Summary
Gene amplification is defined as a copy number (CN) increase in a restricted region of a chromosome arm, and is a mechanism for acquired drug resistance and oncogene activation. In multiple myeloma (MM), high CNs of genes in a 1q12~23 amplicon have been associated with disease progression and poor prognosis. To investigate the mechanisms for gene amplification in this region in MM, we performed a comprehensive metaphase analysis combining G-banding, fluorescence in situ hybridization, and spectral karyotyping in 67 patients with gain of 1q. In six patients (9%), evidence for at least one breakage-fusion-bridge (BFB) cycle was found. In three patients (4%), extended ladders of 1q12~23 amplicons were identified. Several key structures that are predicted intermediates in BFB cycles were observed, including: equal-spaced organization of amplicons, inverted repeat organization of amplicons along the same chromosome arm, and deletion of sequences distal to the amplified region. The 1q12 pericentromeric heterochromatin region served as both a recurrent breakpoint as well as a fusion point for sister chromatids, and ultimately bracketed both the proximal and distal boundaries of the amplicon. Our findings provide evidence for a novel BFB mechanism involving 1q12 pericentromeric breakage in the amplification of a large number of genes within a 1q12~23 amplicon.
Keywords: Multiple myeloma, cytogenetics, gene amplification, pericentromeric heterochromatin, breakage-fusion-bridge cycles
Introduction
Gene amplification is defined as a copy number (CN) increase in a restricted region of a chromosome arm in which multiple copies of an amplicon are accumulated in cancer cells (Albertson, 2006). In multiple myeloma (MM) and B-cell lymphomas, disease progression is characterized in part by the clonal progression of complex cytogenetic aberrations of the proximal region of chromosome 1q. The amplification of CKS1B and the proximal 1q21 region in MM has been reported by fluorescence in situ hybridization (FISH) in about 40% of patients with newly diagnosed MM and in 70% of patients at relapse (Hanamura et al, 2006; Chang et al, 2006a). Recently, the integration of data from various types of high resolution genomic profiles including array comparative genomic hybridization (aCGH), gene expression profiling (GEP), and global single nucleotide polymorphism (SNP) arrays have all provided molecular evidence for the importance of genes in the proximal 1q in MM (Carrasco et al, 2006; Walker et al, 2006; Shaughnessy et al., 2007; Zhan et al., 2007; Chng et al; 2007; Decaux et al, 2009). Furthermore, both aCGH and GEP studies have identified relevant sub-classifications of MM based on the co-expression of a unique cluster of genes on chromosome 1 (Carrasco et al, 2006; Walker et al, 2006; Shaughnessy et al., 2007; Zhan et al., 2007). These studies have defined a region of proximal 1q with a marked enrichment of genes showing a gain and/or amplification which spans approximately a region of 10-15 Mb corresponding to a 1q21-23 amplicon in MM (Carrasco et al, 2006). This region contains a large number of possible candidate genes which show amplification and/or deregulated expression important in myeloma pathogenesis; these genes include MUC1, MCL1, PDZK1, IL6R, BCL9, CKS1B, PSMD4, UBAP2L, and UBE2Q1, among others (Treon et al, 2000; Zhang et al, 2002; Inoue et al, 2004; Shaughnessy, 2005; Chang et al 2006b; Hanamura et al, 2006; Carrasco et al, 2006; Walker et al, 2006; Shaughnessy et al, 2007; Zhan et al, 2007; Fabris et al, 2007; Decaux et al, 2008).
We have previously described a mechanism for the low level increase of CN involving jumping whole-arm translocations of 1q (JT1q) (Sawyer et al., 1998a). The JT1q produces an increase in CN for all the genes on the 1q, generally in the range of 3-4 in affected cells. FISH studies have provided evidence for a heterogeneous set of breakpoints within the 1q12~24 region (Le Baccon et al, 2001; Lestou et al, 2002; Itoyama et al, 2002; Sawyer et al, 2005). The involvement of pericentromeric heterochromatin in amplifications including both direct and inverted duplications, as well as triplications involving a number of different genes in the region, have been reported by several groups (Le Baccon et al, 2001; Lestou et al, 2002; Itoyama et al, 2002; Sawyer et al, 2005). Unfortunately, the origin of these focal pericentromeric amplification events has remained obscure, probably due to the large number of secondary chromosomal aberrations that mask the very early events of gene amplification.
A well-recognized pathway for intrachromosomal gene amplification is the breakage-fusion-bridge (BFB) cycle mechanism which results in megabase-long inverted repeats in a restricted region of a chromosome arm (Alberston, 2006). At least two alternative chromosomal mechanisms have been demonstrated for the initiation of BFB mechanism first described by McClintock (1951). In the telomere fusion model, the triggering mechanism is damaged or shortened telomeres which cause the end-to-end fusion of chromosomes. The telomeric fusion event produces an unstable dicentric chromosome, which is pulled apart during the next anaphase causing a double strand break (DSB) distal to the gene under selection. If the broken chromosome ends are not healed by a functional telomere or translocated to another chromosome, the sister chromatids may fuse, causing the cell to enter into repeated rounds of BFB cycles. The BFB cycles generate amplicons organized as inverted repeats (De Lange, 2005). An alternative initiation mechanism for BFB cycles has been demonstrated whereby common fragile sites (CFS) are activated by exposure to cytotoxic drugs, hypoxia, and deregulation of nucleotide pools (Toledo et al, 1992; Ma et al, 1993; Kuo et al, 1994; Coquelle et al, 1997, 1998; Durkin and Glover, 2007). CFS have been reported as hotspots for deletions, translocations, increased rates of sister chromatid exchange (SCE), and intrachromosomal gene amplification (Yunis and Soreng, 1984; Glover and Stein, 1987). Disrupted or delayed DNA replication causes the DSB in the CFS mechanism, ultimately resulting in BFB cycles and the same type of unstable chromosome intermediates as seen in the telomere model (Toledo et al, 1992; Ma et al, 1993; Kuo et al, 1994; Coquelle et al, 1997, 1998; Ciullo et al, 2002; Hellman et al, 2002; Miller et al, 2006).
Evidence for the origin of BFB cycles in primary tumours is extremely rare and to our knowledge has not been reported in patient samples of MM or other haematological malignancies. Here we provide evidence for intrachromosomal gene amplification of a 1q12~23 amplicon in which pericentromeric breakage mediates the progression of the BFB cycles. Surprisingly, it was the consistent secondary DSBs occurring in the 1q12 heterochromatin that ultimately defined both the proximal and distal boundaries of the amplicon. The finding of pericentromeric heterochromatin serving as a site-specific breakpoint in BFB cycles suggests that some type of epigenetic instability in this region may play a role in gene amplification. The identification of mechanisms involved in the origin of specific amplicons has both diagnostic and prognostic significance.
