Abstract
The neural crest (NC) is a population of multipotent stem cell-like progenitors that arise at the neural plate border in vertebrates and migrate extensively before giving rise to diverse derivatives. A number of components of the neural crest gene regulatory network (NC-GRN) are used reiteratively to control multiple steps in the development of these cells. It is therefore important to understand the mechanisms that control the distinct function of reiteratively used factors in different cellular contexts, and an important strategy for doing so is to identify and characterize the regulatory factors they interact with. Here we report that the LIM adaptor protein, LMO4, is a Slug/Snail interacting protein that is essential for NC development. LMO4 is expressed in NC forming regions of the embryo, as well as in the central nervous system and the cranial placodes. LMO4 is necessary for normal NC development as morpholino-mediated knockdown of this factor leads to loss of NC precursor formation at the neural plate border. Misexpression of LMO4 leads to ectopic expression of some neural crest markers, but a reduction in the expression of others. LMO4 binds directly to Slug and Snail, but not to other components of the NC-GRN and can modulate Slug-mediated neural crest induction, suggesting a mechanistic link between these factors. Together these findings implicate LMO4 as a critical component of the NC-GRN and shed new light on the control of Snail family repressors.
Keywords: Xenopus, Neural crest, LIM, Snail, Slug
Introduction
Neural crest cells are multipotent cells unique to vertebrates that give rise to a diverse array of derivative cell types including elements of the craniofacial skeleton, melanocytes, and neurons and glia of the peripheral nervous system (Le Douarin and Kalcheim, 1999; Hall, 1999; Heeg-Truesdell and LaBonne, 2004). Neural crest precursors first appear at the edges of the neural plate via an inductive event that requires integration of information from multiple signaling pathways including bone morphogenic protein (BMP), Wnt, fibroblast growth factor (FGF) and Notch (Heeg-Truesdell and LaBonne, 2004; Knecht and Bronner-Fraser, 2002; LaBonne and Bronner-Fraser, 1998). These upstream signals induce the expression of components of the neural crest gene regulatory network (NC-GRN) at the neural plate border, including Slug, Snail, FoxD3 and Sox8/9 (Betancur et al., 2010; Gammill and Bronner-Fraser, 2003; Heeg-Truesdell and LaBonne, 2004; Sauka-Spengler and Bronner-Fraser, 2008). Together these transcription factors function to specify and maintain the precursor population, and promote the migration and further development of the neural crest cell population. However, the unique roles of individual components of this gene regulatory network remain poorly understood. Interestingly, many members of the NC-GRN, including Slug/Snail, play multiple temporally distinct roles during neural crest development (Taylor and LaBonne, 2007). The mechanisms underlying the regulation of their function that allow such reiterative usage have yet to be elucidated.
Snail family transcription factors make essential contributions to numerous steps of vertebrate development, including mesoderm formation (Alberga et al., 1991; Nieto et al., 1994), and neural crest development (del Barrio and Nieto, 2002; LaBonne and Bronner-Fraser, 2000; Taylor and LaBonne, 2007), and their misregulation is closely associated with metastasis in cancer (Fujita et al., 2003; Hajra et al., 2002; Vernon and LaBonne, 2006). Snail was first characterized in Drosophila where it is required for the proper formation of mesoderm and repression of neuroectodermal genes such as single-minded and rhomboid (Alberga et al., 1991; Hemavathy et al., 1997). Fly embryos homozygous null for Snail exhibit defects in the invagination of the presumptive mesoderm and retraction of the germ band (Grau et al., 1984; Hemavathy et al., 2000a; Nusslein-Volhard et al., 1984). Snail also regulates the behavior of the mesoderm; for example it has been shown to be required for pulsed contractions and apical actinomysin meshwork assembly (Martin et al., 2009). During mesoderm invagination in Drosophila, pulsed actinomyosin meshwork contractions and a ratchet-like stabilization of cell shape by Twist have been proposed to drive apical constrictions (Martin et al., 2010).
In vertebrates, Snail family proteins include Snail (Snail1), Slug (Snail2), and Smuc (Snail3), and their role in mesoderm development has been conserved (Hemavathy et al., 2000a; Nieto, 2002; Sefton et al., 1998). Like Drosophila Snail, these proteins possess four to five c-terminal C2H2 zinc fingers which mediate binding to target promoters, and which may also play protein–protein interaction roles (Nieto, 2002). Slug and Snail appear to have highly overlapping functions, but are distinguished by the presence of a conserved 29-amino acid motif in the former that has been termed the “Slug domain” which is of unknown function (Cano et al., 2000; Grimes et al., 1996; Hemavathy et al., 2000a; Sefton et al., 1998). Vertebrate Snail family proteins also possess an N-terminal SNAG domain that is absent in Drosophila Snail. This domain is thought to mediate transcriptional repression via recruitment of co-repressor complexes (Grimes et al., 1996; Heeg-Truesdell and LaBonne, 2004; Hemavathy et al., 2000b; Peinado et al., 2004). Drosophila Snail has multiple binding sites for the co-repressor CtBP that confer this function (Hemavathy et al., 2000a; Nibu et al., 1998). Studies in human tumor cells suggest that the SNAG domain, while necessary, may not be sufficient for transcriptional repression (Grimes et al., 1996; Hemavathy et al., 2000b; Nakayama et al., 1998). The most widely studied Snail target in tumor cells is E-cadherin. The E-cadherin promoter contains three tandem Ebox consensus sequences and its down-regulation via these sites is thought to be a central event in tumor progression (Batlle et al., 2000; Cano et al., 2000; Llorens et al., 1998; Perl et al., 1998).
In Xenopus, Slug and Snail expression at the lateral edges of the newly induced neural plate is among the earliest known responses to neural crest inducing signals (Essex et al., 1993; Mayor et al., 1995; Spokony et al., 2002). Slug/Snail function is essential at these early stages for establishing neural crest precursors, and if their activity is blocked the expression of neural plate markers extends into neural crest forming regions. Later in neural crest development, Slug/Snail function is essential for neural crest precursor cells to delaminate from the neural epithelium and gain migratory ability (LaBonne and Bronner-Fraser, 2000). While Snail protein regulation is widely studied in human tumor cells, much less is known about the regulation of these important factors in the neural crest, including the proteins that interact with Slug/Snail to modulate their function. We have previously shown that the F-box protein Ppa binds the N-terminus of Slug and Snail and targets these proteins to the ubiquitin–proteasome system (UPS) for degradation (Vernon and LaBonne, 2006). UPS-mediated control of the threshold level of Slug/Snail in neural crest cells is an important determinant of whether they will impart stem cell properties or mediate morphological/behavior changes in these cells.
Another way in which the functional output of DNA binding transcription factors can be modulated is via regulation of the macromolecular regulatory complexes that they assemble. Adaptor proteins are frequently involved in formation of large multi-protein complexes that influence transcription, and one such factor that has been found to bind to Slug/Snail in neural crest cells is Ajuba LIM (Langer et al., 2008). Ajuba LIM can interact with Snail in the nucleus via its LIM domains and enhance SNAG-mediated repression, in part by recruiting HDAC proteins (Hou et al., 2008; Langer et al., 2008). LIM domains contain two tandem repeated zinc fingers that appear to function primarily in protein–protein interactions. They are found in a wide variety of proteins present in the nucleus or cytoplasm, or that shuttle between these two cellular compartments (Bach, 2000).
Here we report that the LIM domain Only (LMO) protein LMO4 is an essential regulator of neural crest development. LMO family proteins possess two closely spaced LIM domains for protein–protein interactions, but lack DNA-binding or catalytic domains (Bach, 2000; Grutz et al., 1998), and are a family distinct from the Ajuba LIM factors. LMO proteins (LMO1-4) have been implicated in both positive and negative control of transcription depending upon their binding partners and cellular context (Bach, 2000; Grutz et al., 1998; Kudryavtseva et al., 2003; Lu et al., 2006; Novotny-Diermayr et al., 2005; Sum et al., 2002; Wang et al., 2004; Wang et al., 2007). Here we show that LMO4 is expressed in both premigratory and migratory neural crest cells in Xenopus, and that morpholino-mediated depletion of LMO4 leads to profound defects in neural crest formation. We provide evidence that one way LMO4 regulates neural crest development is through binding to Slug/Snail proteins, and that its function here is distinct from that of Ajuba LIM. Together our findings shed new light on the regulation of neural crest precursor formation, as well as on the control of Snail-family transcriptional repressors.
Materials and methods
DNA constructs
A partial LMO4 cDNA was isolated in an in situ based expression screen of a stage 13–17 Xenopus cDNA library. A full-length clone was obtained from the NiBB (accession number NP-001087890). The encoded protein is 98% identical, 99% similar to human LMO4. This is in contrast to a gene previously reported as Xenopus LMO4 (accession number NP-001079179; de la Calle-Mustienes et al., 2003) which encodes a protein that is 79% identical and 87% similar to human LMO4. Truncated forms of LMO4, Slug, and Snail were amplified using low cycle- number PCR and a high fidelity Tgo polymerase (Roche, Dallas, TX, USA) and products were inserted into pCS2-MycC pCS23xFlagC, pCS23xFlagN or pCS2-HA vector as indicated. A full length Human Ajuba cDNA was obtained from Open Biosystems (Image clone 4837383). Morpholino resistant LMO4 was generated by quick-change mutagenesis to introduce four point mutations that did not alter the amino acid sequence but abrogated morpholino hybridization (ATGGTAAACCCCGGA).
