKA also move in concert; a negatively charged functionality flips away and a positively charged residue moves towards the SWNT. The net effect from the combined electrical field of both charged residues is a large decrease in Isd(t), providing excellent signal to noise levels for analysis.
In the absence of binding partners, the PKA apoenzyme adopts an open conformation in which the large lobe (residues 127 to 300) is 1.33 nm away from the smaller lobe (residues 30 to 120, as measured with respect to the α-carbon of S53 and G186).14 This stable, open conformation produces the baseline current observed in Figure 2a. Upon binding to either ATP or Kemptide, PKA moves into an intermediate conformation, in which the two lobes are separated by 1.18 nm or 1.07 nm, respectively.15,16 For either binding partner, the large lobe moves closer to the small one, which is attached to the SWNT conductor. The motions of PKA described by NMR17,18 agree well with the dynamics described here. For example, Veglia, Taylor and co-workers reported that the enzyme transits through three conformational states during catalysis, analogous to the open (apo), intermediate and closed (ternary) states observed using the SWNT device.
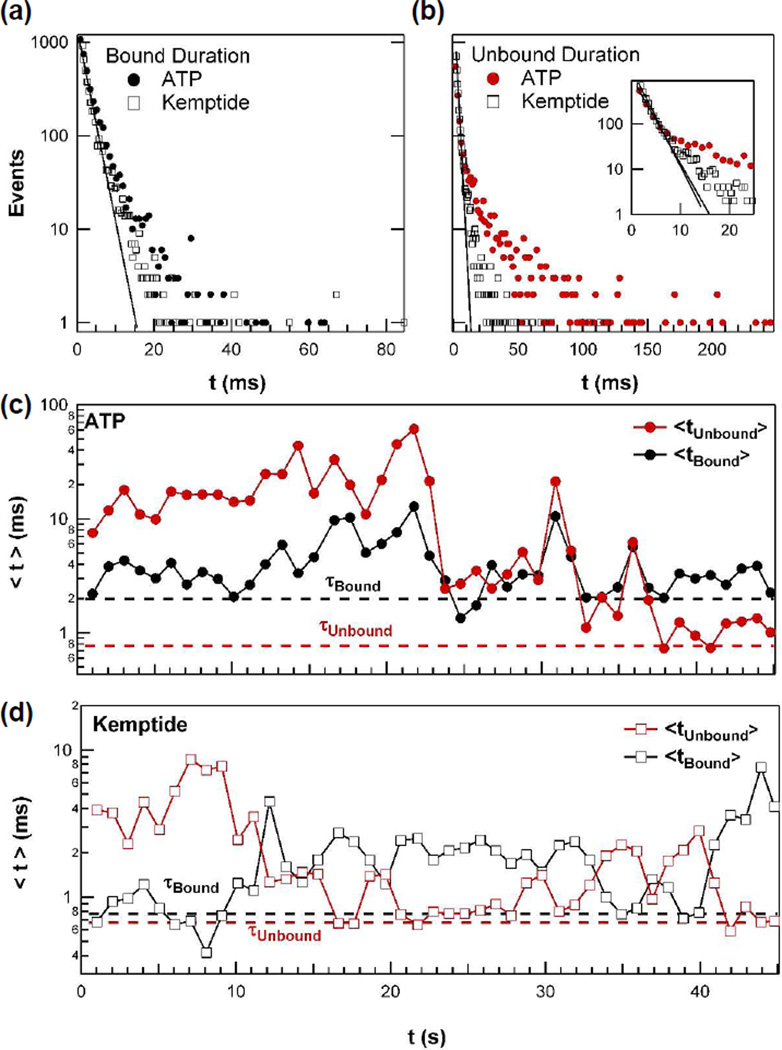
Single molecule measurements offer powerful tools to reveal enzyme variability and dynamic disorder, which are typically hidden during ensemble measurements.19–22 The timing of PKA binding to ATP and Kemptide measured here both exhibited the wide variability associated with the dynamic disorder of the enzyme. Figure 3a shows the distribution of the duration for the enzyme bound state for both ligands individually added to PKA-functionalized nanocircuits; the distributions were constructed from continuous, 45-second subsets of Isd(t) recordings. Figure 3b shows analogous distributions for the waiting times between binding events (i.e., the unbound or open-enzyme state). The slopes of simple, single-exponential fits (solid lines in Figures 3a and b) determine the fitted τ values that represent the majority (95%) of events and determine the most probable on- and off-rates of either ATP or Kemptide.23 However, all four distributions also include long-duration tails that deviate significantly from a single exponential behavior. The waiting time for ATP binding, in particular, exhibits a much longer distribution tail. Furthermore, unlike the near-parity situation for Kemptide on- and off-rates, the PKA-ATP complex has a slower off- than on-rate.
