Abstract
The lipin proteins are evolutionarily conserved proteins with roles in lipid metabolism and disease. There are three lipin protein family members in mammals and one or two orthologs in plants, invertebrates, and single-celled eukaryotes. Studies in yeast and mouse led to the identification of two distinct molecular functions of lipin proteins. Lipin proteins have phosphatidate phosphatase activity and catalyze the formation of diacylglycerol in the glycerol-3-phosphate pathway, implicating them in the regulation of triglyceride and phospholipid biosynthesis. Mammalian lipin proteins also possess transcriptional coactivator activity and have been implicated in the regulation of metabolic gene expression. Here we review key findings in the field that demonstrate roles for lipin family members in metabolic homeostasis and in rare human diseases, and we examine evidence implicating genetic variations in lipin genes in common metabolic dysregulation such as obesity, hyperinsulinemia, hypertension, and type 2 diabetes.
Keywords: triglyceride, obesity, insulin resistance, phosphatidate phosphatase, transcriptional coactivator
THE LIPIN PROTEIN FAMILY
Several important proteins have been identified through the characterization of naturally occurring mutations in humans and in animal models. The family of lipin proteins was identified by elucidation of the gene defect underlying a spontaneous mutation that arose in the BALB/cByJ mouse strain. This mutant mouse had a remarkable lack of adipose tissue as well as other defects in lipid homeostasis, and the mutation was traced to a novel gene that became the founding member of the lipin family, Lpin1 (47). Two additional mammalian genes, Lpin2 and Lpin3, were identified based on sequence similarity (47). This has raised the question of what specific roles each of the three lipin family members play in vivo. Each is expressed with a distinct but overlapping tissue expression pattern. Lpin1 is expressed at highest levels in adipose tissue, skeletal muscle, and testis, and at lower levels in numerous other tissues, including kidney, lung, brain, heart, and liver (47). Lpin2 is prominently expressed in liver and brain, and Lpin3 is expressed in the gastrointestinal tract and liver (14). Evidence available thus far suggests that the three mammalian lipin proteins perform similar molecular functions in different tissues.
Thus far, most studies have focused on the function of the founding member of the lipin family, lipin-1, which therefore constitutes the major subject of this review. In addition, the work of Carman and colleagues on the single lipin homolog present in yeast has contributed important information about the cellular and molecular functions of lipin proteins (23, 24). However, here we largely focus on the mammalian lipin genes and proteins and refer readers elsewhere for excellent reviews on the yeast protein (8, 9).
LIPIN PROTEIN MOLECULAR FUNCTION
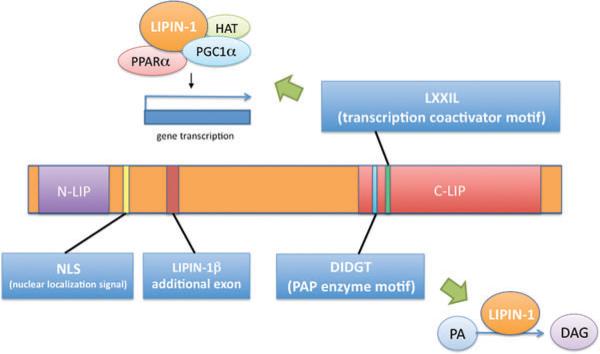
Lipin proteins in all species share two highly conserved regions, termed the N-terminal lipin (N-LIP) and C-terminal lipin (C-LIP) domains, based on their locations at opposite ends of the protein (Figure 1) (47). Lipin proteins in most species also contain at least one nuclear localization sequence. The C-LIP domain contains two known functional motifs: (a) a DIDGT motif, which is present in all species and constitutes the catalytic site for phosphatidate phosphatase (PAP) enzyme activity (24), and (b) an LXXIL motif, which is present in mammalian lipin proteins and is required for activity as a transcriptional coactivator (21). The Mg2+-dependent PAP enzymes catalyze the penultimate step in triglyceride synthesis, converting phosphatidic acid (PA) to diacylglycerol (DAG) (reviewed in 8). The resulting DAG also serves as the immediate precursor of phosphatidylcholine and phosphatidylethanolamine. PAP proteins normally reside in the cytosol and translocate to the endoplasmic reticulum membrane to bind PA and catalyze the enzymatic reaction (4). Although all three mammalian lipin proteins have the DIDGT motif and demonstrable PAP activity, lipin-1 exhibits higher PAP specific activity than lipin-2 and lipin-3 as assessed by an in vitro assay (14). The DIDGT motif is absolutely required for PAP enzyme activity, but mutation of this motif does not affect transcriptional coactivator function (21, 24).
Figure 1.
Mammalian lipin protein functional domains. Key lipin protein motifs are labeled at their relative position along the length of the lipin protein. N-terminal lipin (N-LIP) and C-terminal lipin (C-LIP) domain regions are highly conserved across evolution from yeast to mammals. The lipin-1β-specific region is derived from alternative splicing of the lipin-1 gene. DIDGT is the motif required for PAP enzyme activity and conversion of phosphatidate (PA) to diacylglycerol (DAG). LXXIL is the transcriptional coactivator motif required for interaction of lipin-1 with other proteins and subsequent effects on gene expression. HAT, histone acetyltransferase; NLS, nuclear localization signal; PGC1α, PPARγ coactivator 1α; PPARα, peroxisome proliferator activated receptor α.
A role for lipin protein transcriptional coactivator activity has been demonstrated most convincingly thus far for mouse lipin-1. Lipin-1 coactivator activity was first discovered by Finck et al. (21) in studies of the hepatic response to fasting in mice deficient for the peroxisome proliferator activated receptor γ coactivator-1α (PGC-1α) transcriptional coactivator. Lipin-1 is required for the induction of peroxisome proliferator-activated receptor (PPAR) α expression during fasting: It associates with the PPARα promoter nuclear receptor response element and coactivates the PPARα promoter with PGC-1α. This coactivation is enhanced upon fasting and probably involves an indirect association between lipin-1 and the PPARα gene promoter since lipin-1 has no DNA binding domain. Lipin-1 also coactivates PPARα and PGC-1α to upregulate fatty acid uptake and oxidation, TCA cycle, and mitochondrial metabolism genes (21). The PAP DIDGT motif is not required for this coactivation activity, but interestingly, mutation of the LXXIL coactivation motif abolishes both coactivator and PAP activity (21). There is also some evidence that lipin-1 coactivates mitochondrial biogenesis genes in response to endurance exercise (26). Lipin-1 coactivator activity has also been linked to the suppression of very-low-density lipoprotein secretion in mouse liver, although no effects on the levels of related gene expression were detected (11).
The yeast lipin homolog, known as Pah1p, has been shown to have a role in phospholipid biosynthetic gene expression during expansion of the nuclear membrane (52). However, unlike mammalian lipin-1, this transcriptional activity appears not to be separable from the PAP activity of Pah1p (23), indicating that the observed effects on gene expression in yeast and mouse may occur by different mechanisms. Little is known about whether other lipin proteins have physiological roles in gene regulation. Lipin-2 exhibits similar transcriptional coactivator activity as lipin-1 in a cell-based assay (16), but a role for this activity in vivo has not been demonstrated.
LIPIN-1 DEFICIENCY IN THE MOUSE: IMPAIRED LIPID METABOLISM IN LIVER, ADIPOSE TISSUE, AND PERIPHERAL NERVE
Spontaneous mutations that cause impaired lipin-1 function in the mouse have been instrumental in both the identification of the Lpin1 gene and analysis of the physiological role of lipin-1. The original mutant phenotype, named fatty liver dystrophy ( fld), was first characterized by Langner et al. (32, 33) and was later found to be due to a gene rearrangement and null mutation in Lpin1 (47). A second, independent mouse mutation that causes virtually the same phenotype as the null mutation resulted from a point mutation at a conserved amino acid in the N-LIP domain (47). Recently, a chemical mutagenesis screen identified a point mutation in the C-LIP domain that resulted in a protein with attenuated function and a modest phenotype with effects primarily in the peripheral nerve (17).