Patients and Methods
Patient sample selection was based on the presence of an abnormal karyotype with a suspected 1q aberration by G-banding identified in routine studies, and availability of ample archived methanol:acetic acid fixed cytogenetic cell pellet for FISH analysis. Sixty-seven patients with 1q aberrations were selected for screening of CN for CKS1B using a two-color FISH technique on metaphase chromosome spreads. Metaphase chromosomes were prepared from bone marrow, fine needle aspirates, and plural effusion specimens processed for routine chromosome studies by direct harvest, 24-, 48-, and 72-h cell cultures. Cell cultures were harvested for G-band preparations as previously described (Sawyer et al, 1985). Karyotype designations were made according to the International System for Human Cytogenetic Nomenclature (ISCN 2009). The Institutional Review Board of the University of Arkansas for Medical Sciences approved the research studies, and all subjects provided written informed consent approving the use of their samples for research purposes.
Fluorescence In Situ Hybridization (FISH) and Spectral Karyotyping (SKY)
Probes used to demark the centromeric and pericentromeric region included alpha sat (1p11.1-1q11.1), and satII/III (1q12) (Vysis, Downers Grove, IL ), respectively. Human BAC (bacterial artificial chromosome) clones were allocated according to publication of the human genome resource of the National Centre for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/projects/genome/guide/human/). To verify the specificity of the probes and confirm identification of the chromosome bands, we hybridized the probes to metaphase cells from peripheral-blood lymphocytes of normal donors. BAC clones RP11-153J19 (1p12), RP11-307C12 (CKS1B; 1q21), RP11-550P17 (1q22), RP11-375F2 (1q24.2) ,RP11-177M16 (1q25.1), RP11 -57D16 (1q25.2), and RP11-32D17 (1q31) were purchased from BACPAC Resources (Oakland, CA). BAC DNA was prepared with a NucleoBond plasmid purification kit (BD Biosciences Clontech, Palo Alto, CA). The FISH probes were labeled by a nick translation reaction to incorporate Spectrum-red/green-dUTP (Vysis, Downers Grove, IL) into BAC DNA. Briefly, 1 μg of BAC DNA was mixed with 10 μM of Spectrum-red dUTP or -green dUTP, 5 μM of dTTP, and 15 μM each of dATP, dCTP, and dGTP in 1x nick translation buffer and nick translation enzymes (Vysis, Downers Grove, IL). The reaction was carried out at 15°C for 2 h followed by ethanol precipitation, then suspended in 100 μl of DNA in situ hybridization solution (DAKO Co., Carpinteria, CA). Human Cot-1 DNA (Invitrogen Corp., Carlsbad, CA) was also added to block repetitive DNA sequences. The SKY probe mixture and hybridization reagents were prepared by Applied Spectral Imaging (Carlsbad, CA). Both FISH and SKY procedures were performed as previously described (Sawyer et al, 1998b; 2005) Image acquisition for FISH and SKY was performed using a SD200 Spectracube (Applied Spectral Imaging, Inc., Carlsbad, CA) mounted on a Zeiss Axioplan II microscope (Gottingen, Germany). DAPI (4′,6-diamidino-2-phenylindole) images were captured and then inverted and enhanced by SKYView software to produce G-band-like patterns on the chromosomes. Original magnification of all G-band and FISH images was 1000x. Original magnification for SKY images was 630x.
Results
Composite karyotype designations for G-bands and FISH CN for satII/III and CKS1B probes are provided in Table S1. All patients showed complex karyotypes by G-banding, and 47 patients showed only whole-arm unbalanced jumping translocations (JT1q) and/or isochromosome 1q. Twenty patients showed both proximal duplications/triplications and/or JT1qs (#s 4, 5, 6, 12, 16, 18, 24, 26, 28, 30, 31, 39, 43, 49, 56, 60, 63, 64, 66, 67). FISH analysis showed one patient (1%) with two copies of CKS1B, thirty-nine patients (58%) showed 3-4, 16 patients (23%) with 5-6, and 11 (16%) with a CN of 7-9 (#s 3,5,26,28,39,49,58,60,64,65,66) (Table S1). Six patients (#s 26,28,57,63,64, 66) were of particular interest because they demonstrated both a higher CN and the pattern of two red (sat II/II) signals flanking two green (CKS1B) signals, indicating an inverted duplication (Table 1). Five of these six patients also showed multiple copies of inverted duplications, suggesting that the possible origin of the inverted duplications were BFB cycles. In an effort to further characterize the origin for the focal amplifications of 1q12~23 in these six patients, we expanded the analysis by increasing the number of cells analyzed by FISH from 20 to 200 cells, and utilized additional BAC FISH probes for the proximal 1q, including RP11-550P17 (1q22), RP11-375F2 (1q24.2),RP11-177M16 (1q25.1), RP11-57D16 (1q25.2) (Table 1).
Table 1.
Modal chromosome number and copy number for FISH BAC probes in 1q12~23 region.
| Patient # |
Modal Chromo some # |
sat II/III (1q12) |
RP11- 307C12 (1q21) |
RP11- 550P17 (1q22) |
RP11-375F2 (1q24.2) |
RP11- 177M16 (1q25.1) |
RP11- 57D16 (1q25.2) |
|---|---|---|---|---|---|---|---|
| 26 | 45 | 7 | 7 | 7 | 7 | ||
| 28 | 50~51 | 4 | 9 | 4 | 4 | ||
| 57 | 53~55 | 4 | 6 | 6 | 6 | 2 | 2 |
| 63 | 49~51 | 6 | 6 | 4 | 4 | 3 | 3 |
| 64 | 54~56 | 7 | 7 | 7 | 5 | 3 | 3 |
| 66 | 54~55 | 4 | 9 | 2 | 2 | 2 | 2 |
Strikingly, after expanded analysis, in three of the six patients (#s 66, 64, 28) we identified similar types of unstable intermediates demonstrating ladder-like structures of inverted duplications (Figures 1-3). The ladders showed equal-spaced organization of the region containing copies of probes for CKS1B bracketed by copies of satII/III on each end. For example, Patient # 66 shows expansion of CN of CKS1B from four, to eight, to 16, and finally 18 copies in an inverted duplication pattern on the same chromosome arm (Figure 1). In this patient the initiating breakpoint for the BFB cycles was localized between probes for CKS1B (1q21) and RP11-550P17 (1q22), apparently resulting from an unbalanced translocation of 1q with chromosome 15. Interestingly, the inverted duplication in this patient consistently showed doublets of CKS1B, a pattern that suggests the initiation breakpoint for the BFB cycles is in close proximity to CKS1B. This type of doubling of CN for a gene under selection has been demonstrated previously in rodent cell lines and indicates that the doubling occurs as a result of a very short distance between the initiation breakpoint and the gene under selection (Coquelle et al, 1997). Moreover, the chromosomes 1 in this case also demonstrated isodicentric intermediates with eight copies of CKS1B (Figure 1B), and the expected next step of eight copies with the subsequent deletion of 1p (Figure 1C). When this chromosome underwent another cycle it produced unstable isodicentrics with 16 copies of CKS1B (Figure 1 D&E) corresponding to the expected intermediates in the expansion of the amplicon by BFB cycles. The breakpoints between the centromeres of the isodicentrics occurred in or just adjacent to the duplicated pericentromeric regions. In some metaphase chromosomes the sister chromatids show separation except at the fusion of the distal ends of the chromatids involving the satII/III sequences; this is the only point of attachment between the chromatids other than at the centromere (Figure 1F).