Figure 3.
Dynamic disorder of PKA observed by single molecule measurements with either ATP or Kemptide. (a) Distributions of the duration of the enzyme bound conformation for ATP or Kemptide. (b) Distributions of the waiting times for ATP and Kemptide binding, with the shortest waiting times shown more clearly in the inset. In (a) and (b), single-exponential line fits determining τ are shown as solid lines. (c) Variation in the 1-second arithmetic mean values <t> of ATP binding kinetics. These values were calculated by averaging the indicated one second of data. The corresponding τ values from the exponential fit are indicated by dashed lines for comparison. (d) The analogous variation in Kemptide binding kinetics.
The unusually long events in the distribution tails exert a disproportionate impact on the enzyme’s activities. While most of the distribution has a mean duration τ, an arithmetic average of the entire distribution produces a longer average duration <t>. In binding Kemptide, for example, <t> is more than 2τ. The long-duration events have similar effects on the statistical variances, σ2. Because of these substantial differences, τ and <t> are tabulated independently in Table 1 with their corresponding standard deviations. The practical consequence of the longer <t> values is a reduction of the average ligand cycling rates by 59 to 66%. Even though the faster, maximum rate 1/τ is the probabilistically favored value from cycle to cycle, the reduced rate 1/<t> is the one accessible to ensemble measurements because it includes all excursions in the kinetics.
Consequently, one would not expect rates measured through ensemble techniques to agree with rates determined by molecular dynamics simulations or NMR spectroscopy. PKA is known to have a very dynamic structure with binding sites that allosterically couple with remote loops and subdomains.17,18 Apparently, these interactions can result in a broad range of observable rate constants; such dynamic disorder is immediately evident to single molecule techniques.
Figures 3c and 3d illustrate this dynamic disorder resulting in a temporal variation of PKA-ATP or PKA-Kemptide complex stability. These graphs plot the second-by-second variation in the average times for <tbound> and <tunbound>, throughout the same 45-second intervals. During some seconds, very few long-duration events occur, and <t> approaches τ. During others, <t> grows much longer. The rate-limiting waiting time for ATP binding extended over the widest range, varying over two orders of magnitude from one second to another (Figure 3c). Furthermore for the PKA-ATP complex, <tunbound> and <tbound> follow each other weakly, with a statistical correlation of r = 0.71 (Figure S4). A positive correlation suggests a common mechanism for both steps, such as a conformational change distant from the ATP binding site, that increases or decreases both <tunbound> and <tbound>.
Compared to ATP binding, PKA-Kemptide binding statistics show different trends (Figure 3d). For Kemptide, the binding times <tunbound> and <tbound> were confined to a narrower range of 0.7 ms to 10 ms and tended to move in opposite directions. In fact, ln(<tunbound>) and ln(<tbound>) are anti-correlated with r = −0.84 (Figure S4). This strong, logarithmic anti-correlation is similar to the behavior of slow, viscoelastic motions.24 Here, the binding and unbinding rates are affected by one or more conformational motions, which increase the affinity of PKA for Kemptide, stabilizing the PKA-Kemptide complex and favoring its formation. Altering such conformational motions through binding by regulatory binding partners likely provides the basis for sensitive regulation of PKA activity, as such conformational changes will dramatically favor either the Kemptide bound or unbound states. Furthermore, ATP binding cannot provide a basis for regulated enzyme activity, as such conformational changes affect the stability of both bound and unbound PKA in a correlated manner.
