Abstract
People with mood disorders often have disruptions in their circadian rhythms. Recent molecular genetics has linked circadian clock genes to mood disorders. Our objective was to study two core circadian clock genes, CRY1 and CRY2 as well as TTC1 that interacts with CRY2, in relation to depressive and anxiety disorders. Of these three genes, 48 single-nucleotide polymorphisms (SNPs) whose selection was based on the linkage disequilibrium and potential functionality were genotyped in 5910 individuals from a nationwide population-based sample. The diagnoses of major depressive disorder, dysthymia and anxiety disorders were assessed with a structured interview (M-CIDI). In addition, the participants filled in self-report questionnaires on depressive and anxiety symptoms. Logistic and linear regression models were used to analyze the associations of the SNPs with the phenotypes. Four CRY2 genetic variants (rs10838524, rs7121611, rs7945565, rs1401419) associated significantly with dysthymia (false discovery rate q<0.05). This finding together with earlier CRY2 associations with winter depression and with bipolar type 1 disorder supports the view that CRY2 gene has a role in mood disorders.
Introduction
Circadian rhythms are about 24-hour oscillations in physiological processes that allow humans to anticipate routine changes in the environment. The rhythms are generated by the internal clock, the principal one being located in the suprachiasmatic nucleus of the anterior hypothalamus in the brain. At the molecular level, the transcription of a network of genes is switched on and off following the approximate 24-hour pattern. A feedback loop is initiated by binding of CLOCK protein [1] with aryl hydrocarbon receptor nuclear translocator-like (ARNTL) protein [2]. The heterodimer then activates the transcription of a large set of genes and among them the period (PER1, PER2, PER3) and cryptochrome (CRY1, CRY2) genes. Again, the encoded proteins form heterodimers (PER-with-CRY dimers) which translocate back to the nucleus and inhibit the CLOCK-with-ARNTL mediated transcription, thereby repressing their own transcription [3]. In addition, nuclear receptors together with their co-activators and co-repressors feed back to ARNTL [4].
The cryptochrome genes may be of key importance here, because of their unique role in the core of these feedback loops [3], [5], [6]. The nuclear ratio of CRY1 to CRY2 proteins controls the period of the circadian clock [5] and the effects of CRY2 on circadian regulation are sensitive to the dosage of CRY1 [7]. Findings from knock-out mice suggest that without CRY2 the circadian period is lengthened similar to “evening owls”, whereas without CRY1 it is shortened [8], [9]. Furthermore, CRY2 appears to play a critical role in tuning the suprachiasmatic nucleus to 24 hour cycle by opposing and titrating the decelerating action of CRY1 [7].
“Evening owls” are known to have increased risk for anxiety and depressive symptoms [10], [11] that may develop into depressive disorders [12], [13]. Furthermore, dysfunction of signaling pathways involving CRY1 and CRY2 might lead into depressive behaviors as follows. Interaction of CRY1 and CRY2 proteins directly with the Gsα subunit of heterotrimeric G protein [14] and with adenylyl cyclase [15] reduces the accumulation of cyclic adenosine monophosphate in response to G protein-coupled receptor activation and inhibits downstream reactions [15], [16]. If this inhibition is not appropriate, the inhibitory tone on cyclic adenosine monophosphate signaling is overruled, leading to the activation of cascades downstream, such as cyclic adenosine monophosphate response element-binding protein activity, and to the increase in depressive-like behaviors [17].
Disrupted or misaligned circadian rhythms and circadian gene expression are often found in major depressive disorder and bipolar disorder, with or without a seasonal pattern [18], [19]. Since the original finding [20], there is a growing evidence that genetic variations in the core circadian clock genes associate with mood and anxiety disorders [21]. In addition, these genetic variations associate with sleep and metabolic disorders [22] that are often comorbid conditions with mood and anxiety disorders. With the focus on the cryptochrome genes, earlier studies have indicated CRY1 genetic variants in major depressive disorder [23] and CRY2 genetic variants in seasonal affective disorder, winter depression in particular, and in bipolar type 1 disorder [24], [25].
The aim of the current paper is to study the associations of CRY1, CRY2 and TTC1 genes in depressive and anxiety disorders. In addition to the two cryptochrome genes, TTC1 was selected in order to analyze epistasis, if any, between the TTC1 and CRY2 genes, because the TTC1 protein interacts physically with CRY2 [26]. The present work extends the earlier studies by deeper linkage disequilibrium (LD) coverage of the genes, by including potential functional variants, by using a large sample derived randomly from the general population, by assessment based on both a diagnostic interview and self-reports, and by analysis of diverse phenotype information. Here we report a significant association of CRY2 genetic variants with dysthymia.
Materials and Methods
Ethics Statement
The ethics committees of the National Public Health Institute and the Helsinki and Uusimaa Hospital District accepted the study protocol and its ethical approval, and all participants provided a written informed consent.
Subjects
This study was part of a nationwide health interview and examination survey, Health 2000. The survey included a health status examination and the Munich-Composite International Diagnostic Interview (M-CIDI) [27]. M-CIDI is a valid and reliable instrument for the assessment of depressive, anxiety and alcohol use disorders, yielding diagnoses according to Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV). In addition, participants gave venous blood samples for DNA extraction and filled in two sets of questionnaires. The sample for our current study from the Health 2000 participants aged 30 years and older (n = 8028) included 5910 individuals (3283 women, 2627 men) who had given blood samples, taken part to the M-CIDI interview and filled in the self-report on seasonal changes in mood and behavior. The study design and methods have been described in detail elsewhere (http://www.terveys2000.fi/indexe.html).
Phenotypes
Altogether 12 phenotypes related to mental well-being (diagnoses according to DSM-IV and psychometric scales) were analyzed (Table 1). Mental disorders analyzed in the current study included major depressive disorder, dysthymia, depressive disorders (major depressive disorder, dysthymia), panic disorder, anxiety disorders (panic disorder w/o agoraphobia, generalized anxiety disorder, social phobia, agoraphobia), and the comorbid conditions of depressive, anxiety and alcohol use disorders (alcohol dependence and abuse), as assessed with the M-CIDI. The controls did not have any diagnosis of mental disorders nor met any sub-threshold criteria as assessed with the M-CIDI.