The lipin-1-deficient fld mutant mouse has been extensively studied. The name “fatty liver dystrophy” reflects the presence in neonatal mice of an enlarged, triglyceride-rich fatty liver and the development in adult mice of a dystrophic movement (i.e., neuropathy) characterized by whole-body tremor and apparent weakness in the hind limbs (32). In addition, neonatal fld mice have hypertriglyceridemia, altered hepatic lipid metabolism gene expression, and decreased lipoprotein lipase activity in adipose tissue. The fatty liver and hypertriglyceridemia spontaneously resolve around the time of weaning, although this appears to be a genetically programmed event rather than a result of weaning (32). The two most notable phenotypes in adult fld mice are the absence of functional white and brown adipose tissue throughout the body (51) and the progressively worsening neuropathy, which is caused by impaired formation and maintenance of the myelin sheath in peripheral nerves (33). Analysis of PAP activity in tissues of fld mice revealed that lipin-1 accounts for virtually all of the PAP activity in white and brown adipose tissue, the endoneurium of sciatic nerve, skeletal muscle, heart, and lung, and contributes a large proportion of the PAP activity in liver, brain, and kidney (14, 25, 44). Male mice are infertile, perhaps related to high expression levels of lipin-1 in testis, although an analysis of the PAP activity in fld testis has not been reported.
The lack of lipin-1 PAP activity appears to contribute to the pathology observed in tissues of fld mice by at least two mechanisms. First, the impaired triacylglycerol (TAG) synthesis may directly affect the normal function of tissues that store lipid for local and/or systemic use. Second, PAP deficiency may lead to an accumulation of the enzyme substrate, PA, which acts as a lipid signaling molecule and may lead to dysregulation of downstream signaling pathways (6). There is evidence that both of these mechanisms contribute to the effects of lipin-1 deficiency in tissues such as peripheral nerve and adipose tissue. In sciatic nerve, lipin-1 deficiency causes lipoatrophy of adipocytes that occur in the epineurium as well as impaired lipid synthesis in the endoneurium (56). Furthermore, PA accumulates in the endoneurium due to lack of lipin-1 PAP function (44). PA was shown in vitro to have a direct effect on Schwann cells, reducing both myelination and expression of a marker of differentiated Schwann cells. In addition, PA was shown to activate the MEK-Erk signaling pathway in cultured mouse Schwann cells, a mechanism that was shown to be important for the demyelinating action of PA (44).
In adipose tissue, the loss of lipin-1 PAP activity and resulting impairment in triglyceride synthesis clearly contribute to the failure of adipose tissue in fld mice to store lipids and the resulting lipodystrophy (14, 51). Both white and brown adipose tissue depots in fld mice exhibit an 80% reduction in mass compared to wild-type mice, leading to a 25% reduction in body weight. This defect cannot be overcome by feeding a high-fat diet, which results in virtually no weight gain in fld mice (48). At the microscopic level, fld adipose tissue appears poorly differentiated, with cells having a fibroblastic appearance and sparse, small lipid droplets. This reflects the fact that lipin-1 expression in differentiating preadipocytes is required for normal expression of adipogenic transcription factors, including PPARγ and CAAT/enhancer binding protein (C/EBP) α, which are induced prior to triglyceride synthesis (48). Thus, the lack of lipin-1 in preadipocytes has effects beyond impaired TAG accumulation and may indicate a requirement for lipin-1 in the activation of gene expression during early adipocyte differentiation (46) or aberrant cellular function resulting from the accumulation of PA in adipocytes (44).
The lack of lipin-1-mediated TAG synthesis in fld adipose tissue has important systemic metabolic consequences. Fat stores play a critical role as a sink for excess dietary fatty acids and as a fuel source during fasting and starvation. Wild-type mice utilize glucose in the fed state; during the fasted state, they switch initially to glycogen stores in liver and muscle and then to triglyceride stores in adipose tissue. The lack of fat stores in the fld mouse prevents it from making use of the normal fuel choice program during fasting. To compensate, lipin-1-deficient mice store excess glycogen during the feeding period to use as a sustained fuel source during fasting (59). To spare glucose and allow its storage as glycogen, the fld mice also induce hepatic fatty acid synthesis to supply muscle and other tissues during the feeding period. The enhanced oxidation of fatty acids as fuel in peripheral tissues is a likely mechanism for the lack of fatty liver in the adult fld mice.
Another consequence of lipin-1 deficiency in metabolic tissues is severely impaired glucose homeostasis. The fld mouse exhibits impaired glucose clearance and elevated nonfasting and glucose-stimulated insulin levels, indicating insulin resistance (51, 59). Analysis of hepatic glucose production revealed that fld mice have normal insulin sensitivity in the liver but impaired glucose uptake in peripheral tissues such as adipose tissue and skeletal muscle (59). This is consistent with studies of primary cells isolated from fld and wild-type mice. Cells from both sources were responsive to serum as indicated by formation of elongated actin stress fibers, but only wild-type cells responded to insulin as evidenced by membrane ruffling (31). Furthermore, in human primary adipocytes, lipin-1 mRNA levels exhibit a strong positive correlation with basal and insulin-stimulated glucose transport and with expression of the insulin-sensitive glucose transporter 4 (55). Despite the fact that fld mice have a nonatherogenic plasma lipid profile, they are more susceptible than wild-type mice to diet-induced atherosclerosis (51). Although the cause is unknown, it is interesting to speculate that the increased atherosclerosis is influenced by the insulin-resistant state and/or impaired adipose tissue function and adipokine production, both of which are risk factors for cardiovascular disease (38).
LIPIN-1 EXPRESSION LEVELS INFLUENCE ADIPOSITY, ENERGY METABOLISM, AND INSULIN SENSITIVITY
Lipin-1 represents a link between opposite extremes of adiposity, with lipin-1 deficiency causing lipodystrophy and enhanced expression causing obesity. This was demonstrated first in the mouse, in two transgenic (Tg) mouse models that overexpress lipin-1 in either mature adipocytes (aP2-lipin-1 Tg mice) or skeletal muscle (Mck-lipin-1 Tg mice). Increased lipin-1 expression in either adipose tissue or muscle led to obesity, in stark contrast to the lipodys-trophy observed in lipin-1 deficient mice (49). However, as described below, the mechanism underlying obesity and the resulting effects on glucose homeostasis differed depending on whether lipin-1 expression was enhanced in adipose tissue or muscle.
Transgenic mice with increased lipin-1 expression in adipose tissue have normal weight when fed a chow diet but accumulate twice as much fat mass as wild-type mice when fed a high-fat diet (49). Muscle-specific expression of the lipin-1 transgene leads to increased body weight on chow as well as on high-fat diets, and accumulation of four times the fat mass of wild-type mice after eating the high-fat diet. Although food intake is normal for the transgenics, increased feed conversion efficiency was observed in both transgenic strains, consistent with the increased weight gain. Additionally, the extreme weight gain of Mck-lipin-1 Tg mice on both chow and high-fat diets was characterized by a 15% reduction in energy expenditure, reduced body temperature, and reduced fatty acid oxidation in muscle (49). Interestingly, the energy balance parameters in Mcklipin-1 Tg mice are the opposite of those in fld mice, which exhibit increased energy expenditure, elevated body temperature, and increased fatty acid oxidation in muscle. It was demonstrated that these aspects of the fld phenotype can be largely attributed to the lack of lipin-1 specifically in muscle, since expression of the Mck-lipin-1 transgene in fld mice normalized these energy parameters but did not restore adipose tissue function (49).
