Abstract
The recent advances in the biological research have produced new biological disciplines with clinical applications in medicine and cardiology. The integration of multilevel biological data and the connection with the clinical practice reveal the potential of personalized medicine and nanotechnology with future implications for prognosis, diagnosis and management. In the post-genomic time period the new disciplines, systems biology, synthetic biology and translational medicine are emerging as significant research areas in biology and medicine with extension in the field of clinical medicine and cardiology. These disciplines, with their predictive, preventive and therapeutic potential, are formulating the concept of personalized management, with patient’s energetic involvement and participation in the diagnosis and treatment. Personalized medicine and cardiology, using biomarkers as health and disease indicators, encourage drug development and direct towards a better molecular comprehension of disease processes.
Keywords: systems biology, synthetic biology, translational cardiology, personalized cardiology, review
The term ‘biology’ includes the notion of function or purpose for the living systems which differentiates the science of biology from the other natural sciences of physics and chemistry1. The nonlinear interactions of the biological processes assign a restrictive role to classical biology and do not permit biology to explain the behavior of the components, or determine ‘how’ these components are interconnected, and therefore to interpret and understand the living world2. This led to the next step, to the molecular biology, which characterizes the structure of the biological material and delineates the mechanism of interconnections between molecular elements. Molecular biology was successful in identifying structure and pathways, but proved ineffective in the experimental prediction of biological events. Despite the progress of classical and molecular biology, there are objections to the belief that by studying separately the molecules, molecular biology can explain the complete biological phenomenon. The new biology should decode the functional interconnection of molecules, pathways and networks, and define biological concepts that are effective to explain physiological phenomena, diseased states and genetic and systems adaptation. Human diseases are complex states consisting of various inputs with positive and negative feedback mechanisms.
Systems biology
According to the classical reductionist strategy, only molecular or genetic diversions are responsible for abnormal biological conduct or genesis of pathological situations. In contrast, the philosophy of systems biology is based on the assumption that disturbances in complex biological systems and diseases are directed by the functional impact of integrated networks of molecules, genes or metabolites3. The new approach to understand the biological systems is to see them as a whole instead of explaining them with the reductionist point of view. The systems biology methodology decodes the way that the various biological components and networks are organized, combined and control each other. Also, it is important to recognize the emergent properties of the integrated biological network and their role in the diagnosis and therapy of different clinical situations4. Systems biology is a novel approach to decode the hidden information from genetic and molecular networks and to make significant steps towards drug development process, clinical medicine and personalized therapeutic interventions5-8.
Physiologists regard the term ‘systems biology’ as redundant because biology is basically integrative and physiology has been interested in systems description for a long time9. This position is supported by integrative physiologists who employ the new techniques and modalities used by systems biologists to study complex biological processes, like cardiac remodeling and heart failure10. These physiologists go a step further and advocate that systems biology should be integrated into physiology to create “Integrative Physiology 2.0”. The above opinion is not supported by the recent advances in the fields of cellular molecular networks and disease modeling, from the holistic understanding of the biological functions to personalized medicine.
In the field of complex cardiac system, systems biology integrates the information taken from the available multiple databases and produces experimental or computational models11. The top-down approach for multilevel biological analysis is the classical physiological approach12, while in systems biology three ways of analysis are described: bottom-up, middle-out and top-down13. As an example, according to systems biology approach the biological and clinical analysis of heart failure could be based in two biological systems of interaction: the functional composition (bottom-up direction) and functional decomposition (top-down direction)4.
In the existing and ever-growing number of public databases for systems biology, there are some inconsistencies in nomenclature and differences in conceptual understanding and terminology advanced by various research groups14,15.
Synthetic biology
The term ‘synthetic’ biology was originally invented as a scientific approach to overcome the limits of natural evolution and as a link between functional and evolutionary biology16. The main purpose of synthetic biology is to produce new functional modules from the combined action of different components. Synthetic biology studies cellular behavior, and constructs biological systems and cellular circuits with cells being build module by module in the bottom-up direction17, 18. This way, the role of synthetic biology is expanded and related to systems biology. Systems biology approach constructs modules and networks and gives an explanation for their building and for their emergent properties. Synthetic biology uses the same building blocks, modules and networks, to construct stable and robust biological circuits and networks19, and enables systems biology to break up the complex assembly and composition of cellular systems20. Synthetic biology creates biological networks in order to understand or redesign living complex systems, and to assist biotechnology industry in fundamental research for the improvement of human health, welfare and environment21. A recent article gives a new framework for synthetic biology using a Bayesian model selection and emphasizes the difference between inference (reconstruct the system with the observed data) and design (construct the system with the desired data)22.
Synthetic biology, after the first successes of constructing synthetic gene networks, is producing increasingly complex biological circuits and therapies for a variety of diseases23. Biotechnology companies are using its potential to reduce research time and cost for the production of chemicals, pharmaceutical substances, food ingredients and health care products24.
Translational cardiology
The term ‘evidence based’ medicine and/or ‘evidence based’ cardiology was established as a central concept in medical treatment during the last two decades of the 20th century. This concept meant to ask a proof for a specific treatment after statistical confirmation of its efficiency during execution of prospective experimental protocols. The emerging field of ‘translational’ medicine is related to ‘evidence based’ medicine, and as a concept refers to the translation of the experimental findings obtained in the bench of the research laboratory to terms of clinical practice25. There is a close relationship between medicine, medical technology and society at large, with nanotechnology as an example of emerging and innovative technology with social implications.