Embryo manipulations
Pigmented and albino embryos were obtained using standard methods and staged according to Nieuwkoop and Faber (1994). mRNA for injection was in vitro transcribed using Message Machine (Ambion, Rockville, MD, USA). LMO4 translation blocking morpholino (5′CCTCTTACCTCAGTTACAATTTATA 3′) was obtained from Gene Tools, LLC (Philomath, OR, USA) and injected in a single animal blastomere at the 8-cell stage. β-galactosidase mRNA was co-injected as a lineage tracer and visualized using the Red Gal substrate (Roche, Dallas, TX, USA). Animal pole explants were isolated from stages 8–9 embryos and cultured in 1× MMR containing 50-μg/ml gentamycin until sibling embryos reached the noted stage. For in situ hybridization, embryos were fixed for 1X MEMFA, and stored dehydrated in 100% methanol. In situ hybridization was carried out using digoxigenin or fluorescene-labeled probes (Roche) as previously described (Bellmeyer et al., 2003). Alkaline phosphatase detection was carried out using BMPurple (Roche, Dallas, TX, USA) substrate. Results shown are representative of at least three independent experiments.
Immunoprecipitation, western blot analysis
For immunoprecipitations, embryos were collected at stage 10.5 unless otherwise noted and lysed in 1XPBS +1% NP40 containing protease inhibitors, (aprotinin, leupeptin and PMSF). Cleared embryo lysates were incubated with α-Flag (Sigma Aldrich, St. Louis, MO, USA) diluted with RIPA buffer for 2 h on ice, followed by a 2-hour incubation with protein A Sepharose beads (Sigma Aldrich, St. Louis, MO, USA). After washing, proteins were resolved by SDS-PAGE and immunoblotting was performed using α-Flag (1:3000) (Sigma Aldrich), α-Myc (1:3000) (Santa Cruz Biotechnology) or α-HA (1:2000), gift of R. Lamb (Northwestern University, Evanston, IL, USA) antibodies as indicated. Labeled proteins were detected using HRP-conjugated secondary antibodies and enhanced chemiluminescence (Fisher, Pittsburg, PA, USA). All results shown are representative of at least three independent experiments.
Proliferation and cell death assays
For phosphohistone H3 detection, injected embryos were fixed in formaldehyde at stage 13/14 and processed for β-gal activity. α-phosphohistone H3 antibody (Upstate Biotechnology) was used at a concentration of 5 μg/ml; α-rabbit IgG conjugated with alkaline phosphatase (Boehringer Mannheim) was used at a dilution of 1:1000 and was detected with BM purple. TUNEL staining was carried out as previously described (Bellmeyer et al., 2003). Briefly, fixed embryos were rehydrated in PBT and washed in TdT buffer (Gibco) for 30 min. End labeling was carried out at RT overnight in TdT buffer containing 0.5 μM digoxygenin-dUTP (Boehringer Mannheim) and 150 U/ml TdT (Gibco). Embryos were washed at 65 °C in PBS/1 mM EDTA, and detection of the digoxygenin epitope was carried out as for in situs. At neural plate stages only approximately 50% of control or MO-injected embryos show any endogenous TUNEL staining, therefore Dnmt3b1 was used to induce death as a positive control for TUNEL staining.
GST pulldown assay
Slug-GST protein was expressed in BL21 E. coli, sonicated, purified with Glutathione agarose (Sigma Aldrich, St. Louis, MO, USA). SDS-PAGE and protein induction verified by coomassie staining LMO4 was transcribed and translated in vitro using the TNT quick-coupled transcription/translation system (Promega Madison, WI, USA) in the presence of [35S] methionine. 8% of reaction mixture was kept as input and the remaining protein incubated with glutathione-beads-bound GST fusion proteins for 2 h at 4 °C in 1%NP40/PBS in a 500 μl reaction volume. Pull downs were washed 3 times with RIPA, bound proteins released by boiling in SDS sample buffer, resolved by SDS-PAGE, and detected using autoradiography.
Results
LMO4 is expressed in the neural crest
LMO4 was identified in an expression profile screen in Xenopus designed to uncover novel components of the Neural Crest Gene Regulatory Network (NC-GRN, data not shown). LMO4 is a small (19 kDa) adaptor protein comprised of two tandem LIM domains connected by a small linker region, and short N and C terminal ends (Grutz et al., 1998; Kenny et al., 1998) (Fig. 1A). To better characterize a potential role for LMO4 in regulating neural crest development, the expression of this factor was examined using whole mount in situ hybridization. LMO4 is expressed maternally, and at early cleavage stages LMO4 transcripts are detected throughout the animal hemisphere of the embryo (Fig. 1B I–III). Expression in the animal pole ectoderm continues through the onset of gastrulation (Fig. 1B IV and not shown). By neural plate stages (stage 13/14) LMO4 expression becomes restricted to neural crest and placode forming regions of the ectoderm, including in the transverse neural folds, and portions of the neural plate (Fig. 1B V, VI). By stage 17, however, ectodermal LMO4 expression is primarily restricted to neural crest forming regions, and expression is also observed in the paraxial mesoderm at these stages (Fig. 1B IX, X). Expression of LMO4 in neural crest cells is maintained as these cells start to migrate (Fig. 1B XI, XII). At stage 24 expression is also noted in the otic vesicle and in the tail bud (Fig. 1B XIII). At stage 28 when LMO4 expression is seen in the neural crest that has populated the branchial arches, expression in the tail bud and otic vesicle is still observed (Fig. 1B XIV). Overall, the expression of LMO4 suggests potential functions for this adaptor protein in the neural crest, in other ectodermally derived cell types including the cranial placodes, and in the paraxial mesoderm.
Fig. 1.
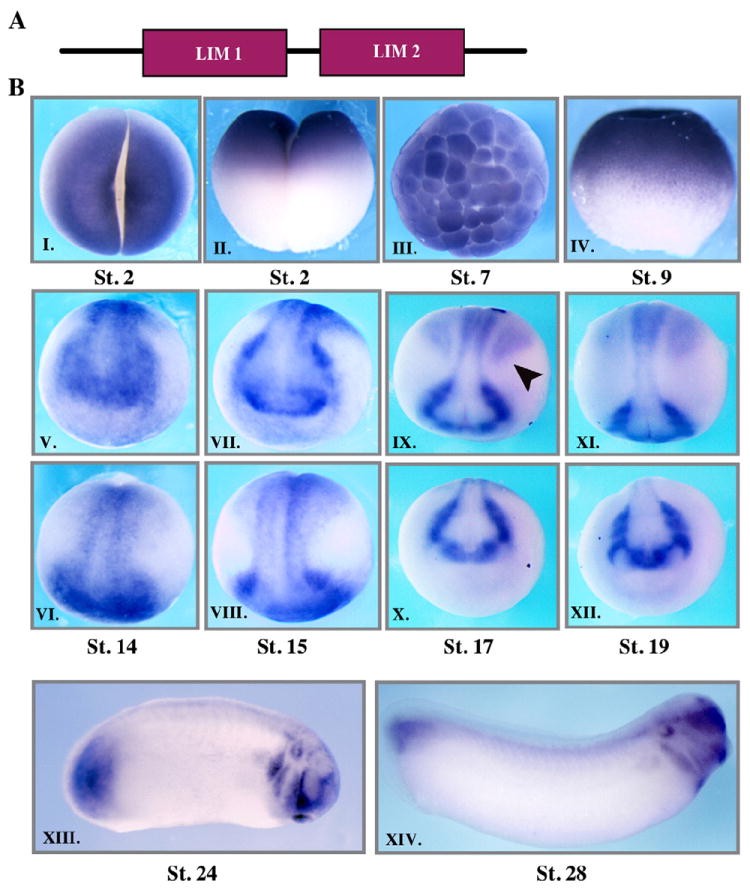
LMO4 is expressed in the neural crest. Schematic diagram showing LMO4 protein domains. (B) Whole mount in situ hybridization examining LMO4 expression in early Xenopus embryos. LMO4 expression in neural crest forming regions is apparent by stage 14, and is maintained in premigratory and migratory neural crest cells. LMO4 expression is also seen in the neural plate, placodal regions, and paraxial mesoderm at neurula stages (stages 14–19). At stage 24 LMO4 is expressed in the otic vesicle and tailbud in addition to migrating neural crest cells. At stage 28 LMO4 expression is seen in post-migratory neural crest cells in the branchial arches as well as in the tailbud, otic vesicle and somites.
LMO4 is required for neural crest development
The expression of LMO4 in neural crest forming regions of the early embryo is consistent with a role for this adaptor protein in the development of these cells. To determine if LMO4 is required for the formation of neural crest precursor cells, or the subsequent development of these cells, a translation blocking morpholino-targeting LMO4 was generated. Effective “knock down” of a c-terminally epitope tagged LMO4 by this morpholino was verified by western blot (Fig. 2F). Embryos were then injected with this morpholino, together with β-galactosidase as a lineage tracer, into one cell at the 8-cell stage to target neural crest and to avoid effects on the mesoderm, where LMO4 is also expressed. Embryos were cultured to early neurula stages when the expression of a number of components of the NC-GRN was examined by in situ hybridization (Fig. 2A).
Fig. 2.