The effects of enhanced lipin-1 expression levels on glucose homeostasis diverged in adipose tissue and muscle-specific lipin-1 transgenic strains. Despite their increased fat mass, aP2-lipin-1 Tg mice exhibited greater insulin sensitivity, whereas Mck-lipin-1 Tg mice had increased insulin resistance compared to wild-type mice (49). The insulin resistance in Mck-lipin-1 Tg mice is likely secondary to the profound obesity, although it is not possible to discount a direct effect due to specific alterations in muscle lipid metabolism. It is, however, counterintuitive that aP2-lipin-1 Tg mice would exhibit improved glucose homeostasis, which was associated with enhanced lipogenic gene expression in fat, and a 60% increase in triglyceride content per cell in adipose tissue (46, 49). This may reflect more efficient fatty acid trapping and incorporation into adipose tissue triglycerides, thereby preventing lipid deposition in muscle, liver, and other tissues, which could compromise insulin action. Furthermore, the induction of endogenous lipin-1 expression may be a positive adaptive response in insulin-sensitive states, as lipin-1 expression in adipose tissue is induced by insulin-sensitizing drugs such as thiazolidinediones and harmine (20, 57, 60). Levels of lipin-1 expression in human adipose tissue are also positively correlated with insulin sensitivity in humans, as described in a later section.
REGULATION OF LIPIN-1 EXPRESSION AND ACTIVITY
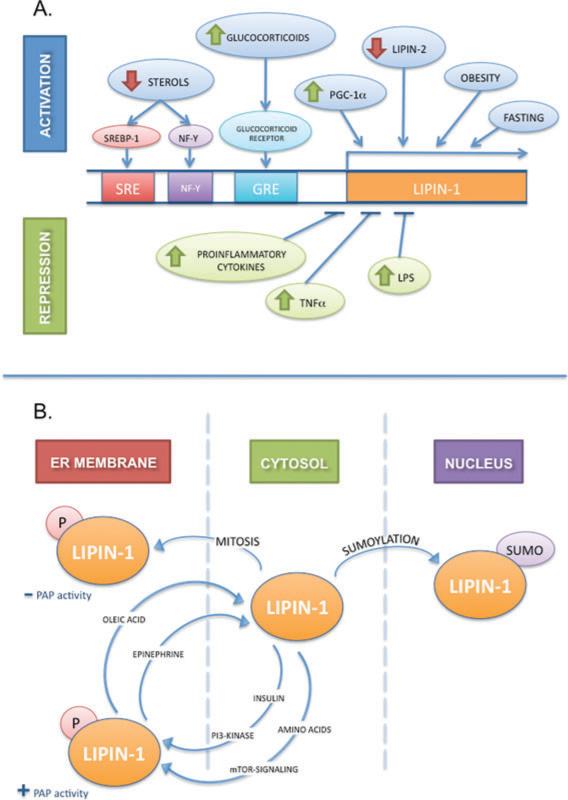
Lipin-1 action is regulated at multiple levels, including gene transcription, alternative mRNA splicing, protein phosphorylation, and subcellular protein localization. This complex array of regulatory mechanisms suggests that the levels and activity of this protein are modulated to adjust to changing physiological conditions. These are summarized in Figure 2 and described below.
Figure 2.
Regulation of lipin-1 gene expression, protein modification, and subcellular localization. (A) Regulation of lipin-1 gene transcription. Diagrammatic view of lipin-1 gene promoter region with known regulatory elements is shown. Conditions known to activate lipin-1 mRNA expression are illustrated in the upper part of the panel. Sterol depletion produces an increase in lipin-1 mRNA expression through sterol regulatory element-binding protein-1 (SREBP-1) and nuclear factor-Y (NF-Y) acting upon a sterol response element (SRE) and an NF-Y response element, respectively. Glucocorticoids activate lipin-1 expression through a glucocorticoid response element (GRE). Obesity and fasting, conditions associated with increased glucocorticoid levels, induce lipin-1 gene expression. Lipin-1 expression is also enhanced by overexpression of PGC-1α or depletion of lipin-2. Conditions known to repress lipin-1 mRNA expression are shown in the lower part of the panel. Proinflammatory cytokines, lipopolysaccharide, and TNFα downregulate lipin-1 expression. (B) Regulation of lipin-1 posttranslational modification and subcellular localization. Both insulin and amino acids induce lipin-1 phosphorylation and translocation to the endoplasmic reticulum (ER) membrane to perform its enzymatic function. This phosphorylation is dependent on the phosphoinositide (PI)3-kinase and mTOR signaling pathways. Oleic acid and epinephrine reverse this phosphorylation, sending lipin-1 back to the soluble fraction. Mitosis has also been shown to induce lipin-1 phosphorylation and translocation, but the enzyme does not become phosphatidate phosphatase (PAP) active. Finally, sumoylation of lipin-1 induces translocation of the protein from the cytosol to the nucleus. See text for reference citations.
Lipin-1 Transcriptional Regulation
Even before the molecular identification of the lipin proteins, it was known that hepatic PAP activity is increased in fasting, diabetes, and in response to glucocorticoids (5). Consistent with these early observations, lipin-1 is regulated at the transcriptional level by glucocorticoids in adipocytes and hepatocytes (40, 63). In primary rat and mouse hepatocytes, lipin-1 mRNA levels are induced by treatment with the synthetic glucocorticoid, dexamethasone, and this effect is augmented by cAMP or glucagon and diminished by insulin (40). In adipocytes, lipin-1 expression is dramatically induced during differentiation, and this was found to be due to stimulation by dexamethasone present in the differentiation cocktail (63). In vivo, conditions associated with increased glucocorticoid levels, such as obesity or fasting, also lead to increased lipin-1 mRNA in adipose tissue (63) and in liver (21). Glucocorticoid regulation is mediated by a glucocorticoid response element within the Lpin1 promoter region, which was shown to bind the glucocorticoid receptor upon addition of dexamethasone (63). Fasting-induced induction of lipin-1 in liver is dependent on the presence of PGC-1α, a protein whose expression is stimulated by dexamethasone (21). An interesting link between cellular cholesterol levels and triglyceride biosynthesis was demonstrated by the determination that sterol depletion in human hepatoblastoma cells induces lipin-1 mRNA expression (28). This induction is regulated by the sterol regulatory response element binding protein 1 (SREBP1) and nuclear factor Y (NF-Y), which bind to sequence motifs upstream of the LPIN1 gene.
Negative regulators of lipin-1 gene expression include cytokines and other members of the lipin family. It is known that inflammation and sepsis lead to the release of free fatty acids from adipose tissue. Consistent with this, inflammatory conditions appear to suppress lipin-1 gene expression, potentially contributing to the reduction in fatty acid utilization for TAG synthesis in these conditions. In mouse adipose tissue, lipopolysaccharide suppressed lipin-1 mRNA and protein levels, and this effect was partially ameliorated in mice lacking tumor necrosis factor (TNF) α, a cytokine implicated in obesity and insulin resistance (37). Indeed, lipin-1 is directly suppressed by TNF-α treatment of cultured adipocytes, and this effect is prevented by inhibition of Jak2 signaling (37, 54). There is also evidence in cultured cells that lipin-1 and lipin-2 may negatively regulate one another. In HeLa cells, siRNA knockdown of either lipin-1 or lipin-2 caused an increase in mRNA and protein levels of the other lipin (22). In 3T3-L1 cells, silencing of lipin-1 led to higher lipin-2 expression levels. However, the effects of these changes on PAP activity are not straightforward, as silencing of lipin-1 dramatically reduces PAP activity in HeLa cells, whereas silencing of lipin-2 increases PAP activity (22). The interplay between different lipin family members is an intriguing area that will need to be confirmed in vivo.