Table 1. Number of subjects in mental health related phenotypes (DSM-IV based diagnoses and quantitative scores on psychometric scales) analyzed in the study.
| All | Women | Men | ||||
| Phenotypes analyzed | Cases | Controls | Cases | Controls | Cases | Controls |
| Depressive disorders | 354 | 3871 | 242 | 2135 | 112 | 1736 |
| Major depressive disorder | 267 | 3871 | 183 | 2135 | 84 | 1736 |
| Dysthymia | 136 | 3871 | 90 | 2135 | 46 | 1736 |
| Anxiety disorders | 221 | 3871 | 137 | 2135 | 84 | 1736 |
| Panic disorder | 106 | 3871 | 71 | 2135 | 35 | 1736 |
| Comorbid depressive disorders | 145 | 3871 | 84 | 2135 | 61 | 1736 |
| Comorbid anxiety disorders | 117 | 3871 | 63 | 2135 | 54 | 1736 |
| Comorbid alcohol use disorders | 105 | 3871 | 38 | 2135 | 67 | 1736 |
| BDI | 5724 | 3153 | 2571 | |||
| GHQ | 5811 | 3211 | 2600 | |||
| MBI | 3379 | 1745 | 1634 | |||
| GSS | 5633 | 3096 | 2537 | |||
BDI; Beck Depression Inventory.
GHQ; General Health Questionnaire.
MBI; Maslach Burnout Inventory.
GSS; Global Seasonality Score.
In addition to attending the diagnostic interview, the participants completed the following four self-reports that were also analyzed: 1) a modified 21-item Beck Depression Inventory (BDI; [28]), as adapted and validated for the Finnish population [29], 2) the 16-item Maslach Burnout Inventory (MBI; [30]); 3) the 12-item General Health Questionnaire (GHQ; [31]), and 4) a modified [32] 7-item Seasonal Pattern Assessment Questionnaire (SPAQ; [33]). These questionnaires give quantitative information about depressive and anxiety symptoms.
Gene and SNP Selection
Three genes of interest were selected: CRY1, CRY2 and TTC1. The SNP selection was based on phase 3 data of the HapMap database (http://www.hapmap.org/) and done using the Tagger program included in the Haploview 4.1 software [34]. Information about the LD within the chosen areas of the genome was used to select an optimal set of SNPs capturing most of the genetic variation. Areas of the genome covered were the genes and 10 kb of their 5′ and 3′ flanking regions. For CRY1, this area of interest was 122 kb (chr12∶105,899–106,021 kb, NCBI36/hg18 assembly) in size, whereas for CRY2 it was 56 kb (chr11∶45,815–45,871 kb) and for TTC1 77 kb (chr5∶159,358–159,435 kb).
The HapMap database included 21, 34 and 15 SNPs having a minor allele frequency (MAF) >5% in the European population (CEU and TSI) for the areas containing the CRY1, CRY2 and TTC1 genes, respectively. The aim was to capture all these SNPs by setting the limit for the pair-wise r2 to ≥0.9. Based on this criterion, ten CRY1 SNPs, ten CRY2 SNPs and nine TTC1 SNPs were decided to be genotyped and all, except one (TTC1 rs3733868), were successfully included in the genotyping multiplexes. One SNP (CRY1 rs11113153) failed in the genotyping phase.
In addition to the aforementioned SNPs, potentially functional variants, as many as possible, were included in the genotyping multiplexes. These 21 additional SNPs (8 for CRY2, 11 for CRY1 and 2 for TTC1) were selected using Pupasuite [35], Variowatch [36], dbSMR [37] and miRNASNP [38] databases. Table 2 presents all the 48 SNPs that were successfully genotyped in this study and their selection criteria.
Table 2. Successfully genotyped SNPs, their selection criteria, allele and genotype frequencies, and Hardy-Weinberg equilibrium p-values.
| Gene | SNP | BP | A1 | A2 | MAF | A1A1 | A1A2 | A2A2 | HWE-P | Selection criteria |
| TTC1 | rs6878309 | 159358102 | C | T | 0.09 | 57 (0.01) | 990 (0.17) | 4800 (0.82) | 0.44 | LD |
| TTC1 | rs1402024 | 159361425 | G | A | 0.02 | 1 (0) | 177 (0.03) | 5636 (0.97) | 1 | LD |
| TTC1 | rs41275305 | 159370449 | G | C | 0 | 0 (0) | 31 (0.01) | 5815 (0.99) | 1 | non-synonymous coding variant |
| TTC1 | rs10515804 | 159380041 | T | C | 0.3 | 531 (0.09) | 2468 (0.43) | 2797 (0.48) | 0.71 | LD |
| TTC1 | rs2176830 | 159380714 | A | G | 0.07 | 32 (0.01) | 756 (0.13) | 5057 (0.87) | 0.48 | LD |
| TTC1 | rs12520927 | 159410735 | G | 0 | 0 (0) | 0 (0) | 5854 (1) | 1 | non-synonymous coding variant | |
| TTC1 | rs3733869 | 159422128 | T | C | 0.3 | 522 (0.09) | 2430 (0.42) | 2866 (0.49) | 0.83 | LD |
| TTC1 | rs6861719 | 159422825 | T | C | 0.23 | 295 (0.05) | 2066 (0.36) | 3448 (0.59) | 0.55 | LD |
| TTC1 | rs1106055 | 159424098 | A | G | 0.32 | 607 (0.1) | 2517 (0.43) | 2674 (0.46) | 0.7 | LD |