Translational cardiology transfers knowledge, from basic research and pre-clinical studies, to the clinical cardiology through well executed clinical trials. Thus, translational cardiology transfers the pre-clinical research from the field of cell-based cardiac tissue repair into early-phase clinical trials in patients with acute myocardial infarction or refractory myocardial ischemia. At the present time, cell priming, bio-nanotechnology and tissue engineering are coming up as valuable techniques for ischemic tissue repair and cell-based therapy application in clinical cardiology26.
As an example, is the translation of S100A1-based research from initial clinical observations in heart failure syndrome, over basic research experiments, back to the clinical setting on the verge of clinical trials27. The loss of cardiomyocyte Ca (2+) cycling integrity is significant for the development and progression of heart failure syndrome. The cardiomyocyte EF-hand Ca (2+) sensor protein S100A1 is a regulator both of sarcoplasmic reticulum, sarcomere and mitochondrial function, and probably the S100A1 gene therapy has a therapeutic potential for heart failure patients25. Another important translational study demonstrates the preclinical feasibility of long-term therapeutic effectiveness of cardiac AAV9-S100A1 gene therapy in a preclinical model of heart failure and opens the possibility for a clinical trial of S100A1 gene therapy for human heart failure28.
A. Personalized cardiology
Personalized cardiology is an ambitious target of systems biology, synthetic biology and translational cardiology, and intends to modify the practice of medicine producing a more predictive, preventive and individualized cardiology. The current approach to human disease is founded on reductionist principles of experimentation and analysis but holistic systems biology proposes a new method for diagnosis and therapy29. The individualized or personalized treatment is an application of the holistic systems biology that is based on modern molecular medicine and use of the large data stores for complex diseases (Figure 1).
Figure 1. Integration of clinical and ‘omics’ information, necessary for personalized cardiology.
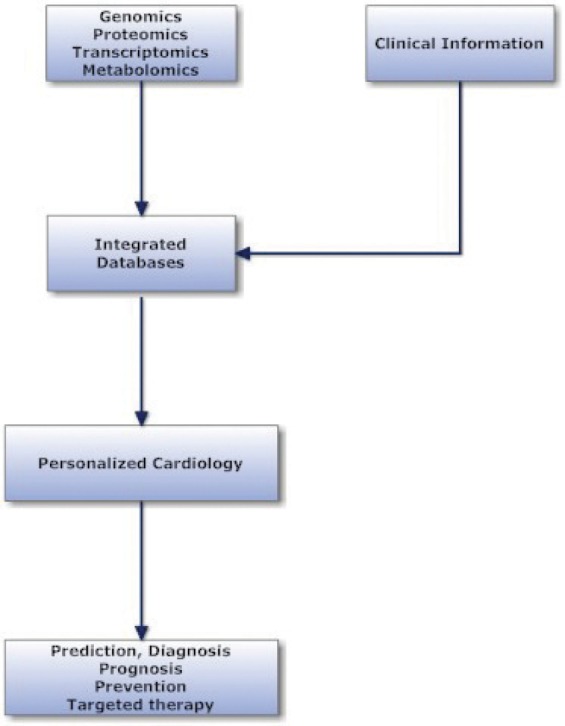
Genetic and epigenetic abnormalities or different environmental circumstances modify the human cardiovascular disease and produce various phenotypes. Therefore, the term of personalized cardiology refers to the prevention, diagnosis and therapy of cardiovascular diseases based on individual genomic, proteomic and metabolomic data30. The clinical practice is individualized especially in complicated cardiac states with multi-organ involvement and in acute heart failure syndromes, with neurohormonal adaptations, abnormal haemodynamics, and variant clinical picture including acute or chronic renal dysfunction31.
Also, the various diagnostic techniques in clinical practice should be expanded and adapted to a more individualized cardiology. A more personalized echocardiography should be advanced to real-time data acquisition from tissue and fluid motion, with improvements in 3D echocardiography, tissue Doppler and exercise stress echocardiography32.
The screening for changes in proteome and metabolome is a useful technique to detect specific biomarkers in coronary heart disease and heart failure. New cardiovascular biomarkers are under investigation for their diagnostic, prognostic and therapeutic value in patients with acute coronary syndrome (ACS) or after percutaneous coronary intervention (PCI). Thus, promising biomarkers like cardiac troponin (cTN), high sensitive cardiac troponin (hscTn), natriuretic peptides (NPs) and some future biomarkers like copeptin, choline and lipoprotein-associated phospholipase A2 (LP-PLA2), will improve the diagnostic and risk stratification processes and soon will be introduced into routine clinical practice33.The identification of all responsible genes for coronary artery disease remains elusive while the identification of variants to disease process is considered as a challenge34,35.
Genomics and Proteomics
Genomic and proteomic advances in the recent years have increased our understanding of cardiovascular diseases. A Mendelian mode of monogenic transmission to an offspring has been demonstrated in a variety of cardiovascular diseases, like hypertrophic cardiomyopathy36, Marfan syndrome37, long QT syndrome38 and arrhythmogenic right ventricular cardiomyopathy39. It appears that there is a genetic ground to more complex cardiovascular diseases like atherosclerosis and heart failure. These complex diseases without monogenic transmission, demonstrate a genotype with interdependency and interactions between various genes, and reciprocal activity between genes and environment.
