Abstract
Takayasu arteritis (TAK) is an autoimmune systemic vasculitis of unknown etiology. Although previous studies have revealed that HLA-B∗52:01 has an effect on TAK susceptibility, no other genetic determinants have been established so far. Here, we performed genome scanning of 167 TAK cases and 663 healthy controls via Illumina Infinium Human Exome BeadChip arrays, followed by a replication study consisting of 212 TAK cases and 1,322 controls. As a result, we found that the IL12B region on chromosome 5 (rs6871626, overall p = 1.7 × 10−13, OR = 1.75, 95% CI 1.42–2.16) and the MLX region on chromosome 17 (rs665268, overall p = 5.2 × 10−7, OR = 1.50, 95% CI 1.28–1.76) as well as the HLA-B region (rs9263739, a proxy of HLA-B∗52:01, overall p = 2.8 × 10−21, OR = 2.44, 95% CI 2.03–2.93) exhibited significant associations. A significant synergistic effect of rs6871626 and rs9263739 was found with a relative excess risk of 3.45, attributable proportion of 0.58, and synergy index of 3.24 (p ≤ 0.00028) in addition to a suggestive synergistic effect between rs665268 and rs926379 (p ≤ 0.027). We also found that rs6871626 showed a significant association with clinical manifestations of TAK, including increased risk and severity of aortic regurgitation, a representative severe complication of TAK. Detection of these susceptibility loci will provide new insights to the basic mechanisms of TAK pathogenesis. Our findings indicate that IL12B plays a fundamental role on the pathophysiology of TAK in combination with HLA-B∗52:01 and that common autoimmune mechanisms underlie the pathology of TAK and other autoimmune disorders such as psoriasis and inflammatory bowel diseases in which IL12B is involved as a genetic predisposing factor.
Introduction
Takayasu arteritis (TAK [MIM 207600]) is an autoimmune systemic vasculitis that was first reported from Japan.1 It is estimated that TAK affects around 0.004% of the population in Japan, especially young women aged between 15 and 35. Although TAK was originally thought to affect individuals of mainly Asian origin, individuals with TAK have been identified worldwide, though with lower prevalence compared to Asia.2 TAK is characterized by the involvement of large arteries, especially the aorta and its large branches, and is grouped into “vasculitis affecting large vessels” according to the Chapel Hill classification.3 Individuals with TAK develop a wide range of symptoms such as fatigue, syncope, and lowering of vision in addition to its characteristic complications including aortic regurgitation (AR), pulselessness, and difference of blood pressure between right and left upper limbs. Previous studies have revealed that genetic components are involved in the pathogenesis of TAK, and HLA-B∗52:01 is so far the only established genetic factor across the world.4–7 Other genetic components especially outside of the HLA locus have not been confirmed to date. Establishment of association with non-HLA regions would lead to a deeper understanding of the basics of TAK pathology and the development of a novel therapy for this vasculitis. Here, we performed a genome-scanning study of TAK to identify the genetic predisposing factors for TAK.
Subjects and Methods
Study Subjects
A total of 379 TAK cases and 1,985 controls were enrolled in this study. All the cases were diagnosed based on the criteria of American College of Rheumatology8 or guideline provided by Japanese Circulation Society.9 The control subjects were collected as a part of the Nagahama Prospective Genome Cohort for Comprehensive Human Bioscience (The Nagahama Study), a community-based prospective multiomics cohort study conducted by Kyoto University.10 This study was approved by the local ethical committees at each institution, and written informed consent was obtained from each subject involved in the study.
Genome Scanning
Illumina Infinium Human Exome BeadChip arrays (Illumina) were used for genome scanning of the cases and the controls. The genome scanning was conducted in Center for Genomic Medicine, Kyoto University Graduate School of Medicine.
Quality Control of Genome Scanning
Polymorphisms showing success rates less than 0.95 in either cases or controls, departure from Hardy-Weinberg equilibrium (HWE) (p < 1.0 × 10−5), or minor allele frequencies less than 0.05 in both cases and controls were excluded from the analysis. Subjects who showed success rates less than 0.95 or evidence of relatedness with other subjects were also excluded. Kinship between study subjects were estimated by PLINK.11 Quantile-quantile plot (QQ plot) was used to assess the population stratification of the study. Because 1,827 markers over 24,487 were located in the HLA locus in which polymorphisms are very closely linked with each other, the 22,660 markers in the non-HLA regions were used for QQ plot.