Alternative Lpin1 mRNA Splicing
Lipin-1 is expressed as two distinct isoforms, known as lipin-1α and lipin-1β, which represent different splice variants from the Lpin1 gene (27, 46). Lipin-1β contains an additional 33 amino acid residues that are not present in lipin-1α (Figure 1). Lpin1 splicing appears to exhibit tissue-specific regulation, as some tissues (brain, spleen, and preadipocytes) have a predominance of lipin-1α whereas others (liver, heart, kidney, and mature adipocytes) have a preponderance of lipin-1β (46). Little is known about the regulation of lipin-1 mRNA splicing to produce the two isoforms, but some clues have been gleaned from the study of lipin-1 expression during adipocyte differentiation in vitro. Lipin-1α is the major mRNA present in undifferentiated preadipocytes, and upon treatment with a differentiation cocktail, lipin-1β is dramatically induced and constitutes the predominant form in mature adipocytes (46). Glucocorticoids may influence splicing, as lipin-1β is induced to a greater extent than lipin-1α in response to dexamethasone (63). Notably, the lipin-1α and lipin-1β proteins have different properties, with lipin-1α exhibiting a greater frequency of nuclear localization than lipin-1β (3, 46). Furthermore, complementation of lipin-1-deficient preadipocytes with each isoform independently showed that lipin-1α expression leads to the induction of adipogenic transcription factors, whereas lipin-1β leads to increased expression of lipogenic genes (46). The lipin-1 transgenic mouse models that were described in a previous section both expressed the lipin-1β isoform (49); it would be interesting to determine if similar physiological effects are obtained with the lipin-1α isoform.
Lipin-1 Protein Modification and Subcellular Localization
Most enzymes in the glycerolipid biosynthetic pathway reside within the membrane of the endoplasmic reticulum (12). In contrast, it was demonstrated some time ago that PAP activity resides in the cytosol until it is recruited to the endoplasmic reticulum (ER) membrane in a transient manner in response to fatty acids or other stimuli (reviewed in 50). PAP phosphorylation was implicated in the subcellular localization of PAP activity, since the phosphatase inhibitor okadaic acid can displace it from the ER membrane. Lipin-1 phosphorylation was first demonstrated by Lawrence and colleagues, who showed that insulin or amino acid treatment of adipocytes caused the appearance of multiple lipin-1 electrophoretic variants that could be collapsed into a common form by phosphatase treatment (27). Lipin-1 phosphorylation by insulin is dependent upon phosphatidylinositol-3-kinase activity and the mammalian target of rapamycin (mTOR) signaling pathways. At least 15 serine and threonine phosphorylation sites have been identified along the length of the lipin-1 protein, including serine 106, which is a key insulin-stimulated phosphorylation site (25). Lipin-1 dephosphorylation occurs in response to oleic acid and epinephrine (25). Importantly, lipin-1 phosphorylation/dephosphorylation by metabolic regulators appears not to affect the intrinsic PAP activity, but rather influences subcellular localization of the protein. Thus, phosphorylation by insulin causes translocation of lipin-1 protein and PAP activity from the soluble to the microsomal fraction, whereas dephosphorylation by oleic acid or epinephrine has the opposite effect (25).
Studies in HeLa cells showed that both lipin-1 and lipin-2 are phosphorylated during mitosis, at cyclin-dependent kinase (Cdk1) phosphorylation consensus sites (22). In contrast to the changes in phosphorylation status by metabolic regulators described above, mitotic phosphorylation was found to decrease PAP activity, whereas dephosphorylation was found to increase it. In addition, phosphorylation changed the affinity of the lipin proteins for membranes in HeLa cells: Phosphorylated lipin-1 and phosphorylated lipin-2 were found to be enriched over dephosphorylated forms in the soluble fraction of HeLa cells (22). The phosphatase Dullard has been shown to dephosphorylate lipin-1 (30), but the physiological role of Dullard in regulation of lipin-1 activity is not clear.
In addition to the regulation of cytosolic/ER membrane localization of lipin-1 by phosphorylation, other protein modifications may modulate cytosolic/nuclear localization. In neuronal cells, lipin-1α nuclear localization is regulated by sumoylation at two lysine residues occurring within sumoylation consensus sites (35). Intact sumoylation sites are required for transcriptional coactivator activity of lipin-1α in cultured neuronal cells, suggesting that sumoylation provides a mechanism to regulate nuclear localization and transcriptional activity of lipin-1α in the brain. The sumoylation of lipin-1α occurs at sites that are distinct from the nuclear localization signal, suggesting that changes in protein conformation may function in combination with the nuclear localization site to optimize nuclear translocation.
ALTERED LIPIN-1 GENE EXPRESSION AND METABOLISM IN HUMANS
Studies of lipin-1 regulation in humans have uncovered significant associations between variations in lipin-1 mRNA expression levels and various metabolic traits. Consistent with studies in mouse models described above, multiple studies found that human adipose tissue lipin-1 expression is positively correlated with insulin sensitivity, as measured by glucose tolerance, euglycemic-hyperinsulinemic clamp, HOMA-IR, and intramyocellular lipids (13, 15, 42, 53, 55, 60). Furthermore, upregulation of lipin-1 by treatment with thiazolidinedione antidiabetic drugs was correlated with improved insulin sensitivity (60). In a unique study, lipin-1β expression was increased in liver and adipose tissue after weight-reduction surgery, and insulin sensitivity was improved (13). Adipose tissue lipin-1β expression levels were reduced in patients with polycystic ovary syndrome, which is characterized by insulin resistance (43).
The relationship between lipin-1 levels and adiposity is not as clear-cut in humans as was observed in mouse models. As described earlier, in the mouse, extreme differences in lipin-1 levels in adipose tissue positively correlate with adiposity, with lipin-1-deficient mice having virtually no adipose tissue mass, and lipin-1 transgenics having enhanced adipose tissue accumulation. In man, a different relationship between lipin-1 expression and adiposity [typically measured as body mass index (BMI)] has been observed. Although one study reported no correlation (53), others have reported a negative correlation between adipose tissue lipin-1 expression levels and BMI (10, 42, 43), percent body fat, fat cell size, fasting free fatty acid levels, and other measures of adipose tissue mass (15), and metabolic syndrome (55). One explanation for the different relationships between lipin-1 and adiposity in mouse and man is that forced lipin-1 expression via transgene leads to continuous PAP activity and increased TAG synthesis in the mouse, whereas the expression of endogenous human lipin-1 is regulated such that when a certain degree of adiposity is reached, expression levels are attenuated. Interestingly, in addition to correlations with metabolic traits, lipin-1 mRNA expression levels are also highly correlated with expression levels of genes involved in several metabolic processes, including PPARα and other genes involved in fatty acid oxidation, adiponectin, glucose transporter 4, and genes involved in lipid biosynthesis (10, 15, 55).
The relationship between lipin-1 levels and lipodystrophy that occurs secondary to human immunodeficiency virus (HIV) infection and antiretroviral drug treatment has also been investigated. Lipin-1 expression levels in adipose tissue were compared between HIV patients who have developed lipodystrophy and those who have not. Consistent with a role for lipin-1 in maintaining adipose tissue, HIV patients with lipodystrophy exhibited decreased lipin-1 mRNA levels in adipose tissue compared to patients without lipodystrophy (34). In addition, lipin-1 levels in adipose tissue were positively correlated with percentage limb fat and negatively correlated with percentage truncal fat. Finally, lipin-1 expression was negatively correlated with inflammatory cytokine expression in abdominal subcutaneous fat in two independent studies (34, 42). However, one of these groups found no correlation between lipin-1 expression and HIV-associated lipodystrophy syndrome (42). Discrepancies between the two studies may be due to many factors, including differences in the stage of lipodystrophy progression among subjects.
