Abstract
The larvae of the wax moth, Galleria mellonella , have been used experimentally to host a range of bacterial and fungal pathogens. In this study we evaluated the suitability of G . mellonella as an alternative animal model of Salmonella infection. Using a range of inoculum doses we established that the LD50 of Salmonella Typhimurium strain NCTC 12023 was 3.6 × 103 bacteria per larva. Further, a set of isogenic mutant strains depleted of known virulence factors was tested to identify determinants essential for S . Typhimurium pathogenesis. Mutants depleted of one or both of the type III secretion systems encoded by Salmonella Pathogenicity Islands 1 and 2 showed no virulence defect. In contrast, we observed reduced pathogenic potential of a phoQ mutant indicating an important role for the PhoPQ two-component signal transduction system. Lipopolysaccharide (LPS) structure was also shown to influence Salmonella virulence in G . mellonella . A waaL (rfaL) mutant, which lacks the entire O-antigen (OAg), was virtually avirulent, while a wzz ST/wzz fepE double mutant expressing only a very short OAg was highly attenuated for virulence. Furthermore, shortly after infection both LPS mutant strains showed decreased replication when compared to the wild type in a flow cytometry-based competitive index assay. In this study we successfully established a G . mellonella model of S . Typhimurium infection. By identifying PhoQ and LPS OAg length as key determinants of virulence in the wax moth larvae we proved that there is an overlap between this and other animal model systems, thus confirming that the G . mellonella infection model is suitable for assessing aspects of Salmonella virulence function.
Introduction
Salmonella enterica is one of the most prevalent bacterial pathogens worldwide. The impressive success of this pathogen can be attributed to its great versatility in surviving within the environment and the capacity to infect a wide range of host organisms [1]. Key to these processes are the array of factors encoded within the genome, which facilitate host colonization and the development of disease [2]. In order to identify novel virulence determinants that represent putative targets for drug development, researchers have designed numerous models of infection based on cell cultures or entire host organisms. Murine models have been used extensively to study the interactions occurring between host and a variety of bacterial pathogens, and the infection of mice with Salmonella enterica serovar Typhimurium ( S . Typhimurium ) has been used for decades as a disease model of human typhoid fever. Subsequent refinements to this system allowed for the induction of Salmonella -mediated colitis by pretreatment of these animals with streptomycin [3], while genetic modifications of the host have resulted in models that are a much closer representation of Salmonella Typhi infection in humans [4,5]. Although these model systems help to improve our understanding of Salmonella pathogenesis, infection studies using mammals are often time consuming and require expensive experimental setup. Furthermore, animal numbers must be kept to a minimum and extensive demands have to be fulfilled to meet the high standards given by law and ethics. Therefore, alternative in vivo models that avoid these limitations would benefit our ability to analyze the complex interplay between a pathogen and a multi-cellular host organism.
In recent years, substantial progress has been made in developing invertebrate-based model systems for studying bacterial and fungal infections [6–8]. It is worth stressing that a suitable surrogate for mammals ought to give comparable results if the gene or pathway of interest is of any importance for human infection. In the case of the soil nematode Caenorhabditis elegans, it has been demonstrated that an alternative sigma factor of Pseudomonas aeruginosa is required for virulence in this invertebrate and mice alike [9]. In addition, Salmonella virulence determinants that similarly affect C. elegans intestinal cells and mammalian epithelial cells have also been described [10]. A key virulence factor of Salmonella is the lipopolysaccharide (LPS) or endotoxin, a complex protruding structure connected to the outer membrane of Gram-negative bacteria via a phospholipid anchor, the Lipid A. Attached to this are repetitive sequences of sugar moieties constituting the core oligosaccharide and the O-antigen (OAg), a polysaccharide structure of variable length. Although LPS is highly immunogenic and activates innate immune defense pathways within the host it also mediates complement resistance and interferes with phagocytosis, and is therefore a prerequisite for full virulence in mice [11,12]. Using a nematode model of infection, Aballay et al. showed that an intact bacterial cell envelope containing full-length LPS is required to establish persistent Salmonella infection of the C. elegans intestine [13]. Similarly, expression of OAg by the apathogenic E. coli strain, K-12, rendered this organism fully virulent in the same model, underlining the importance of this surface structure to the colonization of an invertebrate host [14].
Another well-established alternative model organism is the greater wax moth, Galleria mellonella . G . mellonella larvae offer considerable benefits as an infection model as they are easy to handle, rapidly generate data and can be studied in large numbers while keeping experiments cost-effective. Most importantly, and in contrast to many other invertebrate models, including C. elegans, analyses can be performed at 37° C, a temperature optimal for the vast majority of human pathogens. Moreover, injection of bacteria into the larval hemolymph enables the application of a defined bacterial dose, an advantage over the imprecise infection of C. elegans, which relies on the worms freely grazing on a lawn of bacteria to establish colonization. A further advantage of this model is the innate immune response of G . mellonella , which shares a high degree of homology with the mammalian system [15]. For example, hemocytes, which produce a robust oxidative burst, behave similarly to phagocytic cells in response to bacterial infection [16]. This, amongst other conserved features of anti-microbial action, might explain the positive correlation between data obtained from Galleria and mice infections for both eukaryotic pathogens, such as Candida albicans [17] or Aspergillus fumigatus [18], as well as for different prokaryotes. Hence, the G . mellonella model has been successfully applied to assess the virulence of a variety of bacteria, including: Listeria monocytogenes [19], Francisella tularensis [20], Burkholderia cepacia complex [21], P. aeruginosa [22,23], Cryptococcus neoformans [24], Enterococcus faecium [25], Legionella pneumophila [26] and various corynebacteria [27]. In contrast, data regarding the pathogenicity of S . Typhimurium for G . mellonella is scarce. Although Krustak and colleagues initially described the cellular response of the larvae upon Salmonella challenge and bacteria-mediated lysis of the hemocytes, a detailed examination of the responsible determinants was not pursued [28]. Thus, the mechanism behind Galleria-Salmonella interaction remains obscure. Here, we set out to establish a G . mellonella -based model system for studying Salmonella virulence, which could be used to generate indicatory data prior to comprehensive mammalian studies.
Materials and Methods
Cloning
All bacterial strains and plasmids used in this study are listed in Table 1. An overview about the oligonucleotides used is given in Table 2. The gene for Superfolder GFP (SFGFP) [29] was synthesized codon-optimized for expression in S. enterica (Geneart, Regensburg, Germany). The sfgfp gene was amplified by PCR using primers SmaI-RBS-SFGFP-for and SFGFP-NcoI-rev and the product was cloned via SmaI/NcoI in the similarly-digested pWRG15 [30], yielding pWRG81. The EM7 promoter (Life Technologies) was amplified by PCR from pGEN-luxCDABE [31] using primers EcoRI-EM7-for and EM7-rev and then cloned into pWRG81 via EcoRI/SmaI, yielding pWRG167. The photo-stable TagRFP variant, TagRFP-T [32], was synthesized codon-optimized for expression in S. enterica (Geneart). The gene encoding TagRFP-T was amplified by PCR using primers XbaI-TagRFP-for and TagRFP-HindIII-rev and the product was cloned via XbaI/HindIII into the similarly-digested pFPV25.1 [33], yielding pWRG435.