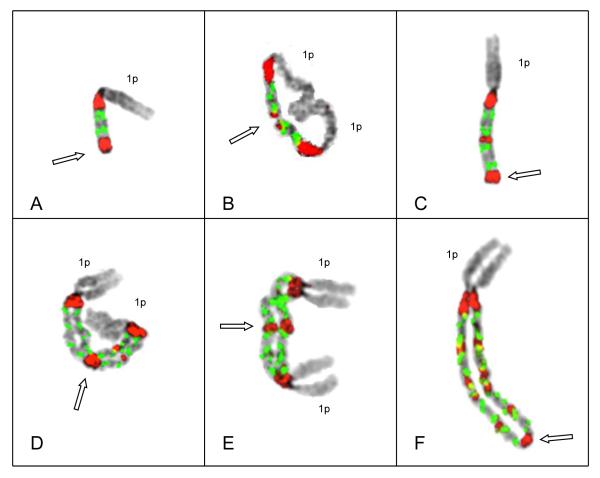
Figure 1.
Representative metaphase chromosomes demonstrating key structures in the progression of intrachromosomal amplification of 1q12~23. Probes for satII/III (red) and CKS1B (green) are shown on inverted DAPI images of chromosomes depicting G-band-like patterns. Patient # 66 demonstrated doublets (two green signals) of CKS1B (see text for explanation) in all cells analyzed. (A) Chromosome 1 in an early stage of amplification, shows four copies of CKS1B bracketed by two copies of satII/III and deletion of chromosome 1q distal to the amplicon. Open arrow demarks the point of sister chromatid fusion (SCF) at distal end. (B) Isodicentric chromosome 1 with two copies of 1p and two copies of the amplicon, both bracketed by satII/III yielding a total of 8 copies of CKS1B. Open arrow indicates the SCF point on the distal 1q (demarked by the satII/III signals) around which 1q hinges in an anaphase bridge. (C) Derivative chromosome 1 showing one copy of 1p and two copies of the amplicon. This chromosome demonstrates the deletion of the 1p distal to the two amplicons (open arrow), and the SCF point for the next step of cycle (open arrow). (D & E) Examples of isodicentric chromosomes 1 from different cells, both showing four amplicons, yielding 16 copies of CKS1B. Note a copy of the short arm of chromosome 1 on each end and the large middle satII/III signals (open arrow) in these chromosomes demarks the point of SCF. (F) Extended ladder-like structure consisting of five copies of the amplicon containing doublets of CKS1B (CN of 18) and one copy of 1p. In this chromosome the lack of sister chromatid cohesion is apparent except at the distinct SCF at the most distal copy of satII/III (open arrow).
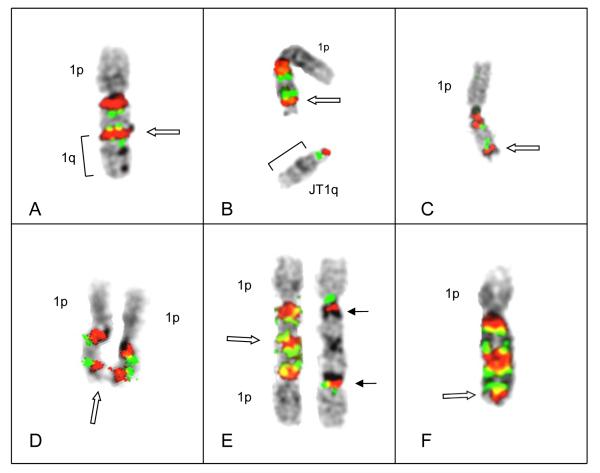
Figure 3.
(A) Patient # 28 demonstrated pericentromeric decondensation in both chromosomes 1, each with a single copy of CKS1B. The chromosome 1 on the left has a normal 1p, while chromosome1 on right has a visible interstitial deletion of 1p. (B) An inverted duplication subsequently occurred in the normal chromosome 1 yielding three copies of CSK1B, associated with two decondensed regions of 1q12 sat II/II. Note the decondensation and near-breakage (solid arrows) in proximal satII/III signal, and decondensation in distal satII/III signal (open arrow). (C) Chromosome 1 with breakage in distal satII/III signal. The satII/III region (open arrow) shows red signal on both sides of thread-like unstained chromatin which appears to break away and result in either a JT1q (bracket) or possibly a micronucleus. (D) Chromosome 1 showing inverted duplication, with two copies of CKS1B bracketed by satII/III, and the loss of an entire 1q distal to the inversion (open arrow), as in Patient #64 (Fig 2C) (E&F) Examples of chromosomes 1 showing ladder-like structures each showing two amplicons bracketed by satII/III signals. Note deletion of 1q distal to the amplicons (open arrows).
Patients # 64 (Figure 2) and # 28 (Figure 3) demonstrated that BFB cycles can occur secondary to an existing inverted duplication on 1q. Chromatid misalignments and unequal sister chromatid exchanges are known to result in direct duplications or, alternatively, a reverse sister chromatid exchange which can produce a large symmetric or asymmetric inverted duplication (Ma et al, 1993). In these cases the chromosome 1 shows three copies of CKS1B and two copies of satII/III resulting from the inversion (Figure 2A). As the progression begins, breakage in the distal satII/III signal results in a jumping translocation of the 1q to a non-homologous chromosome (Figure 2B) or, alternatively, the loss of the whole 1q as a micronucleus. In Patient # 64 the truncated inverted dup of 1q undergoes SCF at the distal satII/III signal (Figure 2C), resulting in an unstable intermediate isodicentric with two copies of 1p and four copies of CKS1B (Figure 2D). The FISH confirmation of isodicentric structure of these unstable intermediates was demonstrated by the rehybridization of this same isodicentric chromosome (Figure 2E). This chromosome demonstrated four copies of CKS1B and three copies of satII/III (on the left), while on the same chromosome (on right) rehybridized with probes for the centromere (alpha sat in red and RP11-153J19 for 1p12 in green) confirms the presence of two centromeric regions and two copies of 1p. Finally, the loss of one of the two copies of the 1p occurs leaving the two copies of the amplicon associated with a single copy of 1p (Figure 2F).
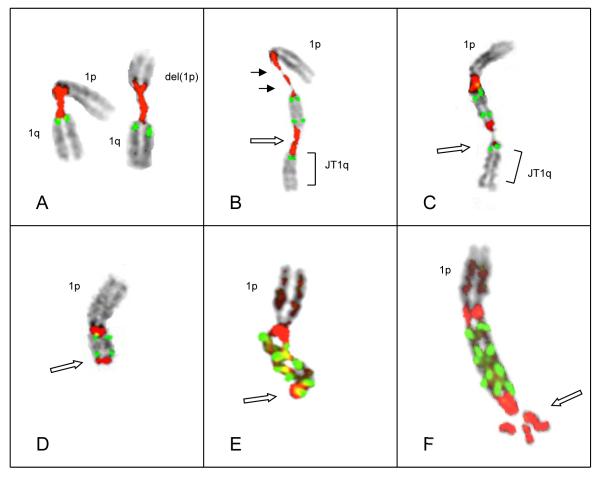
Figure 2.