Replication Study
The SNPs with p values less than 1.0 × 10−5 in the genome scanning were selected for the replication study. Because the association found in the HLA-B region (MIM 142830) was largely attributable to HLA-B∗52:01, rs9263739, a proxy of HLA-B∗52:01, was selected as a representative of the HLA locus. In the replication study, case samples were genotyped by Taqman Assay (Applied Biosystems) and control genotypes were extracted from array data (Table 1).
Table 1.
Summary of Study Subjects
| Case | Control | |
|---|---|---|
| Genome Scanning | ||
| Number | 167 | 663 |
| Agea | 45.7 ± 15.2 | 53.5 ± 13.5 |
| Female ratio | 0.92 | 0.74 |
| Age at onseta | 30.5 ± 14.5 | NA |
| Genotyping | Illumina Infinium Human-Exome BeadChip | Illumina Infinium Human-Exome BeadChip |
| Subjects with clinical information | AR:87; CRP:89 | NA |
| Institutions | Kyoto University; Tokyo Women’s Medical University | Kyoto University |
| Replication Study | ||
| Number | 212 | 1,322 |
| Agea | 46.6 ± 17.6 | 53.3 ± 13.4 |
| Female ratio | 0.94 | 0.62 |
| Age at onseta | 27.0 ± 11.8 | NA |
| Genotyping | Taqman assay | Illumina Infinium Human Omni 2.5-4 BeadChip, Illumina Infinium Human Omni 2.5-8 BeadChip |
| Subjects with clinical information | AR:102; CRP:None | NA |
| Institutions | Tokyo Medical and Dental University; Kyoto University; Niigata University | Kyoto University |
Abbreviations are as follows: NA, not applicable; AR, aortic regurgitation; CRP, C-reactive protein.
Mean ± standard deviation (SD).
Combined Study and Association Study for Genotypes
Association studies of genotypes were performed by chi-square test based on 2 × 2 contingency tables. Combined study of the two studies was performed by inverse-variance method, assuming a fixed-effects model from the effect size (logarithm of odds ratio [OR]) in each study. A significant level for detecting susceptibility genes was set as 2.0 × 10−6, which was obtained by Bonferroni’s correction. A stringent cut-off level of 5.0 × 10−8 was also applied to assess overall significance.
Imputation of Genotypes
Mach dat2 software12 was used for imputation of the whole genomes based on the results of genome scans with the use of the East Asian panel of HapMap phase II data as reference. SNPs with low imputation scores (Rsq < 0.3) were excluded from the analysis.
Calculation of Linkage Disequilibrium
LD between SNPs in the Illumina Infinium Human Exome BeadChip was assessed based on the genome-scanning data. HapMap project phase II data was used when SNPs were not contained in the array. LD between HLA-B∗52:01 and SNPs was calculated by combining our previous HLA-genotyping data of the 173 TAK cases (C.T., unpublished data) by WAKFlow system (Wakunaga Pharmaceutical) with the genome-scanning data.
Estimation of Interaction
We used the method for evaluation of interaction proposed by Andersson et al.13 Gene-gene interaction was defined as departure from additivity of two loci and measured by three indices based on calculation of relative risk (RR); relative excess risk due to interaction (RERI), attributable proportion (AP), and synergy index (SI). We considered an interaction as significant only when both RERI and AP were different from 0 and additionally SI was more than 1. The very low prevalence of TAK justifies to approximate OR by RR. For instance, when we assessed the interaction between rs9263739 and rs6871626 through these three indices, the subjects were classified into four groups: negative for both rs9263739 T allele and rs6871626 A allele, positive for rs9263739 T allele and negative for rs6871626 A allele, negative for rs9263739 T allele and positive for rs6871626 A allele, and positive for both rs9263739 T allele and rs6871626 A allele. Logistic models were used to calculate the indices.
In Silico Analysis of Association between the Gene Expression and rs6871626
We used two methods to assess the effect of rs6871626 on the IL12B (MIM 161561) expression. Gene expression data for IL12B in lymphoblastoid cells were obtained from GEO database (accession number GSE6536)14 and analyzed for association with genotypes of rs6871626 obtained from HapMap project. Genevar software was used for analyzing the IL12B expression in adipose and skin in association with the rs6871626 genotypes.15 Associations between genotypes and gene expression were evaluated by a linear regression analysis.