LIPIN-2 AND LIPIN-3 EXPRESSION AND PHYSIOLOGY
The two remaining lipin family members, lipin-2 and lipin-3, are much less well characterized than lipin-1. Lipin-2 exhibits PAP activity at about one-quarter the level of lipin-1-specific activity (14). It also exhibits transcriptional coactivator activity in combination with PPARα and PGC-1α that is comparable to lipin-1 coactivator activity in an in vitro assay (16). Lipin-2 is expressed at high levels in the liver, kidney, and brain, and is the predominant lipin family member expressed in red blood cells, bone marrow, thymus, and spleen, although at levels that are much lower than those in liver (16). Lipin-2 expression, like lipin-1, is elevated in the liver during fasting (16, 21). However, unlike lipin-1, lipin-2 does not appear to be regulated by PGC-1α (21). Very little is known about the regulation of lipin-2 expression or activity. During differentiation of cultured adipocytes, lipin-2 exhibits a pattern opposite to that of lipin-1, with expression in preadipocytes that is suppressed by differentiation to mature adipocytes (22). Similar to lipin-1, lipin-2 is phosphorylated during mitosis on Cdk1 phosphorylation consensus sites, but the physiological significance is unknown (22).
At present, lipin-3 is the least well-characterized lipin family member. In vitro, lipin-3 exhibits PAP activity at about 20% the level of lipin-1 specific activity (14). In vivo, expression is detected in the gastrointestinal tract and liver (14, 40). Hepatic lipin-3 expression levels are generally much lower than those for lipin-1 or lipin-2, but lipin-3 expression is substantially upregulated in the liver of lipin-1-deficient mice and may partially compensate for PAP activity in this tissue (14).
GENETIC MUTATIONS AND VARIANTS IN LIPIN GENES AND HUMAN DISEASE
The three human lipin proteins exhibit 44% to 48% amino acid identity with the mouse proteins and are encoded by the genes LPIN1, LPIN2, and LPIN3. Studies of LPIN1 and LPIN2 have identified both common and rare genetic variants that are associated with human disease (Table 1).
Table 1.
Lipin gene mutations and polymorphisms associated with human disease
|
LPIN1 mutations | |||
|---|---|---|---|
| Phenotype | Genetic change/locus | Details about change/locus | Reference |
| Recurrent acute myoglobinuria | Homozygous 643 G>T (E215X) | Premature stop codon | 62 |
| Homozygous 1162 C>T (R388X) | Premature stop codon | ||
| 2398 C>T (R800X)/genomic deletion of exons 18 and 19 | Premature stop codon/deletion | ||
| Homozygous 297+2 T>C | Activates cryptic splice site in exon 1; last 106 bp of exon 1 are deleted | ||
| 1259+2 T>C/genomic deletion of exons 18 and 19 (heterozygous) | Skips exon 8/deletion | ||
| Statin-induced myopathy | 2306 A>G (Glu769Gly) | Mutation of a highly conserved amino acid; impaired PAP activity in yeast | 62 |
| LPIN1 polymorphisms | |||
| Body mass index | |||
| rs13412852 | Noncoding SNP | 18 | |
| (Association found in lean men only) | rs893346 A/G, rs2577262 A/G, rs2716610 C/T | Noncoding SNPs | 53 |
| (Association found in obese men only) | rs11693809 C/T, rs10192566 C/G, rs2278513 C/T, rs2577262 A/G | Noncoding SNPs | 53 |
| Haplogroup encompassing SNPs rs33997857, rs6744682, rs6708316 | One exonic SNP leads to a codon change (V494M) and two are intronic; one risk and two protective haplotypes identified | 58 | |
| Insulin levels | |||
| Higher serum insulin | rs11693809 T allele (SNP) | Noncoding SNP | 53 |
| Higher fasting insulin (generation specific) | rs2716609 C allele (SNP) | Noncoding SNP | 36 |
| Waist circumference | |||
| (Generation specific) | rs2716609 C allele (SNP) | Noncoding SNP | 36 |
| Haplogroup encompassing SNPs rs33997857, rs6744682, rs6708316 | One exonic SNP leads to a codon change (V494M) and two are intronic; one risk and two protective haplotypes identified | 58 | |
| Blood pressure | |||
| Lower mean systolic blood pressure (in men only) | rs10495584 G allele (SNP) | Not predicted to be pathogenic, but in high LD with functional SNP rs11524, in which the major allele forms an exonic splicing silencer sequence | 45 |
| Hypertension | D2S0949i (microsatellite) | LPIN1 is the nearest gene to this significantly associated microsatellite | 61 |
| Lower blood pressure | Haplogroup encompassing SNPs rs33997857, rs6744682, rs6708316 | One exonic SNP leads to a codon change (V494M) and two are intronic; one risk and two protective haplotypes identified | 58 |
| Other metabolic phenotypes | |||
| Higher resting metabolic rate (generation specific) | rs2577262 G allele (SNP) | Noncoding SNP | 36 |
| rs2577256 C allele (SNP) | Noncoding SNP | ||
| Metabolic syndrome factor score and A1C levels | Haplogroup encompassing SNPs rs33997857, rs6744682, rs6708316 | One exonic SNP leads to a codon change (V494M) and two are intronic; one risk and two protective haplotypes identified | 58 |
| Type 2 diabetes | Haplotype ATCCG | Haplotype is rare among the Chinese population studied | 10 |
| Improved response to rosiglitazone treatment in type 2 diabetic patients | rs10192566 G allele (SNP) | Noncoding SNP | 29 |
| Elevated hepatic lipase activity (in women only) | rs10192566 G allele (SNP) | Noncoding SNP | 53 |
| LPIN2 mutations | |||
| Majeed syndrome | Homozygous 2201C>T (S734L) | Mutates an amino acid conserved not only across lipins in other species but also in all | 19 |
| HAD family proteins | |||
| Homozygous 540–541delAT | Frameshift mutation; premature stop codon | 19 | |
| Homozygous 2327+1G>C | Mutation of splice site causes an R776S change and a premature stop codon | 1 | |
| Psoriasis | 991G>T (A331S) | All coding changes | 41 |
| 1043C>T (P348L) | |||
| 1159A>G (K387E) | |||
| 1510C>T (L504F) | |||
| 1801G>A (E601K) | |||
| LPIN2 polymorphisms | |||
| Type 2 diabetes and fat distribution | rs3745012 C allele (SNP) | Located within the LPIN2 3′ UTR | 2 |
Abbreviations: HAD, haloacid dehalogenase; LD, linkage disequilibrium; SNP, single nucleotide polymorphism; UTR, untranslated region.
Lipin Gene Mutations and Rare Disease
A striking finding is that mutations in LPIN1 and LPIN2 cause rare recessive Mendelian diseases. Inactivating mutations in LPIN1 cause recurrent episodes of myoglobinuria in childhood (62). The first mutation identified was a nonsense mutation with a recessive inheritance pattern in a single family with three affected children. An accumulation of lysophospholipids and a slight elevation of phosphatidate were present in the muscle of one of the affected children, possibly contributing to the myoglobinuria. Five additional nonsense or frame-shift LPIN1 mutations were identified upon sequencing 22 other unrelated patients suffering from recurrent rhabdomyolysis. None of these mutations were detected in control individuals of similar ethnic background. The affected patients had normal fat distribution and average weights, suggesting that there may be compensation for lipin-1 deficiency in human adipose tissue, although none of these patients have yet been studied as adults. LPIN1 mutations have not been detected in cases of human lipodystrophy (7, 18), but it should be noted that the patients analyzed did not exhibit the full array of symptoms that characterize lipin-1 deficiency in the mouse, including peripheral neuropathy, male infertility, and neonatal dyslipidemia and fatty liver.