In the majority of the patients with coronary artery disease there is genotypic heterogeneity, and therefore the clinical appearance has a multifactorial origin as the result of many genes action with minimal individual effects. Recently, many genome-associated studies have replicated a novel gene marker on chromosome locus 9p21 which is related to non-coding RNA gene and needs further research. Genetic information acquired from single nucleotide polymorphisms (SNPs) or haplotypes of genes related to atherothrombotic cardiovascular process, is expected to improve prediction and management of coronary artery disease. In the near future, a more personalized approach, that integrates clinical data with environmental and genomic risks, is anticipated to clarify risk stratification and early clinical intervention in high-risk persons40.
Proteome includes all existent proteins in a cell or tissue, and proteomics is the study of the proteome encoded by the genome. In contrast to the genome, proteome is not static and adapts to cellular circumstances and environmental conditions. The existing 30000 human genes are responsible for the construction of one million proteins41, but there is a protein diversity due to alternative splicing, multiple transcription start sites, changes in pre-messenger RNA, polyadenylation and post-translational modification of proteins42. The liquid chromatography-mass spectroscopy technique is a sophisticated invention that could evaluate complex biological material, fluid and tissue proteomes, enclosing many biological molecules like proteins or lipids43-45.
The proteomic cardiac studies receive significant assistance from online databases of human cardiac proteins and from international organizations that provide standards of data being discovered in proteomic studies30. In the online databases are included the HSC-2DPAGE46, HEART-2DPAGE47 and HP-2DPAGE48, which are displaying more than 6000 myocardial proteins. Two known international organizations are publishing standards of proteomic data for diagnosis, therapy, prevention and research: the Human Proteome Organization49 that provides guidelines for proteomic research and the Proteomics Division at the National Heart, Lung and Blood Institute50.
Proteomic studies have recognized changes in many protein groups that are related to cytoskeletal, sarcomeric and extracellular matrix construction and function of the myocardium, to mitochondrial metabolism pathways in preconditioning, to calcium control mechanisms and to redox regulation51,52.
Pharmacological targeted therapy is the main field of clinical implementation of genomic and proteomic technological advances after the identification of specific molecular targets in disordered biochemical pathways. The use of anticoagulant drugs is an example of personalized medicine in the practice of modern cardiology53.Warfarin is used extensively in clinical medicine and cardiology, but it has a small therapeutic window with an uncertainty in dosing and inconsistency in patient’s reaction. The responsiveness to the warfarin depends on polymorphisms in genes with an impact to metabolism (CYP2C9) and to pharmacodynamic response (VKORC1). Two allelic variants, CYP2C9*2 and CYP2C9*3, are associated with impaired hydroxylation of S-warfarin and inefficient warfarin metabolism54. Individuals having one or more CYP2C9 variant alleles require a low warfarin dose and have an increased risk of bleeding. Also, variants in the gene VKORC1, that encodes vitamin K epoxide reductase complex 1, are associated with reduced expression of VKORC1 and lower response to warfarin. The VKORC1 haplotypes can explain differences in dose requirements while the molecular mechanism of this response is regulated at the transcriptional level55.
In interventional cardiology and in patients with ACS, it is essential to individualize the antiplatelet therapy. Particularly in patients with ACS, who are submitted to PCI and stent implantation, the use of dual antiplatelet therapy with aspirin and clopidogrel, a P2Y12 receptor antagonist, is considered as the mainstay of the management for preventing future cardiovascular events. The object of many recent trials is the assessment of the relationship between the genetic variation in cytochrome P450 (CYP) isoenzymes and the pharmacokinetic response of the clopidogrel. There is significant variability in the antiplatelet effect between patients due to drug interactions, clinical factors, and the presence of CYP2C19 loss of function alleles that obstruct the metabolism of clopidogrel to its active form56. There is 3-fold increase in .stent thrombosis among patients with the CYP2C19*2 genotype who were treated with clopidogrel57. Therefore, in patients with acute coronary syndromes, genotyping for a CYP2C19 loss of function variant could be considered58.
Also, the pharmacodynamic response to clopidogrel is variable due to impaired activity of CYP3A4 enzyme. Clopidogrel therapy is ineffective in 10-30% of the treated patients while the resistance and suboptimal response to aspirin is about 5.5-9.5% and 23.8% accordingly59,60. Probably, platelet function tests measuring the effect of clopidogrel on the P2Y12 receptor and upcoming clinical trials would change our approach to antiplatelet therapy and lead to a more personalized cardiology61.
Three hundred patients were evaluated after PCI for changes of the clopidogrel platelet reactivity (PR) and its relationship with genotype and clinical outcomes. In these patients, the PR decreased from baseline to one month while the genotype (gene polymorphisms, CYP2C19*2, *17, CYP3A5*3, and ABCB1) influenced approximately 18% of this trend62,63.
The beta-blockers are an important group of drugs for heart failure patients. Genetic variants of the beta1-adrenergic receptor are considered therapeutic beta-blocker targets. In humans, polymorphisms at amino acid residue 389 (Arg/Gly) of the beta1-adrenergic receptors, predisposes to heart failure due to hyperactive signaling programs guiding to ventricular dysfunction64. The homozygosity for Arg389 was characterized by left ventricular functional improvement when these patients with heart failure were treated with carvedilol64. Homozygotes with Arg389 treated with bucindolol, had a 38% reduction in mortality, and 34% reduction in mortality or hospitalization compared with placebo65.