Associations between Genotypes and Clinical Phenotypes of TAK
Data of age at onset were analyzed for the association with the susceptibility alleles. AR, ischemic heart disease, and pulmonary infarction were selected for the association with genotypes as representative complications of TAK because cardiovascular event was the major cause of death in TAK individuals16 and it was previously demonstrated that these phenotypes were associated with HLA-B∗52:01,17 suggesting that genetic backgrounds were at least partly responsible for these clinical manifestations. Data of the clinical manifestations were collected in Kyoto University Hospital or Tokyo Medical and Dental University by medical doctors who were blinded to genotype data reviewing clinical charts. Although AR evaluated by transthoracic echocardiography or angiography was positive for 44% of cases, other complications were found in less than 16%. Only AR was analyzed because of lack of power for other manifestations. Data for severity of AR assessed by the three categories18 (mild, moderate, and severe) were also collected. C-reactive protein (CRP) was focused on as a biomarker reflecting disease activity. We calculated time-averaged CRP and dosage of prednisolone. Individuals who had visited hospitals for less than 500 days were excluded from the analysis of CRP. The associations between genotypes and clinical phenotypes were assessed by logistic regression analysis for existence of AR or linear regression analysis for severity of AR, time-averaged CRP, and age at onset. Time-averaged CRP was analyzed in condition with time-averaged dosage of prednisolone alone or in combination with rs3093059 genotypes in the CRP (MIM 123260) region. Associations between genotypes and clinical manifestations with p values less than 0.05 were regarded as significant.
Statistical Analysis
Statistical analyses were performed by PLINK v.1.07, R statistical software, or SPSS v.18.0.
Results
A summary of basic information of the subjects in our study is shown in Table 1. DNA samples from 167 cases and 663 healthy controls were genome scanned with the use of Illumina Human-Exome arrays containing 247,730 SNPs. One sample of the TAK cases and six samples in controls with success rates of less than 0.95 or with evidence of relatedness with other subjects (PI_HAT > 0.2 calculated by PLINK, see Subjects and Methods) were excluded from further analysis. The genotyping revealed that more than 80% of the markers in the array were monomorphic and 9% of the markers showed low minor allele frequency (<0.05) in the Japanese population, respectively. A total of 24,487 markers remained after filtering of SNPs that showed success rates of less than 0.95, deviation from HWE (p < 1 × 10−5) in either cases or controls, or minor allele frequencies of less than 0.05 in both cases and controls. The mean success rate of individuals was 0.999 after filtering.
Association studies were performed by chi-square test to compare allele frequencies between cases and controls. Population stratification was evaluated by QQ plot. The results indicated a lambda value of 1.05 in the QQ plot, indicating no excess population stratification in our study. Manhattan plot revealed that a region on chromosome 5 as well as the HLA locus showed significant associations that satisfied the genome-wide significant threshold obtained by Bonferroni’s correction (p = 2.0 × 10−6; Figure 1). The associations were also confirmed by the imputed results (Figure S1 available online). rs4947248 in the HLA-B region, which is a known susceptibility gene to TAK, showed the strongest association (p = 5.1 × 10−9, OR = 2.17, 95% CI 1.67–2.82). rs9263739, a proxy of HLA-B∗52:01 (r2 = 0.94), similarly showed a significant association (p = 8.0 × 10−9, OR = 2.30, 95% CI 1.72–3.07; Table 2) and in moderate LD with rs4947248 (D’ = 0.95, r2 = 0.58). Because rs4947248 did not show evidence of an independent association from rs9263739 in logistic regression analysis (p = 0.04), we assumed that the top association in the HLA locus was attributable to HLA-B∗52:01. rs6871626 in the IL12B region on chromosome 5 also showed a significant association (p = 1.8 × 10−7, OR = 1.90, 95% CI 1.49–2.42; Table 2 and Figure 2A). Four other loci showed suggestive associations in our study (p < 5.0 × 10−5; Table 2). No departure from HWE was observed for these six SNPs (p ≥ 0.041).
Figure 1.