In addition to the mutations causing lipin-1 deficiency described above, LPIN1 missense mutations were identified in two of six patients who developed myopathy while taking statin drugs, a serious side effect that occurs in a small percentage of patients (62). The association of LPIN1 mutations with statin-induced myopathy is intriguing but awaits confirmation in larger numbers of subjects.
Mutations in the LPIN2 gene cause a rare recessive Mendelian disease known as Majeed syndrome (1, 19, 39). Majeed syndrome is characterized by chronic recurrent multifocal osteomyelitis, an autoinflammatory condition that usually occurs sporadically. Other symptoms include congenital dyserythropoietic anemia and an inflammatory dermatitis. These phenotypes may be related to the loss of lipin-2 in erythrocytes and lymphocytes, where lipin-2 seems to be the major lipin expressed (16). Three affected families have been described: Two involve mutations that introduce a premature stop codon, and one is a single missense mutation at a serine residue in the C-LIP domain that is conserved in all mammalian lipin-1, lipin-2, and lipin-3 proteins (1, 19, 39). Mutation of the serine residue abolishes the PAP activity of lipin-2 (and also of lipin-1) but does not impair the association of lipin-2 with microsomal membranes or its transcriptional coactivator function (16). Several distinct missense mutations in lipin-2 have been associated with psoriasis, although the functional consequences of these amino acid substitutions on lipin-2 protein function have not been evaluated (41).
Genetic Variations in Lipin Genes Associated with Common Disease
Based on the established roles of lipin-1 as a determinant of adiposity, lipid metabolism, and insulin sensitivity, several studies have analyzed LPIN1 genetic variants for association with these and related metabolic traits (see Table 1). Several different variants in noncoding and intronic regions, as well as one exonic variant that leads to an amino acid substitution, have been associated with BMI and/or waist circumference in four different populations (18, 36, 53, 58). LPIN1 associations with insulin levels or type 2 diabetes, or with blood pressure (45, 58, 61), have also been reported in multiple populations. Other metabolic traits associated with LPIN1 polymorphisms or haplotypes include resting metabolic rate, hepatic lipase activity, overall metabolic syndrome factor score, and improvement of type 2 diabetes in response to rosiglitazone (10, 29, 36, 53, 58). In some cases, the LPIN1 genetic associations were significant only in one sex, suggesting that interactions may occur with other sex-specific biological factors.
A genomewide scan of type 2 diabetic patients led investigators to a significant common variant of LPIN2 (2). Refinement of the signal at 18p11 revealed a single nucleotide polymorphism within the 3′ untranslated region of LPIN2 that is significantly associated with BMI and fat distribution, glucose, and insulin levels. The polymorphism alters a putative micro-RNA nucleation site, but experimental assays failed to demonstrate a significant difference in lipin-2 mRNA levels in nucleated blood cells from homozygotes compared to heterozygotes for the associated allele. It will be important to assess the association of LPIN2 polymorphisms with similar metabolic traits in additional populations.
SUMMARY AND FUTURE DIRECTIONS
Lipin proteins are present in organisms ranging from yeast to man and play a fundamental role in cellular metabolism. All lipin proteins appear to function as PAP enzymes that play a key role in glycerolipid biosynthesis. The mammalian lipins may also have roles in the regulation of gene expression through activity as transcriptional coactivators. Lipin-1 and lipin-2 are necessary for normal physiological function, as evidenced by severe disease phenotypes in mice and humans with inactivating mutations in the corresponding genes. A particularly exciting finding is that common polymorphisms in both the LPIN1 and LPIN2 genes may influence important metabolic traits that underlie prevalent diseases such as increased adiposity, hypertension, psoriasis, insulin sensitivity, and response to commonly used antidiabetic drugs. Further studies in mouse models and human genetic variants will be instrumental to fully elucidate the role of lipin family members in maintenance of metabolic homeostasis in health and disease.
PAP: phosphatidate phosphatase
PA: phosphatidic acid
PGC-1α: peroxisome proliferator activated receptor γ coactivator-1α
fld: fatty liver dystrophy
TAG: triacylglycerol
Tg: transgenic
BMI: body mass index
SUMMARY POINTS.
Lipin proteins are phosphatidate phosphatase enzymes that provide diacylglycerol for synthesis of triacylglycerol and phospholipids. Mammalian lipin proteins also exhibit transcriptional coactivator activity.
The three mammalian lipin proteins—lipin-1, lipin-2, and lipin-3—appear to have nonredundant physiological roles, as evidenced by mutations in mice and humans.
Lipin-1, the founding member of the lipin family, is critical for lipid homeostasis in adipose tissue, skeletal muscle, liver, and peripheral nerve in mouse. In humans, lipin-1 deficiency is associated with childhood myopathy.
Lipin-1 gene expression levels are regulated by glucocorticoids, sterols, fasting, and lipin-2 levels and are correlated with insulin sensitivity in humans.
Lipin-1 activity is regulated by protein modification (phosphorylation, sumoylation) and subcellular localization.
Lipin-2 deficiency in humans causes Majeed syndrome, an inflammatory condition associated with osteomyelitis and anemia.
Common polymorphisms in human LPIN1 and LPIN2 genes are associated with traits underlying common metabolic diseases, including body mass index, hypertension, insulin resistance, and diabetes.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health grants HL28481 and HL90553 (K.R.) and T32HG002536 (L.C.).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Al-Mosawi ZS, Al-Saad KK, Ijadi-Maghsoodi R, El-Shanti HI, Ferguson PJ. A splice site mutation confirms the role of LPIN2 in Majeed syndrome. Arthritis Rheum. 2007;56:960–64. doi: 10.1002/art.22431. [DOI] [PubMed] [Google Scholar]
- 2.Aulchenko YS, Pullen J, Kloosterman WP, Yazdanpanah M, Hofman A, et al. LPIN2 is associated with type 2 diabetes, glucose metabolism, and body composition. Diabetes. 2007;56:3020–26. doi: 10.2337/db07-0338. [DOI] [PubMed] [Google Scholar]
- 3.Bou Khalil M, Sundaram M, Zhang HY, Links PH, Raven JF, et al. The level and compartmentalization of phosphatidate phosphatase-1 (lipin-1) control the assembly and secretion of hepatic VLDL. J. Lipid Res. 2009;50:47–58. doi: 10.1194/jlr.M800204-JLR200. [DOI] [PubMed] [Google Scholar]
- 4.Brindley DN. Intracellular translocation of phosphatidate phosphohydrolase and its possible role in the control of glycerolipid synthesis. Prog. Lipid Res. 1984;23:115–33. doi: 10.1016/0163-7827(84)90001-8. [DOI] [PubMed] [Google Scholar]
- 5.Brindley DN. Phosphatidate phosphohydrolase activity in the liver. In: Brindley DN, editor. Phosphatidate Phosphohydrolase. Vol. 1. CRC Press; Boca Raton, FL: 1988. pp. 21–77. [Google Scholar]