| TTC1 | rs7715826 | 159425636 | T | C | 0.02 | 5 (0) | 254 (0.04) | 5595 (0.96) | 0.22 | LD |
| CRY2 | rs7121611 | 45820718 | A | T | 0.46 | 1218 (0.21) | 2883 (0.5) | 1711 (0.29) | 0.96 | LD |
| CRY2 | rs7121775 | 45820899 | C | T | 0.27 | 384 (0.07) | 2326 (0.4) | 3100 (0.53) | 0.06 | LD |
| CRY2 | rs61884508 | 45821508 | G | T | 0.02 | 1 (0) | 241 (0.04) | 5600 (0.96) | 0.52 | Pupasuite OregannoFilter TFBS |
| CRY2 | rs75065406 | 45821518 | T | C | 0.04 | 13 (0) | 421 (0.07) | 5414 (0.93) | 0.11 | TFBS, MAF |
| CRY2 | rs3747548 | 45825589 | A | C | 0 | 0 (0) | 1 (0) | 5847 (1) | 1 | Pupasuite non-synonymous & VarioWatch |
| CRY2 | rs10838524 | 45826753 | G | A | 0.48 | 1337 (0.23) | 2897 (0.5) | 1579 (0.27) | 0.92 | LD & Lavebratt et al. |
| CRY2 | rs2292913 | 45834105 | T | C | 0.05 | 18 (0) | 590 (0.1) | 5233 (0.9) | 0.7 | LD & splice site |
| CRY2 | rs7945565 | 45835568 | G | A | 0.46 | 1213 (0.21) | 2890 (0.5) | 1695 (0.29) | 0.79 | Pupasuite Triplex |
| CRY2 | rs1401419 | 45836315 | G | A | 0.46 | 1211 (0.21) | 2909 (0.5) | 1681 (0.29) | 0.48 | Pupasuite Triplex |
| CRY2 | rs72902437 | 45838834 | C | T | 0.03 | 2 (0) | 313 (0.05) | 5499 (0.95) | 0.45 | Pupasuite Triplex |
| CRY2 | rs35488012 | 45845804 | G | 0 | 0 (0) | 0 (0) | 5854 (1) | 1 | Variowatch synonymous | |
| CRY2 | rs7123390 | 45847994 | A | G | 0.29 | 431 (0.07) | 2445 (0.42) | 2915 (0.5) | 0.01 | LD & Lavebratt et al. |
| CRY2 | rs4755345 | 45848084 | A | G | 0.05 | 18 (0) | 598 (0.1) | 5229 (0.89) | 0.8 | LD |
| CRY2 | rs17787136 | 45851212 | G | C | 0.28 | 409 (0.07) | 2385 (0.41) | 3014 (0.52) | 0.03 | Pupasuite TFBS |
| CRY2 | rs10838527 | 45859770 | G | A | 0.12 | 89 (0.02) | 1236 (0.21) | 4509 (0.77) | 0.67 | LD & Lavebratt et al. |
| CRY2 | rs2292910 | 45860189 | A | C | 0.34 | 650 (0.11) | 2707 (0.47) | 2455 (0.42) | 0.02 | LD & dbSMR miRNA target site |
| CRY2 | rs3824872 | 45862181 | T | G | 0.25 | 372 (0.06) | 2173 (0.37) | 3276 (0.56) | 0.65 | LD & Lavebratt et al. |
| CRY2 | rs1554338 | 45863406 | G | A | 0.05 | 14 (0) | 528 (0.09) | 5289 (0.91) | 0.77 | LD |
| CRY1 | rs4964513 | 105899888 | C | T | 0.12 | 84 (0.01) | 1224 (0.21) | 4492 (0.77) | 0.95 | LD |
| CRY1 | rs714359 | 105902975 | A | G | 0.22 | 276 (0.05) | 2006 (0.35) | 3516 (0.61) | 0.67 | LD |
| CRY1 | rs12821586 | 105904582 | A | G | 0.11 | 72 (0.01) | 1138 (0.19) | 4629 (0.79) | 0.84 | LD |
| CRY1 | rs2287161 | 105905270 | C | G | 0.5 | 1408 (0.24) | 2930 (0.51) | 1461 (0.25) | 0.43 | Soria et al. & Utge et al. & Pupasuite triplex |
| CRY1 | rs8192441 | 105909584 | C | A | 0.01 | 1 (0) | 136 (0.02) | 5712 (0.98) | 0.56 | miRNASNP miRNA target site |
| CRY1 | rs3741892 | 105911293 | C | G | 0.49 | 1395 (0.24) | 2937 (0.51) | 1477 (0.25) | 0.4 | Pupasuite Triplex |
| CRY1 | rs10861688 | 105918178 | T | C | 0.17 | 164 (0.03) | 1642 (0.28) | 4011 (0.69) | 0.82 | LD |
| CRY1 | rs10861697 | 105943792 | C | G | 0.49 | 1352 (0.23) | 2923 (0.5) | 1521 (0.26) | 0.48 | Pupasuite Triplex |
| CRY1 | rs2078074 | 105960936 | C | T | 0.42 | 1017 (0.18) | 2815 (0.49) | 1930 (0.33) | 0.87 | Pupasuite Transfac |
| CRY1 | rs59790130 | 105964433 | T | C | 0.06 | 26 (0) | 692 (0.12) | 5131 (0.88) | 0.58 | Pupasuite Transfac |
| CRY1 | rs10437895 | 105964954 | C | T | 0.49 | 1398 (0.24) | 2936 (0.5) | 1482 (0.25) | 0.46 | Pupasuite Transfac |
| CRY1 | rs10746077 | 105965682 | A | G | 0.42 | 1027 (0.18) | 2832 (0.49) | 1960 (0.34) | 0.96 | Pupasuite Transfac |
| CRY1 | rs11613557 | 105966445 | T | C | 0.06 | 26 (0) | 692 (0.12) | 5130 (0.88) | 0.58 | Pupasuite Triplex |
| CRY1 | rs2888896 | 105970712 | T | C | 0.42 | 1019 (0.18) | 2830 (0.49) | 1947 (0.34) | 0.87 | LD |
| CRY1 | rs11113179 | 105976915 | T | C | 0.08 | 39 (0.01) | 833 (0.14) | 4942 (0.85) | 0.53 | LD & Utge et al. |
| CRY1 | rs10746083 | 105978532 | T | C | 0.49 | 1391 (0.24) | 2941 (0.51) | 1481 (0.25) | 0.37 | Pupasuite Triplex |
| CRY1 | rs4964518 | 105990347 | T | C | 0.07 | 30 (0.01) | 778 (0.13) | 5027 (0.86) | 1 | LD |
| CRY1 | rs7294758 | 105991959 | A | T | 0.01 | 0 (0) | 97 (0.02) | 5758 (0.98) | 1 | Pupasuite Triplex |
| CRY1 | rs17289712 | 105993098 | G | A | 0.05 | 6 (0) | 524 (0.09) | 5308 (0.91) | 0.07 | LD |
| CRY1 | rs10778528 | 105998092 | G | T | 0.48 | 1358 (0.23) | 2926 (0.5) | 1533 (0.26) | 0.62 | LD |
BP; Base pair position based on NCBI36/hg18 build.