Additionally, we note that <tbound> and <tunbound> varied widely enough to reverse their relative durations. Generally, the bound state of PKA interacting with either ATP or Kemptide was much shorter-lived than the unbound state; thus, the transduced Isd(t) signal looked like the pattern of excursions depicted in Figures 2b and 2c. Approximately 25% of the time, however, the bound and unbound lifetimes became comparable, and the resultant Isd(t) showed two-level switching with essentially symmetric timing. In rarer situations, the bound state became much longer than the unbound state. During these periods, the excursions in Isd(t) become positive spikes corresponding to a short-lived, apo PKA conformation. Examples of each case are shown for PKA binding to Kemp-tide in Fig. S5. Fortunately, continuous single-molecule monitoring provided long-duration Isd(t) records in which the two states could be unambiguously identified throughout the experiment.
The simultaneous binding of both ATP and Kemptide allows PKA to assume a fully closed conformation that completes formation of the catalytically functional, ternary complex. Once in this conformation, the enzyme can transfer the gamma phosphate of ATP to the Kemptide substrate.2 Experimentally, the addition of both ATP and Kemptide to the PKA-conjugated SWNT produced three-level Isd(t) fluctuations shown in Figure 2d. The baseline and intermediate states are identical to those described above for single ligand binding events, and these intermediate states are joined by a new, quasi-stable current level that was 20 nA, or 42%, below the baseline current. This amplified signal is consistent with the fully closed enzyme conformation; such a conformation has been observed previously by NMR and X-ray crystallography.17,25 In this conformation, the large lobe approaches even more closely to the SWNT conductor, thus altering the electronic signal even more than upon ATP or Kemptide binding. Figure 2e shows a high resolution portion of just 0.2 s of data, and illustrates excellent separation of individual events for statistical analysis.
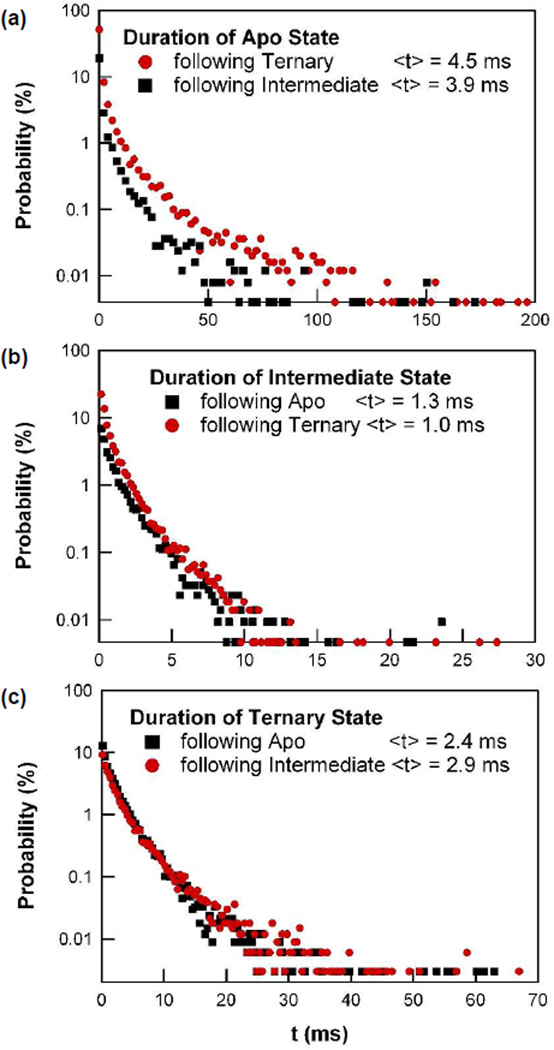
Figure 4 shows probability distributions and mean values for the durations of the apo, intermediate, and fully closed PKA conformations. The mean durations are 4.4, 1.1, and 2.6 ms for the apo, intermediate, and closed conformations, respectively. Such durations are naturally influenced by the concentrations of ATP and Kemptide present, which were 2 mM and 100 µM, respectively. For each conformation, we separately plot two distributions to account for the possibility that the duration of the conformation depends on the previous state of the enzyme. For example, Figure 4a shows the apoenzyme conformation. The entire distribution is stretched to longer durations when the open state follows the closed state instead of the intermediate one. A physical explanation of this increase could be that the preceding closed state has resulted in a successful catalytic event, after which product release slows the enzyme as it begins a new cycle; furthermore, the divergence between the distributions of the apoenzyme state resulting from either the ternary or the intermediate states is consistent with a different conformation required for catalysis, than for mere binding. The intermediate conformation (Figure 4b) has a much smaller, but still significant dependence on the previous conformational state. Transitions through the intermediate conformation are 30% longer, on average, when they follow the closed rather than the open conformation. The duration of the ternary complex has essentially no such dependence. All six probability distributions are stretched exponentials, indicating varying kinetics and possibly a variety of hidden states to which Isd(t) may not be sensitive; for example, a slow step might be required before product release.