Table 1. Bacterial strains and plasmids used in this study.
| Salmonella strain | Relevant characteristic(s) | Source or Reference |
|---|---|---|
| NCTC 12023 | wild type, NalS, isogenic to ATCC 14028 | NCTC, Colindale, UK |
| MvP724 | Δwzz ST FRT Δwzz fepE FRT | [42] |
| MvP818 | ΔinvC FRT | [60] |
| MvP1036 | ΔwaaL (rfaL) FRT | [61] |
| MvP1213 | ΔfliI FRT | M. Hensel, unpublished |
| P2D6 | ssaV::mTn5, Kmr | [50] |
| WRG6 | ΔphoQ | [62] |
| WRG107 | ΔinvC FRT ssaV::mTn5, Kmr | This study |
| Plasmids | ||
| p3313 | PrfaD::rfaDFCL in pWSK29, Apr | [61] |
| p3390 | wzz ST and wzz fepE in pWSK29, Apr | [42] |
| pFPV25.1 | PrpsM::gfpmut3a in pFPV25, Apr | [33] |
| pWSK29 | Low copy-number vector, Apr | [63] |
| pWRG81 | sfgfp in pWRG15 [30], Apr | This study |
| pWRG103 | PphoP::phoPQ in pWSK29, Apr | [62] |
| pWRG167 | PEM7::sfgfp in pWRG81, Apr | This study |
| pWRG435 | PrpsM::tagrfp-t in pFPV25, Apr | This study |
Table 2. Oligonucleotides used in this study.
| Oligonucleotide | Sequence (5’→3’), restriction sites underlined |
|---|---|
| EcoRI-EM7-for | CAGG A A T T CATCCGCGGCCGCGTTTAAAC |
| EM7-rev | AGAGGATCC C C G G GTACCAC |
| SFGFP-NcoI-rev | ATAC C A T G GTTATTATTTATACAGTTCATCCATG |
| SmaI-RBS-SFGFP-for | ATCC C C G G GAAAGAGGAGAAAAGTATGCGCAAAGGCGAAGAACTG |
| TagRFP-HindIII-rev | GCGA A G C T TATTATTTATACAGTTCATCCATGC |
| XbaI-TagRFP-for | GCGT C T A G ATTTAAGAAGGAGATATACATATGGTGAGCAAAGGCGAAGAACTG |
| SmaI-RBS-SFGFP-for | ATCC C C G G GAAAGAGGAGAAAAGTATGCGCAAAGGCGAAGAACTG |
| TagRFP-HindIII-rev | GCGA A G C T TATTATTTATACAGTTCATCCATGC |
| XbaI-TagRFP-for | GCGT C T A G ATTTAAGAAGGAGATATACATATGGTGAGCAAAGGCGAAGAACTG |
Infection model
G . mellonella larvae were obtained from Reptilienkosmos.de (Schwalmtal, Germany) and selected for no visible signs of melanization, pupation, or any other illnesses before being infected. At the time of infection each larva weighed approximately 250 mg. Bacterial strains were grown overnight and the indicated amount of bacteria, suspended in 5 µl Dulbecco’s PBS (PAA, Pasching, Austria), was injected into the hindmost left proleg of the larvae using a 10 µl syringe (Hamilton, Bonaduz, Switzerland). As controls, non-injected and PBS-injected animals were included in each experiment. Groups of 16 identically treated larvae were placed in a 100 mm petri dish and incubated at 37° C. Individual larvae were scored regularly over the course of the experiment for signs of melanization and for viability using their reflex in response to contact. Data are expressed as percent survival, and are the representative results of at least three independent experiments.
Flow cytometry
Larvae were first homogenized in 2 ml PBS using a sterile pipette tip. After vigorous pipetting, each lysate was cleared of coarse debris by centrifugation (500 x g, 10 min). A 1:100 dilution of the supernatant in 1% paraformaldehyde in PBS was then analyzed using a FACSAria III (BD, Heidelberg, Germany) cell sorter equipped with 488 nm and 561 nm lasers. Two-color analyses according to Bumann [34] were applied to discriminate between cellular debris and Salmonella expressing the fluorescent proteins SFGFP and TagRFP-T, respectively. For SFGFP detection, 488 nm excitation in combination with two detectors and the two filters, 502LP+530/30 and 555LP+586/15, was used. For TagRFP-T detection, 561 nm excitation in combination with two detectors and the two filters, 582/15 and 685LP+710/50, was used. Data acquisition was performed using FACS DIVA v 6.1.3 software (BD).
Data analysis
Data were analyzed and plotted using Prism 5 software (GraphPad Software Inc., La Jolla, CAL, USA). Flow cytometry data was analyzed and plotted using FlowJo v 9.4.7 (Tree Star Inc., Ashland, OR, USA).
Results
Infection of G . mellonella with S . Typhimurium and determination of LD50
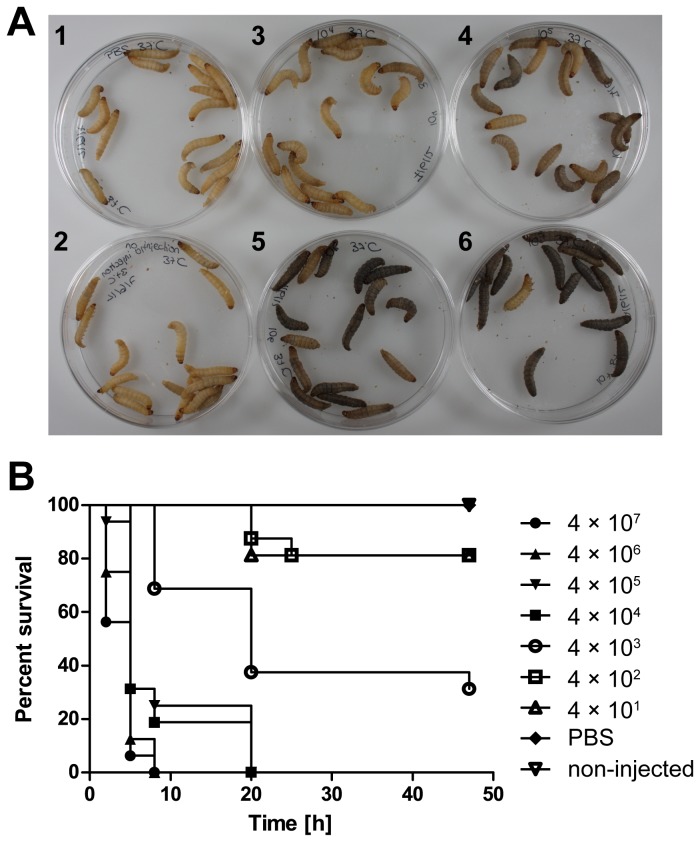
In order to investigate the pathogenic potential of Salmonella towards arthropods, we studied the susceptibility of G . mellonella larvae to infection with S . Typhimurium strain NCTC 12023. To determine the parameters necessary for establishing infection by the pathogen, larvae were challenged with bacterial loads ranging from 40 to 4 × 107 bacteria. Incubation was carried out at 37° C, the optimum temperature for Salmonella growth and mammalian physiology, and the larvae were monitored for up to 50 h to assess physical conditions, such as melanization and rates of survival. Melanization, a process resulting in the systemic deposition of the pigment melanin, is part of the innate immune response of Arthropoda to parasite challenge and can be used as an effective measure to evaluate insect health. Following infection with Salmonella , melanin production by G . mellonella became apparent only 2.5 h after injection of 4 × 105 to 4 × 107 bacteria (Figure 1A). Moreover, synthesis of melanin was triggered in a dose-dependent manner, eventually resulting in 100% dark-colored larvae at 20 h post inoculation using 4 × 107 bacteria, as compared to moderate melanization in only 38% of larvae that received 4 × 104 or fewer bacteria (data not shown). Non-injected, or PBS-injected larvae controls showed no signs of melanization (Figure 1A). In order to reflect a more physiological temperature for G . mellonella , infected larvae were incubated also at 27° C. Similar results to those described above were obtained under these conditions, although the progression of melanin production was notably slower (data not shown).