A1; Minor allele.
A2; Major allele.
MAF; Minor allele frequency.
A1A1, A1A2, A2A2; genotype counts and frequencies (%).
HWE-P; Hardy-Weinberg equilibrium p-value.
LD; Linkage disequilibrium.
TFBS, Transcription factor binding site.
Genotyping
Genomic DNA was isolated from the whole blood according to standard procedures. DNA samples were available from 5910 subjects for this study. The SNPs were genotyped using the Sequenom MassARRAY system and the iPLEX Gold Single Base Extension chemistry (Sequenom, San Diego, CA, USA) in a multiplex format [39]. This method has excellent success (>95%) and accuracy (100%) rates [40]. For the genotyping quality control purposes, both positive (CEPH) and negative water controls were included in each 384-plate. Genotyping was performed blind to phenotypic information.
173 individuals were removed from the statistical analyses due to a high missing genotype rate (i.e. >0.1). The total genotyping rate in the remaining individuals was 0.9987. In addition, the following five SNPs were removed because of the minor allele frequency of <0.01: rs41275305, rs12520927, rs3747548, rs35488012, rs7294758. Finally, there were 5737 individuals and 43 SNPs for the statistical analyses.
Statistical Analyses
Statistical analyses were performed with the PLINK software [41], release v1.07. The additive model was calculated using logistic and linear regression models controlling for age and sex. In addition, dominant and recessive models were calculated and the analyses were performed in both sexes separately. For continuous phenotypes 10,000 permutations were used to produce empirical p-values in order to relax the assumption of normality. The results were corrected for multiple testing across all the tests (SNPs, phenotypes, gender, genetic models) by calculating false discovery rate (FDR) q-values [42] using R software (http://www.r-project.org/). The q-values of <0.05 were considered significant.
Haploview software [34] was used to define haplotype blocks by the confidence interval method. Each haplotype in the formed blocks was analyzed by PLINK software using linear and logistic regression, additive model and controlling for age and sex. Sexes were also analyzed separately. Continuous phenotypes were permuted 10,000 times. Interactions between CRY2 and TTC1 (SNP × SNP epistasis) were tested and calculated using the PLINK software.
Results
The genotype and allele frequencies and Hardy-Weinberg equilibrium (HWE) estimates are shown in Table 2 (see Table S1 for the genotype counts by phenotype). The results of the association analyses where q<0.15 are presented in Table 3 and the remaining results of the association analyses in Table S2. Four CRY2 SNPs showed evidence of association with dysthymia using the additive model (rs10838524 risk allele G, p = 0.000020, OR = 1.75; rs7121611 risk allele A, p = 0.000022, OR = 1.74; rs7945565 risk allele G, p = 0.000039, OR = 1.71; rs1401419 risk allele G, p = 0.000041, OR = 1.71). CRY2 SNP rs10838524 showed evidence of association also using the dominant model (p = 0.000059, OR = 3.27) and the additive model for women (p = 0.000026, OR = 1.97). The CRY1 and TTC1 genetic variants studied showed no evidence of association with any of the phenotypes analyzed. No epistasis between CRY2 and TTC1 SNPs was observed.
Table 3. Results from the single SNP association analyses (q<0.15).
| Phenotype | Model | Gene | SNP | A1 | Odds ratio | L95 | U95 | P-value | Q-value |
| Dysthymia | ADD | CRY2 | rs10838524 | G | 1.75 | 1.35 | 2.27 | 2.00E-05 | 0.04* |
| Dysthymia | ADD women | CRY2 | rs10838524 | G | 1.97 | 1.44 | 2.71 | 2.60E-05 | 0.04* |
| Dysthymia | DOM | CRY2 | rs10838524 | G | 3.27 | 1.84 | 5.83 | 5.90E-05 | 0.04* |
| Dysthymia | DOM women | CRY2 | rs10838524 | G | 5.11 | 2.22 | 11.77 | 0.00013 | 0.07 |
| Dysthymia | ADD | CRY2 | rs1401419 | G | 1.71 | 1.32 | 2.21 | 4.10E-05 | 0.04* |
| Dysthymia | ADD women | CRY2 | rs1401419 | G | 1.82 | 1.33 | 2.48 | 0.00018 | 0.07 |
| Dysthymia | DOM | CRY2 | rs1401419 | G | 2.67 | 1.59 | 4.46 | 0.0002 | 0.07 |
| Dysthymia | DOM women | CRY2 | rs1401419 | G | 3.20 | 1.65 | 6.23 | 0.00061 | 0.14 |
| Depressive disorders | DOM | CRY2 | rs1401419 | G | 1.62 | 1.23 | 2.13 | 0.00063 | 0.14 |
| Dysthymia | ADD | CRY2 | rs3824872 | T | 0.52 | 0.37 | 0.74 | 0.00024 | 0.08 |
| Dysthymia | ADD | CRY2 | rs7121611 | A | 1.74 | 1.35 | 2.24 | 2.20E-05 | 0.04* |
| Dysthymia | ADD women | CRY2 | rs7121611 | A | 1.86 | 1.36 | 2.53 | 9.40E-05 | 0.06 |
| Dysthymia | DOM | CRY2 | rs7121611 | A | 2.74 | 1.63 | 4.58 | 0.00013 | 0.07 |
| Depressive disorders | DOM | CRY2 | rs7121611 | A | 1.66 | 1.26 | 2.18 | 0.00033 | 0.1 |
| Dysthymia | DOM women | CRY2 | rs7121611 | A | 3.30 | 1.7 | 6.42 | 0.00043 | 0.12 |
| Dysthymia | ADD | CRY2 | rs7945565 | G | 1.71 | 1.33 | 2.21 | 3.90E-05 | 0.04* |
| Dysthymia | ADD women | CRY2 | rs7945565 | G | 1.82 | 1.33 | 2.49 | 0.00017 | 0.07 |
| Dysthymia | DOM | CRY2 | rs7945565 | G | 2.68 | 1.6 | 4.49 | 0.00018 | 0.07 |
| Depressive disorders | DOM | CRY2 | rs7945565 | G | 1.65 | 1.25 | 2.17 | 0.00042 | 0.12 |
| Dysthymia | DOM women | CRY2 | rs7945565 | G | 3.22 | 1.65 | 6.26 | 0.00058 | 0.14 |
ADD; Additive model.