Figure 4.
Probability distributions of the duration spent in each of the following three PKA conformations: (a) the unbound apo state, (b) the intermediate state with one binding partner, and (c) the ternary, fully closed state. For each state, two distributions are shown to distinguish the direction of conformational change. Only the apo state in (a) has a significant dependence on its previous conformational state.
The high quality signal shown in Figure 2e allow determination of a wide variety of statistics, and Table 2 shows mean durations of the final state and the transition probability matrix amongst each of the possible conformations. Beginning in the open conformation, transitions directly to the ternary complex were observed 73% of the time, while transitions to the intermediate state only occurred in the remaining 27%. Of course, every formation of the ternary complex must occur via the intermediate bound state; however, the binding of Kemptide and ATP are known to be highly cooperative17, and therefore occur nearly simultaneously on the time scale of the electronic measurements. The measurements here were performed with 20 µs time resolution, during which diffusion-limited transport and near-simultaneous binding could very well have occurred in too few data points to be resolvable.
Table 2.
Matrix of transition probabilities and mean durations.
| Final State | ||||||
|---|---|---|---|---|---|---|
| Low | Middle | High | ||||
|
Initial state |
P | <t> | P | <t> | P | <t> |
| Low | 45% | 1.0 ms | 55% | 4.5 ms | ||
| Middle | 71% | 2.9 ms | 29% | 3.9 ms | ||
| High | 73% | 2.5 ms | 27% | 1.3 ms | ||
Complex transition probabilities between more than just two conformations are interesting to complete the portrait of PKA activity. For example, the mean time from the beginning of one closure to the next via the intermediate configuration determines the effective catalytic turnover rate, assuming successful phosphorylation takes place each time the enzyme accesses the fully closed conformation and then returns to the apo state. In a single data set, we observed 20,000 such events and were able to determine a mean turnover rate of 107 s−1.