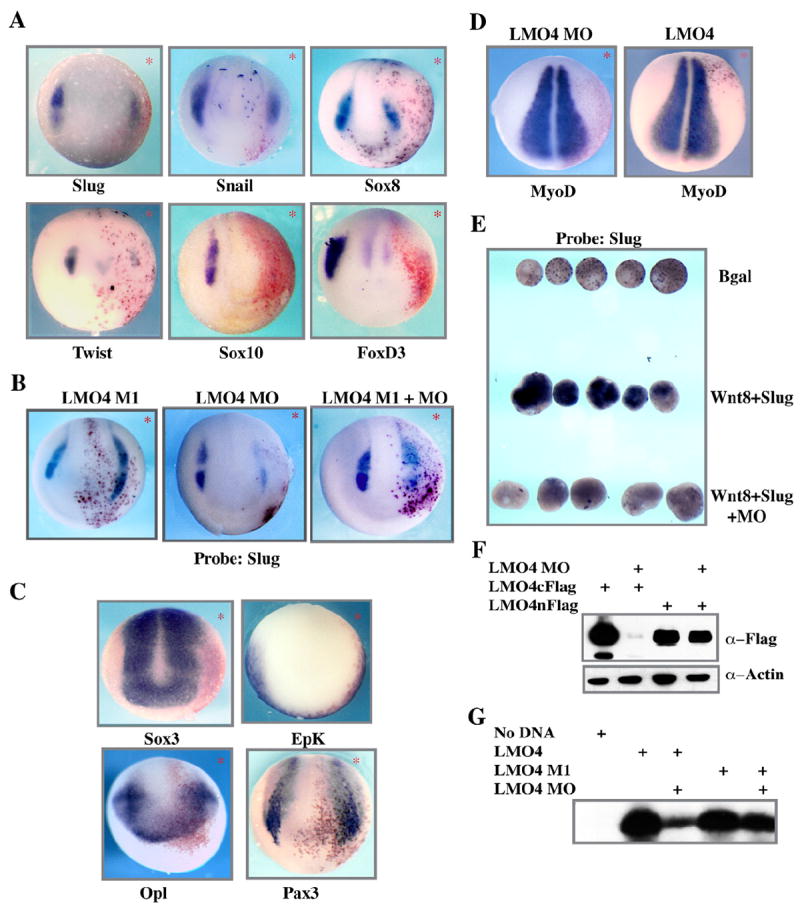
LMO4 is required for neural crest formation. (A) Whole mount in situ hybridization of embryos co-injected in one animal micromere at the eight-cell stage with LMO4 morpholino and β-gal as a lineage tracer. Embryos were examined at mid-neurula stage (St17) with neural crest markers Slug, Snail, Sox8, Twist, Sox10, FoxD3. * indicates side of injection which is also denoted by red gal staining. (B) Effects of LMO4 depletion can be rescued with a morpholino resistant form of LMO4 (LMO4 M1). Whole mount in situ hybridization injected probed for neural crest marker Slug. (C) Whole mount in situ of stage 13 LMO4 depleted embryos probed for neural plate marker Sox3, epidermal marker Epk, and neural plate border markers Opl, and Pax3. (D) In situs of LMO4 MO injected embryos probed for mesoderm marker MyoD. Normal expression of mesodermal markers indicates that effects of LMO4 depletion on the neural crest are not a consequence of mesodermal defects. (E) Animal cap assay demonstrating that mesoderm independent induction of neural crest by Wnt/Slug is blocked by LMO4 depletion. (F) Western blot analysis validating the knockdown of LMO4 immunoblotting with antibodies against the tagged protein. Actin is used as a loading control. (G) Western blot of in vitro translated (IVT) LMO4 proteins demonstrating that the mutant form is resistant to translation blocking morpholino.
The expression of several NC-GRN factors, including Twist, Sox10 and FoxD3, was completely abolished in LMO4 depleted embryos. The effects of LMO4 depletion on other essential regulators of neural crest development, including Slug, Snail, Sox9 and Sox8 were comparatively less severe, although their expression was still significantly reduced (Fig. 2A; Slug: 75%, n=364; Snail: 61%, n=109; Sox8: 85%, n=59; Twist: 77%, n=39; Sox10: 83%, n=426; FoxD3: 70%, n=279; Sox9: 71%, n=176, not shown). Interestingly, the factors affected more severely by LMO4 depletion were those that have a somewhat later onset of expression, suggesting that LMO4 may play a greater role in the maintenance of neural crest precursor cells than in their initial specification. Importantly, the effects of LMO4 depletion on neural crest formation can be rescued by a mutated form of LMO4 (LMO4M1) that cannot be targeted by the morpholino, demonstrating the specificity of the observed phenotype (Figs. 2B, F). Co-injection of LMO4 M1 mRNA resulted in enhanced rather than inhibited, Slug expression (Fig. 2B; LMO4M1 injected: 66% enhanced, n=65; LMO4 MO: 67% inhibited, n=42; co-injected: 87% rescued, n=78). Together, these results indicate that LMO4 is required for the establishment of the neural crest precursor pool at the neural plate border. We also examined the effects of LMO4 depletion on other ectodermally derived cell types. Morpholino injected embryos exhibited a loss of epidermal keratin (EpK) expression (Fig. 2C). By contrast, expression of both the neural plate border maker, Pax3, and of Opl, which is expressed primarily in placodal regions, was modestly but consistently expanded following LMO4 depletion, whereas expression of Sox3, which is expressed throughout the neural plate, was largely unchanged. (Fig. 2C; Epk: 80%, n=69; Sox3: 83% n=181; Opl: 73%, n=35; Pax3: 74% n=84).
Morpholino injections were carried out at the 8-cell stage to target the ectoderm and avoid depleting LMO4 in the mesoderm. Nevertheless, we examined the expression of mesodermal markers in LMO4 depleted embryos in order to rule out the possibility that loss of neural crest gene expression was an indirect consequence of defects in mesoderm formation. We found that expression of both MyoD and alpha-actin was unchanged in LMO4 MO injected embryos (Fig. 2D and data not shown; MyoD: 95% n=80; actin: 86% n=50). We next examined whether the requirement for LMO4 during neural crest induction could be demonstrated under conditions where mesoderm is not present. We have previously demonstrated that neural crest can be induced in animal pole ectoderm (“animal caps”) by a combination of Slug and Wnt expression (LaBonne and Bronner-Fraser, 1998). Accordingly, embryos were injected at the 2-cell stage with mRNAs encoding both Slug and Wnt8, in the presence or absence of LMO4 MO. Animal caps were isolated at stage 9 and cultured to stage 17 when they were examined by in situ hybridization. We found that co-injection of the LMO4 morpholino inhibited Wnt/Slug-mediated neural crest induction (Fig. 2E). Together these results demonstrate that ectodermally expressed LMO4 is required for neural crest induction.
Misexpression of LMO4 interferes with neural crest development
Having established that LMO4 is required for neural crest formation, we next wanted to better understand its possible function in these cells. Toward this end, we examined the consequences of ectopic LMO4 expression for the development of the neural crest and other ectodermally derived cell types. mRNA encoding LMO4 was injected into one cell at the 2-cell stage. β-galactosidase was co-injected as a lineage tracer, and injected embryos were allowed to develop to neurula stages for analysis by in situ hybridization. LMO4 misexpression led to striking increases in Slug and Snail expression (Fig. 3A; Slug: 76%, n=370; Snail: 71%, n=197). Ectopic expression of these markers was noted not only adjacent to endogenous neural crest forming regions, but also in the neural plate and in large expanses of the non-neural ectoderm (presumptive epidermis). These are regions of the ectoderm that are generally refractory to ectopic expression of neural crest markers, even in response to neural crest inducing factors such as Wnts or noggin. This suggests a novel or direct mechanism for LMO4 dependent expression of these factors. To determine if LMO4 was capable of directly activating Slug expression in “naïve” ectoderm, animal caps were isolated from LMO4 injected embryos and cultured to neurula stages where they were examined by in situ hybridization using a probe for Slug. β-galactosidase mRNA was used as negative control and Wnt8/Noggin co-injection as a positive control. As previously described (LaBonne and Bronner-Fraser, 1998), the combination of Wnt8 and noggin induced strong expression of Slug throughout the animal cap. By contrast, LMO4 injected caps failed to express Slug, indicating that it must cooperate with other factors not present in animal caps in order to induce expression of this marker (Figs. 3Ba–c).
Fig. 3.
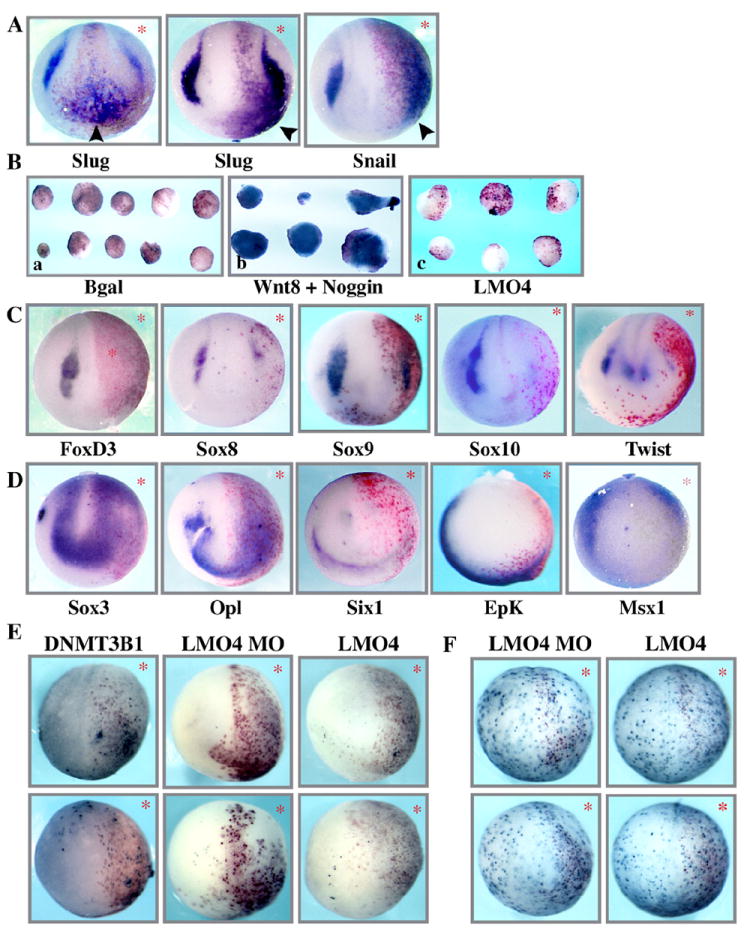
Excess LMO4 interferes with neural crest formation. (A) Whole mount in situ hybridization of embryos injected in one cell at two-cell stage with LMO4 and β-gal lineage tracer. Embryos were examined at mid-neurula stage (St. 17) with neural crest markers Slug and Snail, which display significant ectopic expression. B) Animal cap assay demonstrating that, in contrast to Wnt/noggin, LMO4 cannot induce Slug expression in isolated ectoderm. (C) Whole mount in situ hybridization of embryos injected in one cell at two-cell stage with mRNA encoding LMO4 and β-gal. Embryos were examined at mid-neurula stage (St. 17) with neural crest markers FoxD3, Sox8, Sox9, Sox10 and Twist expression of which, in contrast to Slug and Snail, were all inhibited by LMO4 misexpression. (D) In situ hybridization of stage 13 embryos injected with LMO4 and β-gal probed for neural plate marker Sox3, placodal markers Opl and Six1, epidermal marker Epk, and neural plate border markers Msx1 (St. 13). The expression of Slug and Snail is massively expanded while expression of other markers is inhibited. (E) TUNEL staining of stage 15 embryos injected with LMO4 MO, mRNA encoding LMO4, or apoptosis inducing factor DNMT3B1 (as a positive control). No significant changes in cell death were noted following either LMO4 up or down regulation. (F) phospho Histone H3 staining of stage 15 embryos injected with LMO4 MO or LMO4 mRNA. No significant changes in cell proliferation were noted following either LMO4 up or down regulation. * indicates injected side of embryo which is also marked by red gal staining.