Figure 1. Dose-dependent killing of G. mellonella by S. Typhimurium NCTC 12023 WT.
G. mellonella larvae were infected with increasing bacterial loads, ranging from 40 to 4 × 107 bacteria, and incubated for up to 50 h at 37° C. (A) Deposition of melanin at 2.5 h p. i. was dependent on the number of bacteria injected (1) PBS control; 2) non-injected control; 3) 4 × 104; 4) 4 × 105; 5) 4 × 106; 6) 4 × 107). (B) The percent of larvae surviving was assessed after injecting different doses of S. Typhimurium WT as indicated. Increasing amounts of dark-colored and/or dead larvae were obtained in a dose-dependent manner. PBS: buffer control. Data as shown are the representative results of three independent experiments, for which similar outcomes were obtained.
Following the onset of an immune response, larvae rapidly succumbed to Salmonella infection in a dose-dependent manner (Figure 1B). As seen for the deposition of melanin, PBS-injected or non-injected controls showed no symptoms, and 100% of control larvae survived (Figure 1A and 1B). Dose-dependent evaluation of Salmonella inoculi yielded an LD50 value of 3.6 × 103 bacteria at 25 h post infection. Thus, further experiments were all performed using an inoculum of 4 × 104 bacteria per larvae, which were followed for up to 48 h, to ensure that any minor differences in the pathogenic potential of the various Salmonella strains investigated would be observed (see below). As the onset of pupation occurred naturally after 40-48 h of incubation all experiments were concluded 48 h after bacterial challenge, at which point larvae were counted as either dead or alive.
It is widely accepted that changes occur in virulence factor expression by S . Typhimurium as it transits from logarithmic growth into stationary phase [35]. To evaluate the contribution of pre-induced virulence factors important for early colonization events such as invasion, adhesion or motility, we additionally compared the effects of using S . Typhimurium grown to late exponential or stationary phase for infection of G . mellonella . However, we did not observe any significant difference in the survival rates of larvae inoculated with either late-log invasive bacteria (3.5 h of sub-cultivation), or Salmonella that were grown overnight (20 h) to stationary phase (data not shown).
Investigation of major virulence factors in the Galleria model
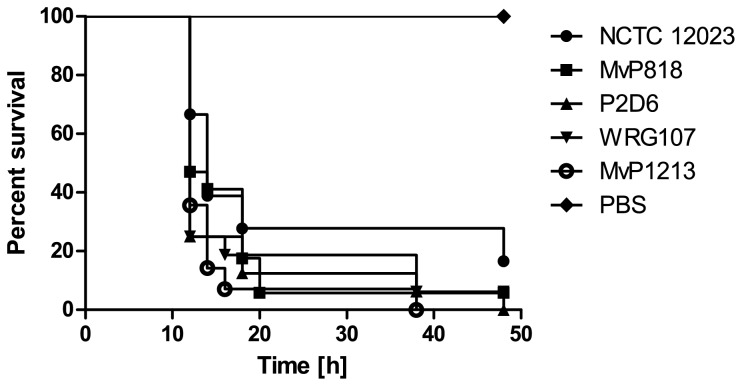
The genus Salmonella possesses a variety of proteins, commonly referred to as virulence factors that are required for bacterial survival and/or proliferation within the host organism. A vast number of studies have identified various open reading frames (ORFs) that are transcribed from defined genomic regions known as Salmonella pathogenicity islands (SPI), which are essential for these host–pathogen interactions [2]. For instance, SPI-1 and SPI-2, each encoding genes for a type III secretion system (T3SS), were shown to be necessary for in vitro and in vivo infection of cell lines, C. elegans, mice or calves [10,36–38]. In order to investigate their contribution to Galleria infection, G . mellonella larvae were injected with S . Typhimurium mutant strains deficient in either a specific virulence factor, or an entire pathogenicity island (Table 1). Deletion of fliI, the gene encoding the flagella apparatus-associated ATPase, did not influence infectivity of the respective mutant strain MvP1213 (Figure 2). Likewise, strains disrupted in SPI-1, SPI-2 or both pathogenicity islands showed no attenuation when compared to the wild type (WT) strain NCTC 12023 (Figure 2).
Figure 2. Survival of G. mellonella infected with S. Typhimurium NCTC 12023 WT or mutants deficient for known virulence factors.
G. mellonella larvae were infected with 4 × 104 S. Typhimurium NCTC 12023 WT and incubated for 48 h at 37° C. Likewise, larvae were challenged with the same amount of Salmonella mutant strains lacking a functional T3SS encoded by pathogenicity islands SPI-1 (MvP818), SPI-2 (P2D6), SPI-1 plus SPI-2 (WRG107) or the flagellar export ATPase gene, fliI (MvP1213). PBS injections were included as a negative control. Data as shown are the representative results of three independent experiments, for which similar outcomes were obtained.
Assessing the impact of the PhoPQ regulon on Galleria infection
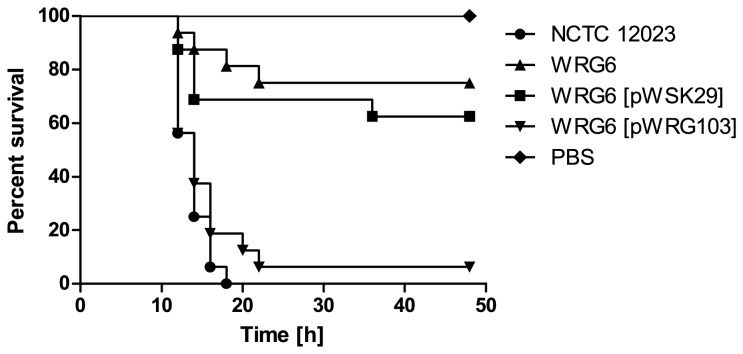
Previous studies have demonstrated that Salmonella of various serovars are capable of infecting the soil nematode C. elegans, where they reside in the lumen of the worm intestinal tract [13]. The ability to sense changes in the environmental surroundings with the two-component system PhoPQ is therefore a prerequisite for successful colonization by S . Typhimurium . PhoPQ has been shown to contribute to infectivity of this bacterium towards C. elegans, macrophages and mice alike [13,39]. Hence, we determined rates of survival of the G . mellonella larvae upon infection with a phoQ deletion mutant strain of S . Typhimurium . Deletion of the sensor kinase significantly reduced bacterial virulence in the Galleria model (Figure 3). While no larvae survived infection by WT bacteria, up to 75% of the arthropods were still alive at 24 h post challenge with the phoQ mutant strain WRG6 (Figure 3). This defect could be compensated for by trans-complementation experiments, whereby episomal introduction of phoPQ using the low-copy number vector pWRG103 resulted in the death of 94% of the larvae 24 h after challenge (Figure 3). By confirming that deletion of known virulence determinants, such as the conserved signaling pathway PhoPQ, attenuates the virulence of S . Typhimurium in this model these results support the utility in using G . mellonella as a model system for studying Salmonella pathogenesis.
Figure 3. Survival of G. mellonella infected with S. Typhimurium NCTC 12023 WT and a ΔphoQ mutant strain.