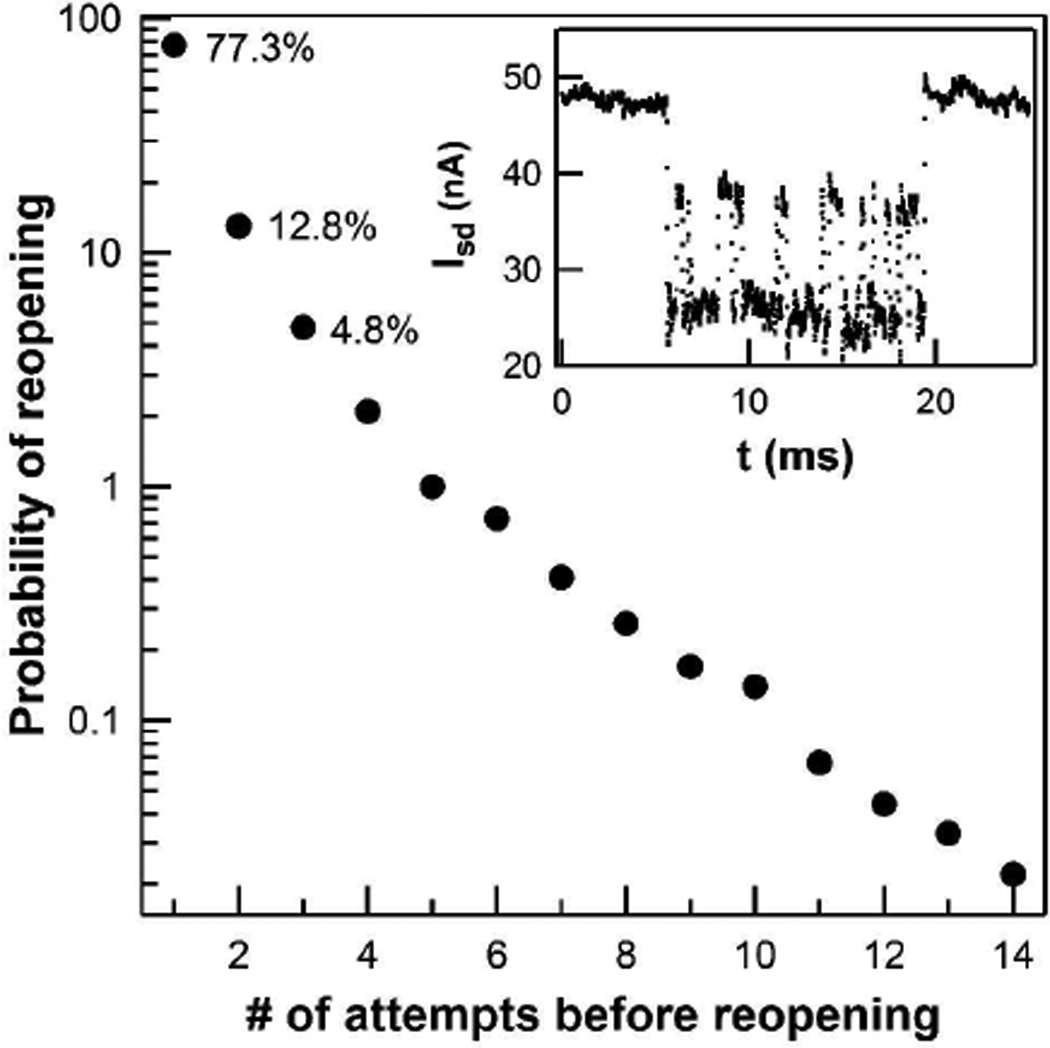
As with the other rates described above, however, the mean value concealed the true variability of the processes observed. The mean turnover rate for 10 s intervals ranged broadly from 13 to 230 s−1, which overlaps with the ensemble-measured value of 21 s−1.6 The vast majority (77%) of turnovers occurred via a straightforward sequence of open-intermediate-closed-open conformations and with a rate of 155 s−1. A minority (13%) of turnovers visited the closed state twice before opening, leading to a slower rate of 91 s−1. And finally, the remaining 10% of turnovers cycled multiple times between the closed and intermediate conformations before finally opening with product release with a turnover rate of 37 s−1. We interpret these sequences as failed attempts to phosphorylate the Kemptide, which remained bound while PKA repeatedly closed upon it. Thus, we determine PKA to have a 77% probability of success during the first closure and only a ~50% probability of catalysis during each subsequent attempt (Figure 5). This relatively low probability leads to occasional sequences of enzyme motion in which 10 or more closures are observed before substrate release (Figure 5, inset).
Figure 5.
Probability of re-opening, as indicated by the return to the apo conformation. Each time PKA reaches the ternary complex, the protein can either return to the open or the intermediate state. As shown here, the enzyme typically (77.3% of the time) accesses the ternary complex only once before reopening and returning to the apo state; less commonly, multiple transitions from the intermediate to the ternary state were observed with the percentages shown. Inset: Isd(t) trace shows an example of multiple transitions between the intermediate and closed conformations before finally re-opening.