To further investigate the functional consequences of LMO4 misexpression we examined its effects on the expression of a number of other key components of the NC-GRN. Strikingly, in contrast to what was observed for Slug and Snail, the expression of FoxD3, Sox8/9/10 and Twist was all inhibited by LMO4 misexpression (Fig. 3C; FoxD3: 78%, n=286; Sox8: 90%, n=43; Sox9: 92% n=38; Sox10: 88%, n=430; Twist: 81%, n=32). This was a surprising and intriguing finding as experimental manipulations that affect the formation of the neural crest generally have similar consequences for most markers of this cell type, and it suggests that LMO4 may play multiple mechanistically distinct roles in the formation of the neural crest.
We next examined the effects of LMO4 misexpression on the formation of other ectodermally-derived cell types. We found that the expression domains of the neural plate marker Sox3 and the neural plate border/placodal marker Opl were significantly expanded in LMO4 injected embryos. By contrast, the expression of Epidermal Keratin, a marker of presumptive epidermis, was significantly reduced, as was the placodal marker Six1 (Fig. 3D). Given the divergent effects on the expression of definitive neural crest markers and placodal makers, we also examined the effects of LMO4 misexpression on factors that broadly define the neural plate border region at early neurula stages (Sauka-Spengler and Bronner, 2010; Sauka-Spengler and Bronner-Fraser, 2008; Taylor and LaBonne, 2007). Expression of Pax3 and Msx1 was significantly inhibited following LMO4 misexpression, suggesting that inappropriate expression of this LIM adaptor protein is incompatible with normal formation of the neural plate border and its derivative cell types (Fig. 3D and data not shown; Sox3: 60% expanded, n=81; Opl: 49% expanded, n=35; Six1: 79% reduced, n=19; Epidermal Keratin: 95% reduced, n=40; Msx1: 60% reduced, n=40; Pax3: 74% reduced, n=23).
As the expression of a number of neural plate border and neural crest markers was lost or diminished following LMO4 depletion and/or LMO4 misexpression, we next examined whether their loss could be attributed to an increase in the number of apoptotic cells, although the continued presence of numerous β-galactosidase expressing cells on the injected sides of these embryos suggested that this was unlikely. Embryos injected either with LMO4 MO, or mRNA encoding LMO4, were allowed to develop to mid neurula stages (stages 15–17) when the extent of apoptosis was assessed using whole-mount TUNEL staining. DNMT3B1, which potently induces cell death, was used as a positive control for apoptosis as healthy neurula stage embryos display very few apoptotic nuclei. Importantly, no significant difference in the numbers of TUNEL-positive nuclei was observed on the LMO4-depleted or LMO4 injected side of these embryos when compared with the control sides of the same embryos (Fig. 3E; DNMT3B1 injected: 100% increased, n=7, LMO4 MO injected: 100% no effect, n=42, LMO4 injected: 100% no effect, n=52). This suggests that the loss of gene expression reflects an alteration in cell specification as opposed to the death of specific populations of cells. In an analogous set of experiments, we asked if the increase seen in the expression of some genes such as Slug, Snail, Sox3 and Opl could reflect enhanced proliferation of certain populations of cells. To determine this we examined the numbers of cells immunoreactive for the mitotic marker phospho-histone H3 on the LMO4-depleted or LMO4 injected side of embryos. Again, no apparent difference was noted in the number of mitotic cells when compared to the control side of the embryos, indicating that the observed phenotypes are not mediated by altered levels of cell cycle progression (Fig. 3F; LMO4 MO injected: 100% no effect, n=49; LMO4 injected: 100% no effect, n=40).
LMO4 specifically interacts with Slug and Snail
Together, the above experiments demonstrate that LMO4 is essential for neural crest precursor formation, and that the expression of this factor has dramatic and complex consequences for the patterning of the early embryonic ectoderm. In order to further understand the mechanisms underlying the function of this LIM domain containing adaptor protein, we first examined whether it could physically interact with components of the NC-GRN and thus have a potential role in modulating their function. To this end, embryos were co-injected at the 2-cell stage with mRNA encoding a flag-tagged LMO4 protein plus myc-tagged forms of the neural crest regulatory proteins Slug, Snail, Sox10, FoxD3 or Twist. Embryos were cultured to late blastula stages when LMO4 was immunoprecipitated using α-flag antibodies and interacting factors were detected by western blot analysis using α-myc antibodies. LMO4 displayed robust interactions with both Slug and Snail, but failed to interact with other NC-GRN components in this assay (Fig. 4A), suggesting that it may serve as a specific adaptor for Slug/Snail family proteins in regulating neural crest formation. In order to determine if the interaction between LMO4 and Slug/Snail is direct, we generated Slug protein in bacteria as a GST fusion and LMO4 protein in reticulocyte lysates. The ability of LMO4 to interact with Slug was then investigated using GST-pull down assays. LMO4 bound strongly to GST-Slug but not to equivalent amounts of GST alone, indicating that the interaction between these two factors is direct (Fig. 4B). Together these findings suggest that the requirement for LMO4 in neural crest formation is based at least in part on its ability to directly bind Slug/Snail proteins and serve as a scaffold for the further recruitment of co-regulatory factors.
Fig. 4.
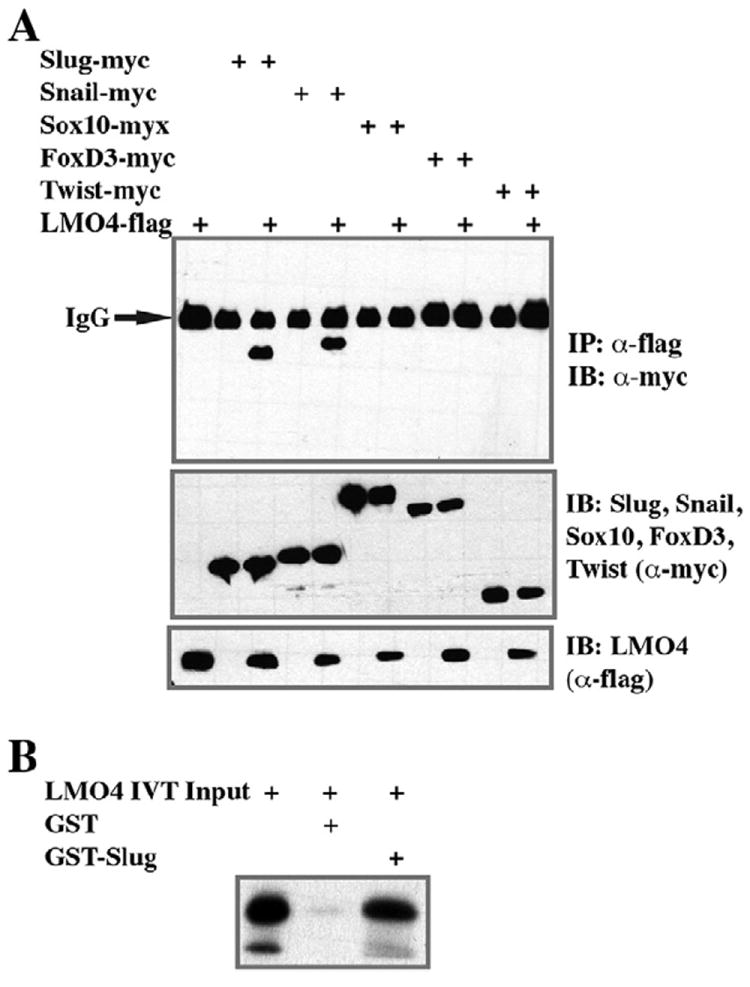
LMO4 forms a complex with Slug and Snail. (A) Co-immunoprecipitation (IP) assay probing the ability of LMO4 to interact with neural crest regulatory factors. Embryos were injected with mRNA encoding flag-tagged LMO4 and indicated myc-tagged neural crest transcription factors. Whole embryo lysates were prepared at stage 10.5, immunoprecipitated with α-flag antibodies, resolved by SDS page and subjected to western analysis using α-myc antibody. LMO4 interacts strongly with Slug and Snail but not Sox10, FoxD3 or Twist. (B) GST pull-down assay demonstrating that in vitro translated LMO4 protein directly interacts with GST-Slug, but not with GST alone.
Interaction between LMO4 and Slug requires both LIM domains and the Slug N-terminus
To better understand the functional consequences of LMO4’s interaction with Slug/Snail family factors, we investigated which domains of each factor were required for this interaction. A panel of LMO4 deletions was constructed in which only one LIM domain was present (LIMN or LIMC) or in which the N-and C-terminal extensions were deleted (LMO4DNC). In addition, mutant forms of Slug and Snail were generated in which the SNAG domain, or entire N-terminus of the proteins had been deleted (Fig. 5A). Co-immunoprecipitation assays were carried out using full length LMO4 and the three LMO4 deletion mutants to determine which proteins retained their ability to bind full length Slug and Snail. We found that deletion of the N- and C-terminal linker regions had no effect on LMO4 binding to either Slug or Snail. By contrast, deletion of either LIM domain severely diminished this interaction, indicating that both LIM domains play required roles in mediating this interaction (Fig. 5B).
Fig. 5.