Survival of G. mellonella larvae was monitored for 48 h at 37° C after injection of Salmonella WT and the ΔphoQ deletion strain WRG6. Complementation of WRG6 was carried out in trans by transformation with the low-copy number plasmid pWRG103 expressing phoQ, or empty vector pWSK29. PBS: buffer control. Experiments were performed in triplicate using 16 larvae per group.
LPS is required for full virulence of S . Typhimurium in the Galleria infection model
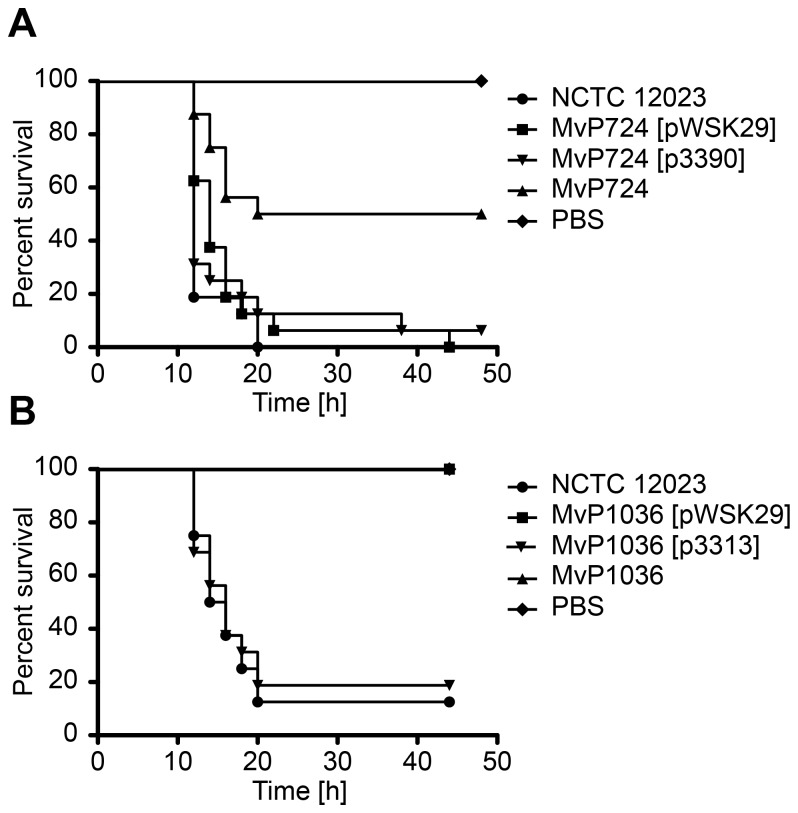
PhoPQ signaling comprises a complex regulatory network that influences numerous cellular processes, including outer membrane modifications [40]. Because the LPS of Salmonella is detrimental to bacterial survival within host cells [12,41,42], we next analyzed whether truncation of this structure impacted Salmonella -induced killing of G . mellonella . Larvae were injected with the wzz ST/wzz fepE double knockout mutant MvP724, which lacks the enzymes responsible for synthesizing long and very long modal length OAg, respectively [12,43]. Disruption of both ORFs results in a strain expressing a relatively short OAg (S–OAg). G . mellonella survival increased significantly upon truncation of the OAg (Figure 4A), while complementation of wzz ST/wzz fepF in trans restored pathogenicity of the resulting S . Typhimurium strain to WT levels (Figure 4A). However, it must be noted that introducing the empty vector, pWSK29, used for in trans complementation also reconstituted virulence, albeit to a lesser extent than plasmid p3390, a pWSK29 derivative harboring both ORFs of wzz ST and wzz fepE (Figure 4A).
Figure 4. Effect of deletion of LPS-modifying enzymes on Salmonella-mediated killing of G. mellonella.
Survival of G. mellonella was assessed after injection of 4 × 104 S. Typhimurium NCTC 12023 WT or mutant derivatives lacking different LPS-modifying regulators and enzymes. (A) Comparison of WT with mutant strain MvP724, which lacks the regulators of gene expression wzz ST /wzz fepE. For complementation purposes, plasmid p3390 was transformed into MvP724, from which both wzz ST and wzz fepE were expressed under control of the wzz ST promoter. Plasmid pWSK29 was transformed into the mutant strain as a vector-only control. (B) Impact of the O-antigen ligase, WaaL, on bacterial-mediated killing of G. mellonella larvae. Infection by the ΔwaaL mutant MvP1036, with or without the empty vector (pWSK29), and its complemented derivative MvP1036 (p3313), was performed alongside WT injection at a total amount of 4 × 104 bacteria per larvae. PBS: buffer control. Data as shown are the representative results of three independent experiments, for which similar outcomes were obtained.
The contribution of LPS to Salmonella virulence was also examined by challenging G . mellonella larvae with the waaL (rfaL) mutant strain, MvP1036, which lacks the immunoresponsive oligosaccharide chain. As depicted in Figure 4B, deletion of the entire OAg completely abolished infectivity of this S . Typhimurium derivative. Twenty hours after inoculation only 12% of WT-infected larvae had survived bacterial challenge, whereas all larvae infected with the waaL mutant strain remained alive (Figure 4B). Complementation with long to very long modal lengths of OAg introduced by the low-copy number vector p3313 restored pathogenicity of mutant strain MvP1036 to 92% that of WT levels (Figure 4B). This was independent of carrying the empty vector alone, as transformation of the waaL mutant strain MvP1036 with pWSK29 did not influence virulence (Figure 4B).
Taken together, these data clearly demonstrate that Salmonella require an intact LPS with a specific modal length to resist the immune response, persist, and most likely multiply within the gastrointestinal tract of the arthropod G . mellonella .
Competitive index analysis shows a role for LPS in bacterial replication within G . mellonella
Salmonella OAg length significantly contributes to G . mellonella mortality as shown above. To elucidate this effect in more detail we set out to establish a competitive index (C. I.) [44] model of G . mellonella infection, which would allow the relative fitness of a mutant strain to be assessed in comparison to the WT within an individual larva at much earlier time points than used previously. Two fluorescent marker proteins, GFP and RFP, were chosen to discriminate between mutant and WT strains in the co-infection. This methodology has been successfully used in a previous study analyzing the capability of different Salmonella mutants to replicate within cultured cells [45]. Since it is known that expression of fluorescent markers can significantly attenuate Salmonella virulence both in vitro and in vivo [46], we first tested whether WT bacteria expressing either GFP or RFP exhibited reduced virulence in the G . mellonella model when compared to a non-tagged WT strain. In single infections using 4 × 104 bacteria per larva, no differences in pathogenicity were observed between the non-tagged WT, or WT strains expressing either SFGFP (from plasmid pWRG167) or TagRFP-T (pWRG435) following 48 h of infection (Figure 5A).