Representative examples of chromosomes 1 in Patient # 64. (A) Chromosome 1 with an inverted duplication of 1q12~23 and whole 1q distal to the duplication. This chromosome 1 shows three copies of CKS1B, two inside the inverted duplication and one in the normal position. (B) Example of a cell where the entire 1q distal to the inverted duplication has broken off (open arrow), forming a jumping 1q (JT1q)(bracket). (C) Example of another intermediate der(1) following the loss of the whole 1q distal to the amplicon. Breakage in the distal satII/III sequences (open arrow) results in a truncated der(1) with an inverted duplication and deletion of 1q similar to that seen in Patient #66 (Fig 1A). (D) Isodicentric chromosome 1 showing two copies of 1p and two copies of the amplicon (CKS1B CN of 4) bracketed by satII/III signals. Open arrow indicates the point of SCF at the distal satII/III signal. (E) Hybridization of an isodicentric chromosome 1 showing two copies of the amplicon with four copies of CKS1B bracketed by satII/III (Left). On right, rehybridization of the same chromosome 1 with alpha sat 1 (red) to demonstrate two copies of chromosome 1 centromere and BAC probe RP11-153J19 (1p12 in green) just distal to the centromere confirming the intermediate isodicentric structure with two copies of the centromeric region and 1p. (F) Chromosome 1 demonstrating the loss of one of the copies of the short arm of chromosome 1p, distal to two copies of the amplicon (open arrow).
The third patient (#28) showing the amplicon ladders demonstrated decondensation of 1q12 pericentromeric heterochromatin in both copies of chromosome 1, each showing only a single copy of CKS1B (Figure 3A). A subclone was identified with an inverted duplication in the proximal region of the chromosome 1 which showed the normal 1p (with breakpoints in the inversion between F2 and M16). This inverted duplication resulted in three copies of CKS1B (Figure 3B) two of which were bracketed by decondensed regions of satII/III sequences. The distal satII/III sequences demonstrated decondensation (Figure 3B), resulting in breakage in the most distal satII/III sequences (Fig 3C) and the formation of a truncated 1q (Figure 3D) similar to those seen in Patients #66 and #64 (Figures 1A and 2C, respectively). The fusion of sister chromatids at the distal satII/III and progression through further cells cycles ultimately produced amplified chromosomes (Figure 3E and F) similar to chromosomes 1 in Patients #66 and #64 (Figures 1F and 2F, respectively).
The amplicon ladders in Patients # 66, 64 and 28 were transient intermediates which subsequently underwent unbalanced translocations to non-homologous chromosomes or were lost as acentric fragments of amplicons (Figure 4). Unbalanced translocations between the most distal satII/III sequences and non-homologous chromosomes were documented in two of the patients (#s 66, 64) (Figure 4A-D). In Patient # 66, a non-homologous chromosome fused with the donor chromosome 1 at the distal pericentromeric region producing an unstable fusion dicentric (Figure 4A). This unstable intermediate chromosome demonstrated the predicted breakage between the centromeres, leaving a shortened amplicon ladder (two copies) on the donor chromosome 1 and a jumping segmental translocation (two copies) on the recipient chromosome (Figures 4B). This same translocation process was seen in Patient #64 as the distal 1q fused with chromosome 21p (Figure 4C) creating the unstable fusion dicentric and ultimately breaking between the centromeres translocating an amplicon to 21p (Figure 4D). These unbalanced translocations are the type predicted to occur in both the telomere fusion and CFS models for gene amplification. Further evidence for the breakage-prone nature of satII/III sequences is the presence of acentric amplicon segments found in some cells (not shown). These segments presumably occurred by simultaneous breakage in two different pericentromeric junctions, thus yielding unstable acentric amplicon fragments. These acentric fragments would be subsequently lost as micronuclei with high CNs of CKS1B (Figure 4 E-F).
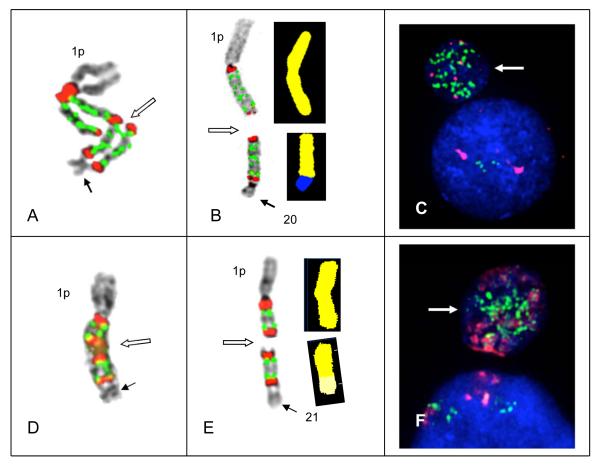
Figure 4.
Secondary aberrations including unbalanced translocations and/or the formation of micronuclei subsequently results in the masking of amplicon ladders. Translocation of intact copies of amplicons to non-homologous chromosomes and loss of amplicons by micronucleus formation was found in both Patients # 66 and 64. (A) In Patient #66 an amplified chromosome 1 demonstrating a ladder of four amplicons yielding 16 copies of CKS1B showing a small non-homologous chromosome (solid arrow) fused at the most distal satII/III signal. Breakage is occurring in the satII/III signals in one chromatid (open arrow) midway between the centromeres of the two chromosomes. This unstable dicentric is the putative precursor to chromosomes shown in (B). (B) An unbalanced translocation of two amplicons in patient # 66 is characterized by both FISH (left) and SKY (right) in the same cell. FISH hybridization (on left) shows two copies of the amplicon remaining on donor chromosome 1, while a section of 1q also containing two copies of the amplicon is translocated to a section of receptor chromosome 20 (on right) identified by SKY. Breakage in the middle of the amplicon ladder between the centromeres of this unstable dicentric ladder resulted in eight copies of CKS1B remaining on the der(1), while 8 copies were translocated to der(20). (C) Chromosome 1 from Patient # 64 showing chromosome 21 fused to distal satII/III signal with two copies of the amplicon (CKS1B CN 4). Open arrow indicates the putative breakage point between the centromeres of this dicentric which would result in the chromosomes shown in (D). FISH signals (left) show one amplicon copy remains on the donor chromosome 1 and one copy of the amplicon is translocated to the receptor chromosome 21. SKY (right) confirms one copy of the amplicon is translocated to the chromosome 21. (E and F) DAPI counterstaining demonstrates micronuclei (arrows) from Patients # 66 (E) and # 64 (F) showing high CNs of CKS1B in micronuclei, presumably resulting from the loss of acentric copies of amplicons (not shown).