DOM; Dominant model.
A1; Tested allele (minor allele).
L95, U95; Lower and upper bounds of 95% confidence interval for odds ratio.
Significant association.
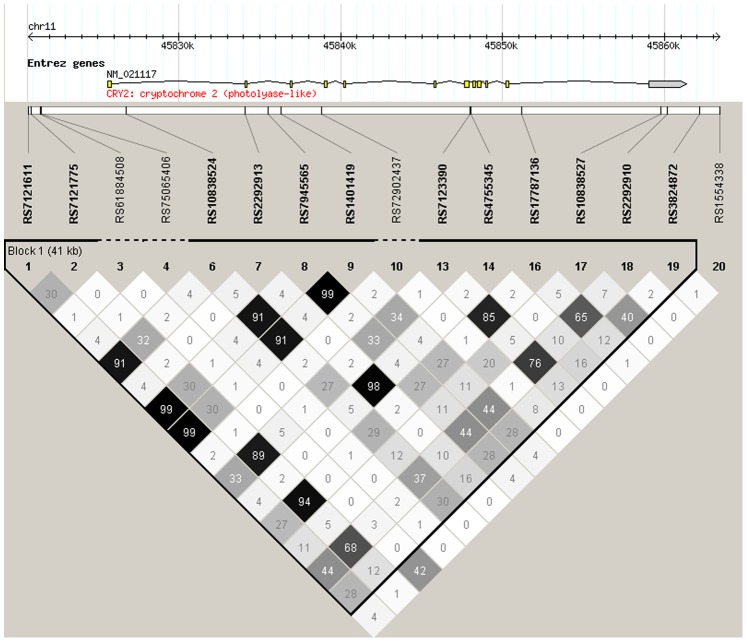
Haploview produced two haplotype blocks for CRY1 (Figure 1), one for CRY2 (Figure 2) and one for TTC1 (Figure 3). The haplotype analyses supported the association of CRY2 with dysthymia, as the ATTCGCGGTGGCACG haplotype containing the risk alleles A, G, G and G of SNPs rs7121611, rs10838524, rs7945565 and rs1401419, respectively, showed association with dysthymia in all cases (p = 0.00034, OR = 1.58) and among women (p = 0.0011, OR = 1.66). Table 4 presents the haplotype associations with p-values (or empirical p-values) of <0.010.
Figure 1. The analyzed CRY1 SNPs in this study, their location and the haplotype block structure of the area formed based on our sample showing r2 values.
The confidence interval algorithm implemented in the Haploview program was used to construct the haplotype blocks.
Figure 2. The analyzed CRY2 SNPs, their location and the haplotype block structure constructed using the Haploview program showing r2 values.
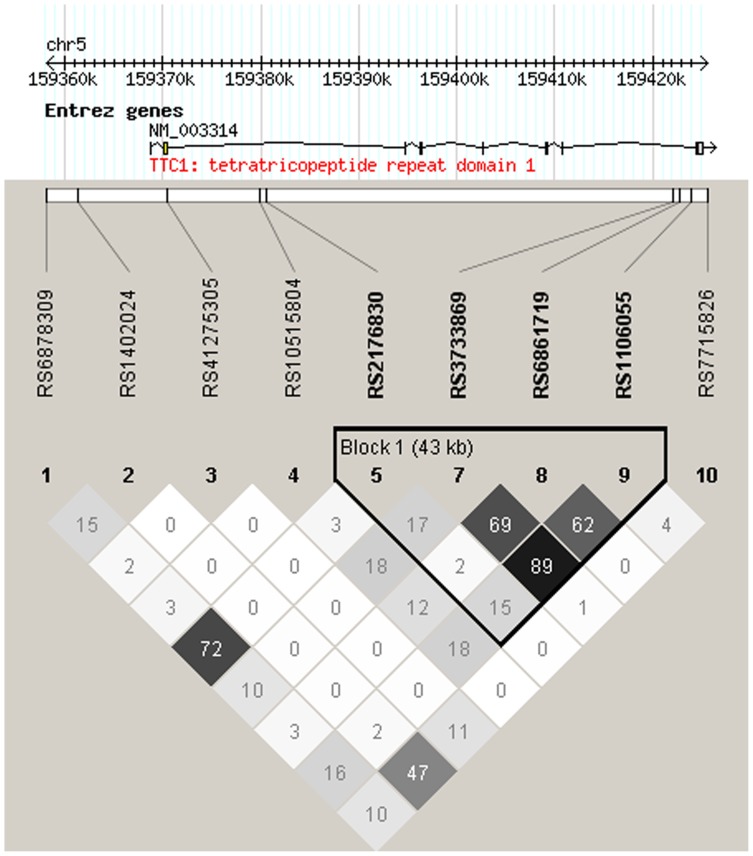
Figure 3. The analyzed TTC1 SNPs, their location and the haplotype block structure constructed using the Haploview program showing r2 values.