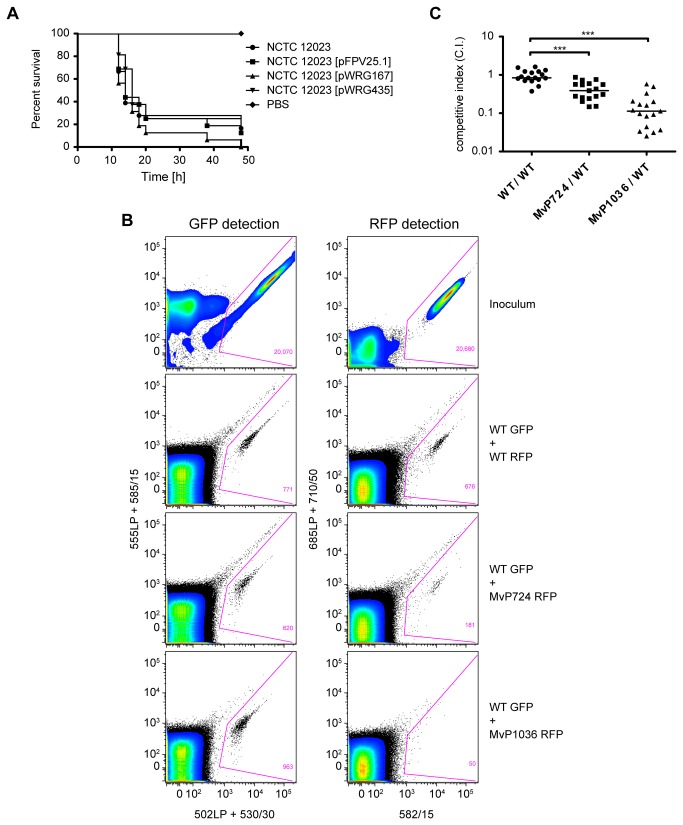
Figure 5. Impact of successive truncation of the O-antigen of Salmonella in the pathogenicity of these bacteria in competitive index experiments.
G. mellonella larvae were injected with a suspension containing a total of 4 × 104 bacteria. A 1:1 mixture of S. Typhimurium WT and each LPS-truncated mutant was used to inoculate the larvae, which were incubated for 6 h before larval lysates were analyzed by flow cytometry for bacterial quantification. Expression of either GFP from pFPV25.1 or pWRG167, or TagRFP‑T from pWRG435 was used to discriminate between WT and mutant strains, respectively. (A) The influence of gfp and rfp expression on bacterial-mediated killing of G. mellonella larvae was assessed using S. Typhimurium NCTC 12023 WT with or without the expression plasmids. PBS: buffer control. (B) Gating strategy and examples of bacterial cell counts as measured from homogenized larva tissue after co-infection with Salmonella WT and Δwzz ST /wzz fepE mutant (MvP724) or ΔwaaL mutant (MvP1036). (C) Competitive index analysis of mixed infections involving WT/WT (gfp/rfp-tagged WT strain), MvP724/WT (rfp-tagged Δwzz ST /wzz fepE mutant and gfp-expressing WT) and MvP1036/WT (rfp-containing ΔwaaL deletion mutant and gfp-tagged WT). One data point represents one individual larva. Horizontal bars indicate the median of data from three independent experiments. Statistical analysis by one-tailed Mann-Whitney ranked-sum test was done by comparing C.I. data of different co-infections as depicted. ***, P <0.0001.
For each C. I. experiment, larvae were inoculated with 2 × 104 each of the differentially tagged bacteria and followed for 6 h. We then used flow cytometry to determine the ratios between GFP-tagged WT bacteria and TagRFP‑T-tagged mutant strain in the inoculum, and in larval lysates 6 h post-infection. To discriminate between cellular debris of larval origin and the fluorescently labeled bacteria, samples were subjected to two-color flow cytometry for GFP as well as for RFP detection [34]. The gating strategy used for sample analysis is shown in Figure 5B. To calculate the C. I., the total numbers of fluorescent bacteria within these gates were used. Co-infection with differentially labeled WT Salmonella resulted in relatively equal amounts of GFP- and RFP-expressing bacteria in both the inoculum and larval lysates (mean C. I. = 0.962), proving that WT bacteria expressing either GFP or RFP are equally virulent in this model (Figure 5C). A co-infection using the RFP-tagged wzz ST/wzz fepE double knockout mutant strain MvP724 together with GFP-tagged WT resulted in a mean C. I. of 0.43, indicating that significantly fewer mutant bacteria were present after 6 h of infection (Figure 5C). The colonizing ability of the RFP-labeled waaL mutant, MvP1036, at this time was also reduced, showing a six-fold reduction in bacterial numbers when compared to a GFP-labeled WT strain (mean C. I. = 0.165) (Figure 5C).
Taken together, these results confirm our observations from the single infection experiments, with the calculated C. I. reporting attenuation for both the waaL and the wzz ST/wzz fepE Salmonella mutants just 6 h after infection of G . mellonella larvae.
Discussion
Galleria mellonella is susceptible to infection with S . Typhimurium strain NCTC 12023
The greater wax moth Galleria mellonella has been utilized as a novel model host for determining the virulence of a variety of bacterial pathogens. However, to date, data on infection of this caterpillar by the genus Salmonella has remained scarce. We herein demonstrate that these larvae are susceptible to Salmonella infection in a dose-dependent manner. Injection of about 3,600 S . Typhimurium NCTC 12023 cells was sufficient to cause the death of 50% (LD50) of the challenged larvae within 25 h of inoculation. LD50 scores for different pathogens are reported in the range from as little as 1 to exceeding 7 × 104 bacteria per larva, depending on the experimental conditions used. While P. aeruginosa strain PA14 or selected strains of the B. cepacia complex were reported to exhibit an LD50 of less than 10 bacterial cells [21–23], Acinetobater baumanii , Group A Streptococci (GAS) and L. monocytogenes required 2 × 104, 105 and 106 bacteria to cause 50% of the larvae to succumb to infection, respectively [19,47,48]. Our finding that G . mellonella is susceptible to S . Typhimurium infection indicates that this organism can be used as a novel model system for studying the pathogenic mechanisms of Salmonella .
SPI-1 and SPI-2 are dispensable for G . mellonella infection by S . Typhimurium
In order to examine the virulence potential of certain pathogens in various hosts, researchers utilize deletion mutants with interruptions in known and putative virulence factors to study the role these specific determinants play in establishing and progression of disease. For example, depleting P. aeruginosa of its T3SS or important effector proteins such as the phospholipase ExoU renders this organism avirulent in a Galleria model killing assay [23]. Similarly, Tenor and colleagues also used Salmonella knockout strains to demonstrate the importance of both effector proteins and the entire SPI-1 in establishing intestinal colonization of C. elegans [10]. Deletion of one or two of the SPI-encoded T3SSs of S . Typhimurium also has a substantial impact on virulence during infection of mice [49,50]. Conversely, in this study we showed that the deletion of either SPI-1 or SPI-2 did not affect Salmonella virulence in a Galleria model of infection (Figure 2). However, since we only focused on data obtained by end point experiments, we cannot rule out a role for these two SPI-encoded T3SSs in pathogenicity of S . Typhimurium against G . mellonella at earlier time points of infection.
S . Typhimurium requires PhoQ signaling and intact LPS for successful infection of G . mellonella
Although some established Salmonella virulence determinants do not appear essential for the successful colonization of the Galleria host, our study revealed that certain signal transduction pathways and outer membrane structures are important for larval infection. Deletion of phoQ, which encodes the membrane-spanning sensor kinase of the PhoP-PhoQ two-component system, severely attenuated virulence of S . Typhimurium NCTC 12023 in G . mellonella larvae (Figure 3). When ambient Mg2+-levels drop, as occurs in the transition from the environment to the host cell, PhoQ undergoes spontaneous autophosphorylation. Subsequent transfer of a phosphate residue to the response regulator, PhoP, leads to activation of gene transcription, stimulating a complex network of regulatory processes [51]. This PhoPQ regulatory system is essential for pathogenicity not only in Salmonella [39], but impacts the virulence of Y. pestis [52], Shigella flexneri [53] and the plant pathogen Erwinia carotovora subsp. carotovora [54]. Since all of these organisms thrive in quite distinct habitats, it is likely that this two-component system functions in the sensing of an appropriate environmental niche for colonization. Although Salmonellae usually reside inside a specialized vacuole within the host cell, it remains to be determined whether or not S . Typhimurium actively invades and replicates inside the hemocytes of G . mellonella .