Taken together the findings in this study showed a recurring pattern of 1q12~23 amplification (Figures 1, 2, 3). Importantly, two types of initiating breaks for the BFB amplification were demonstrated. In the classic model for BFB cycles, a DSB occurs in a region distal to the gene under selection, as in Patient # 66. However, in the other two patients (#s 64 and 28) the initiating DSB occurred in the distal copy of the 1q12 heterochromatin of an existing inverted duplication. Breakage in 1q12 pericentromeric heterochromatin has not been reported in the initiation of BFB cycles. Importantly, all three patients (#s 66, 64, 28) showed that the breakpoints within the pericentromeric heterochromatin ultimately set both the centromeric and telomeric boundaries of the 1q12~23 amplicon. The breakage and fusion occurring within the satII/III sequences accounts for the regular pattern within ladder-like expansions and provides evidence for a mechanism which mediates the progression of the BFB cycles.
Discussion
Gene amplification is process in which multiple copies of an amplicon are accumulated in cells. In MM and aggressive B-cell lymphomas disease progression is often characterized by the clonal progression of aberrations of the proximal region of chromosome 1q. The molecular characterization of the 1q21~23 amplicon in MM has shown that the amplicon spans approximately 10-15 Mb and contains a marked enrichment in overexpressed genes (Carrasco et al, 2006; Walker et al, 2006; Shaughnessy et al, 2007; Zhan et al, 2007; Chng et al, 2007). In this study we set out to identify the chromosomal mechanisms behind the amplification of CKS1B and other genes in this region. Multiple contiguous copies of inverted duplications of the kind seen in these patients is thought to represent the type of intrachromosomal amplification which originates by BFB cycles. In the patients with the amplicon expansions described here, several of the key structures of BFB cycles predicted by both the telomere fusion and CFS models were identified. Alternatively, there were also other potential sources for proximal 1q aberrations seen here. Segmental duplications (SD), also known as low copy repeats, are submicroscopic structural variations in the human genome that have been identified adjacent to 1q12 heterochromatin (Horvath et al, 2001; Bailey et al, 2002). These regions have been implicated as rearrangement hotspots in both constitutional and cancer-related chromosome aberrations, including tandem duplications/deletions, non homologous translocations, and inversions. Just distal to the region of SDs, are at least two breakpoints of potential interest, including an aphidicolin-type fragile site FRA1F (1q21.1) (Glover et al, 1984) and a jumping translocation breakpoint (JTB), mapped to 1q21 (Hatakeyama et al, 1999). In this and previous studies of the proximal 1q21~23 region, the breakpoints have been found to be heterogeneous (Le Baccon et al, 2001; Lestou et al, 2002; Itoyama et al, 2002; Sawyer et al, 2005), and to our knowledge, no one has identified a breakpoint associated with BFB cycle amplification in this region.
The only recurring breakpoint associated with the BFB cycles in this study involved the 1q12 pericentromeric heterochromatin. Previous studies of MM and other B-cell haematological malignancies have also shown a clustering of breakpoints in the 1q12 pericentromeric region, suggesting a role for this breakpoint in a wide range of neoplasms (Sawyer et al, 1998a; Le Baccon et al, 2001; Lestou et al, 2002; Itoyama et al, 2002; Sawyer et al, 2005). We have previously speculated that hypomethylation of the pericentromeric heterochromatin in 1q12 was a possible cause for the decondensation and JT aberrations (Sawyer et al, 1998a). Genome-wide hypomethylation occurs in many human cancers and has been shown to induce tumours in mice, however it is unclear whether this epigenetic change is the cause or consequence of tumourigenesis (Gaudet et al, 2003). Both gene mutations and chemotherapeutic agents can affect methyltransferase gene function and can induce genome-wide hypomethylation which results in genomic instability. Paediatric patients with Immunodeficiency Centromeric Instability and Facial Anomalies syndrome (ICF), have a defect in DNA methyltransferase 3B (Xu et al, 1999) and develop the same types of striking 1q12 aberrations seen in MM. These include 1q12 pericentromeric decondensation, whole arm unbalanced translocations of chromosomes 1 and 16, multiradial chromosomes 1, and micronuclei of chromosome 1 (Sawyer et al, 1998a; 2005). Chemotherapeutic agents, such as 5-azacytidine, which inhibit methyltransferases are known to specifically induce hypomethylation of the 1q12 pericentromeric DNA. In metaphase chromosomes, 5-azacytidine causes a characteristic pattern of pericentromeric decondensation, somatic associations and multibranched chromosomes 1 (Schmid et al, 1984; Guttenback and Schmid, 1994; Satoh et al, 2004). 5-azacytidine has also been shown to induce aberrant mitosis and subsequent endoreduplication and diplochromosomes in Chinese hamster cells (Mateos et al, 2005). The cause of BFB cycles seen here is not known, however an accumulation of epigenetic modifications occurring during tumour progression may somehow relate to the breakage of a 5-azacytidine fragile site FRA1J (Sutherland et al, 1985) located in the 1q12 pericentromeric heterochromatin. In addition to global hypomethylation, other factors, such as hyperacetlyation and cellular stress induced by the tumour microenvironment are also potential sources for epigenetic modification and subsequent amplification during tumour progression (Coquelle et al, 1988).
Identification of key intermediates predicted by the BFB cycles model
Here we document the key structures of chromosome 1 in MM that are predicted as unstable intermediates in the BFB models for gene amplification. The most striking chromosomal intermediates resulting from BFB cycles are the ladder-like structures composed of equally spaced inverted duplications of 1q12~23 (Figures 1-3). The equal spacing of amplicons occurring on the same chromosome arm is a cardinal feature of BFB cycles in both the telomere fusion and CFS models. At the sites of the DSBs it is predicted that the broken chromatids will undergo SCF. Although the first DSB occurred in the 1q21~23 region (either from an unbalanced translocation or previous inversion), in the next mitosis and all subsequent mitotic cells identified, the breakpoints occurred in or adjacent to the 1q12 pericentromeric region. Remarkably, even in the most highly amplified chromosomes, the terminal FISH signals of the satII/III indicated the fusion of the sister chromatids (Figure 1 F).
According to the BFB models, SCFs occurring from a DSB in the region 1q21~23 should lead to the formation of isodicentric chromosomes 1 in the next mitosis. Patients # 66 and 64 (Figures 1 & 2) provide striking examples of the predicted intermediates of BFB cycles. As previously mentioned, breakpoints in the amplicon ladders during the BFB cycles cluster in the satII/III heterochromatin between the centromeres of the isodicentric chromosomes 1. Therefore, the satII/III sequences set the original centromeric boundary of the 1q12~23 amplicon, and after the first BFB cycle also set telomeric boundaries of the amplicon by bracketing the amplicon. It has previously been demonstrated that the boundaries of an amplicon can be defined by a fragile site (Coquelle et al,1997; Cillo et al, 2002; Hellamn et al, 2002; Miller et al, 2006). If one considers the boundaries of the amplicon as a marker of the original DNA DSB initiating the amplification process, then the pericentromeric heterochromatin could potentially habour a fragile site activated in the late progression of MM. Finally, as predicted by the models for BFB cycles, the 1q showed a consistent deletion distal to the pericentromeric breakpoints (Figures 1-3).