The angiotensin-converting enzyme (ACE) deletion allele (ACE-D) is associated with increased renin-angiotensin-aldosterone system (RAAS) activation and in patients with systolic dysfunction was associated with a significantly poorer transplant-free survival66. The use of higher doses of ACE inhibitors reduced the impact of the ACE-D allele in patients with systolic dysfunction, while the advantages of using beta-blockers and high-dose ACE inhibitors seem to be the greatest for DD patients67.
Genetically based individual differences, are considered as major determinants of left ventricular remodeling. The application of molecular imaging techniques, to decipher the cellular and molecular mechanisms of the left ventricular remodeling, contributes to a personalized assessment and follow-up in patients with heart failure68.
‘Theranostics’ is the scientific field that combines diagnostic methodology and therapy and incorporates the entities of personalized medicine, pharmacodiagnostics, integrated medicine and nanotechnology. The development of molecular-imaging techniques, like MRI and optical imaging, is based on nanoparticles, while drug-delivery approaches are important in order to understand the biomedical processes and therapies at a molecular level69. The molecular imaging technique is targeting molecular and cellular sites designed to visualize disease-associated molecules and cells, to assess disease progression and to evaluate the in vivo molecular effects of drugs70. The single nucleotide polymorphism (SNP) mapping technique is effective to detect genes important for coronary artery disease genesis and progression, and advances the concept of personalized medicine 71,72.
Trancriptomics
Transcriptomics is the study of the transcriptome, which includes the complete set of mRNA transcripts in the cell generated by the genome, and reflects the genes that are expressed at any given time. In contrast to the genome, which is fixed for a given cell line, the transcriptome can change with external environmental conditions. The microarray analysis is a technology that enables the quantification of many thousands of mRNA transcripts, detects new molecular abnormalities, produces new clinical biomarkers, and explores drug efficacy73. In human heart failure, transcriptional regulation and transcriptome variability was studied with microarray analysis74,75. Gene expression and transcription analysis in human heart failure was portrayed in ischemic and nonischemic cardiomyopathy76, and in patients supported with left ventricular assist devices77. The integration of clinical assessment (NYHA class) with T cell receptor signaling gene expression was proposed as a model to predict survival of heart failure patients78.
MicroRNAs (miRNAs, miRs) are a class of small (22-nucleotide) noncoding RNAs, post-transcriptional regulators of gene expression, which can link to messenger RNA transcripts79. The microRNAs are an endogenous class of small RNA molecules that negatively regulate gene expression and mediate post-transcriptional repression (inhibit translation) or mRNA degradation80. The miR-133 and miR-1 are expressed in cardiac and skeletal muscle, and regulate myogenesis, cardiac development, cardiac performance and cardiomyocyte hypertrophy, while other microRNAs participate on the myocardial growth, electrical balance and angiogenesis78. Probably, future experimental and clinical research on microRNAs will contribute to sudden cardiac death prevention and heart failure treatment targeting cardiac fibrosis, hypertrophy, stem cell differentiation, cardiomyocyte survival, apoptosis and myocardial failure through modulation of cardiac microRNAs81,82.
Genes were identified indicating the important role of chemokines, cell-extracellular matrix and lipoprotein alterations in the pathophysiology of acute myocardial infarction83. Also, were identified genes preferably expressed in atrial cardiomyocytes and proposed to be tested as potential biomarkers for atrial stress84.
Metabolomics
A number of small molecules named metabolites reside in the human cells or tissues with a significant biological effect on health and disease. There is speculation about the exact number of metabolites normally existing in human cells, but it is calculated to be a few thousands. The discipline of metabolomics studies the biological impact of metabolites under normal circumstances and during the state of a disease, and identifies novel biomarkers and new drugs85. In transgenic mouse model, are described experimental methods able to determine genes, proteins and metabolites involved in the three processes of atherosclerosis: lipid metabolism, inflammation, and tissue changes86.
Three complementary approaches are used for metabolic research: metabolomic fingerprinting (metabolites altered in a disease), metabolomic profiling (metabolites that participate in a targeted pathway), and metabolomic footprinting (monitoring metabolites that are secreted or fail to be taken up by a cell or tissue)87.
The main objective of metabolomics studies is to identify and intervene in specific locations of metabolic pathways with advantageous effect in early detection, metabolic individuality, exact diagnoses and ultimate halt of disease processes. In patients with primary dilated cardiomyopathy, 61 metabolites were found to be significantly different between people with primary dilated cardiomyopathy and control individuals88. This metabolomic profiling identifies biomarkers of primary dilated cardiomyopathy that probably have protective or harmful effects on cardiac structure and function.
Conclusions
In this paper an overview is presented of the new disciplines of systems biology, synthetic biology and translational medicine with a focus on cardiology. These new fields of knowledge integrate data from physiological measurements, genetic and molecular networks, and clinical findings. The recent advances in these disciplines improve the status of current practice of medicine and cardiology, and guide to a more personalized cardiology with personal participation in diagnostic, preventive and therapeutic decisions.
Conflict of Interest Statement:
The corresponding author confirms that there are no financial or other relations that could lead to a conflict of interest.