In Salmonella , PhoPQ controls the expression of approximately 1% of all ORFs, many of which are involved in remodeling the bacterial cell envelope [39,55]. Included in this series of modifications is the incorporation of aminoarabinose into Lipid A, which reduces the negative net charge of the LPS and confers increased resistance to cationic antimicrobial peptides (CAMPs) such as polymyxin [40]. Moreover, PhoQ can be directly activated by CAMPs further contributing to the recognition of these antimicrobial molecules by Salmonella [56]. This might, at least in part, explain the reduced pathogenicity of S. Typmiurium phoQ mutants in our Galleria model. Since these larvae are capable of synthesizing a range of antimicrobial substances [57], the inability of these strains to acquire a resistant phenotype could severely impact bacterial viability inside the hemocoel.
Our investigations showed that truncation of the OAg had a dramatic effect on the ability of S . Typhimurium to kill the Galleria larvae. Shortening OAg chain length by deleting the regulatory genes wzz ST and wzz fepE, reduced the pathogenic potential of the resultant mutant strain to 50% that of WT levels after 20 h of infection (Figure 4A). This effect was even more pronounced following disruption of the OAg liagse, WaaL, which rendered the mutant strain completely avirulent in our Galleria model of infection (Figure 4B). Truncation of the OAg has also been implicated in increased neutrophil-mediated killing, complement-mediated susceptibility, and phagocytosis of Salmonella by RAW264.7 macrophages [12,42,58]. Furthermore, Murray and colleagues demonstrated that a wzz ST/wzz fepE double mutant of S . Typhimurium was highly attenuated in competition with the WT following intraperitoneally injection of mice, as well as in single infection analysis following oral administration [12]. This is consistent with our findings, whereby C. I. experiments clearly showed that the double mutant and the waaL deletion strain were outcompeted by WT bacteria in our G . mellonella model of infection. Interestingly, truncation of the LPS of P. aeruginosa results in increased secretion of effector proteins by the T3SS, and hence enhances lung injury [59]. However, this was not shown to be the case in S . Typhimurium , where reduced modal repeats within the LPS had no impact on T3SS function or intracellular replication by the bacteria [42]. Hölzer et al. identified another side effect of LPS truncation in Salmonella , in that the respective waaL mutant had impaired motility due to malfunction of the flagella assembly [42]. It is therefore feasible that the loss of virulence of the waaL strain in the Galleria model seen here is the result of the reduced ability of the bacteria to disseminate evenly throughout the larvae. However, as we did not observe attenuation of a non-motile fliI mutant strain, which lacks the flagella-associated ATPase, the mechanism through which loss of waaL attenuates virulence remains to be determined.
The presence of Salmonella -LPS is known to confer protection against the early epithelial immune response both in vivo and in vitro [41]. Likewise, intact LPS is required for the establishment of persistent infections, and causes gonadal cell death in C. elegans intestines [13]. In this C. elegans model of infection, the phenotypes of two distinct LPS mutants of Salmonella were comparable to that of the phoPQ deficient strain. We were able to replicate these findings using similar phoQ- and LPS- mutant strains in our Galleria model of Salmonella infection. Thus, prospective examinations using this novel model system should include detailed analysis of LPS-modifications in a PhoQ-dependent manner. This might reveal a positive correlation between PhoQ activation, signaling and subsequent LPS modification, which is important for Salmonella pathogenicity in various host backgrounds.
In conclusion, we demonstrated that G . mellonella represents a valuable alternative model for the investigation of Salmonella virulence determinants. The identification of LPS OAg length and PhoQ as factors important for the colonization of G . mellonella proves that there are an overlapping functions between virulence factors important for Salmonella pathogenesis in the wax moth, as well as in mammals.
Acknowledgments
We thank Evelyn Skiebe and Gottfried Wilharm for their help in setting up the G . mellonella infection model in our laboratory. We also thank Michael Hensel for providing different Salmonella mutant strains and plasmids p3313 and p3390. We want to thank Emily Hart for critical reading of the manuscript.
Funding Statement
This work was supported in part by DFG (www.dfg.de) grant GE2533/1-1 and an intramural research grant of the RKI (www.rki.de) to RGG. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Haraga A, Ohlson MB, Miller SI (2008) Salmonellae interplay with host cells. Nat Rev Microbiol 6: 53-66. doi:10.1038/nrmicro1788. PubMed: 18026123. [DOI] [PubMed] [Google Scholar]
- 2. Gerlach RG, Hensel M (2007) Salmonella pathogenicity islands in host specificity, host pathogen-interactions and antibiotics resistance of Salmonella enterica . Berl Münch Tierärztl Wochenschr 120: 317-327. PubMed: 17715824. [PubMed] [Google Scholar]
- 3. Barthel M, Hapfelmeier S, Quintanilla-Martínez L, Kremer M, Rohde M et al. (2003) Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71: 2839-2858. doi:10.1128/IAI.71.5.2839-2858.2003. PubMed: 12704158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Libby SJ, Brehm MA, Greiner DL, Shultz LD, McClelland M et al. (2010) Humanized nonobese diabetic-scid IL2rγ null mice are susceptible to lethal Salmonella Typhi infection. Proc Natl Acad Sci U S A 107: 15589-15594. doi:10.1073/pnas.1005566107. PubMed: 20713716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Song J, Willinger T, Rongvaux A, Eynon EE, Stevens S et al. (2010) A mouse model for the human pathogen Salmonella Typhi. Cell Host Microbe 8: 369-376. doi:10.1016/j.chom.2010.09.003. PubMed: 20951970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cotter G, Doyle S, Kavanagh K (2000) Development of an insect model for the in vivo pathogenicity testing of yeasts. FEMS Immunol Med Microbiol 27: 163-169. doi:10.1111/j.1574-695X.2000.tb01427.x. PubMed: 10640612. [DOI] [PubMed] [Google Scholar]
- 7. Mellies JL, Lawrence-Pine ER (2010) Interkingdom signaling between pathogenic bacteria and Caenorhabditis elegans . Trends Microbiol 18: 448-454. doi:10.1016/j.tim.2010.07.002. PubMed: 20667738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Waterfield NR, Sanchez-Contreras M, Eleftherianos I, Dowling A, Yang G et al. (2008) Rapid Virulence Annotation (RVA): identification of virulence factors using a bacterial genome library and multiple invertebrate hosts. Proc Natl Acad Sci U S A 105: 15967-15972. doi:10.1073/pnas.0711114105. PubMed: 18838673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hendrickson EL, Plotnikova J, Mahajan-Miklos S, Rahme LG, Ausubel FM (2001) Differential roles of the Pseudomonas aeruginosa PA14 rpoN gene in pathogenicity in plants, nematodes, insects, and mice. J Bacteriol 183: 7126-7134. doi:10.1128/JB.183.24.7126-7134.2001. PubMed: 11717271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tenor JL, McCormick BA, Ausubel FM, Aballay A (2004) Caenorhabditis elegans-based screen identifies Salmonella virulence factors required for conserved host-pathogen interactions. Curr Biol 14: 1018-1024. doi:10.1016/j.cub.2004.05.050. PubMed: 15182677. [DOI] [PubMed] [Google Scholar]