A Model for Intrachromosomal 1q12~23 Amplification in MM
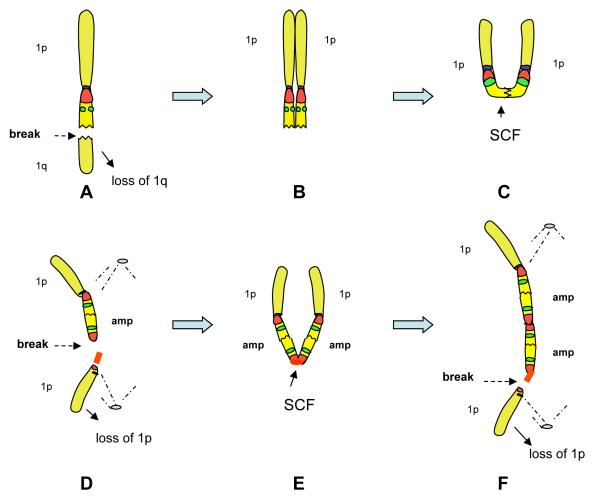
We propose a possible model for 1q12~23 amplification in MM (Figure 5). In a subset of patients, a DSB occurs distal to CKS1B in the 1q21~23 region (Figure 5A) or, alternatively, a DSB occurs in the distal pericentromeric region of an existing inverted duplication (not shown), leading to a deletion of 1q distal to the breakpoint. Following replication (Figure 5B), the deleted 1q can undergo a SCF at the DSB, resulting in an isodicentric chromosome 1 in the next mitosis (Figure 5C). The isodicentric chromosome 1 will break between the centromeres, within or adjacent to one of the pericentromeric regions. This results in one daughter cell with a chromosome 1 consisting of a 1p and totally deleted 1q (not selected for), and another daughter cell with a chromosome 1 consisting of a 1p and a single copy of the 1q12~23 amplicon (Figure 5D). Following the next round of replication, the distal pericentromeric region serves as a point of SCF (Figure 5E), forming another isodicentric chromosome 1, this time containing two copies of the amplicon. In the next anaphase this chromosome breaks, resulting in a chromosome 1 with one copy of 1p associated with two linear copies of the amplicon (Figure 5F). This process is repeated in subsequent cell cycles with the heterochromatin acting as a site specific point of the SCF, and also as the breakpoint in the anaphase isodicentrics. In this BFB cycle mechanism, the 1q12 pericentromeric heterochromatin ultimately sets both the proximal and distal boundaries of the amplicon. Secondary aberrations subsequently mask the presence of BFB cycles by unbalanced translocations of segments of the amplicon ladders to non-homologous chromosomes (Figure 4A-D) in the form of jumping segmental translocations, or acentric fragments of amplicons lost from the cell as micronuclei (Figure 4 E&F).
Figure 5.
A possible model depicting the key steps and intermediate rearrangements of chromosome 1 in BFB cycles. As in the FISH images, red represents satII/III while CKS1B is green. Chromosome 1 can initially undergo a double strand break (DSB) in the region 1q21~23 (A) or, alternatively, a DSB in the distal pericentromeric copy of a chromosome 1 with preexisting direct or inverted duplication of 1q12~23 (not depicted). (B) The DSB in 1q12~23 can result in a sister chromatid fusion (SCF) at the DSB, which in turn will result in the formation of an isodicentric chromosome 1 (C). In the next mitosis, the isodicentric 1 will break between the centromeres, in or immediately adjacent to one of the satII/III regions. The breakage between the centromeres results in the loss of one chromosome 1p arm (D), and therefore a deletion distal to the 1q12 amplicon (amp). Following replication, a SCF takes place in the distal pericentromeric sequences, resulting in another isodicentric chromosome 1, which contains two copies of the amp (E). In the next anaphase this chromosome again breaks in one of the pericentromeric regions between the centromeres resulting in a chromosome 1 with two copies of the amplicon thus beginning the process of enlarging the amplicon ladder (F). This process may be repeated resulting in larger amplicon ladders or may subsequently be disrupted and/or masked by secondary aberrations.
The clonal evolution of tumour cell populations in MM is manifested by increasing genomic instability and during the later stages disease progression this can result in a significant increase in the CN of a number of genes in the 1q12~23 amplicon. The combined findings of the clustering of breakpoints in 1q12 heterochromatin, SCFs at the satII/III breakpoints, isodicentric chromosomes 1, and ladder-like inverted duplications provide evidence for the BFB cycles involving the 1q12~23 region. The key intermediates characterized here were all punctuated by the progression of pericentromeric decondensation, breakage, and/or fusions involving 1q12 satII/III sequences. As predicted by the BFB models, copies of the amplicons were located on the chromosome 1q arm bearing the original copies of the genes, and were organized as megabase-long inverted repeats. These findings provide evidence for 1q12 pericentromeric instability as a mediating breakpoint for the progression of BFB cycles in MM. The possibility that epigenetic modifications to heterochromatin may be involved in the origin of specific amplicons has both diagnostic and prognostic significance, and can also lead to the possibility of targeted therapies.
Supplementary Material
Acknowledgements
This work was supported in part by National Cancer Institute (P01 Grant CA55819 (JDS and BB) .
Footnotes
Authorship Contribution: JRS conceptualized project, interpreted data, and prepared the manuscript; ET prepared FISH probes, ET, MK, CS, LG, GS, CS, performed G-band cytogenetic analysis, RLB performed FISH and SKY, BB enrolled patients, JDS participated in data interpretation and analysis. All authors have read and approved the final manuscript.
Conflict of interest disclosure The authors declare no competing financial interest.