References
- 1.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402(Supp):C47–52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 2.Boogerd FC, Bruggeman fj, Hofmeyr J-H S, Westerhoff HV. Systems Biology, Philosophical Foundations. Elsevier BV. 2007 [Google Scholar]
- 3.Louridas GE, Kanonidis IE, Lourida KG. Systems biology in heart diseases. Hippokratia. 2010;14:10–16. [PMC free article] [PubMed] [Google Scholar]
- 4.Louridas GE, Lourida KG. A conceptual paradigm of heart failure and systems biology approach. Intern J Cardiol. 2011;doi:10.1016/j.ijcard.2011.07.014 doi: 10.1016/j.ijcard.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 5.Van der Greef J, Hankemeier T, McBurney RN. Metabolomics-based systems biology and personalized medicine: moving towards n=1 clinical trials? Pharmacogenomics. 2006;7:1087–1094. doi: 10.2217/14622416.7.7.1087. [DOI] [PubMed] [Google Scholar]
- 6.Barabasi AL, Oltvai ZN. Network biology: Understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 7.WenJun Zhang. Network Biology: an exciting frontier science. Network Biology. 2011;1:79–80. [Google Scholar]
- 8.Ferrarini A. Some steps forward in semi-quantitative networks modeling. Network Biology. 2011b;1:72–78. [Google Scholar]
- 9.Joyner MJ. Physiology: alone at the bottom, alone at the top. J Physiol. 2011;589:1005. doi: 10.1113/jphysiol.2010.203893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuster DW, Merkus D, van der Velden J, Verhoeven AJ, Duncker DJ. “Integrative Physiology 2.0”: integration of systems biology into physiology and its application to cardiovascular homeostasis. J Physiol. 2011;589:1037–1045. doi: 10.1113/jphysiol.2010.201533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shreenivasaiah PK, Rho S, Kim T, Kim DH. An overview of cardiac systems biology. J Mol Cell Cardiol. 2008;44:460–469. doi: 10.1016/j.yjmcc.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Hester RL, Iliescu R, Summers R, Coleman TG. Systems biology and integrative physiological modeling. J Physiol. 2011;589:1053–1060. doi: 10.1113/jphysiol.2010.201558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohl P, Crampin EJ, Quinn TA, Noble D. Systems biology: an approach. Clin Pharmacol Ther. 2010;88:25–33. doi: 10.1038/clpt.2010.92. [DOI] [PubMed] [Google Scholar]
- 14.Ng A, Bursteinas B, Gao Q, Mollison E, Zvelebil M. Resources for integrative systems biology: from data through databases to networks and dynamic system models. Briefings in Bioinformatics. 2006;7:318–330. doi: 10.1093/bib/bbl036. [DOI] [PubMed] [Google Scholar]
- 15.Stein LD. Integrating biological databases. Nat Rev Genet. 2003;4:337–345. doi: 10.1038/nrg1065. [DOI] [PubMed] [Google Scholar]
- 16.Morange M. Synthetic biology: a bridge between functional and evolutionary biology. Biological Theory. 2009;4:368–377. [Google Scholar]
- 17.Nandagopal N, Elowitz MB. Synthetic biology: integrated gene circuits. Science. 2011;333:1244–1248. doi: 10.1126/science.1207084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwille P. Bottom-up synthetic biology: engineering in a tinkerer’s world. Science. 2011;333:1252–1254. doi: 10.1126/science.1211701. [DOI] [PubMed] [Google Scholar]
- 19.Chen BS, Chang CH, Lee HC. Robust synthetic biology design: Stochastic game theory approach. Bioinformatics. 2009;25:1822–1830. doi: 10.1093/bioinformatics/btp310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinkhabwala A, Guet CC. Uncovering cis-regulatory codes using synthetic promoter shuffling. PloS One. 2008;3:e2030. doi: 10.1371/journal.pone.0002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alberts B. Editorial: A grand challenge in biology. Science. 2011;333:1200. doi: 10.1126/science.1213238. [DOI] [PubMed] [Google Scholar]
- 22.Barnes CP, Silk D, Sheng X, Stumpf MP. Bayesian design of synthetic biological systems. Proc Natl Acad Sci USA. 2011 Aug 29;[Epub ahead of print] doi: 10.1073/pnas.1017972108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruder WC, Lu T, Collins JJ. Synthetic biology moving into the clinic. Science. 2011;333:1248–1252. doi: 10.1126/science.1206843. [DOI] [PubMed] [Google Scholar]
- 24.Erickson B, Singh R, Winters P. Synthetic biology: regulating industry uses of new biotechnologies. Science. 2011;333:1254–1256. doi: 10.1126/science.1211066. [DOI] [PubMed] [Google Scholar]
- 25.Woolf SH. The meaning of translational research and why it matters. JAMA. 2008;299:211–213. doi: 10.1001/jama.2007.26. [DOI] [PubMed] [Google Scholar]