- 11. Fierer J, Guiney DG (2001) Diverse virulence traits underlying different clinical outcomes of Salmonella infection. J Clin Invest 107: 775-780. doi:10.1172/JCI12561. PubMed: 11285291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Murray GL, Attridge SR, Morona R (2003) Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol Microbiol 47: 1395-1406. doi:10.1046/j.1365-2958.2003.03383.x. PubMed: 12603743. [DOI] [PubMed] [Google Scholar]
- 13. Aballay A, Drenkard E, Hilbun LR, Ausubel FM (2003) Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway. Curr Biol 13: 47-52. doi:10.1016/S0960-9822(02)01424-0. PubMed: 12526744. [DOI] [PubMed] [Google Scholar]
- 14. Browning DF, Wells TJ, França FL, Morris FC, Sevastsyanovich YR et al. (2013) Laboratory adapted Escherichia coli K-12 becomes a pathogen of Caenorhabditis elegans upon restoration of O antigen biosynthesis. Mol Microbiol 87: 939-950. doi:10.1111/mmi.12144. PubMed: 23350972. [DOI] [PubMed] [Google Scholar]
- 15. Champion OL, Cooper IA, James SL, Ford D, Karlyshev A et al. (2009) Galleria mellonella as an alternative infection model for Yersinia pseudotuberculosis . Microbiology 155: 1516-1522. doi:10.1099/mic.0.026823-0. PubMed: 19383703. [DOI] [PubMed] [Google Scholar]
- 16. Bergin D, Reeves EP, Renwick J, Wientjes FB, Kavanagh K (2005) Superoxide production in Galleria mellonella hemocytes: identification of proteins homologous to the NADPH oxidase complex of human neutrophils. Infect Immun 73: 4161-4170. doi:10.1128/IAI.73.7.4161-4170.2005. PubMed: 15972506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brennan M, Thomas DY, Whiteway M, Kavanagh K (2002) Correlation between virulence of Candida albicans mutants in mice and Galleria mellonella larvae. FEMS Immunol Med Microbiol 34: 153-157. doi:10.1111/j.1574-695X.2002.tb00617.x. PubMed: 12381467. [DOI] [PubMed] [Google Scholar]
- 18. Slater JL, Gregson L, Denning DW, Warn PA (2011) Pathogenicity of Aspergillus fumigatus mutants assessed in Galleria mellonella matches that in mice. Med Mycol 49 Suppl 1: S107-S113. doi:10.3109/13693786.2010.494255. PubMed: 20950221. [DOI] [PubMed] [Google Scholar]
- 19. Mukherjee K, Altincicek B, Hain T, Domann E, Vilcinskas A et al. (2010) Galleria mellonella as a model system for studying Listeria pathogenesis. Appl Environ Microbiol 76: 310-317. doi:10.1128/AEM.01301-09. PubMed: 19897755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aperis G, Fuchs BB, Anderson CA, Warner JE, Calderwood SB et al. (2007) Galleria mellonella as a model host to study infection by the Francisella tularensis live vaccine strain. Microbes Infect 9: 729-734. doi:10.1016/j.micinf.2007.02.016. PubMed: 17400503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Seed KD, Dennis JJ (2008) Development of Galleria mellonella as an alternative infection model for the Burkholderia cepacia complex. Infect Immun 76: 1267-1275. doi:10.1128/IAI.01249-07. PubMed: 18195031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jander G, Rahme LG, Ausubel FM (2000) Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J Bacteriol 182: 3843-3845. doi:10.1128/JB.182.13.3843-3845.2000. PubMed: 10851003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miyata S, Casey M, Frank DW, Ausubel FM, Drenkard E (2003) Use of the Galleria mellonella caterpillar as a model host to study the role of the type III secretion system in Pseudomonas aeruginosa pathogenesis. Infect Immun 71: 2404-2413. doi:10.1128/IAI.71.5.2404-2413.2003. PubMed: 12704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mylonakis E, Moreno R, El Khoury JB, Idnurm A, Heitman J et al. (2005) Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Immun 73: 3842-3850. doi:10.1128/IAI.73.7.3842-3850.2005. PubMed: 15972469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lebreton F, Le Bras F, Reffuveille F, Ladjouzi R, Giard JC et al. (2011) Galleria mellonella as a model for studying Enterococcus faecium host persistence. J Mol Microbiol Biotechnol 21: 191-196. doi:10.1159/000332737. PubMed: 22286046. [DOI] [PubMed] [Google Scholar]
- 26. Harding CR, Schroeder GN, Reynolds S, Kosta A, Collins JW et al. (2012) Legionella pneumophila Pathogenesis in the Galleria mellonella Infection Model. Infect Immun 80: 2780-2790. doi:10.1128/IAI.00510-12. PubMed: 22645286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ott L, McKenzie A, Baltazar MT, Britting S, Bischof A et al. (2012) Evaluation of invertebrate infection models for pathogenic corynebacteria. FEMS Immunol Med Microbiol 65: 413-421. doi:10.1111/j.1574-695X.2012.00963.x. PubMed: 22443092. [DOI] [PubMed] [Google Scholar]
- 28. Kurstak E, Goring I, Vega C (1969) Cellular defense in an Arthropod in response to infection with a Salmonella typhimurium strain. Antonie Van Leeuwenhoek 35: 45-51. doi:10.1007/BF02219115. PubMed: 4891326. [DOI] [PubMed] [Google Scholar]
- 29. Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS (2006) Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24: 79-88. doi:10.1038/nbt1172. PubMed: 16369541. [DOI] [PubMed] [Google Scholar]
- 30. Wille T, Blank K, Schmidt C, Vogt V, Gerlach RG (2012) Gaussia princeps Luciferase as a Reporter for Transcriptional Activity, Protein Secretion, and Protein-Protein Interactions in Salmonella enterica Serovar Typhimurium. Appl Environ Microbiol 78: 250-257. doi:10.1128/AEM.06670-11. PubMed: 22020521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lane MC, Alteri CJ, Smith SN, Mobley HL (2007) Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc Natl Acad Sci U S A 104: 16669-16674. doi:10.1073/pnas.0607898104. PubMed: 17925449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL et al. (2008) Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods 5: 545-551. doi:10.1038/nmeth.1209. PubMed: 18454154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Valdivia RH, Falkow S (1996) Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol Microbiol 22: 367-378. doi:10.1046/j.1365-2958.1996.00120.x. PubMed: 8930920. [DOI] [PubMed] [Google Scholar]
- 34. Bumann D (2002) Examination of Salmonella gene expression in an infected mammalian host using the green fluorescent protein and two-colour flow cytometry. Mol Microbiol 43: 1269-1283. doi:10.1046/j.1365-2958.2002.02821.x. PubMed: 11918812. [DOI] [PubMed] [Google Scholar]
- 35. Ibarra JA, Knodler LA, Sturdevant DE, Virtaneva K, Carmody AB et al. (2010) Induction of Salmonella pathogenicity island 1 under different growth conditions can affect Salmonella-host cell interactions in vitro . Microbiology 156: 1120-1133. doi:10.1099/mic.0.032896-0. PubMed: 20035008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Galán JE, Curtiss R 3rd (1989) Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci U S A 86: 6383-6387. doi:10.1073/pnas.86.16.6383. PubMed: 2548211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morgan E, Campbell JD, Rowe SC, Bispham J, Stevens MP et al. (2004) Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol Microbiol 54: 994-1010. doi:10.1111/j.1365-2958.2004.04323.x. PubMed: 15522082. [DOI] [PubMed] [Google Scholar]