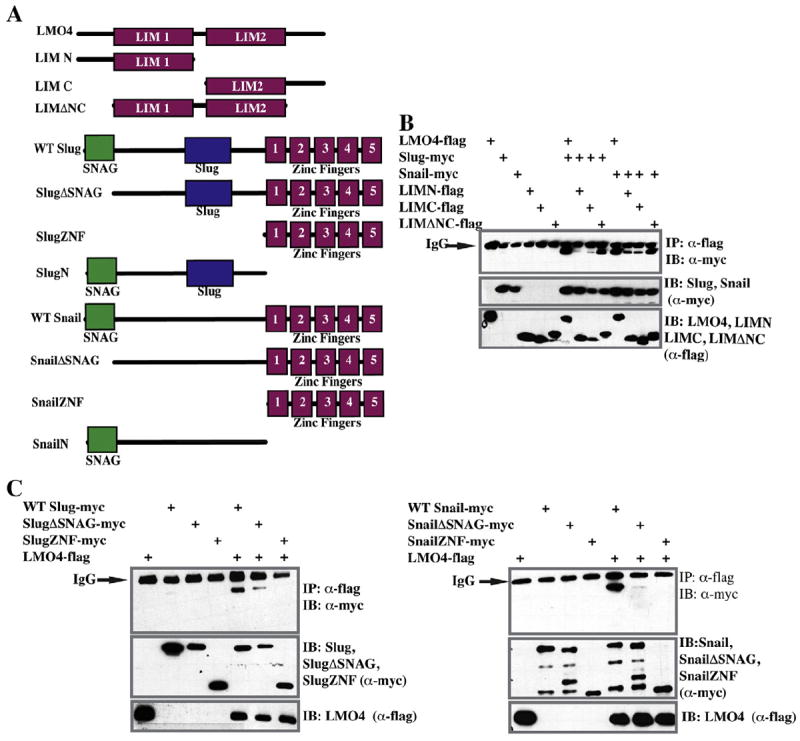
The LIM domains of LMO4 and the N-terminus of Slug are necessary for interaction. (A) Schematic of showing deletion mutants of LMO4, Slug, or Snail used in this study. (B) Co-immunoprecipitation (IP) assay probing the LMO4 domains required for interaction with Slug and Snail. Embryos were injected with mRNA encoding flag-tagged LMO4 constructs and myc-tagged Slug or Snail. α-flag IP followed by α-myc western shows that both LIM domains are required for robust interaction with Slug and Snail. (C) Co-immunoprecipitation assay using flag-tagged LMO4 and myc-tagged forms of either Slug (left panel) or Snail (right panel). Deletion of the Snag domain has a greater effect on the ability of Snail to interact with LMO4. (D) Co-immunoprecipitation assays comparing the ability of the full length or N-terminus of Slug (left panel) or Snail (right panel) to interact with LMO4, HDAC or Ajuba. Both Slug and Snail interact comparably with all three co-regulatory factors, but the Snag domain containing N-terminus is not sufficient for this interaction. Lower molecular weight bands on Snail immunoblot are common Snail degradation products.
The ability of the Slug/Snail deletion mutants to interact with LMO4 was similarly interrogated. Full length LMO4 was co-expressed with either full length Slug/Snail, or forms of these proteins in which either their SNAG domain, or their entire N-terminus had been deleted, and interactions were assessed using co-immunoprecipitation assays. A Snail protein in which the SNAG domain was absent was no longer able to interact with LMO4, indicating that this repressor domain is essential to that interaction (Fig. 5C, right panel). Interestingly, deletion of the Slug SNAG domain diminished but did not fully abrogate that interaction. Only when the entire N-terminus of Slug was absent was the interaction with LMO4 lost (Fig. 5C, left panel). Together these findings suggest that there may be sequence differences in the residue used by Slug and Snail to recruit LMO4, and raise the possibility that there might be previously unrecognized functional differences between these two closely related transcriptional repressors.
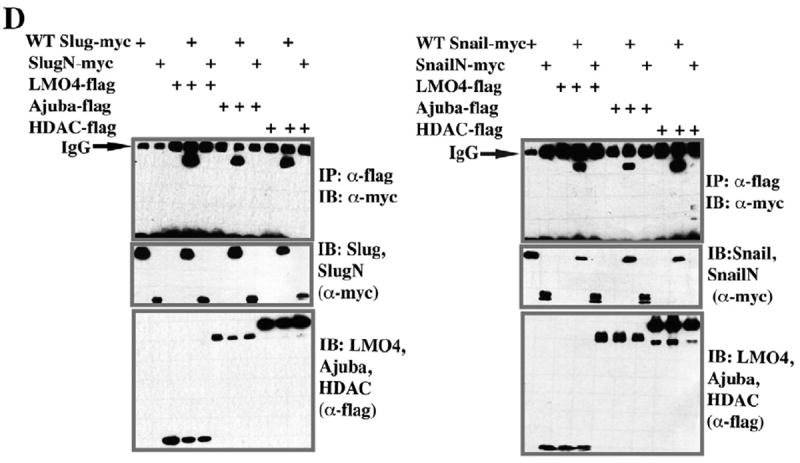
Another LIM domain containing protein, Ajuba LIM, has previously been shown to interact with Snail via its LIM domains, in a manner dependent on Snail’s SNAG domain, and is proposed to play a central role in SNAG-mediated HDAC recruitment (Langer et al., 2008). We therefore asked if the N-terminus of either Slug or Snail was sufficient to recruit LMO4, Ajuba, or HDAC1. Surprisingly, co-immunoprecipitation assays indicated that the SNAG domain containing N-termini of Slug or Snail is not sufficient for binding LMO4, Ajuba, or HDAC1 (Fig. 5D). Thus, while the SNAG domain may play an important role in recruiting the transcriptional repression machinery to Snail (and to a lesser extent Slug), there must also be residues in the c-terminus of the proteins that are essential to this role.
Slug and Snail display differences in their abilities to recruit Ajuba and HDAC
It had previously been proposed that Ajuba and HDAC are recruited to Snail family proteins via the SNAG domain. Our finding that neither the Slug nor Snail N-terminus is sufficient to recruit these factors provides the first evidence that sequences outside the Nterminus are also required. Given our finding that Slug and Snail exhibit differences in the degree to which the SNAG domain is required to recruit LMO4, we asked if they also behaved differently with respect to their recruitment of Ajuba or HDAC. To test this we utilized full length Slug or Snail, the zinc finger regions of these factors alone, or forms of the proteins in which their SNAG domains had been deleted. Co-immunoprecipitation assays were used to determine the ability of these proteins to bind Ajuba or HDAC1. As we had found for LMO4 (Fig. 5C), and consistent with previously published findings (Langer et al., 2008), we found that deletion of the Snail SNAG domain led to loss of Ajuba recruitment (Fig. 6A). In marked contrast to these findings, a mutant Slug protein missing the SNAG domain retained its ability to interact with Ajuba. Moreover, we found that the C-terminal zinc finger domain of Slug was sufficient for Ajuba recruitment (Fig. 6A). When we examined the ability of Slug and Snail mutants to interact with HDAC1 we found analogous results (Fig. 6B)—the zinc finger domain of Slug, but not Snail, was sufficient for HDAC1 binding. The findings indicate that there are differences in how Slug and Snail assemble transcriptional co-regulatory complexes that may have important functional consequences. These results also indicate that although they are both LIM domain containing adaptor proteins, Ajuba and LMO4 display differences in how they interface with Snail family proteins.
Fig. 6.
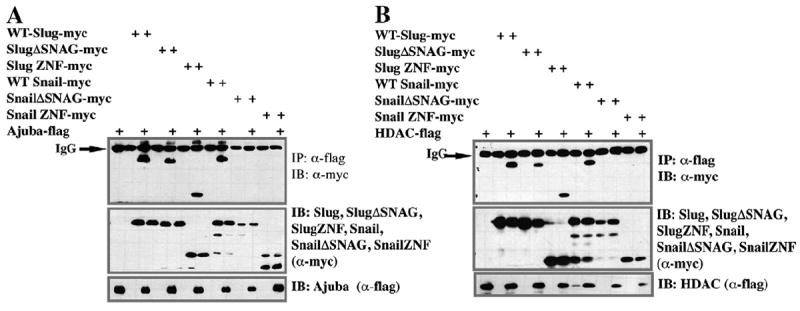
Slug and Snail utilize different domains to recruit Ajuba and HDAC. (A–B) Co-immunoprecipitation (IP) assay comparing the protein domains of (myc-tagged) Slug and Snail required for interaction with flag-tagged HDAC1 (A) or Ajuba (B). Deletion of the SNAG domain of Snail but not Slug, leads to loss of interaction with both HDAC1 and Ajuba. Conversely, the zinc finger domain of Slug, but not Snail, is sufficient for both interactions. These findings reveal novel differences in how Slug and Snail interaction with transcriptional co-regulatory factors.
LMO4 can interact with Ajuba but not HDAC1
Because LMO4 and Ajuba are both LIM domain containing adaptor proteins that can interact with Snail family transcriptional repressors and potentially modulate their function, we asked if these two factors could themselves interact. LMO4 was co-expressed with either Slug (as a positive control) or with Ajuba. Co-immunoprecipitation assays demonstrated strong binding between LMO4 and Ajuba (Fig. 7A). We further asked if LMO4, like Ajuba, could interact with HDAC1. Although both Slug and Ajuba strongly interact with HDAC1 when co-expressed in early embryos, interaction between LMO4 and HDAC1 was at best very weak (Fig. 7B) and could not be detected in some experiments. This data indicate that LMO4 and Ajuba are functionally distinct adaptor proteins, with Ajuba but not LMO4 capable of mediating interaction between Slug/Snail proteins and HDAC1, thus facilitating transcriptional repression. Since LMO4 and Ajuba can interact, but only Ajuba interacts with HDAC1, we next examined the consequences of LMO4 co-expression on the ability of Ajuba to recruit HDAC1. For these experiments we utilized an HA tagged form of LMO4, myc-tagged Ajuba, and flag-tagged HDAC1 in three way co-immunoprecipitation assays. Co-expression of LMO4 was found to interfere with the interaction between Ajuba and HDAC1, suggesting a possible competition for Ajuba binding by HDAC1 and LMO4 (Fig. 7B).