- 26.Tongers J, Losordo DW, Landmesser U. Stem and progenitor cell-based therapy in ischaemic heart disease: promise, uncertainties, and challenges. Eur Heart J. 2011;32:1197–1206. doi: 10.1093/eurheartj/ehr018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rohde D, Brinks H, Ritterhoff J, Qui G, Ren S, Most P. S100A1 gene therapy for heart failure: a novel strategy on the verge of clinical trials. J Mol Cell Cardiol. 2011;50:777–784. doi: 10.1016/j.yjmcc.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 28.Pleger ST, Shan C, Ksienzyk J, Bekeredjian R, Boekstegers P, Hinkel R, et al. Cardiac AAV9-S100A1 gene therapy rescues post-ischemic heart failure in a preclinical large animal model. Sci Transl Med. 2011;3:92ra64. doi: 10.1126/scitranslmed.3002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loscalzo J, Barabasi A-L. Systems biology and the future of medicine. WIREs Syst Biol Med. 2011;DOI:10.1002/wsbm.144. doi: 10.1002/wsbm.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ouzounian M, Lee DS, Gramolini AO, Emili A, Fukuoka M, Liu PP. Predict, prevent and personalize: Genomic and proteomic approaches to cardiovascular medicine. Can J Cardiol. 2007;23(Suppl A):28A–33A. doi: 10.1016/s0828-282x(07)71003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aspromonte N, Cruz DN, Valle R, Ronco C. Management and monitoring of haemodynamic complications in acute heart failure. Heart Fail Rev. 2011; Feb 8;[Epub ahead of print] doi: 10.1007/s10741-011-9229-3. [DOI] [PubMed] [Google Scholar]
- 32.Shizukuda Y, Bhatti S, Munjal J, Hu YL, Harrelson A. Personalized echocardiography: clinical applications of advanced echocardiography and future directions. Future Cardiol. 2010;6:833–844. doi: 10.2217/fca.10.103. [DOI] [PubMed] [Google Scholar]
- 33.Searle J, Danne O, Muller C, Mockel M. Biomarkers in acute coronary syndrome and percutaneous coronary intervention. Minerva Cardioangiol. 2011;59:203–224. [PubMed] [Google Scholar]
- 34.Rosenson RS. New technologies personalize diagnostics and therapeutics. Curr Atheroscler Rep. 2010;12:184–186. doi: 10.1007/s11883-010-0103-x. [DOI] [PubMed] [Google Scholar]
- 35.Padmanabhan S, Hastie C, Prabhakaran D, Dominczak AF. Genomic approaches to coronary artery disease. Indian J Med Res. 2010;132:567–578. [PMC free article] [PubMed] [Google Scholar]
- 36.Geisterfer-Lowrance AA, Kass S, Tanigawa G, Vosberg HP, McKenna W, Seidman CE, et al. A molecular basis for familial hypertrophic cardiomyopathy: A beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 37.Kaartinen V, Warburton D. Fibrillin controls TGF-beta activation. Nat Genet. 2003;33:331–332. doi: 10.1038/ng0303-331. [DOI] [PubMed] [Google Scholar]
- 38.Modell SM, Lehmann MH. The long QT syndrome family of cardiac ion channelopathies: A HuGE review. Genet Med. 2006;8:143–155. doi: 10.1097/01.gim.0000204468.85308.86. [DOI] [PubMed] [Google Scholar]
- 39.Calkins H. Arrhythmogenic right ventricular dysplasia/cardiomyopathy. Curr Opin Cardiol. 2006;21:55–63. doi: 10.1097/01.hco.0000198984.70884.4d. [DOI] [PubMed] [Google Scholar]
- 40.Lee S, Shin D, Jang Y. Personalized medicine in coronary artery disease: insights from genomic research. Korean Circ J. 2009;39:129–137. doi: 10.4070/kcj.2009.39.4.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Humphery-Smith I. A human proteome project with a beginning and an end. Proteomics. 2004;4:2519–2521. doi: 10.1002/pmic.200400866. [DOI] [PubMed] [Google Scholar]
- 42.Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 43.Lane CS. Mass spectrometry-based proteomics in the life sciences. Cell Mol Life Sci. 2005;62:848–869. doi: 10.1007/s00018-005-5006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Listgarten J, Emili A. Statistical and computational methods for comparative proteomic profiling using liquid chromatography-tandem mass spectrometry. Mol Cell Proteomics. 2005;4:419–434. doi: 10.1074/mcp.R500005-MCP200. [DOI] [PubMed] [Google Scholar]
- 45.Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452:571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 46.Evans G, Wheeler CH, Corbett JM, Dunn MJ. Construction of HSC-2DPAGE: A two- dimensional gel electrophoresis database of heart proteins. Electrophoresis. 1997;18:471–479. doi: 10.1002/elps.1150180322. [DOI] [PubMed] [Google Scholar]
- 47.Pleissner KP, Sander S, Oswald H, Regitz-Zagrosek V, Fleck E. The construction of the World Wide Web-accessible myocardial two-dimensional gel electrophoresis protein database ‘HEART-2DPAGE’: A practical approach. Electrophoresis. 1996;17:1386–1392. doi: 10.1002/elps.1150170818. [DOI] [PubMed] [Google Scholar]
- 48.Muller EC, Thiede B, Zimny-Arndt U, Scheler C, Prehm J, Muller-Werdan U, et al. High-performance human myocardial two-dimensional electrophoresis database: Edition 1996. Electrophoresis. 1996;17:1700–1712. doi: 10.1002/elps.1150171107. [DOI] [PubMed] [Google Scholar]
- 49.HUPO (Human Proteome Organization) http://www.hupo.org.
- 50.NHLBI Proteomics. http://www.nhlbi-proteomics.org.