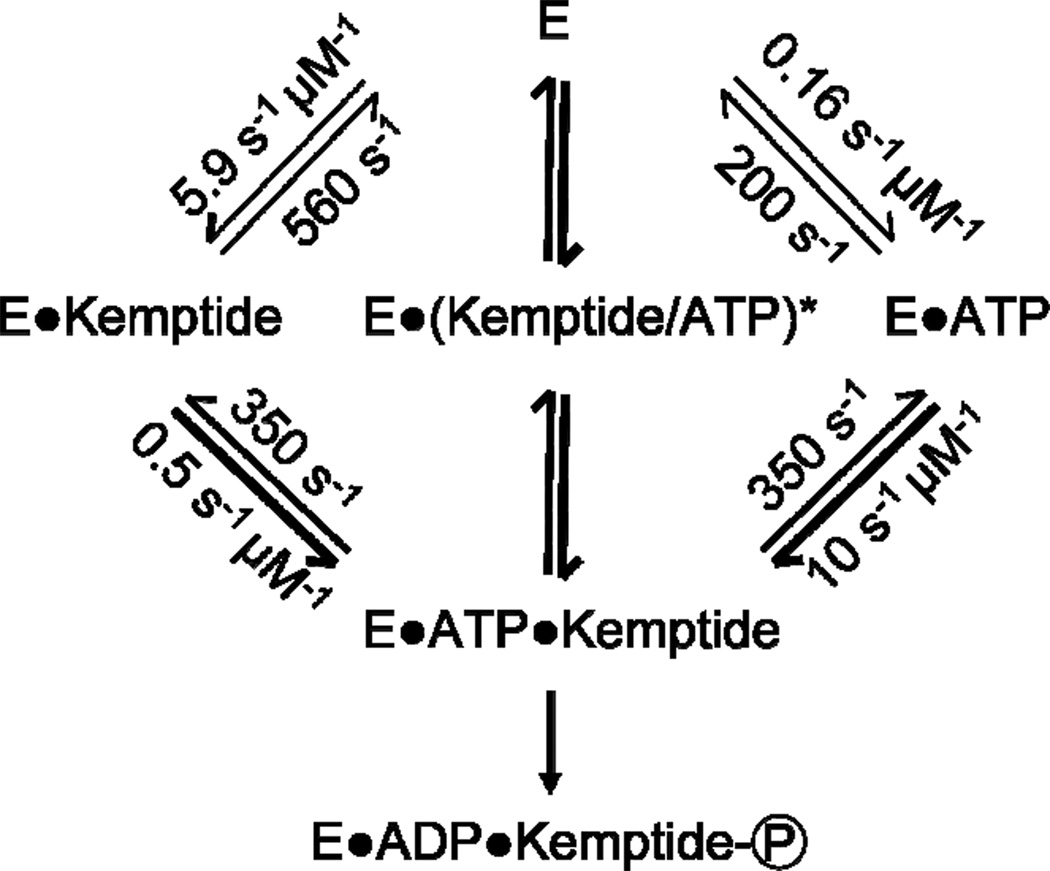
A minimal kinetic scheme consistent with this data is shown in Figure 6. If the enzyme fails to catalyze the phosphorylation reaction each time it cycles between fully closed and the apo state, the turnover rates and efficiency could be even lower. The reported rates, thus, represent an upper bound on the catalytic efficiency of PKA. Transition probabilities highlighted as arrow thicknesses in Figure 6 again emphasize the enzyme’s strongly cooperative formation of the ternary complex.
Figure 6.
A kinetic scheme for the PKA-catalyzed phosphoryla-tion of Kemptide with rates measured by a single PKA functionalized SWNT device. Second order rate constants for complex formation were calculated by dividing the observed rates by the appropriate substrate concentrations. The thickness of the lines reflects the transition probabilities from Table 2 with thicker lines indicating a higher probability. As described in the text, the intermediate marked with an asterisk is unobserved; formation and dissociation of the ternary complex by this route occurred with rates of 230 and 410 s−1, respectively.
In the perfect enzyme described by Knowles, every substrate binding event results in catalysis.26 Conversely, the single molecule examination of PKA dramatically illustrates an imperfect enzyme, as the conformational vicissitudes described above undermine the catalytic efficiency of PKA. Each failure to access the fully closed configuration necessary state for catalysis wastes an opportunity for substrate-to-product conversion. In comparison, the enzyme hexokinase catalyzes a similar phosphoryl transfer reaction to a primary hydroxyl functionality, yet exhibits far higher catalytic efficiency (2 × 109 M−1s−1 for hexokinase vs. 5–6 × 106 M−1s−1 for PKA).27,28 The efficiency of hexokinase is consistent with its role as catalyst for a key step in metabolism. PKA’s vacillation from the intermediate conformation reflects the evolution of signaling kinases like PKA to provide a regulatable molecular switch, rather than evolution to optimize catalytic efficiency.
A final notable aspect of the Isd(t) signals in Figure 2 is a difference in the widths of their distributions. Individual peaks in the Isd(t) histograms were fit to Gaussian functions to quantitatively compare these widths. The baseline current centered around ΔIsd = 0 had the narrowest distribution σo = 1.03± 0.06 nA, and we found this width to be independent of whether binding partners were present (e.g., Figure 2b, 2c or 2d) or not (Figure 2a). The smaller, secondary peaks associated with PKA binding to ATP or Kemptide both had widths of approximately 1.5σo. The distribution of the ternary complex had the lowest mean value but the largest width of nearly 2σo. This trend is opposite to the behavior of the SWNT’s 1/f noise, which normally decreases proportionally to the mean current. Thus, the increased width of ΔIsd is most likely associated with the attached PKA and the motions of its surface charges, which are more stochastic upon ligand binding. In other words, the enzyme accesses a broader range of conformational motions during formation of the ternary complex from the intermediate conformation.