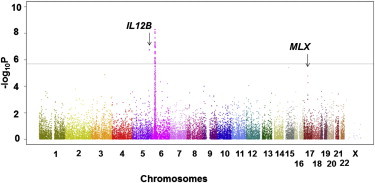
Manhattan Plot of Genome Scanning
The horizontal line indicates the significant level based on Bonferroni’s correction. The HLA locus on chromosome 6 and the IL12B region on chromosome 5 reached the significant level.
Table 2.
Results of Association Studies for TAK Susceptibility
| SNP | Chr | Position | Gene | Ref(A1) | Var(A2)a |
Genome Scan |
Replication |
Meta-analysis |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CaseA2freq | ContA2freq | p | CaseA2freq | ContA2freq | p | p | OR (95% CI) | ||||||
| rs10934853 | 3 | 129521063 | EEFSEC | A | C | 0.59 | 0.45 | 1.3 × 10−5 | 0.52 | 0.47 | 0.066 | 2.6 × 10−5 | 1.40 (1.20–1.64) |
| rs6871626 | 5 | 158759370 | IL12B | C | A | 0.53 | 0.37 | 1.8 × 10−7 | 0.53 | 0.39 | 1.1 × 10−7 | 1.7 × 10−13 | 1.75 (1.42–2.16) |
| rs9263739 | 6 | 31219335 | CCHCR1 | C | T | 0.27 | 0.14 | 8.0 × 10−9 | 0.30 | 0.14 | 6.0 × 10−15 | 2.8 × 10−21 | 2.44 (2.03–2.93) |
| rs1570843 | 6 | 84577239 | RIPPLY2 | C | T | 0.62 | 0.50 | 4.6 × 10−5 | 0.54 | 0.51 | 0.19 | 3.1 × 10−4 | 1.34 (1.14–1.57) |
| rs12102203 | 15 | 49578851 | DMXL2 | G | A | 0.64 | 0.49 | 3.8 × 10−6 | 0.53 | 0.54 | 0.71 | 0.0081 | 1.24 (1.06–1.46) |
| rs665268 | 17 | 37975555 | MLX | A | G | 0.58 | 0.44 | 1.7 × 10−5 | 0.49 | 0.42 | 0.0032 | 5.2 × 10−7 | 1.50 (1.28–1.76) |
Abbreviations are as follows: chr, chromosome; ref, reference allele; var, variant allele; CaseA2freq, variant allele frequency in cases; ContA2freq, variant allele frequency in controls; OR, odds ratio; CI, confidence interval. Positions are according to National Center for Biotechnology Information (NCBI) build 36.
Risk alleles for TAK based on the results of the genome scanning are set as variant alleles.
Figure 2.
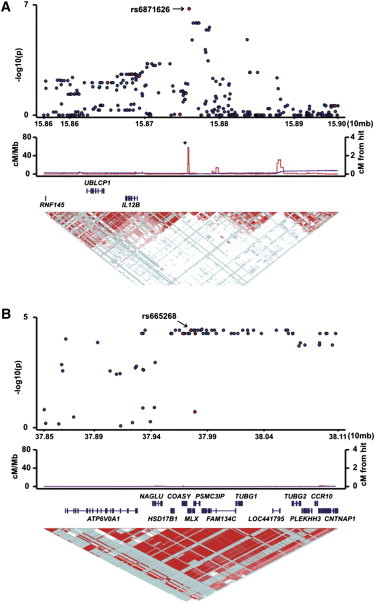
Associations of the IL12B and MLX Regions with the Susceptibility to TAK
Associations of SNPs in the (A) IL12B and (B) MLX regions in the genome scanning are plotted according to the position of the markers. Red circles indicate results of the current genome scanning. Blue circles indicate results of the imputation analysis based on the current results. The middle panel indicates recombination rates. The lower panel indicates LD of markers. Asterisk indicates a recombination hot spot in the IL12B region.
A replication study was performed with the use of DNA samples from 212 cases and 1,322 controls. The six SNPs with p values less than 5.0 × 10−5 in the genome scanning were genotyped in the replication study. rs9263739 was selected as a representative of the associations in the HLA locus. As a result, the significant associations of TAK with rs6871626 and rs665268 in the MLX (MAX dimerization protein [MIM 602976]) region on chromosome 17 as well as rs9263739 were replicated (p = 1.1 × 10−7, 0.0032, and 6.0 × 10−15, respectively; Table 2, Figures 2A and 2B). The suggestive association on chromosome 15 (Figures S1 and 1) was not replicated. Again, no departure from HWE was observed (p ≥ 0.11).