Fig. 7.
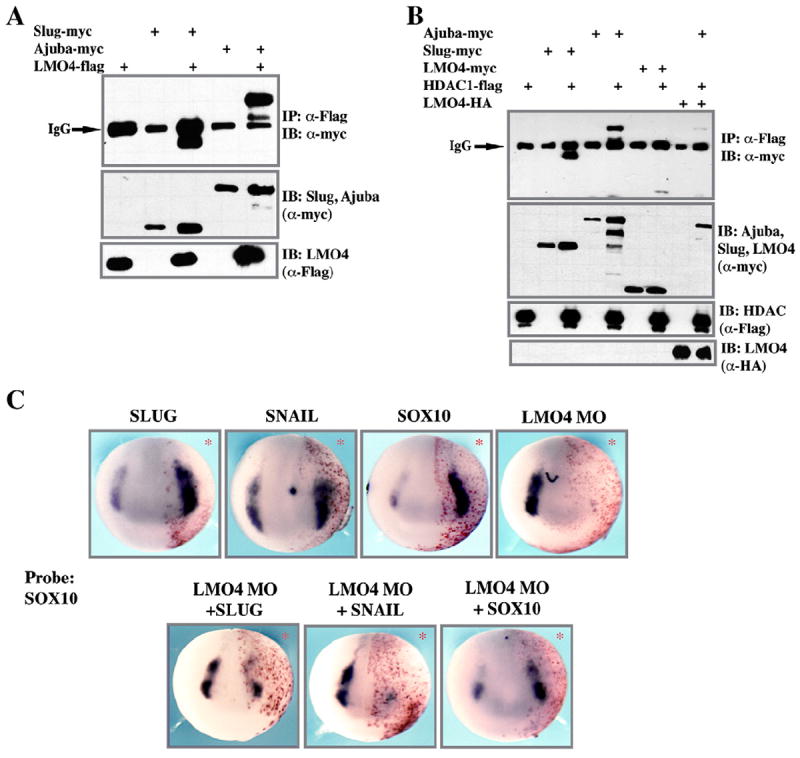
Functional roles for LMO4 in neural crest induction. (A) Co-IP assay demonstrating interaction between two LIM domain containing adaptor proteins, LMO4 and Ajuba. (B) Co-IP assay demonstrating that HDAC interacts strongly with Ajuba but not LMO4, indicating that the functions of these two adaptor proteins can be distinguished. Co-expression of LMO4 abrogates the interaction between Ajuba and HDAC, most likely by competing for Ajuba binding. (C) LMO4 is necessary for Slug/Snail-mediated, but not Sox10-mediated, neural crest induction. Whole mount in situ hybridization of embryos co-injected in one cell at the eight-cell stage with LMO4 morpholino and β-gal lineage tracer alone or in the presence of mRNA encoding Slug, Snail or Sox10. Embryos were examined at mid-neurula stages for expression of neural crest marker Sox10. Whereas all three NC regulatory factors can induce ectopic Sox10 expression, only Sox10 can do so in LMO4 depleted cells. These findings point to a more direct the role for LMO4 in the function of Slug and Snail.
LMO4 is essential for Slug/Snail-mediated neural crest induction
Together the above findings indicate that LMO4 is an essential regulator of neural crest development. They further demonstrate that LMO4, while functionally distinct from the distantly related LIM adaptor protein Ajuba, can also interact with and potentially modulate the function of Slug/Snail family proteins. We therefore hypothesized that the requirement for LMO4 in the establishment of the neural crest precursor pool might stem from its ability to interact with Slug and Snail and contribute to their function. If this hypothesis is correct, then we might expect that loss of LMO4 would have more severe consequences for Slug/Snail-mediated induction of neural crest markers than for neural crest induction by other components of the NC-GRN. To test this hypothesis we compared the effects of LMO4 depletion on Slug vs. Sox10-mediated induction of Sox10. Overexpression of Slug, Snail or Sox10 strongly induces ectopic expression of this marker (Fig. 7C; Slug injected: 83% expanded n=36; Snail injected: 71% expanded, n=84; Sox10 injected: 80% expanded, n=91). Conversely, LMO4-depleted embryos display a severe loss of Sox10 expression (Fig. 7C LMO4 MO: 85% reduced, n=100). When co-injected with LMO4 MO, neither Slug nor Snail can induce ectopic Sox10 expression (LMO4 MO+Slug: 89% reduced, n=55; LMO4 MO+Snail: 89% reduced, n=90). By contrast, Sox10 induces strong expression of Sox10 even in LMO4 depleted embryos (LMO4 MO+Sox10: 87% increased, n=46). These findings demonstrate that LMO4 depletion impacts Slug/Snail function to a far greater extent than it does Sox10 function. Together these data support a model in which the requirement for LMO4 in the neural crest is a least partially linked to the ability of this adaptor protein to modulate Slug/Snail function.
Discussion
A gene regulatory network underlying the formation, migration and differentiation of neural crest cells has begun to be delineated (Betancur et al., 2010; LaBonne and Bronner-Fraser, 1999; Sauka-Spengler and Bronner-Fraser, 2008). A central challenge to understanding complex developmental processes such as neural crest development on a systems-wide level is to understand how the function of each protein in the network is controlled, often by post-translational modification and/or by interacting factors. In this study we report on a novel component of the NC-GRN, the LIM adaptor protein LMO4. We show that LMO4 is expressed in neural crest forming regions, and that both depletion of LMO4 and LMO4 misexpression, causes defects in the formation of neural crest precursors and in the patterning of the early embryonic ectoderm more broadly. As LMO proteins lack DNA binding domains, we asked if this adaptor protein could interact with previously identified components of the NC-GRN. We demonstrate that LMO4 strongly and directly interacts with Slug/Snail family proteins and is required for their function.
LMO4 is a member of the LMO protein family, a group of four nuclear LIM Only factors (designated LMO1-4) that consist almost entirely of LIM domains (Kenny et al., 1998; Yu et al., 2008). LMO4 consist of two-tandem LIM domains connected by a small linker region and short N-terminal (22 residues) and C-terminal (25 residues) ends. LMO proteins localize to the nucleus and can bind with high affinity to the widely expressed nuclear LIM interactor (NLI, also known as CLIM or LIM domain binding protein LDB1) (Kenny et al., 1998; Mizunuma et al., 2003). Other characterized LMO-interacting proteins include Deaf-1, GET-1, HEN-1, BMP7, and SMAD proteins, and LMO factors have been found to participate in both transcriptional activation and repression (Bach, 2000; Grutz et al., 1998; Kudryavtseva et al., 2003; Lu et al., 2006; Novotny-Diermayr et al., 2005; Sum et al., 2002; Wang et al., 2004; Wang et al., 2007). Within the highly homologous LMO family, LMO4 is the most divergent and is the least well studied factor. It has been reported to be expressed in the thymus, brain, skin, pituitary gland, nervous system as well as the neural crest (Bach, 2000; Lane et al., 2002; McCollum et al., 2007; Setogawa et al., 2006; Sum et al., 2005). Interestingly, LMO4 is also highly expressed in breast cancer cells and at locations of active mesenchymal–epithelial interactions (Lu et al., 2006). Recently it has been shown that LMO4 can act as a co-activator for neurogenin 2 in the developing mouse cortex (Asprer et al., 2011). A role for a Xenopus LMO4-related protein in mesoderm patterning has previously been reported (de la Calle-Mustienes et al., 2003). However there is considerable sequence divergence between that protein (currently annotated as Xenopus laevis LMO4 gene 2) and Xenopus laevis LMO4 gene 1, which we report here. Importantly, LMO4 gene 1 is most closely related to human LMO4 (98% identical, 99% similar) and is therefore likely to be the true LMO4 orthologue in Xenopus. Moreover, the reported expression pattern for the LMO4 gene 2 differs significantly from the expression of LMO4 (LMO4 gene 1) we report here, with no expression in neural crest forming regions until fairly late stages (stage 19) (de la Calle-Mustienes et al., 2003).
Our finding that LMO4 specifically interacts with Slug and Snail, but not with other components of the NC-GRN, provides a potential mechanism for its requirement during the formation of neural crest precursor cells. Slug/Snail factors play a central role in the regulation of neural crest development. These closely related zinc finger transcriptional repressors are among the first factors expressed in response to neural crest inducing signals, and their function is required for the formation of the neural crest precursor cell population (Essex et al., 1993; LaBonne and Bronner-Fraser, 2000; Mayor et al., 1995). Snail family proteins also play a second, temporally distinct, role in the EMT/onset of migration of neural crest cells (Batlle et al., 2000; Cano et al., 2000; Hemavathy et al., 2000a; LaBonne and Bronner-Fraser, 2000). EMT is a highly conserved cellular process that is critical for numerous stages of embryonic development as well as for the metastasis of epithelially derived cancer (Peinado et al., 2007; Tucker, 2004; Yang and Weinberg, 2008). Snail proteins are among the core regulatory factors that control both developmental and pathological EMTs (Alves et al., 2009; Cano et al., 2000; Hemavathy et al., 2000a; Tucker, 2004). Invasive tumor cells frequently express high levels of Slug and/or Snail, and this is considered a marker for aggressive disease and poor prognosis (Alves et al., 2009; Shioiri et al., 2006; Uchikado et al., 2011). The mechanisms underlying transcriptional repression and EMT regulation by Snail family proteins are an important topic of investigation. Interestingly, their ability to regulate the stem cell like characteristics in neural crest cells, in addition to EMT, may be mirrored by an ability to promote the formation of cancer stem cells (Dumont et al., 2008; Polyak and Weinberg, 2009; Yu et al., 2007).