- 51.Sawicki G, Jugdutt BI. Detection of regional changes in protein levels in the in vivo canine model of acute heart failure following ischemia-reperfusion injury: Functional proteomics studies. Proteomics. 2004;4:2195–2202. doi: 10.1002/pmic.200300746. [DOI] [PubMed] [Google Scholar]
- 52.Arrell DK, Elliott ST, Kane LA, Guo Y, Ko YH, Pedersen PL, et al. Proteomic analysis of pharmacological preconditioning: Novel protein targets converge to mitochondrial metabolism pathways. Circ Res. 2006;99:706–714. doi: 10.1161/01.RES.0000243995.74395.f8. [DOI] [PubMed] [Google Scholar]
- 53.Kamali F, Pirmohamed M. The future prospects of pharmacogenetics in oral anticoagulation therapy. Br J Clin Pharmacol. 2006;61:746–751. doi: 10.1111/j.1365-2125.2006.02679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aithal GP, Day CP, Kesteven PJ, Daly AK. Association of polymorphisms in the cytochrome P450CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet. 1999;353:717–719. doi: 10.1016/S0140-6736(98)04474-2. [DOI] [PubMed] [Google Scholar]
- 55.Rieder MJ, Reiner AP, Cage BF, Nickerson DA, Eby CS, McLeod HL, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 56.Brandt T, Close SL, Iturria SJ, Payne CD, Farid NA, Ernest CS 2nd, et al. Common polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. J Thromb Haemost. 2007;5:2429–2436. doi: 10.1111/j.1538-7836.2007.02775.x. [DOI] [PubMed] [Google Scholar]
- 57.Montalescot G, Hulot JS, Collet JP. Stent thrombosis: who’s guilty? Eur Heart J. 2009;30:2685–2688. doi: 10.1093/eurheartj/ehp436. [DOI] [PubMed] [Google Scholar]
- 58.Wright RS, Anderson JL, Adams CD, Bridges CR, Casey DE Jr, Ettinger SM, et al. 2011 ACCF/AHA focused update incorporated into the ACC/AHA 2007 Guidelines for the management of patients with unstable angina/non-ST-elevation myocardial infarction. J Am Coll Cardiol. 2011;57:e215–367. doi: 10.1016/j.jacc.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 59.Podolec J, Gajos G, Budziaszek L, Czaniecka M, Kleczkoska A, Zmudka K. New approach to interventional cardiology treatment with personalized medicine. Przegl Lek. 2008;65:850–857. [PubMed] [Google Scholar]
- 60.Geisler T, Gawaz M. Individualized antiplatelet therapy: what can a clinical score contribute? Hamostaseologie. 2009;29:360–367. [PubMed] [Google Scholar]
- 61.Price MJ, Barker CM. Functional testing methods for the antiplatelet effect of P2Y12 receptor antagonists. Biomark Med. 2011;5:43–51. doi: 10.2217/bmm.11.1. [DOI] [PubMed] [Google Scholar]
- 62.Campo G, Parrinello G, Ferraresi P, Lunghi B, Tebaldi M, Miccoli M, et al. Prospective evaluation of on-clopidogrel platelet reactivity over time in patients treated with percutaneous coronary intervention. J Am Coll Cardiol. 2011;57:2474–2483. doi: 10.1016/j.jacc.2010.12.047. [DOI] [PubMed] [Google Scholar]
- 63.Angiolillo DJ. Editorial:Unraveling myths of platelet function and genetic testing. J Am Coll Cardiol. 2011;57:2484–2486. doi: 10.1016/j.jacc.2011.01.040. [DOI] [PubMed] [Google Scholar]
- 64.Mialet PJ, Rathz DA, Petrashevskaya NN, Hahn HS, Wagoner LE, Schwartz A, et al. Beta 1-adrenergic receptor polymorphisms confer differential function and predisposition to heart failure. Nat Med. 2003;9:1300–1305. doi: 10.1038/nm930. [DOI] [PubMed] [Google Scholar]
- 65.Liggett SB, Mialet-Perez J, Thaneemit-Chen S, Weber SA, Greene SM, Hodne D, et al. A polymorphism within a conserved beta(1)-adrenergic receptor motif alters cardiac function and beta-blocker response in human heart failure. Proc Natl Acad Sci USA. 2006;103:11288–11293. doi: 10.1073/pnas.0509937103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McNamara DM, Holubkov R, Janosko K, Palmer A, Wang JJ, MacGowan GA, et al. Pharmacogenetic interactions between beta-blocker therapy and the angiotensin-converting enzyme deletion polymorphism in patients with congestive heart failure. Circulation. 2001;103:1644–1648. doi: 10.1161/01.cir.103.12.1644. [DOI] [PubMed] [Google Scholar]
- 67.McNamara DM, Holubkov R, Postava L, Janosko K, MacGowan GA, Mathier M, et al. Pharmacogenetic interactions between angiotensin-converting enzyme inhibitor therapy and the angiotensin-converting enzyme deletion polymorphism in patients with congestive heart failure. J Am Coll Cardiol. 2004;44:2019–2026. doi: 10.1016/j.jacc.2004.08.048. [DOI] [PubMed] [Google Scholar]
- 68.Shirani J, Dilsizian V. Molecular imaging targets of cardiac remodeling. Curr Cardiol Rep. 2009;11:148–154. doi: 10.1007/s11886-009-0022-z. [DOI] [PubMed] [Google Scholar]