A combined study in which the associations in the two studies were integrated by inverse-variance method demonstrated that rs6871626, rs665268, and rs9263739 showed significant associations (p = 1.7 × 10−13, 5.2 × 10−7, and 2.8 × 10−21; OR = 1.75, 1.50, and 2.44; 95% CI 1.42–2.16, 1.28–1.76, and 2.03–2.93, respectively; Table 2) satisfying the significance obtained by Bonferroni’s correction. rs6871626 and rs9263739 satisfied the more stringent, widely accepted genome-wide significance (p = 5.0 × 10−8).
Because it was suggested that genetic components had influence on the manifestations of the disease,17 we analyzed whether the variant of the IL12B region had clinical effects on the disease course or severity. Age at onset was not associated with rs6871626 (p = 0.36), whereas a significant association between rs6871626 and development of AR was observed in a recessive model (p = 0.0046; Figure 3A). Focusing on the cases with AR, a significant association between rs6871626 and severity of AR was observed in the recessive model (p = 0.0018; Figure 3B). Risk allele of rs6871626 (A allele) also demonstrated a significant association with increased level of time-averaged CRP, which was a representative marker of the disease activity (p = 0.021; Figure 3C). The association between rs6871626 and CRP levels was independent from rs3093059 in the CRP region (p = 0.029), which showed the strongest association with circulating CRP levels in Japanese.19 These associations between rs6871626 and clinical manifestations were independent from rs9263739 (conditioned p value of rs6871626 ≤ 0.020). Although rs665268 also demonstrated a significant association with development of AR in a dominant model (p = 0.0089; Figure S2A), the association was not significant in condition with rs9263739 (p = 0.080). No significant associations were observed between rs665268 and other clinical phenotypes (Figures S2B and S2C).
Figure 3.
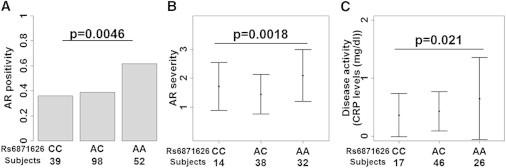
Associations between rs6871626 Genotypes and Clinical Manifestations of TAK
An association between rs6871626 genotypes and (A) development of AR, (B) severity of AR, and (C) time-averaged CRP levels in TAK cases. The p value was calculated by (A) logistic regression analysis, (B) linear regression analysis, and (C) linear regression analysis with time-averaged dosage of prednisolone as covariate. The recessive model is applied to all calculations. Severity of 1 to 3 in AR corresponds to mild, moderate, and severe, respectively. Mean ± SD are indicated for (B) and (C).
Next, we investigated the interaction between the IL12B and HLA-B loci to TAK susceptibility. The risk of TAK in the population positive for both rs6871626 A allele and rs9263739 T allele surpassed the product and sum of the risk in those who were positive for either rs6871626 A allele or rs9263739 T allele alone (Figure 4). The analysis revealed that those who were positive for both had OR of 6.00 (95% CI 4.22–8.55), whereas those who were positive for either rs9263739 T allele or rs6871626 A allele showed OR of 1.80 (95% CI 1.11–2.93) or 1.74 (95% CI 1.23–2.47), respectively. Interaction measures revealed RER of 3.46 (p = 1.4 × 10−5, 95% CI 1.90–5.01), AP of 0.58 (p = 1.0 × 10−12, 95% CI 0.42–0.73), and SI of 3.24 (p = 0.00028, 95% CI 1.72–6.11). This significant interaction between IL12B and HLA-B on TAK susceptibility could be observed in both studies (Table 3). The synergistic interaction effects between rs6871626 and rs9263739 were not evident in the clinical manifestations associated with rs6871626 (Figure S3). When we analyzed the interaction between the MLX and HLA-B regions, we observed suggestive interaction with RER of 1.73, AP of 0.43, and SI of 2.29 (p ≤ 0.027; Figure S4 and Table S1). The associations between the interaction and clinical manifestations were not significant (Figure S5).
Figure 4.