References
- Albertson DG. Gene amplification in cancer. Trends Genetics. 2006;22:447–455. doi: 10.1016/j.tig.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Addams MD, Myers EW, Li PW, Eichler EE. Recent segmental duplication in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Carrasco DR, Tonon G, Huang Y, Zhang Y, Sinha R, Feng B, Stewart JP, Zhan F, Khatry D, Protopopova M, Protopopov A, Sukhdeo K, Hanamura I, Stephens O, Barlogie B, Anderson KC, Chin L, Shaughnessy JD, Jr, Brennan C, Depinho RA. High-resolution genomic profiles defines distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell. 2006;4:313–325. doi: 10.1016/j.ccr.2006.03.019. [DOI] [PubMed] [Google Scholar]
- Chang H, Yeung J, Xu W, Ning Y, Patterson B. Significant increase of CKS1B amplification from monoclonal gammopathy of undetermined significance to multiple myeloma and plasma cell leukaemia as demonstrated by interphase fluorescence in situ hybridization. British Journal of Haematology. 2006a;134:613–615. doi: 10.1111/j.1365-2141.2006.06237.x. [DOI] [PubMed] [Google Scholar]
- Chang H, Qi X, Trieu Y, Xu W, Reader JC, Ning Y, Reece D. Multiple myeloma patients with CKS1B gene amplification have a shorter progression-free survival post-autologous stem cell transplantation. British Journal of Haematology. 2006b;135:486–491. doi: 10.1111/j.1365-2141.2006.06325.x. [DOI] [PubMed] [Google Scholar]
- Chng WJ, Kumar S, Vanwier S, Ahmann G, Price-Troska T, Henderson K, Chung TH, Kim S, Mulligan G, Bryant B, Carpten J, Gertz M, Rajkumar SV, Lacy M, Dispenzieri A, Kyle R, Greipp P, Bergsagel PL, Fonseca R. Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer Research. 2007;67:2982–2989. doi: 10.1158/0008-5472.CAN-06-4046. [DOI] [PubMed] [Google Scholar]
- Ciullo M, Debily MA, Rozier L, Autiero M, Billault A, Mayau V, El Marhomy S, Guardiola J, Bernheim A, Coullin P, Piatier-Tonneau D, Debatisse M. Initiation of the breakage-fusion-bridge mechanism through common fragile site activation in human breast cancer cells: the model of PIP gene duplication from a break at FRA7I. Human Molecular Genetics. 2002;11:2887–2894. doi: 10.1093/hmg/11.23.2887. [DOI] [PubMed] [Google Scholar]
- Coquelle A, Pipiras E, Toledo F, Debatisse M. Expression of fragile sites triggers intrachromosomal mammalian gene amplification and sets boundaries to early amplicons. Cell. 1997;89:215–225. doi: 10.1016/s0092-8674(00)80201-9. [DOI] [PubMed] [Google Scholar]
- Coquelle A, Toledo F, Stern S, Bieth A, Debatisse M. A new role for hypoxia in tumour progression: Induction of fragile site triggering genomic rearrangements and formation of complex DMs and HSRs. Molecular Cell. 1998;2:259–265. doi: 10.1016/s1097-2765(00)80137-9. [DOI] [PubMed] [Google Scholar]
- Decaux O, Lodé L, Magrangeas F, Charbonnel C, Gouraud W, Jézéquel P, Attal M, Harousseau JL, Moreau P, Bataille R, Campion L, Avet-Loiseau H, Minvielle S, Intergroupe Francophone du Myélome Prediction of survival in multiple myeloma based on gene expression profiles reveals cell cycle and chromosomal instability signatures in high-risk patients and hyperdiploid signatures in low-risk patients: A study of the Intergourpe Francophone du Myelome. Journal of Clinical Oncology. 2008;26:4798–4805. doi: 10.1200/JCO.2007.13.8545. [DOI] [PubMed] [Google Scholar]
- Decaux O, Lode L, Magrangeas F, Charbonnel C, Gouraud W, Jézéquel P, Attal M, Harousseau JL, Moreau P, Bataille R, Campion L, Avet-Loiseau H, Minvielle S, Intergroupe Francophone du Myélome Prediction of survival in multiple myeloma based on gene expression profiles reveals cell cycle and chromosomal instability signatures in high-risk patients and hyperdiploid signatures in low-risk patients: A study of the Intergourpe Francophone du Myelome. J Clin Oncol. 2008;26:4798–4805. doi: 10.1200/JCO.2007.13.8545. [DOI] [PubMed] [Google Scholar]
- De Lange T. Telomere-related genome instability in cancer. Cold Spring Harbor Symposium Quantitative Biology. 2005;70:197–204. doi: 10.1101/sqb.2005.70.032. [DOI] [PubMed] [Google Scholar]
- Durkin SG, Glover TW. Chromosome Fragile Sites. Annual Review Genetics. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- Fabris S, Ronchetti D, Agnelli L, Baldini L, Morabito F, Bicciato S, Basso D, Todoerti K, Lombardi L, Lambertenghi-Deliliers G, Neri A. Transcriptional features of multiple myeloma patients with chromosome 1q gain. Leukemia. 2007;21:1113–1116. doi: 10.1038/sj.leu.2404616. [DOI] [PubMed] [Google Scholar]
- Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- Glover TW, Stein CK. Induction of sister chromatid exchanges at common fragile sites. American Journal of Human Genetics. 1987;41:882–890. [PMC free article] [PubMed] [Google Scholar]
- Glover TW, Berger C, Coyle J, Echo B. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Human Genetics. 1984;67:136–142. doi: 10.1007/BF00272988. [DOI] [PubMed] [Google Scholar]
- Guttenbach M, Schmid M. Exclusion of specific human chromosomes into micronuclei by 5-azcytidine treatment of lymphocyte culture. Experimental Cell Research. 1994;211:127–132. doi: 10.1006/excr.1994.1068. [DOI] [PubMed] [Google Scholar]
- Hanamura I, Stewart JP, Huang Y, Zhan F, Santra M, Sawyer JR, Hollmig K, Zangarri M, Pineda-Roman M, van Rhee F, Cavallo F, Burington B, Crowley J, Tricot G, Barlogie B, Shaughnessy JD., Jr. Frequent gain of chromosome band 1q21 in plasma cell dyscrasias detected by fluorescence in situ hybridization: Incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem cell transplantations. Blood. 2006;108:1724–1732. doi: 10.1182/blood-2006-03-009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama S, Osawa M, Omine M, Ishikawa F. JTB: A novel membrane protein gene at 1q21 rearranged in a jumping translocation. Oncogene. 1999;18:2085–2090. doi: 10.1038/sj.onc.1202510. [DOI] [PubMed] [Google Scholar]
- Hellman A, Zlotorynski E, Scherer SW, Cheung J, Vincent JB, Smith JB, Trakhtenbrot L, Kerem B. A role for common fragile site induction in amplification of human oncogenes. Cancer Cell. 2002;1:89–97. doi: 10.1016/s1535-6108(02)00017-x. [DOI] [PubMed] [Google Scholar]
- Horvath JE, Bailey JA, Locke DP, Eichler EE. Lessons from the human genome: transitions between euchromatin and heterochromatin. Human Molecular Genetics. 2001;10:2215–2223. doi: 10.1093/hmg/10.20.2215. [DOI] [PubMed] [Google Scholar]
- Inoue J, Otsuki T, Hirasawa A, Imoto I, Matsuo Y, Shimizu S, Taniwaki M, Inazawa J. Overexpression of PDZK1 within the 1q12-q22 amplicon is likely to be associated with drug-resistance phenotype in multiple myeloma. American Journal of Pathology. 2004;165:71–81. doi: 10.1016/S0002-9440(10)63276-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ISCN . In: An International System for Human Cytogenetic Nomenclature: Recommendations of the International Standing Committee on Human Cytogenetic Nomenclature. Shaffer LG, Slovak ML, Campbell LJ, editors. S Karger; Basel: 2009. [Google Scholar]