- 6.Brindley DN, Pilquil C, Sariahmetoglu M, Reue K. Phosphatidate degradation: phosphatidate phosphatases (lipins) and lipid phosphate phosphatases. Biochim. Biophys. Acta. 2009;1791:956–61. doi: 10.1016/j.bbalip.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao H, Hegele RA. Identification of single-nucleotide polymorphisms in the human LPIN1 gene. J. Hum. Genet. 2002;47:370–72. doi: 10.1007/s100380200052. [DOI] [PubMed] [Google Scholar]
- 8.Carman GM, Han GS. Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem. Sci. 2006;31:694–99. doi: 10.1016/j.tibs.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carman GM, Han GS. Phosphatidic acid phosphatase, a key enzyme in the regulation of lipid synthesis. J. Biol. Chem. 2009;284:2593–97. doi: 10.1074/jbc.R800059200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang YC, Chang LY, Chang TJ, Jiang YD, Lee KC, et al. The associations of LPIN1 gene expression in adipose tissue with metabolic phenotypes in the Chinese population. Obesity (Silver Spring) 2009;18:7–12. doi: 10.1038/oby.2009.198. [DOI] [PubMed] [Google Scholar]
- 11.Chen Z, Gropler MC, Norris J, Lawrence JC, Jr, Harris TE, Finck BN. Alterations in hepatic metabolism in fld mice reveal a role for lipin 1 in regulating VLDL-triacylglyceride secretion. Arterioscler. Thromb. Vasc. Biol. 2008;28:1738–44. doi: 10.1161/ATVBAHA.108.171538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coleman RA, Lee DP. Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 2004;43:134–76. doi: 10.1016/s0163-7827(03)00051-1. [DOI] [PubMed] [Google Scholar]
- 13.Croce MA, Eagon JC, LaRiviere LL, Korenblat KM, Klein S, Finck BN. Hepatic lipin 1beta expression is diminished in insulin-resistant obese subjects and is reactivated by marked weight loss. Diabetes. 2007;56:2395–99. doi: 10.2337/db07-0480. [DOI] [PubMed] [Google Scholar]
- 14.Donkor J, Sariahmetoglu M, Dewald J, Brindley DN, Reue K. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J. Biol. Chem. 2007;282:3450–57. doi: 10.1074/jbc.M610745200. [DOI] [PubMed] [Google Scholar]
- 15.Donkor J, Sparks LM, Xie H, Smith SR, Reue K. Adipose tissue lipin-1 expression is correlated with peroxisome proliferator-activated receptor alpha gene expression and insulin sensitivity in healthy young men. J. Clin. Endocrinol. Metab. 2008;93:233–39. doi: 10.1210/jc.2007-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donkor J, Zhang P, Wong S, O'Loughlin L, Dewald J, et al. A conserved serine residue is required for the phosphatidate phosphatase activity but not the transcriptional coactivator functions of lipin-1 and lipin-2. J. Biol. Chem. 2009;284:29968–78. doi: 10.1074/jbc.M109.023663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douglas DS, Moran JL, Bermingham JR, Jr, Chen XJ, Brindley DN, et al. Concurrent Lpin1 and Nrcam mouse mutations result in severe peripheral neuropathy with transitory hindlimb paralysis. J. Neurosci. 2009;29:12089–100. doi: 10.1523/JNEUROSCI.2029-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fawcett KA, Grimsey N, Loos RJ, Wheeler E, Daly A, et al. Evaluating the role of LPIN1 variation on insulin resistance, body weight and human lipodystrophy in UK populations. Diabetes. 2008;57(9):2527–33. doi: 10.2337/db08-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferguson PJ, Chen S, Tayeh MK, Ochoa L, Leal SM, et al. Homozygous mutations in LPIN2 are responsible for the syndrome of chronic recurrent multifocal osteomyelitis and congenital dyserythropoietic anaemia (Majeed syndrome). J. Med. Genet. 2005;42:551–57. doi: 10.1136/jmg.2005.030759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Festuccia WT, Blanchard PG, Turcotte V, Laplante M, Sariahmetoglu M, et al. Depot-specific effects of the PPARgamma agonist rosiglitazone on adipose tissue glucose uptake and metabolism. J. Lipid Res. 2009;50:1185–94. doi: 10.1194/jlr.M800620-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finck BN, Gropler MC, Chen Z, Leone TC, Croce MA, et al. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 2006;4:199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Grimsey N, Han GS, O'Hara L, Rochford JJ, Carman GM, Siniossoglou S. Temporal and spatial regulation of the phosphatidate phosphatases lipin 1 and 2. J. Biol. Chem. 2008;283:29166–74. doi: 10.1074/jbc.M804278200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han GS, Siniossoglou S, Carman GM. The cellular functions of the yeast lipin homolog PAH1p are dependent on its phosphatidate phosphatase activity. J. Biol. Chem. 2007;282:37026–35. doi: 10.1074/jbc.M705777200. [DOI] [PubMed] [Google Scholar]
- 24.Han GS, Wu WI, Carman GM. The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 2006;281:9210–18. doi: 10.1074/jbc.M600425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris TE, Huffman TA, Chi A, Shabanowitz J, Hunt DF, et al. Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. J. Biol. Chem. 2007;282:277–86. doi: 10.1074/jbc.M609537200. [DOI] [PubMed] [Google Scholar]
- 26.Higashida K, Higuchi M, Terada S. Potential role of lipin-1 in exercise-induced mitochondrial biogenesis. Biochem. Biophys. Res. Commun. 2008;374:587–91. doi: 10.1016/j.bbrc.2008.07.079. [DOI] [PubMed] [Google Scholar]
- 27.Huffman TA, Mothe-Satney I, Lawrence JC., Jr Insulin-stimulated phosphorylation of lipin mediated by the mammalian target of rapamycin. Proc. Natl. Acad. Sci. USA. 2002;99:1047–52. doi: 10.1073/pnas.022634399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishimoto K, Nakamura H, Tachibana K, Yamasaki D, Ota A, et al. Sterol-mediated regulation of human lipin 1 gene expression in hepatoblastoma cells. J. Biol. Chem. 2009;284:22195–205. doi: 10.1074/jbc.M109.028753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang ES, Park SE, Han SJ, Kim SH, Nam CM, et al. LPIN1 genetic variation is associated with rosiglitazone response in type 2 diabetic patients. Mol. Genet. Metab. 2008;95:96–100. doi: 10.1016/j.ymgme.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 30.Kim Y, Gentry MS, Harris TE, Wiley SE, Lawrence JC, Jr, Dixon JE. A conserved phosphatase cascade that regulates nuclear membrane biogenesis. Proc. Natl. Acad. Sci. USA. 2007;104:6596–601. doi: 10.1073/pnas.0702099104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klingenspor M, Xu P, Cohen RD, Welch C, Reue K. Altered gene expression pattern in the fatty liver dystrophy mouse reveals impaired insulin-mediated cytoskeleton dynamics. J. Biol. Chem. 1999;274:23078–84. doi: 10.1074/jbc.274.33.23078. [DOI] [PubMed] [Google Scholar]
- 32.Langner CA, Birkenmeier EH, Ben-Zeev O, Schotz MC, Sweet HO, et al. The fatty liver dystrophy (fld) mutation. A new mutant mouse with a developmental abnormality in triglyceride metabolism and associated tissue-specific defects in lipoprotein lipase and hepatic lipase activities. J. Biol. Chem. 1989;264:7994–8003. [PubMed] [Google Scholar]
- 33.Langner CA, Birkenmeier EH, Roth KA, Bronson RT, Gordon JI. Characterization of the peripheral neuropathy in neonatal and adult mice that are homozygous for the fatty liver dystrophy (fld) mutation. J. Biol. Chem. 1991;266:11955–64. [PubMed] [Google Scholar]