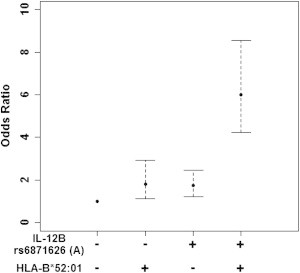
A Synergistic Effect between IL12B and HLA-B∗52:01 on TAK Susceptibility
ORs are shown for the four strata of subjects according to combination of rs6871626 and rs9263739 genotypes. Those who are negative for rs9263739 T allele, a proxy of HLA-B∗52:01, and rs6871626 A allele are used as reference. ORs and 95% CI are indicated.
Table 3.
Synergistic Effects between IL12B and HLA-B∗52:01 in Each Study
| Study |
RERI |
AP |
SI |
|||
|---|---|---|---|---|---|---|
| (95% CI) | p | (95% CI) | p | (95% CI) | p | |
| Genome scanning | 2.90 (0.60–5.20) | 0.014 | 0.50 (0.23–0.78) | 0.00034 | 2.57 (1.08–6.09) | 0.032 |
| Replication study | 3.87 (1.70–6.05) | 0.00049 | 0.62 (0.42–0.81) | 4.7 × 10−10 | 3.76 (1.51–9.32) | 0.0043 |
| Combined study | 3.46 (1.90–5.02) | 1.4 × 10−5 | 0.58 (0.42–0.73) | 1.0 × 10−12 | 3.24 (1.72–6.11) | 0.00028 |
Abbreviations are as follows: RERI, relative excess risk; AP, attributable proportion; SI, synergy index; CI, confidence interval.
IL12B encodes a common subunit of the IL12 and IL23 protein, known as p40. Because previous studies showed that the IL23R/IL12RB2 (MIM 607562/601642) region was associated with Behçet disease20 (MIM 109650), another connective tissue disease where vasculitis is involved in its pathology, we investigated this region for the possible associations in the current study. As a result, no suggestive association was found, either in our study or in the imputed results (Figure S6).
Discussion
This study provides a convincing evidence of associations between non-HLA genes and TAK susceptibility along with a synergistic role of susceptibility genes to TAK. The lack of evidence for associations of non-HLA genes with TAK so far is attributable to the lack of GWASs of TAK performed to date. Low prevalence of this disease had made it difficult to collect DNA samples to obtain sufficient power to detect susceptibility genes and perform a GWAS. Previous studies have revealed that the IL12B region was associated with a wide variety of autoimmune disorders and infectious diseases, including psoriasis21–23 (MIM 177900), ankylosing spondylitis24 (MIM 106300), Crohn disease25 (CD [MIM 266600]), ulcerative colitis26 (UC [MIM 191390]), and leprosy27 (MIM 609888). rs6871626 showed a significant association with UC and leprosy over the genome-wide significance. Notably, rs6871626 A allele is susceptible to UC but protective against leprosy. A previous study from Turkey reported a suggestive association of TAK with rs3212227 in the 3′ UTR of the IL12B region.28 rs3212227 is not in strong LD with rs6871626 in the Japanese population (r2 = 0.11) and in Europeans (r2 = 0.06) because of a recombination hot spot adjacent to rs6871626 (Figure 2A). In fact, an imputed association of rs3212227 with TAK in the current study resulted in only a suggestive association (p = 0.0027). There is a possibility that rs6871626 was responsible for the suggestive association between rs3212227 and TAK reported in the Turkish population. The association between gene expression and SNPs in the IL12B region appears to be complicated and inconsistent across different studies. rs3212227 in the 3′ UTR and rs17860508, an ins/del polymorphism in the promoter region of IL12B, were shown to have potential effects on the gene expression.29,30 However, the previous studies showed that the association patterns varied according to the cell type and the protocol used for stimulation.31–33 No previous report analyzed the effects of rs6871626 on the gene expression of IL12B. Although our in silico analysis failed to show the effects of rs6871626 on IL12B expression (data not shown, see Subjects and Methods), specific cell types or stimulus could lead to a significant association. Because a recent study showed that a haplotype of SNPs in the IL12B region could influence the gene and protein expression of IL12B,22 a combination of rs6871626 and other SNPs in the IL12B region might lead to consistent results.