Because Snail factors can regulate both the acquisition of stem cell like properties (cell fate decisions) and the onset of EMT (cell behavior/morphology), their function must be subject to strict cell context dependent controls. All vertebrate Snail family members possess an NH2-terminal SNAG repression domain that appears to have been acquired at the base of the vertebrates (Hemavathy et al., 2000a). Transcriptional repression by vertebrate Snail family proteins has been reported to require the SNAG domain, which contributes to recruitment of HDAC1 other proteins to assemble a repressor complex (Cano et al., 2000; Comijn et al., 2001; Peinado et al., 2007). The SNAG domain of Snail has been shown to recruit the adaptor protein Ajuba, and it has been suggested that Ajuba functions as an obligate co-repressor for Snail-mediated repression (Ayyanathan et al., 2007; Langer et al., 2008). The Ajuba protein in turn can recruit Prmt5 (protein arginine methyltransferase 5) (Hou et al., 2008). It has been suggested that 14-3-3 proteins may bridge and stabilize Snail–Ajuba complexes and possibly also mediate connections with histone tails, thereby anchoring the complex to chromatin (Hou et al., 2010). It remains unclear, however, how this complex might assemble and be maintained on Snail target genes, what alternative regulatory complexes might also assemble, and whether any of those might carry out distinct functions.
While also a LIM domain containing adaptor protein, Ajuba is structurally and functionally distinct from LMO4 (Kanungo et al., 2000; Langer et al., 2008). Here we show that unlike Ajuba, LMO4 does not bind HDAC1 and therefore does not also function to bridge Slug/Snail recruitment of this component of the transcriptional repression machinery. Indeed, co-expression of LMO4 leads to loss of the interaction between Ajuba and HDAC, suggesting it could play an inhibitory roll in this context. Not surprisingly for an adaptor protein where stoichiometry is expected to play a central role, the amount of LMO4 present is critical for proper neural crest formation, with both LMO4 depletion and LMO4 misexpression causing the inhibition of some neural crest markers. The loss of function phenotype is of course the most critical, as it demonstrates the absolute requirement for LMO4 for the formation of neural crest precursor cells. Of interest is the observation that later expressed components of the NC-GRN are somewhat more sensitive to LMO4 depletion than are some of the earliest components, suggesting that LMO4 might act in concert with one or more of the earliest expressed factors to regulate the expression of downstream genes in the network. This is consistent with the finding that LMO4 directly interacts with Slug and Snail and is required for their function.
Another intriguing finding from this study is that LMO4 induced dramatic expression of Slug and Snail in the neural plate and non-neural ectoderm, while inhibiting the expression of other components of the NC-GRN. This finding suggests that LMO4 misexpression might first play a role in patterning events critical for Slug and Snail expression. Once these proteins are expressed, LMO4 can then physically interact with them in order to modulate their function, leading the observed inhibitory effects on other components of the NC-GRN. It will be important to elucidate the upstream interaction partners for LMO4 that contribute to its ability to dramatically upregulate the expression of Slug and Snail. Additionally, it will be essential to determine which proteins LMO4 recruits to Slug/Snail-dependent transcriptional regulatory complexes on target promoters, and to better understand the provocative difference between Slug and Snail brought to light by this study. Most centrally, however, our findings identify LMO4 as an essential new component of the NC-GRN, and shed novel mechanistic light on the regulation of Snail family transcriptional repressors, which play essential roles in both neural crest development and tumor progression.
Acknowledgments
The authors thank Joe Nguyen, Robert Beal, Deanna Wong and Rob Hartemayer for technical assistance; Allison Harney, Kara Nordin, and members of the lab for helpful discussions. S.O. acknowledges support from T32GM008061, CLIMB, and the Malkin Scholars program of the RHLCCC, S.S acknowledges support from the T32CA009560. This work was supported by NIH RO1CA114058 to CL.
References
- Alberga A, Boulay JL, Kempe E, Dennefeld C, Haenlin M. The snail gene required for mesoderm formation in Drosophila is expressed dynamically in derivatives of all three germ layers. Development. 1991;111:983–992. doi: 10.1242/dev.111.4.983. [DOI] [PubMed] [Google Scholar]
- Alves CC, Carneiro F, Hoefler H, Becker KF. Role of the epithelial–mesenchymal transition regulator Slug in primary human cancers. Front Biosci. 2009;14:3035–3050. doi: 10.2741/3433. [DOI] [PubMed] [Google Scholar]
- Asprer JS, Lee B, Wu CS, Vadakkan T, Dickinson ME, Lu HC, Lee SK. LMO4 functions as a co-activator of neurogenin 2 in the developing cortex. Development. 2011;138:2823–2832. doi: 10.1242/dev.061879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyanathan K, Peng H, Hou Z, Fredericks WJ, Goyal RK, Langer EM, Longmore GD, Rauscher F., Jr The Ajuba LIM domain protein is a corepressor for SNAG domain mediated repression and participates in nucleocytoplasmic Shuttling. Cancer Res. 2007;67:9097–9106. doi: 10.1158/0008-5472.CAN-07-2987. [DOI] [PubMed] [Google Scholar]
- Bach I. The LIM domain: regulation by association. Mech Dev. 2000;91:5–17. doi: 10.1016/s0925-4773(99)00314-7. [DOI] [PubMed] [Google Scholar]
- Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- Bellmeyer A, Krase J, Lindgren J, LaBonne C. The protooncogene c-myc is an essential regulator of neural crest formation in Xenopus. Dev Cell. 2003;4:827–839. doi: 10.1016/s1534-5807(03)00160-6. [DOI] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, Sauka-Spengler T. Assembling neural crest regulatory circuits into a gene regulatory network. Annu Rev Cell Dev Biol. 2010;26:581–603. doi: 10.1146/annurev.cellbio.042308.113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D, van Roy F. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7:1267–1278. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- de la Calle-Mustienes E, Lu Z, Cortes M, Andersen B, Modolell J, Gomez-Skarmeta JL. Xenopus Xlmo4 is a GATA cofactor during ventral mesoderm formation and regulates Ldb1 availability at the dorsal mesoderm and the neural plate. Dev Biol. 2003;264:564–581. doi: 10.1016/j.ydbio.2003.09.002. [DOI] [PubMed] [Google Scholar]
- del Barrio MG, Nieto MA. Overexpression of Snail family members highlights their ability to promote chick neural crest formation. Development. 2002;129:1583–1593. doi: 10.1242/dev.129.7.1583. [DOI] [PubMed] [Google Scholar]
- Dumont N, Wilson MB, Crawford YG, Reynolds PA, Sigaroudinia M, Tlsty TD. Sustained induction of epithelial to mesenchymal transition activates DNA methylation of genes silenced in basal-like breast cancers. Proc Natl Acad Sci U S A. 2008;105:14867–14872. doi: 10.1073/pnas.0807146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essex LJ, Mayor R, Sargent MG. Expression of Xenopus snail in mesoderm and prospective neural fold ectoderm. Dev Dyn. 1993;198:108–122. doi: 10.1002/aja.1001980205. [DOI] [PubMed] [Google Scholar]
- Fujita N, Jaye DL, Kajita M, Geigerman C, Moreno CS, Wade PA. MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell. 2003;113:207–219. doi: 10.1016/s0092-8674(03)00234-4. [DOI] [PubMed] [Google Scholar]
- Gammill LS, Bronner-Fraser M. Neural crest specification: migrating into genomics. Nat Rev Neurosci. 2003;4:795–805. doi: 10.1038/nrn1219. [DOI] [PubMed] [Google Scholar]
- Grau Y, Carteret C, Simpson P. Mutations and chromosomal rearrangements affecting the expression of snail, a gene involved in embryonic patterning in Drosophila melanogaster. Genetics. 1984;108:347–360. doi: 10.1093/genetics/108.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes HL, Chan TO, Zweidler-McKay PA, Tong B, Tsichlis PN. The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol Cell Biol. 1996;16:6263–6272. doi: 10.1128/mcb.16.11.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grutz G, Forster A, Rabbitts TH. Identification of the LMO4 gene encoding an interaction partner of the LIM-binding protein LDB1/NLI1: a candidate for displacement by LMO proteins in T cell acute leukaemia. Oncogene. 1998;17:2799–2803. doi: 10.1038/sj.onc.1202502. [DOI] [PubMed] [Google Scholar]
- Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62:1613–1618. [PubMed] [Google Scholar]
- Hall BK. The Neural Crest in Development and Evolution. Springer; New York: 1999. [Google Scholar]
- Heeg-Truesdell E, LaBonne C. A slug, a fox, a pair of sox: transcriptional responses to neural crest inducing signals. Birth Defects Res C Embryo Today. 2004;72:124–139. doi: 10.1002/bdrc.20011. [DOI] [PubMed] [Google Scholar]
- Hemavathy K, Ashraf SI, Ip YT. Snail/slug family of repressors: slowly going into the fast lane of development and cancer. Gene. 2000a;257:1–12. doi: 10.1016/s0378-1119(00)00371-1. [DOI] [PubMed] [Google Scholar]
- Hemavathy K, Guru SC, Harris J, Chen JD, Ip YT. Human Slug is a repressor that localizes to sites of active transcription. Mol Cell Biol. 2000b;20:5087–5095. doi: 10.1128/mcb.20.14.5087-5095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemavathy K, Meng X, Ip YT. Differential regulation of gastrulation and neuroectodermal gene expression by Snail in the Drosophila embryo. Development. 1997;124:3683–3691. doi: 10.1242/dev.124.19.3683. [DOI] [PubMed] [Google Scholar]