Table 4. Results from the haplotype association analyses (p<0.10).
| Phenotype | Population | NSNP | NHAP | Gene | SNP1 | SNP2 | Haplotype | Frequency | Odds ratio | p-value | Empirical p-value |
| Dysthymia | all | 15 | 10 | CRY2 | rs7121611 | rs3824872 | ATTCGCGGTGGCACG | 0.41 | 1.58 | 0.00034 | |
| Dysthymia | women | 15 | 10 | CRY2 | rs7121611 | rs3824872 | ATTCGCGGTGGCACG | 0.407 | 1.66 | 0.0011 | |
| Depressivedisorders | men | 3 | 4 | CRY1 | rs4964513 | rs12821586 | TAG | 0.217 | 1.67 | 0.0014 | |
| Depressivedisorders | all | 15 | 10 | CRY2 | rs7121611 | rs3824872 | ATTCGCGGTGGCACG | 0.41 | 1.27 | 0.0039 | |
| Depressivedisorders | women | 15 | 10 | CRY2 | rs7121611 | rs3824872 | TTTCATAATGACAAT | 0.0539 | 0.45 | 0.0057 | |
| Depressivedisorders | all | 15 | 10 | CRY2 | rs7121611 | rs3824872 | TTTCATAATGACAAT | 0.0514 | 0.53 | 0.0061 | |
| Dysthymia | all | 15 | 10 | CRY2 | rs7121611 | rs3824872 | TTTCATAATGACAAT | 0.0514 | 0.2 | 0.0065 | |
| Major depressivedisorder | men | 3 | 4 | CRY1 | rs4964513 | rs12821586 | TAG | 0.217 | 1.62 | 0.0082 | |
| GSS | all | 14 | 6 | CRY1 | rs3741892 | rs10778528 | GTGTCTGCCCCCAT | 0.17 | 0.2 | 0.011 | 0.01 |
NSNP; Number of SNPs in this haplotype.
NHAP; Number of common haplotypes (f>0.01).
SNP1; SNP ID of the first SNP (5′).
SNP2; SNP ID of the last SNP (3′).
Discussion
Our main finding was that CRY2 genetic variants associated with dysthymia in our random sample derived from the general population aged 30 years and older. The associations with dysthymia were stronger in the additive than in the dominant model and became weaker when we analyzed only women (n = 88). No association was observed in men. The sample size for dysthymia was limited (n = 128) and the lack of association in men could be due to a small sample size (n = 40). We did not find any significant association of CRY2 with major depressive disorder, whether with a single or recurrent episode, or with anxiety disorders, and there was no significant association of CRY1 or TTC1 with any of the phenotypes assessed.
Of the individual CRY2 SNPs, rs7121611 (upstream), rs10838524 (intron 1), rs7945565 (intron 2) and rs1401419 (intron 2) showed evidence of association with dysthymia, with the A, G, G and G alleles being predisposing, respectively. These associations are supported by the haplotype analysis, as the CRY2 ATTCGCGGTGGCACG haplotype including the individual risk alleles associated with the phenotype. The associated haplotype of CRY2 spans almost the whole gene and the four associated SNPs are located in the first half of the haplotype block (Figure 2). According to transcription factor CHiPSeq from ENCODE (http://genome.ucsc.edu/ENCODE/) SNP rs10838524 is located in a regulatory region and according to Pupasuite software [35] rs7945565 and rs1401419 were predicted to be putative triplex disrupting SNPs which may also be part of regulatory regions for controlling gene expression [43].
Of the four associated CRY2 SNPs, only rs10838524 has previously been linked to mental health disorders. Concerning this SNP, our current results are partly in line with the findings of Lavebratt et al. [24] who found the same risk allele for winter depression in the Finnish subpopulation of their study. The allele frequencies in cases and controls were similar in these two sets, as expected for the controls, since they were drawn from the same Health 2000 cohort. The cases, on the other hand, had been recruited from outpatient services in the capital area [24]. Another Finnish study, using a population based sample also from the Health 2000 cohort, found the same minor allele to weakly associate with depressive disorders in women and in women having depressive disorders with early morning awakening, and the association was supported with haplotype analyses but did not hold after correcting for multiple testing [44]. We also analyzed depressive disorders in women and found similar crude p-value (p = 0.035, Table S2). In the study by Utge et al., the statistical analyses had been done separately in men and women. In our analyses, the associations were stronger when using the dominant model in both sexes together (p = 0.0010), but still the association to depressive disorders did not reach statistical significance after correcting for multiple testing. For the Swedish subpopulation of the winter depression study [24] as well as for a Swedish bipolar study [25], the allele frequencies for CRY2 rs10838524 were reversed and the other allele, i.e. A, was found as the risk allele. Earlier, the A allele has also been associated with a greater degree of chronic course of depressive symptoms in a sample of 35 individuals with major depression or bipolar disorder [45].
Concerning SNP rs3824872, located in the 3′ region of CRY2 (but also in the 5′ region of MAPK8IP1 gene), our study suggests (q = 0.08) that the T allele is the protective one against dysthymia, and a similar (though non-significant) tendency is seen in the Finnish subpopulation of the winter depression study [24], while in the Swedish subpopulation of the same study, the other allele, i.e. A, was the risk allele. With regard to CRY2 rs7123390, the SNP was earlier associated with winter depression in the Finnish subpopulation [24], but in the current study, we did not observe any significant association with this intronic SNP. Moreover, there was no significant association with the 3′UTR CRY2 rs10838527, similar to earlier findings where no association with winter depression was observed among the Finnish subpopulation, whereas this SNP did associate with winter depression among the Swedish subpopulation [24].