- Itoyama T, Nanjungud G, Chen W, Dyomin VG, Teruya-Feldstein J, Jhanwar SC, Zelenetz AD, Chaganti RS. Molecular cytogenetic analysis of genomic instability at the 1q12-22 chromosomal site in B-cell non-Hodgkin lymphoma. Genes Chromosomes & Cancer. 2002;35:318–328. doi: 10.1002/gcc.10120. [DOI] [PubMed] [Google Scholar]
- Kuo MT, Vyas RC, Jiang LX, Hettelman WN. Chromosome breakage at a major fragile site associated with P-glycoprotein gene amplification in multidrug-resistant CHO cells. Molecular Cell Biology. 1994;14:5202–5211. doi: 10.1128/mcb.14.8.5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Baccon P, Leroux D, Dascalescu C, Duley S, Marais D, Esmenjaud E, Sotto JJ, Callanan M. Novel evidence of a role for chromosome 1 pericentric heterochromatin in the pathogenesis of B-cell lymphoma and multiple myeloma. Genes Chromosomes & Cancer. 2001;32:250–264. doi: 10.1002/gcc.1189. [DOI] [PubMed] [Google Scholar]
- Lestou VS, Gascoyne RD, Salski C, Connors JM, Horsman DE. Uncovering novel inter-and intrachromosomal chromosome 1 aberrations in follicular lymphomas by using an innovative multicolor banding technique. Genes Chromosomes & Cancer. 2002;34:201–210. doi: 10.1002/gcc.10069. [DOI] [PubMed] [Google Scholar]
- Ma C, Martin S, Trask B, Hamlin JL. Sister chromatid fusion initiates amplification of the dihydrofolate reductase gene in Chinese hamster cells. Genes and Development. 1993;7:605–620. doi: 10.1101/gad.7.4.605. [DOI] [PubMed] [Google Scholar]
- Mateos S, Domínguez I, Pastor N, Cantero G, Cortés F. The DNA demethylating 5-azaC induces endoreduplication in cultured Chinese hamster cells. Mutation Research. 2005;578:33–42. doi: 10.1016/j.mrfmmm.2005.02.001. [DOI] [PubMed] [Google Scholar]
- McClintock B. Chromosome organization and genic expression. Cold Spring Harbor Symposium Quantitative Biology. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- Miller CT, Lin L, Casper AM, Lim J, Thomas DG, Orringer MB, Chang AC, Chambers AF, Giordano TJ, Glover TW, Beer DG. Genomic amplification of MET with boundaries within fragile site FRA7G and up regulation of MET pathways in esophageal adenocarcinoma. Oncogene. 2006;25:409–418. doi: 10.1038/sj.onc.1209057. [DOI] [PubMed] [Google Scholar]
- National Centre for Biotechnology Information . NCBI; http://www.ncbi.nlm.nih.gov/projects/genome/guide/human/ [Google Scholar]
- Satoh T, Yamamoto K, Miura KF, Sofuni T. Region-specific chromatin decondensation and micronucleus formation induced by 5-azacytidine in human TIG-7 cells. Cytogenetic .& Genome Research. 2004;104:289–294. doi: 10.1159/000077504. [DOI] [PubMed] [Google Scholar]
- Sawyer JR, Waldron JA, Jagannath S, Barlogie B. Cytogenetic findings in 200 patients with multiple myeloma. Cancer Genetics and Cytogenetics. 1985;82:41–49. doi: 10.1016/0165-4608(94)00284-i. [DOI] [PubMed] [Google Scholar]
- Sawyer JR, Tricot G, Mattox S, Jagannath S, Barlogie B. Jumping translocations of chromosome 1q in multiple myeloma: Evidence for a mechanism involving decondensation of pericentromeric heterochromatin. Blood. 1998a;91:1732–1741. [PubMed] [Google Scholar]
- Sawyer JR, Lukacs JL, Munshi N, Desikan KR, Singhal S, Mehta J, Siegel D, Shaughnessy J, Barlogie B. Identification of new nonrandom translocations in multiple myeloma with multicolor spectral karyoytping. Blood. 1998b;91:4269–4278. [PubMed] [Google Scholar]
- Sawyer JR, Tricot G, Lukacs JL, Binz RL, Tian E, Barlogie B, Shaughnessy J., Jr. Genomic instability in multiple myeloma: evidence for jumping segmental duplications of chromosome arm 1q. Genes Chromosomes & Cancer. 2005;42:95–106. doi: 10.1002/gcc.20109. [DOI] [PubMed] [Google Scholar]
- Shmid M, Haaf T, Grunert D. 5-Azacytidine-induced undercondensation in human chromosomes. Human Genetics. 1984;67:257–263. doi: 10.1007/BF00291352. [DOI] [PubMed] [Google Scholar]
- Shaughnessy JD., Jr. Amplification and overexpression of CKS1B at chromosome band 1q21 is associated with reduced levels of p27Kip1 and an aggressive clinical course in multiple myeloma. Hematology. 2005;10:117–26. doi: 10.1080/10245330512331390140. [DOI] [PubMed] [Google Scholar]
- Shaughnessy JD, Jr, Zhan F, Burington BE, Huang Y, Colla S, Hanamura I, Stewart JP, Kordsmeier B, Randolph C, Williams DR, Xiao Y, Xu H, Epstein J, Anaissie E, Krishna SG, Cottler-Fox M, Hollmig K, Mohiuddin A, Pineda-Roman M, Tricot G, van Rhee F, Sawyer J, Alsayed Y, Walker R, Zangari M, Crowley J, Barlogie B. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109:2276–2284. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- Sutherland GR, Parslow MI, Baker E. New classes of fragile sites induced by 5-azacytidine and BrdU. Human Genetics. 1985;69:233–237. doi: 10.1007/BF00293031. [DOI] [PubMed] [Google Scholar]
- Toledo F, Le Roscouet D, Buttin G, Debatisse M. Co-amplified markers alternate in megabase long chromosomal inverted repeats and cluster independently in interphase nuclei at early steps of mammalian gene amplification. EMBO Journal. 1992;11:2665–2673. doi: 10.1002/j.1460-2075.1992.tb05332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treon SP, Maimonis P, Bua D, Young G, Raje N, Mollick J, Chauhan D, Tai YT, Hideshima T, Shima Y, Hilgers J, von Mensdorff-Pouilly S, Belch AR, Pilarski LM, Anderson KC. Elevated soluble MUC1 levels and decreased anti-MUC1 antibody levels in patients with multiple myeloma. Blood. 2000;96:3147–3153. [PubMed] [Google Scholar]
- Walker BA, Leone PE, Jenner MW, Li C, Gonzalez D, Johnson DC, Ross FM, Davies FE, Morgan GJ. Integration of global SNP-based mapping and expression arrays reveals key regions, mechanisms, and genes important in the pathogenesis of multiple myeloma. Blood. 2006;108:1733–1743. doi: 10.1182/blood-2006-02-005496. [DOI] [PubMed] [Google Scholar]
- Xu GL, Bestor TH, Bourc’his D, Hsieh CL, Tommerup N, Bugge M, Hulten M, Qu X, Russo JJ, Viegas-Péquignot E. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- Yunis JJ, Soreng AL. Constitutive fragile sites and cancer. Science. 1984;226:1199–1204. doi: 10.1126/science.6239375. [DOI] [PubMed] [Google Scholar]
- Zhan F, Colla S, Wu X, Chen B, Stewart JP, Kuehl WM, Barlogie B, Shaughnessy JD., Jr. CKS1B, over expressed in aggressive disease, regulates multiple myeloma growth and survival through SKP2- and p27 Kip1-dependent and independent mechanisms. Blood. 2007;109:4995–5001. doi: 10.1182/blood-2006-07-038703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gojo I, Fenton RG. Myeloid cell factor-1 is a critical survival factor for multiple myeloma. Blood. 2002;99:1885–1893. doi: 10.1182/blood.v99.6.1885. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.