The fluctuation magnitudes ΔIsd shown in the y-axis of Figure 2 are sensitive to both the contact resistance and transconductance of a particular SWNT device. Both of these parameters vary from device to device. Thus, to aid direct comparisons within this report, all of the data in Figures 2–4 were collected from a single device. Similar fluctuations have been observed over shorter durations using other PKA-labeled SWNT devices, and past work9 has proven that fluctuation magnitudes ΔIsd can be converted into an effective molecular “gating” (ΔVG) that eliminates device-to-device variability; we have successfully applied this approach to eliminate this variability in >100 devices with different enzymes attached.9 The effective ΔVG relates ΔIsd to the transconductance of the device, and is a measure of the change in electrolyte gate that would reproduce the observed signal.
For the PKA described here, the transition from the open or apoenzyme to the intermediate state produces a ΔIsd that is equivalent to a change in electrolyte gate ΔVG of +87 ± 7 mV for ATP and +65 ± 11 mV for Kemptide. The transition from the open apoenzyme to the fully closed, ternary complex is equivalent to ΔVG = 221 ± 20 mV, an effect that is much greater than the sum of ATP and Kemptide binding. All of the stated error bars are one standard deviation calculated from relatively limited data sets (Figure S6), and they may reduce significantly once data is available from a wider range of SWNT devices. In the meantime, these mean ΔVG values allow the data to be compared to theoretical models and predict how similar PKA-SWNT devices should behave. Other planned experiments will examine perturbation of PKA by inhibitors and varied co-enzyme concentration.
CONCLUSION
In conclusion, SWNT-based devices are proving to be powerful tools for observing single molecule kinetics. Using PKA, we have observed highly variable on- and off-rates for both the substrate and the ATP co-factor, and we have directly observed dynamic interconversion between the intermediate and fully closed conformations. Using 10-minute recordings extending over many thousands of binding events, we have calculated the full transition probability matrix among three possible configurations and the conversion rates among them. The resulting portrait of PKA depicts a highly dynamic enzyme with turnover rates varying over two orders of magnitude. Such abilities befit the function of an enzyme subject to extensive regulatory pressures; as shown by the anti-correlated bound and unbound PKA-Kemptide stabilities changing over time, enzyme regulation could operate on binding to its substrates, but not ATP. Furthermore, the stochastic choreography of PKA motions, with only 77% of events following a set pattern, illustrates the complicated nature of coupling enzyme motion with catalysis.
Supplementary Material
ACKNOWLEDGMENT
This work was supported financially by NIH NCI (R01 CA133592-01) and NSF (DMR-1104629). We thank an anonymous reviewer for helpful comments.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Materials and methods, bulk assays of enzymatic activity, and additional electronic signals and analysis. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Adams JA. Chem. Rev. 2001;101:2271. doi: 10.1021/cr000230w. [DOI] [PubMed] [Google Scholar]
- 2.Johnson DA, Akamine P, Radzio-Andzelm E, Madhusudan, Taylor SS. Chem. Rev. 2001;101:2243. doi: 10.1021/cr000226k. [DOI] [PubMed] [Google Scholar]