- 38. Ochman H, Soncini FC, Solomon F, Groisman EA (1996) Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci U S A 93: 7800-7804. doi:10.1073/pnas.93.15.7800. PubMed: 8755556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miller SI, Mekalanos JJ (1990) Constitutive expression of the phoP regulon attenuates Salmonella virulence and survival within macrophages. J Bacteriol 172: 2485-2490. PubMed: 2185222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Guo L, Lim KB, Gunn JS, Bainbridge B, Darveau RP et al. (1997) Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ . Science 276: 250-253. doi:10.1126/science.276.5310.250. PubMed: 9092473. [DOI] [PubMed] [Google Scholar]
- 41. Duerr CU, Zenk SF, Chassin C, Pott J, Gütle D et al. (2009) O-antigen delays lipopolysaccharide recognition and impairs antibacterial host defense in murine intestinal epithelial cells. PLOS Pathog 5: e1000567 PubMed: 19730692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hölzer SU, Schlumberger MC, Jäckel D, Hensel M (2009) Effect of the O-antigen length of lipopolysaccharide on the functions of Type III secretion systems in Salmonella enterica . Infect Immun 77: 5458-5470. doi:10.1128/IAI.00871-09. PubMed: 19797066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Daniels C, Morona R (1999) Analysis of Shigella flexneri Wzz (Rol) function by mutagenesis and cross-linking: wzz is able to oligomerize. Mol Microbiol 34: 181-194. doi:10.1046/j.1365-2958.1999.01591.x. PubMed: 10540296. [DOI] [PubMed] [Google Scholar]
- 44. Beuzón CR, Holden DW (2001) Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect 3: 1345-1352. doi:10.1016/S1286-4579(01)01496-4. PubMed: 11755424. [DOI] [PubMed] [Google Scholar]
- 45. Segura I, Casadesús J, Ramos-Morales F (2004) Use of mixed infections to study cell invasion and intracellular proliferation of Salmonella enterica in eukaryotic cell cultures. J Microbiol Methods 56: 83-91. doi:10.1016/j.mimet.2003.09.004. PubMed: 14706753. [DOI] [PubMed] [Google Scholar]
- 46. Knodler LA, Bestor A, Ma C, Hansen-Wester I, Hensel M et al. (2005) Cloning vectors and fluorescent proteins can significantly inhibit Salmonella enterica virulence in both epithelial cells and macrophages: implications for bacterial pathogenesis studies. Infect Immun 73: 7027-7031. doi:10.1128/IAI.73.10.7027-7031.2005. PubMed: 16177386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Iwashkiw JA, Seper A, Weber BS, Scott NE, Vinogradov E et al. (2012) Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PLOS Pathog 8: e1002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Olsen RJ, Watkins ME, Cantu CC, Beres SB, Musser JM (2011) Virulence of serotype M3 Group A Streptococcus strains in wax worms (Galleria mellonella larvae). Virulence 2: 111-119. doi:10.4161/viru.2.2.14338. PubMed: 21258213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Coombes BK, Coburn BA, Potter AA, Gomis S, Mirakhur K et al. (2005) Analysis of the contribution of Salmonella pathogenicity islands 1 and 2 to enteric disease progression using a novel bovine ileal loop model and a murine model of infectious enterocolitis. Infect Immun 73: 7161-7169. doi:10.1128/IAI.73.11.7161-7169.2005. PubMed: 16239510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shea JE, Hensel M, Gleeson C, Holden DW (1996) Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium . Proc Natl Acad Sci U S A 93: 2593-2597. doi:10.1073/pnas.93.6.2593. PubMed: 8637919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Groisman EA (2001) The pleiotropic two-component regulatory system PhoP-PhoQ. J Bacteriol 183: 1835-1842. doi:10.1128/JB.183.6.1835-1842.2001. PubMed: 11222580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Oyston PC, Dorrell N, Williams K, Li SR, Green M et al. (2000) The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis . Infect Immun 68: 3419-3425. doi:10.1128/IAI.68.6.3419-3425.2000. PubMed: 10816493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Moss JE, Fisher PE, Vick B, Groisman EA, Zychlinsky A (2000) The regulatory protein PhoP controls susceptibility to the host inflammatory response in Shigella flexneri . Cell Microbiol 2: 443-452. doi:10.1046/j.1462-5822.2000.00065.x. PubMed: 11207599. [DOI] [PubMed] [Google Scholar]
- 54. Flego D, Marits R, Eriksson AR, Kõiv V, Karlsson MB et al. (2000) A two-component regulatory system, pehR-pehS, controls endopolygalacturonase production and virulence in the plant pathogen Erwinia carotovora subsp. carotovora . Mol Plant Microbe Interact 13: 447-455. doi:10.1094/MPMI.2000.13.4.447. PubMed: 10755308. [DOI] [PubMed] [Google Scholar]
- 55. Ernst RK, Guina T, Miller SI (2001) Salmonella typhimurium outer membrane remodeling: role in resistance to host innate immunity. Microbes Infect 3: 1327-1334. doi:10.1016/S1286-4579(01)01494-0. PubMed: 11755422. [DOI] [PubMed] [Google Scholar]
- 56. Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U et al. (2005) Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122: 461-472. doi:10.1016/j.cell.2005.05.030. PubMed: 16096064. [DOI] [PubMed] [Google Scholar]
- 57. Ratcliffe NA (1985) Invertebrate immunity – a primer for the non-specialist. Immunol Lett 10: 253-270. doi:10.1016/0165-2478(85)90100-2. PubMed: 3930392. [DOI] [PubMed] [Google Scholar]
- 58. Okamura N, Spitznagel JK (1982) Outer membrane mutants of Salmonella typhimurium LT2 have lipopolysaccharide-dependent resistance to the bactericidal activity of anaerobic human neutrophils. Infect Immun 36: 1086-1095. PubMed: 7047389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Augustin DK, Song Y, Baek MS, Sawa Y, Singh G et al. (2007) Presence or absence of lipopolysaccharide O antigens affects type III secretion by Pseudomonas aeruginosa . J Bacteriol 189: 2203-2209. doi:10.1128/JB.01839-06. PubMed: 17209027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gerlach RG, Cláudio N, Rohde M, Jäckel D, Wagner C et al. (2008) Cooperation of Salmonella pathogenicity islands 1 and 4 is required to breach epithelial barriers. Cell Microbiol 10: 2364-2376. doi:10.1111/j.1462-5822.2008.01218.x. PubMed: 18671822. [DOI] [PubMed] [Google Scholar]
- 61. Zenk SF, Jantsch J, Hensel M (2009) Role of Salmonella enterica lipopolysaccharide in activation of dendritic cell functions and bacterial containment. J Immunol 183: 2697-2707. doi:10.4049/jimmunol.0900937. PubMed: 19625639. [DOI] [PubMed] [Google Scholar]
- 62. Blank K, Hensel M, Gerlach RG (2011) Rapid and Highly Efficient Method for Scarless Mutagenesis within the Salmonella enterica Chromosome. PLOS ONE 6: e15763. doi:10.1371/journal.pone.0015763. PubMed: 21264289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang RF, Kushner SR (1991) Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli . Gene 100: 195-199. doi:10.1016/0378-1119(91)90366-J. PubMed: 2055470. [PubMed] [Google Scholar]