- 34.Lindegaard B, Larsen LF, Hansen AB, Gerstoft J, Pedersen BK, Reue K. Adipose tissue lipin expression levels distinguish HIV patients with and without lipodystrophy. Int. J. Obes. 2006;31:449–56. doi: 10.1038/sj.ijo.0803434. [DOI] [PubMed] [Google Scholar]
- 35.Liu GH, Gerace L. Sumoylation regulates nuclear localization of lipin-1alpha in neuronal cells. PLoS One. 2009;4:e7031. doi: 10.1371/journal.pone.0007031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loos RJ, Rankinen T, Perusse L, Tremblay A, Despres JP, Bouchard C. Association of lipin 1 gene polymorphisms with measures of energy and glucose metabolism. Obesity (Silver Spring) 2007;15:2723–32. doi: 10.1038/oby.2007.324. [DOI] [PubMed] [Google Scholar]
- 37.Lu B, Lu Y, Moser AH, Shigenaga JK, Grunfeld C, Feingold KR. LPS and proinflammatory cytokines decrease lipin-1 in mouse adipose tissue and 3T3-L1 adipocytes. Am. J. Physiol. Endocrinol. Metab. 2008;295:E1502–9. doi: 10.1152/ajpendo.90323.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lusis AJ, Attie AD, Reue K. Metabolic syndrome: from epidemiology to systems biology. Nat. Rev. Genet. 2008;9:819–30. doi: 10.1038/nrg2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Majeed HA, Al-Tarawna M, El-Shanti H, Kamel B, Al-Khalaileh F. The syndrome of chronic recurrent multifocal osteomyelitis and congenital dyserythropoietic anaemia. Report of a new family and a review. Eur. J. Pediatr. 2001;160:705–10. doi: 10.1007/s004310100799. [DOI] [PubMed] [Google Scholar]
- 40.Manmontri B, Sariahmetoglu M, Donkor J, Khalil MB, Sundaram M, et al. Glucocorticoids and cyclic AMP selectively increase hepatic lipin-1 expression, and insulin acts antagonistically. J. Lipid Res. 2008;49:1056–67. doi: 10.1194/jlr.M800013-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milhavet F, Cuisset L, Hoffman HM, Slim R, El-Shanti H, et al. The infevers autoinflammatory mutation online registry: update with new genes and functions. Hum. Mutat. 2008;29:803–8. doi: 10.1002/humu.20720. [DOI] [PubMed] [Google Scholar]
- 42.Miranda M, Chacon MR, Gomez J, Megia A, Ceperuelo-Mallafre V, et al. Human subcutaneous adipose tissue LPIN1 expression in obesity, type 2 diabetes mellitus, and human immunodeficiency virus–associated lipodystrophy syndrome. Metabolism. 2007;56:1518–26. doi: 10.1016/j.metabol.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 43.Mlinar B, Pfeifer M, Vrtacnik Bokal E, Jensterle M, Marc J. Decreased lipin 1beta expression in visceral adipose tissue is associated with insulin resistance in polycystic ovary syndrome. Eur. J. Endocrinol. 2008;59:833–39. doi: 10.1530/EJE-08-0387. [DOI] [PubMed] [Google Scholar]
- 44.Nadra K, de Preux Charles AS, Medard JJ, Hendriks WT, Han GS, et al. Phosphatidic acid mediates demyelination in Lpin1 mutant mice. Genes Dev. 2008;22:1647–61. doi: 10.1101/gad.1638008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ong KL, Leung RY, Wong LY, Cherny SS, Sham PC, et al. Association of a polymorphism in the lipin 1 gene with systolic blood pressure in men. Am. J. Hypertens. 2008;21:539–45. doi: 10.1038/ajh.2008.21. [DOI] [PubMed] [Google Scholar]
- 46.Péterfy M, Phan J, Reue K. Alternatively spliced lipin isoforms exhibit distinct expression pattern, subcellular localization, and role in adipogenesis. J. Biol. Chem. 2005;280:32883–89. doi: 10.1074/jbc.M503885200. [DOI] [PubMed] [Google Scholar]
- 47.Péterfy M, Phan J, Xu P, Reue K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 2001;27:121–24. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- 48.Phan J, Péterfy M, Reue K. Lipin expression preceding peroxisome proliferator-activated receptor-gamma is critical for adipogenesis in vivo and in vitro. J. Biol. Chem. 2004;279:29558–64. doi: 10.1074/jbc.M403506200. [DOI] [PubMed] [Google Scholar]
- 49.Phan J, Reue K. Lipin, a lipodystrophy and obesity gene. Cell Metab. 2005;1:73–83. doi: 10.1016/j.cmet.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 50.Reue K, Brindley DN. Multiple roles for lipins/phosphatidate phosphatase enzymes in lipid metabolism. J. Lipid Res. 2008;49:2493–503. doi: 10.1194/jlr.R800019-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reue K, Xu P, Wang XP, Slavin BG. Adipose tissue deficiency, glucose intolerance, and increased atherosclerosis result from mutation in the mouse fatty liver dystrophy (fld) gene. J. Lipid Res. 2000;41:1067–76. [PubMed] [Google Scholar]
- 52.Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 2005;24:1931–41. doi: 10.1038/sj.emboj.7600672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suviolahti E, Reue K, Cantor RM, Phan J, Gentile M, et al. Cross-species analyses implicate lipin 1 involvement in human glucose metabolism. Hum. Mol. Genet. 2006;15:377–86. doi: 10.1093/hmg/ddi448. [DOI] [PubMed] [Google Scholar]
- 54.Tsuchiya Y, Takahashi N, Yoshizaki T, Tanno S, Ohhira M, et al. A Jak2 inhibitor, AG490, reverses lipin-1 suppression by TNF-alpha in 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 2009;382:348–52. doi: 10.1016/j.bbrc.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 55.van Harmelen V, Ryden M, Sjolin E, Hoffstedt J. A role of lipin in human obesity and insulin resistance: relation to adipocyte glucose transport and GLUT4 expression. J. Lipid Res. 2007;48:201–6. doi: 10.1194/jlr.M600272-JLR200. [DOI] [PubMed] [Google Scholar]
- 56.Verheijen MH, Chrast R, Burrola P, Lemke G. Local regulation of fat metabolism in peripheral nerves. Genes Dev. 2003;17:2450–64. doi: 10.1101/gad.1116203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waki H, Park KW, Mitro N, Pei L, Damoiseaux R, et al. The small molecule harmine is an antidiabetic cell-type specific regulator of PPARg expression. Cell Metab. 2007;5:357–70. doi: 10.1016/j.cmet.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 58.Wiedmann S, Fischer M, Koehler M, Neureuther K, Riegger G, et al. Genetic variants within the LPIN1 gene, encoding lipin, are influencing phenotypes of the metabolic syndrome in humans. Diabetes. 2008;57:209–17. doi: 10.2337/db07-0083. [DOI] [PubMed] [Google Scholar]
- 59.Xu J, Lee WN, Phan J, Saad MF, Reue K, Kurland IJ. Lipin deficiency impairs diurnal metabolic fuel switching. Diabetes. 2006;55:3429–38. doi: 10.2337/db06-0260. [DOI] [PubMed] [Google Scholar]
- 60.Yao-Borengasser A, Rasouli N, Varma V, Miles LM, Phanavanh B, et al. Lipin expression is attenuated in adipose tissue of insulin-resistant human subjects and increases with peroxisome proliferator-activated receptor gamma activation. Diabetes. 2006;55:2811–18. doi: 10.2337/db05-1688. [DOI] [PubMed] [Google Scholar]
- 61.Yatsu K, Mizuki N, Hirawa N, Oka A, Itoh N, et al. High-resolution mapping for essential hypertension using microsatellite markers. Hypertension. 2007;49:446–52. doi: 10.1161/01.HYP.0000257256.77680.02. [DOI] [PubMed] [Google Scholar]
- 62.Zeharia A, Shaag A, Houtkooper RH, Hindi T, de Lonlay P, et al. Mutations in LPIN1 cause recurrent acute myoglobinuria in childhood. Am. J. Hum. Genet. 2008;83:1–6. doi: 10.1016/j.ajhg.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang P, O'Loughlin L, Brindley DN, Reue K. Regulation of lipin-1 gene expression by glucocorticoids during adipogenesis. J. Lipid Res. 2008;49:1519–28. doi: 10.1194/jlr.M800061-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]