- 3.Manning G, Whyte DB, Martinez R, Hunter T, Su-darsanam S. Science. 2002;298:1912. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 4.Taylor SS, Yang J, Wu J, Haste NM, Radzio-Andzelm E, Anand G. Biochim. Biophys. Acta, Proteins and Prote-omics. 2004;1697:259. doi: 10.1016/j.bbapap.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 5.Kemp BE, Graves DJ, Benjamini E, Krebs EG. J. Biol. Chem. 1977;252:4888. [PubMed] [Google Scholar]
- 6.Adams JA, Taylor SS. Biochemistry. 1992;31:8516. doi: 10.1021/bi00151a019. [DOI] [PubMed] [Google Scholar]
- 7.Choi Y, Moody IS, Sims PC, Hunt SR, Corso BL, Weiss GA, Collins PG. Science. 2012;335:319. doi: 10.1126/science.1214824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi Y, Moody IS, Sims PC, Hunt SR, Corso BL, Seitz DE, Blaszcazk LC, Collins PG, Weiss GA. J. Am. Chem. Soc. 2012;134:2032. doi: 10.1021/ja211540z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi Y, Olsen TJ, Sims PC, Moody IS, Corso BL, Dang M, Weiss GA, Collins PG. Nano Lett. 2013;13:625. doi: 10.1021/nl304209p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson NC, Taylor SS. J. Biol Chem. 1983;258:10981. [PubMed] [Google Scholar]
- 11.Lew J, Taylor SS, Adams JA. Biochemistry. 1997;36:6717. doi: 10.1021/bi963164u. [DOI] [PubMed] [Google Scholar]
- 12.Whitehouse S, Feramisco JR, Casnellie JE, Krebs EG, Walsh DA. J. Biol Chem. 1983;258:3693. [PubMed] [Google Scholar]
- 13.Kong CT, Cook PF. Biochemistry. 1988;27:4795. doi: 10.1021/bi00413a032. [DOI] [PubMed] [Google Scholar]
- 14.Akamine P, Madhusudan, Wu J, Xuong NH, Ten Eyck LF, Taylor SS. J. Mol Biol. 2003;327:159. doi: 10.1016/s0022-2836(02)01446-8. [DOI] [PubMed] [Google Scholar]
- 15.Madhusudan, Trafny EA, Xuong NH, Adams JA, Ten Eyck LF, Taylor SS, Sowadski JM. Protein Sci. 1994;3:176. doi: 10.1002/pro.5560030203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narayana N, Cox S, Nguyen-huu X, Ten Eyck LF, Taylor SS. Structure. 1997;5:921. doi: 10.1016/s0969-2126(97)00246-3. [DOI] [PubMed] [Google Scholar]
- 17.Masterson LR, Mascioni A, Traaseth NJ, Taylor SS, Veglia G. Proc. Nat. Acad. Sci U.S.A. 2008;105:506. doi: 10.1073/pnas.0709214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masterson LR, Cheng C, Yu T, Tonelli M, Kornev A, Taylor SS, Veglia G. Nat. Chem Biol. 2010;6:821. doi: 10.1038/nchembio.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu HP, Xun LY, Xie XS. Science. 1998;282:1877. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 20.Xie XS. Single Molecules. 2001;2:229. [Google Scholar]
- 21.Yang H, Luo GB, Karnchanaphanurach P, Louie TM, Rech I, Cova S, Xun LY, Xie XS. Science. 2003;302:262. doi: 10.1126/science.1086911. [DOI] [PubMed] [Google Scholar]
- 22.Park J, Myong S, Niedziela-Majka A, Lee KS, Yu J, Lohman TM, Ha T. Cell. 2010;142:544. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.For Kemptide, the tails are composed of 5% of all events. For ATP, the tails are composed of 15% of binding times and 32% of unbound times.
- 24.Goychuk I. Phys. Rev E. 2009;80:046125. doi: 10.1103/PhysRevE.80.046125. [DOI] [PubMed] [Google Scholar]
- 25.Zheng JH, Trafny EA, Knighton DR, Xuong NH, Taylor SS, Ten Eyck LF, Sowadski JM. Acta Crystallogr Sect. D: Biol Crystallogr. 1993;49:362. doi: 10.1107/S0907444993000423. [DOI] [PubMed] [Google Scholar]
- 26.Knowles JR. Nature. 1991;350:121. doi: 10.1038/350121a0. [DOI] [PubMed] [Google Scholar]
- 27.Rose IA, O’Connell EL, Litwin S. J. Biol Chem. 1974;249:5163. [PubMed] [Google Scholar]
- 28.Adams JA, McGlone ML, Gibson R, Taylor SS. Biochemistry. 1995;34:2447. doi: 10.1021/bi00008a007. [DOI] [PubMed] [Google Scholar]
- 29.Ni DQ, Shaffer J, Adams JA. Protein Sci. 2000;9:1818. doi: 10.1110/ps.9.9.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grant BD, Adams JA. Biochemistry. 1996;35:2022. doi: 10.1021/bi952144+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.