- 69.Pan D, Caruthers SD, Chen J, Winter PM, SenPan A, Schmieder AH, et al. Nanomedicine strategies for molecular targets with MRI and optical imaging. Future Med Chem. 2010;2:471–490. doi: 10.4155/fmc.10.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Osborn EA, Jaffer FA. The year in molecular imaging. J Am Coll Cardiol Cardiovasc Imaging. 2010;3:1181–1195. doi: 10.1016/j.jcmg.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seo D, Wang T, Dressman H, Herderick EE, Iversen ES, Dong C, et al. Gene expression phenotypes of atherosclerosis. Arterioscler Thromb Vasc Biol. 2004;24:1922–1927. doi: 10.1161/01.ATV.0000141358.65242.1f. [DOI] [PubMed] [Google Scholar]
- 72.Roberts R, Gollob M. Molecular cardiology and genetics in the 21st century-a primer. Curr Probl Cardiol. 2006;31:637–701. doi: 10.1016/j.cpcardiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 73.Heidecker B, Hare JM. The use of transcriptomic biomarkers for personalized medicine. Heart Fail Rev. 2007;12:1–11. doi: 10.1007/s10741-007-9004-7. [DOI] [PubMed] [Google Scholar]
- 74.Kaab S, Barth AS, Margerie D, Dugas M, Gebauer M, Zwermann L, et al. Global gene expression in human myocardium-oligonucleotide microarray analysis of regional diversity and transcriptional regulation in heart failure. J Mol Med. 2004;82:308–316. doi: 10.1007/s00109-004-0527-2. [DOI] [PubMed] [Google Scholar]
- 75.Boheler KR, Volkova M, Morrell C, Garg R, Zhu Y, Margulies K, et al. Sex- and age-dependent human transcriptome variability: Implications for chronic heart failure. Proc Natl Acad Sci USA. 2003;100:2754–2759. doi: 10.1073/pnas.0436564100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kittleson MM, Minhas KM, Irizarry RA, Ye SQ, Edness G, Breton E, et al. Gene expression analysis of ischemic and nonischemic cardiomyopathy: Shared and distinct genes in the development of heart failure. Physiol Genomics. 2005;21:299–307. doi: 10.1152/physiolgenomics.00255.2004. [DOI] [PubMed] [Google Scholar]
- 77.Margulies KB, Matiwala S, Cornejo C, Olsen H, Craven WA, Bednarik D. Mixed messages: Transcription patterns in failing and recovering human myocardium. Circ Res. 2005;96:592–599. doi: 10.1161/01.RES.0000159390.03503.c3. [DOI] [PubMed] [Google Scholar]
- 78.Vanburen P, Ma J, Chao S, Mueller E, Schneider DJ, Liew CC. Blood gene expression signatures associate with heart failure outcomes. Physiol Genomics. 2011;43:392–397. doi: 10.1152/physiolgenomics.00175.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Port JD, Sucharov C. Role of microRNAs in cardiovascular disease: therapeutic challenges and potentials. J Cardiovasc Pharmacol. 2010;56:444–453. doi: 10.1097/FJC.0b013e3181f605b6. [DOI] [PubMed] [Google Scholar]
- 80.Zorio E, Medina P, Rueda J, Millan JM, Arnau MA, Beneyto M, et al. Insights into the role of microRNAs in cardiac diseases: from biological signaling to therapeutic targets. Cardiovasc Hematol Agents Med Chem. 2009;7:82–90. doi: 10.2174/187152509787047676. [DOI] [PubMed] [Google Scholar]
- 81.Wang N, Zhou Z, Liao X, Zhang T. Role of microRNAs in cardiac hypertrophy and heart failure. IUBMB Life. 2009;61:566–571. doi: 10.1002/iub.204. [DOI] [PubMed] [Google Scholar]
- 82.Ikeda S, Pu WT. Expression and function of microRNAs in heart disease. Curr Drug Targets. 2010;11:913–925. doi: 10.2174/138945010791591304. [DOI] [PubMed] [Google Scholar]
- 83.Muller O, Delrue L, Hamilos M, Vercauteren S, Ntalianis A, Trana C, et al. Transcriptional fingerprint of human whole blood at the site of coronary occlusion in acute myocardial infarction. EuroIntervention. 2011;7:458–466. doi: 10.4244/EIJV7I4A75. [DOI] [PubMed] [Google Scholar]
- 84.Maas AH, De Jong AM, Frederiks J, Smit MD, Gouweleeuw L, de Boer RA, et al. Cardiac gene expression profiling-the quest for an atrium-specific biomarker. Neth Heart J. 2010;18:610–614. doi: 10.1007/s12471-010-0844-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Van der Greef J, McBurney RN. Rescuing drug discovery and drug development: in vivo systems pathology and systems pharmacology. Nat Rev Drug Discov. 2005;4:961–967. doi: 10.1038/nrd1904. [DOI] [PubMed] [Google Scholar]
- 86.Oresic M, Clish CB, Davidov EJ, Verheij E, Vogels J, Havekes LM, et al. Phenotype characterization using integrated gene transcript, protein and metabolite profiling. Appl Bioinformatics. 2004;3:205–217. doi: 10.2165/00822942-200403040-00002. [DOI] [PubMed] [Google Scholar]
- 87.Barderas MG, Laborde CM, Posada M, de la Cuesta F, Zubin I, Vivanco F, et al. Metabolomic profiling for identification of novel potential biomarkers in cardiovascular diseases. J Biomed Biotechnol. 2011;doi:10.1155/2011/790132 doi: 10.1155/2011/790132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alexander D, Lombardi R, Rodriguez G, Mitchell MM, Marian AJ. Metabolomic distinction and insights into the pathogenesis of human primary dilated cardiomyopathy. Eur J Clin Invest. 2011;41:527–538. doi: 10.1111/j.1365-2362.2010.02441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