Although preliminary, our analysis has demonstrated that CRY2 associates robustly with dysthymia which is characterized by a chronic course of illness where a depressive episode lasts for two years or longer and often deepens into a major depressive episode. Earlier, CRY2 genetic variants have been associated with winter recurrent depressive disorder [24], and with the rapid cycling of bipolar type 1 disorder where depressive episodes dominate the clinical course of illness [25]. A single CRY1 variant has been associated with major depressive disorder earlier [23] but there is no replication of the finding [21]. Furthermore, it has been previously observed that following the antidepressant sleep deprivation, the expression of CRY2 mRNA increased in controls, whereas no change or response to the treatment was observed in patients with bipolar disorder whose CRY2 mRNA levels were markedly lower than in controls [24]. Moreover in an animal study, CRY2 mRNA expression was abnormal when inbred-strain mice with the intrinsic level of high anxiety were deprived of sleep [46]. Currently, it is however not known, whether the knock-out or the knock-down of CRY2 in the whole organism or specific to a tissue produces any change in anxiety-like or depressive-like behaviors, or how mutated CRY2 proteins influence mood-related behaviors. CRY2 and CRY1 proteins are functionally different [7] which could impact their circadian outputs differently and make a difference in the consequent phenotype. This difference could explain why CRY2, not CRY1, associated with dysthymia in our study. Moreover, it might be that CRY2 variants play a leading role in circadian hierarchy [3] and thereby in the pathogenesis of mood disorders as well. Some circadian clock gene variants seem to have a role in neuropsychiatric disorders in general [47], [48]. Elucidating the mechanisms of action with these genetic variants might therefore benefit the development of treatments for and beyond mood and anxiety disorders [18], [49], [50].
A limitation in our current epidemiological health examination study is that the seasonal pattern, whether in major depressive disorder or bipolar disorder, was not assessed at all. Another limitation is that the diagnoses of bipolar disorders were considered not adequate for analysis. However, it is of note that bipolar type 1 disorder is markedly less frequent in Finland compared with the prevalence in many Western-type societies [51].
To conclude, CRY2 genetic variants associated with dysthymia. This new data, together with the earlier findings, reinforces the involvement of CRY2 in mood disorders.
Supporting Information
Genotype counts and Hardy-Weinberg equilibrium p-values for successfully genotyped SNPs in the analyzed phenotypes.
(XLS)
Results from the single SNP and haplotype association analyses.
(XLS)
Funding Statement
The authors have no support or funding to report.
References
- 1. Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, et al. (1994) Mutagenesis and mapping of a mouse gene, clock, essential for circadian behavior. Science 264: 719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ikeda M, Nomura M (1997) cDNA cloning and tissue-specific expression of a novel basic helix-loop-helix/PAS protein (BMAL1) and identification of alternatively spliced variants with alternative translation initiation site usage. Biochem Biophys Res Commun 233: 258–264. [DOI] [PubMed] [Google Scholar]
- 3. Dardente H, Fortier EE, Martineau V, Cermakian N (2007) Cryptochromes impair phosphorylation of transcriptional activators in the clock: A general mechanism for circadian repression. Biochem J 402: 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ko CH, Takahashi JS (2006) Molecular components of the mammalian circadian clock. Hum Mol Genet 15 Spec No 2: R271–7. [DOI] [PubMed] [Google Scholar]
- 5. Hirota T, Lee JW, St John PC, Sawa M, Iwaisako K, et al. (2012) Identification of small molecule activators of cryptochrome. Science 337: 1094–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Padmanabhan K, Robles MS, Westerling T, Weitz CJ (2012) Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science 337: 599–602. [DOI] [PubMed] [Google Scholar]
- 7. Anand SN, Maywood ES, Chesham JE, Joynson G, Banks GT, et al. (2013) Distinct and separable roles for endogenous CRY1 and CRY2 within the circadian molecular clockwork of the suprachiasmatic nucleus, as revealed by the Fbxl3Afh mutation. J Neurosci 33: 7145–7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, et al. (1999) Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398: 627–630. [DOI] [PubMed] [Google Scholar]
- 9. Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, et al. (1999) Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci U S A 96: 12114–12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hidalgo MP, Caumo W, Posser M, Coccaro SB, Camozzato AL, et al. (2009) Relationship between depressive mood and chronotype in healthy subjects. Psychiatry Clin Neurosci 63: 283–290. [DOI] [PubMed] [Google Scholar]
- 11. Kitamura S, Hida A, Watanabe M, Enomoto M, Aritake-Okada S, et al. (2010) Evening preference is related to the incidence of depressive states independent of sleep-wake conditions. Chronobiol Int 27: 1797–1812. [DOI] [PubMed] [Google Scholar]
- 12. Drennan MD, Klauber MR, Kripke DF, Goyette LM (1991) The effects of depression and age on the horne-ostberg morningness-eveningness score. J Affect Disord 23: 93–98. [DOI] [PubMed] [Google Scholar]
- 13. Gaspar-Barba E, Calati R, Cruz-Fuentes CS, Ontiveros-Uribe MP, Natale V, et al. (2009) Depressive symptomatology is influenced by chronotypes. J Affect Disord 119: 100–106. [DOI] [PubMed] [Google Scholar]
- 14. Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, et al. (2010) Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med 16: 1152–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Narasimamurthy R, Hatori M, Nayak SK, Liu F, Panda S, et al. (2012) Circadian clock protein cryptochrome regulates the expression of proinflammatory cytokines. Proc Natl Acad Sci U S A 109: 12662–12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O’Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH (2008) cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science 320: 949–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Park SK, Nguyen MD, Fischer A, Luke MP, Affar el B, et al. (2005) Par-4 links dopamine signaling and depression. Cell 122: 275–287. [DOI] [PubMed] [Google Scholar]
- 18. McCarthy MJ, Welsh DK (2012) Cellular circadian clocks in mood disorders. J Biol Rhythms 27: 339–352. [DOI] [PubMed] [Google Scholar]
- 19.Li JZ, Bunney BG, Meng F, Hagenauer MH, Walsh DM, et al.. (2013) Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed]
- 20. Johansson C, Willeit M, Smedh C, Ekholm J, Paunio T, et al. (2003) Circadian clock-related polymorphisms in seasonal affective disorder and their relevance to diurnal preference. Neuropsychopharmacology 28: 734–739. [DOI] [PubMed] [Google Scholar]
- 21. Partonen T (2012) Clock gene variants in mood and anxiety disorders. J Neural Transm 119: 1133–1145. [DOI] [PubMed] [Google Scholar]
- 22. Takahashi JS, Hong HK, Ko CH, McDearmon EL (2008) The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat Rev Genet 9: 764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Soria V, Martinez-Amoros E, Escaramis G, Valero J, Perez-Egea R, et al. (2010) Differential association of circadian genes with mood disorders: CRY1 and NPAS2 are associated with unipolar major depression and CLOCK and VIP with bipolar disorder. Neuropsychopharmacology 35: 1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lavebratt C, Sjoholm LK, Soronen P, Paunio T, Vawter MP, et al. (2010) CRY2 is associated with depression. PLoS One 5: e9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sjoholm LK, Backlund L, Cheteh EH, Ek IR, Frisen L, et al. (2010) CRY2 is associated with rapid cycling in bipolar disorder patients. PLoS One 5: e12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhao S, Sancar A (1997) Human blue-light photoreceptor hCRY2 specifically interacts with protein serine/threonine phosphatase 5 and modulates its activity. Photochem Photobiol 66: 727–731. [DOI] [PubMed] [Google Scholar]
- 27. Wittchen HU, Lachner G, Wunderlich U, Pfister H (1998) Test-retest reliability of the computerized DSM-IV version of the munich-composite international diagnostic interview (M-CIDI). Soc Psychiatry Psychiatr Epidemiol 33: 568–578. [DOI] [PubMed] [Google Scholar]
- 28. Beck AT, Ward CH, Mendelson M, Mock J, Erbaugh J (1961) An inventory for measuring depression. Arch Gen Psychiatry 4: 561–571. [DOI] [PubMed] [Google Scholar]
- 29.Raitasalo R (2007) Mielialakysely. suomen oloihin beckin lyhyen depressiokyselyn pohjalta kehitetty masennusoireilun ja itsetunnon kysely. Helsinki: Kela, Sosiaali- ja terveysturvan tutkimuksia 86.