- Hou Z, Peng H, Ayyanathan K, Yan KP, Langer EM, Longmore GD, Rauscher FJ., III The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol. 2008;28:3198–3207. doi: 10.1128/MCB.01435-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Peng H, White DE, Wang P, Lieberman PM, Halazonetis T, Rauscher FJ., III 14-3-3 binding sites in the snail protein are essential for snail-mediated transcriptional repression and epithelial–mesenchymal differentiation. Cancer Res. 2010;70:4385–4393. doi: 10.1158/0008-5472.CAN-10-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanungo J, Pratt SJ, Marie H, Longmore GD. Ajuba, a cytosolic LIM protein, shuttles into the nucleus and affects embryonal cell proliferation and fate decisions. Mol Biol Cell. 2000;11:3299–3313. doi: 10.1091/mbc.11.10.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny DA, Jurata LW, Saga Y, Gill GN. Identification and characterization of LMO4, an LMO gene with a novel pattern of expression during embryogenesis. Proc Natl Acad Sci U S A. 1998;95:11257–11262. doi: 10.1073/pnas.95.19.11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht AK, Bronner-Fraser M. Induction of the neural crest: a multigene process. Nat Rev Genet. 2002;3:453–461. doi: 10.1038/nrg819. [DOI] [PubMed] [Google Scholar]
- Kudryavtseva EI, Sugihara TM, Wang N, Lasso RJ, Gudnason JF, Lipkin SM, Andersen B. Identification and characterization of Grainyhead-like epithelial transactivator (GET-1), a novel mammalian Grainyhead-like factor. Dev Dyn. 2003;226:604–617. doi: 10.1002/dvdy.10255. [DOI] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Neural crest induction in Xenopus: evidence for a two-signal model. Development. 1998;125:2403–2414. doi: 10.1242/dev.125.13.2403. [DOI] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Molecular mechanisms of neural crest formation. Annu Rev Cell Dev Biol. 1999;15:81–112. doi: 10.1146/annurev.cellbio.15.1.81. [DOI] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Snail-related transcriptional repressors are required in Xenopus for both the induction of the neural crest and its subsequent migration. Dev Biol. 2000;221:195–205. doi: 10.1006/dbio.2000.9609. [DOI] [PubMed] [Google Scholar]
- Lane ME, Runko AP, Roy NM, Sagerstrom CG. Dynamic expression and regulation by Fgf8 and Pou2 of the zebrafish LIM-only gene, lmo4. Gene Expr. 2002;Patterns 2:207–211. doi: 10.1016/s1567-133x(02)00061-3. [DOI] [PubMed] [Google Scholar]
- Langer EM, Feng Y, Zhaoyuan H, Rauscher F, Jr, Kroll KL, Longmore GD. Ajuba LIM proteins are snail/slug corepressors required for neural crest development in Xenopus. Dev Cell. 2008;14:424–436. doi: 10.1016/j.devcel.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin N, Kalcheim C. The Neural Crest. Second Edition. Cambridge University Press; Cambridge: 1999. [Google Scholar]
- Llorens A, Rodrigo I, Lopez-Barcons L, Gonzalez-Garrigues M, Lozano E, Vinyals A, Quintanilla M, Cano A, Fabra A. Down-regulation of E-cadherin in mouse skin carcinoma cells enhances a migratory and invasive phenotype linked to matrix metalloproteinase-9 gelatinase expression. Lab Invest. 1998;78:1131–1142. [PubMed] [Google Scholar]
- Lu Z, Lam KS, Wang N, Xu X, Cortes M, Andersen B. LMO4 can interact with Smad proteins and modulate transforming growth factor-beta signaling in epithelial cells. Oncogene. 2006;25:2920–2930. doi: 10.1038/sj.onc.1209318. [DOI] [PubMed] [Google Scholar]
- Martin AC, Gelbart M, Fernandez-Gonzalez R, Kaschube M, Wieschaus EF. Integration of contractile forces during tissue invagination. J Cell Biol. 2010;188:735–749. doi: 10.1083/jcb.200910099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC, Kaschube M, Wieschaus EF. Pulsed contractions of an actin–myosin network drive apical constriction. Nature. 2009;457:495–499. doi: 10.1038/nature07522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor R, Morgan R, Sargent MG. Induction of the prospective neural crest of Xenopus. Development. 1995;121:767–777. doi: 10.1242/dev.121.3.767. [DOI] [PubMed] [Google Scholar]
- McCollum CW, Amin SR, Pauerstein P, Lane ME. A zebrafish LMO4 ortholog limits the size of the forebrain and eyes through negative regulation of six3b and rx3. Dev Biol. 2007;309:373–385. doi: 10.1016/j.ydbio.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizunuma H, Miyazawa J, Sanada K, Imai K. The LIM-only protein, LMO4, and the LIM domain-binding protein, LDB1, expression in squamous cell carcinomas of the oral cavity. Br J Cancer. 2003;88:1543–1548. doi: 10.1038/sj.bjc.6600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama H, Scott IC, Cross JC. The transition to endoreduplication in trophoblast giant cells is regulated by the mSNA zinc finger transcription factor. Dev Biol. 1998;199:150–163. doi: 10.1006/dbio.1998.8914. [DOI] [PubMed] [Google Scholar]
- Nibu Y, Zhang H, Levine M. Interaction of short-range repressors with Drosophila CtBP in the embryo. Science. 1998;280:101–104. doi: 10.1126/science.280.5360.101. [DOI] [PubMed] [Google Scholar]
- Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- Nieto MA, Sargent MG, Wilkinson DG, Cooke J. Control of cell behavior during vertebrate development by Slug, a zinc finger gene. Science. 1994;264:835–839. doi: 10.1126/science.7513443. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin): a systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. Garland Publishing, Inc.; New York: 1994. [Google Scholar]
- Novotny-Diermayr V, Lin B, Gu L, Cao X. Modulation of the interleukin-6 receptor subunit glycoprotein 130 complex and its signaling by LMO4 interaction. J Biol Chem. 2005;280:12747–12757. doi: 10.1074/jbc.M500175200. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E, Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. I. Zygotic loci on the second chromosome. Dev Biol. 1984:267–282. doi: 10.1007/BF00848156. [DOI] [PubMed] [Google Scholar]
- Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2004;24:306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- Perl AK, Wilgenbus P, Dahl U, Semb H, Christofori G. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature. 1998;392:190–193. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner M. Snapshot: neural crest. Cell. 2010;143(486–486):e481. doi: 10.1016/j.cell.2010.10.025. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Sefton M, Sanchez S, Nieto MA. Conserved and divergent roles for members of the Snail family of transcription factors in the chick and mouse embryo. Development. 1998;125:3111–3121. doi: 10.1242/dev.125.16.3111. [DOI] [PubMed] [Google Scholar]
- Setogawa T, Shinozaki-Yabana S, Masuda T, Matsuura K, Akiyama T. The tumor suppressor LKB1 induces p21 expression in collaboration with LMO4, GATA-6, and Ldb1. Biochem Biophys Res Commun. 2006;343:1186–1190. doi: 10.1016/j.bbrc.2006.03.077. [DOI] [PubMed] [Google Scholar]
- Shioiri M, Shida T, Koda K, Oda K, Seike K, Nishimura M, Takano S, Miyazaki M. Slug expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Br J Cancer. 2006;94:1816–1822. doi: 10.1038/sj.bjc.6603193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spokony RF, Aoki Y, Saint-Germain N, Magner-Fink E, Saint-Jeannet JP. The transcription factor Sox9 is required for cranial neural crest development in Xenopus. Development. 2002;129:421–432. doi: 10.1242/dev.129.2.421. [DOI] [PubMed] [Google Scholar]
- Sum EY, O’Reilly LA, Jonas N, Lindeman GJ, Visvader JE. The LIM domain protein Lmo4 is highly expressed in proliferating mouse epithelial tissues. J Histochem Cytochem. 2005;53:475–486. doi: 10.1369/jhc.4A6553.2005. [DOI] [PubMed] [Google Scholar]
- Sum EY, Peng B, Yu X, Chen J, Byrne J, Lindeman GJ, Visvader JE. The LIM domain protein LMO4 interacts with the cofactor CtIP and the tumor suppressor BRCA1 and inhibits BRCA1 activity. J Biol Chem. 2002;277:7849–7856. doi: 10.1074/jbc.M110603200. [DOI] [PubMed] [Google Scholar]
- Taylor KM, LaBonne C. Modulating the activity of neural crest regulatory factors. Curr Opin Genet Dev. 2007;17:326–331. doi: 10.1016/j.gde.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Tucker RP. Neural crest cells: a model for invasive behavior. Int J Biochem Cell Biol. 2004;36:173–177. doi: 10.1016/s1357-2725(03)00243-7. [DOI] [PubMed] [Google Scholar]
- Uchikado Y, Okumura H, Ishigami S, Setoyama T, Matsumoto M, Owaki T, Kita Y, Natsugoe S. Increased Slug and decreased E-cadherin expression is related to poor prognosis in patients with gastric cancer. Gastric Cancer. 2011;14:41–49. doi: 10.1007/s10120-011-0004-x. [DOI] [PubMed] [Google Scholar]
- Vernon AE, LaBonne C. Slug stability is dynamically regulated during neural crest development by the F-box protein Ppa. Development. 2006;133:3359–3370. doi: 10.1242/dev.02504. [DOI] [PubMed] [Google Scholar]
- Wang N, Kudryavtseva E, Ch’en IL, McCormick J, Sugihara TM, Ruiz R, Andersen B. Expression of an engrailed-LMO4 fusion protein in mammary epithelial cells inhibits mammary gland development in mice. Oncogene. 2004;23:1507–1513. doi: 10.1038/sj.onc.1207288. [DOI] [PubMed] [Google Scholar]
- Wang N, Lin KK, Lu Z, Lam KS, Newton R, Xu X, Yu Z, Gill GN, Andersen B. The LIM-only factor LMO4 regulates expression of the BMP7 gene through an HDAC2-dependent mechanism, and controls cell proliferation and apoptosis of mammary epithelial cells. Oncogene. 2007;26:6431–6441. doi: 10.1038/sj.onc.1210465. [DOI] [PubMed] [Google Scholar]
- Yang J, Weinberg RA. Epithelial–mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Yu J, Ohuchida K, Nakata K, Mizumoto K, Cui L, Fujita H, Yamaguchi H, Egami T, Kitada H, Tanaka M. LIM only 4 is overexpressed in late stage pancreas cancer. Mol Cancer. 2008;93 doi: 10.1186/1476-4598-7-93. [DOI] [PMC free article] [PubMed] [Google Scholar]