- 30.Schaufeli W, Leiter M, Maslach C, Jackson S (1996) The maslach burnout inventory - general survey (MBI-GS). In: Maslach C, Jackson S, Leiter M, editors. Maslach Burnout Inventory manual. Palo Alto (CA): Consulting Psychologists Press.
- 31.Goldberg D (1978) Manual of the general health questionnaire. Windsor, UK: NFER (National Foundation for Educational Research) Publishing.
- 32. Rintamaki R, Grimaldi S, Englund A, Haukka J, Partonen T, et al. (2008) Seasonal changes in mood and behavior are linked to metabolic syndrome. PLoS One 3: e1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenthal NE, Bradt GH, Wehr TA (1984) Seasonal pattern assessment questionnaire. Bethesda, Md, National Institute of Mental Health.
- 34. Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. [DOI] [PubMed] [Google Scholar]
- 35. Conde L, Vaquerizas JM, Dopazo H, Arbiza L, Reumers J, et al. (2006) PupaSuite: Finding functional single nucleotide polymorphisms for large-scale genotyping purposes. Nucleic Acids Res 34: W621–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cheng YC, Hsiao FC, Yeh EC, Lin WJ, Tang CY, et al. (2012) VarioWatch: Providing large-scale and comprehensive annotations on human genomic variants in the next generation sequencing era. Nucleic Acids Res 40: W76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hariharan M, Scaria V, Brahmachari SK (2009) dbSMR: A novel resource of genome-wide SNPs affecting microRNA mediated regulation. BMC Bioinformatics 10: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gong J, Tong Y, Zhang HM, Wang K, Hu T, et al. (2012) Genome-wide identification of SNPs in microRNA genes and the SNP effects on microRNA target binding and biogenesis. Hum Mutat 33: 254–263. [DOI] [PubMed] [Google Scholar]
- 39. Jurinke C, van den Boom D, Cantor CR, Koster H (2002) Automated genotyping using the DNA MassArray technology. Methods Mol Biol 187: 179–192. [DOI] [PubMed] [Google Scholar]
- 40. Lahermo P, Liljedahl U, Alnaes G, Axelsson T, Brookes AJ, et al. (2006) A quality assessment survey of SNP genotyping laboratories. Hum Mutat 27: 711–714. [DOI] [PubMed] [Google Scholar]
- 41. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Storey JD (2003) The positive false discovery rate: A bayesian interpretation and the q-value. The Annals of Statistics 31: 2013–2035. [Google Scholar]
- 43. Goni JR, de la Cruz X, Orozco M (2004) Triplex-forming oligonucleotide target sequences in the human genome. Nucleic Acids Res 32: 354–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Utge SJ, Soronen P, Loukola A, Kronholm E, Ollila HM, et al. (2010) Systematic analysis of circadian genes in a population-based sample reveals association of TIMELESS with depression and sleep disturbance. PLoS One 5: e9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fiedorowicz JG, Coryell WH, Akhter A, Ellingrod VL (2012) Chryptochrome 2 variants, chronicity, and seasonality of mood disorders. Psychiatr Genet 22: 305–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wisor JP, Pasumarthi RK, Gerashchenko D, Thompson CL, Pathak S, et al. (2008) Sleep deprivation effects on circadian clock gene expression in the cerebral cortex parallel electroencephalographic differences among mouse strains. J Neurosci 28: 7193–7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Barnard AR, Nolan PM (2008) When clocks go bad: Neurobehavioural consequences of disrupted circadian timing. PLoS Genet 4: e1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Coogan AN, Thome J (2011) Chronotherapeutics and psychiatry: Setting the clock to relieve the symptoms. World J Biol Psychiatry 12 Suppl 140–43. [DOI] [PubMed] [Google Scholar]
- 49.Bunney BG, Bunney WE (2012) Mechanisms of rapid antidepressant effects of sleep deprivation therapy: Clock genes and circadian rhythms. Biol Psychiatry. [DOI] [PubMed]
- 50. Ono D, Honma S, Honma K (2013) Cryptochromes are critical for the development of coherent circadian rhythms in the mouse suprachiasmatic nucleus. Nat Commun 4: 1666. [DOI] [PubMed] [Google Scholar]
- 51. Perala J, Suvisaari J, Saarni SI, Kuoppasalmi K, Isometsa E, et al. (2007) Lifetime prevalence of psychotic and bipolar I disorders in a general population. Arch Gen Psychiatry 64: 19–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genotype counts and Hardy-Weinberg equilibrium p-values for successfully genotyped SNPs in the analyzed phenotypes.
(XLS)
Results from the single SNP and haplotype association analyses.
(XLS)