The associations between rs6871626 and clinical manifestations of TAK suggest the fundamental effects of IL-12p40 protein on TAK progression as well as TAK onset. We found that HLA-B∗52:01 was associated with AR as reported previously (p = 0.00014). This finding supported the accuracy of our data. Although the risk allele of rs6871626 was associated with a significant dose-dependent increase in risk and severity of AR and in circulating CRP levels (p = 0.013, 0.030, and 0.023, respectively), these associations were more evident in a recessive manner. This raises a possibility that those who are homozygous for rs6871626 have strong disease activity that exceeds the additive disease activity of cases with single risk alleles, leading to severe destruction of aortic valve. Genetic variations in IL12B are known to influence the risk of psoriasis21–23 and CD.25 Because ustekinumab, a monoclonal antibody against IL-12p40, is an effective treatment for both diseases,34,35 our findings would raise a possibility of its therapeutic use for TAK by targeting the IL-12/23 pathway. A previous study reported that the level of IL-12 protein was increased in TAK cases compared to healthy populations,36 whereas there have been no reports addressing the circulating levels of IL-23 in TAK cases. IL-12 directly leads to type 1 helper T cell proliferation37 and IL-23 upregulates IL-17 production and supports survival of activated Th17 cells.38 Further analyses addressing circulating T cells in individuals with TAK or cell types infiltrating the artery specimen obtained from cases would provide clues to specify a critical pathway in TAK pathology.
The synergistic effect between rs6871626 and HLA-B∗52:01 was notable. Those carrying both risk alleles had OR of 6.00 in comparison with those not carrying any risk alleles. Combination of rs6871626 and HLA-B∗52:01 showed tendency of severe clinical phenotypes. Thus, we assume that increase of subjects and extraction of subjects who are homozygous for rs6871626 and positive for HLA-B∗52:01 would provide evidence for significant effects of the combination on the disease phenotypes. The synergistic effect of these two loci raises a possibility that immune-related cells that recognize a yet-to-be-determined antigen through HLA-B∗52:01 can be overactivated by IL-12/23 whose expression or function is modulated by rs6871626. In vitro analysis of immune-related cells from cases with TAK or healthy individuals would provide functional evidence of this synergistic role in the TAK pathogenesis.
rs665268 is a missense mutation of MLX that alters the 139th glutamine to arginine (Gln139Arg). MLX is a member of the basic helix-loop-helix leucine zipper (bHLH-Zip) transcription factor family and regulates gene expression by forming heterodimers with Mad protein.39 The 17q21 region, whose associations with other autoimmune diseases including psoriasis40 and CD41 were shown, contains a number of genes including immune-related genes and polymorphisms that are in strong LD with each other (Figure 2B), so the corresponding gene to TAK susceptibility was inconclusive. Because risk allele frequency of rs665268 is comparable to that of rs6871626, the lack of associations between rs665268 and clinical manifestations and the weaker interaction between rs665268 and HLA-B∗52:01 compared to rs6871626 might be a reflection of the milder effect of rs665268 on TAK progression. No interaction was observed between rs665268 and rs6871626 (data not shown).
We set the relatively low cut-off value of imputation score (Rsq ≥ 0.3) in the imputation analysis to increase sensitivity at the expense of specificity, but we failed to find other candidates of susceptibility loci. Another imputation analysis based on the data from the 1000 Genomes Project42 revealed the same signals as the current study (data not shown). However, because the array used in the current study focused on SNPs in exons or nearby genes, it did not fully cover the whole genome with dense markers even in imputation analysis. There is a possibility that other SNPs not tagged by the markers on the array are associated with TAK. When the associations in the HLA locus were conditioned by rs9263739 or rs4947248, the most significantly associated SNPs, suggestive association signals in this locus could still be observed (the smallest p value = 5.5 × 10−5, data not shown). The use of arrays with denser markers especially in intergene regions and using an increased number of cases could lead to the discovery of other susceptibility regions or independent associations in the HLA locus. Considering that both of the non-HLA susceptibility loci to TAK found in the current study are also associated with psoriasis and inflammatory bowel diseases, further analysis of TAK susceptibility genes would reveal other overlapping loci and common autoimmune mechanisms between TAK and other autoimmune diseases. It is feasible to analyze whether these two loci are associated with TAK and whether the interactions are observed in other populations.
Taken together, the current study identified two susceptibility genes to TAK and provided evidence of a common immunological pathway exerted by the IL12B region that is involved in the etiology of TAK and other autoimmune disorders and of its synergistic role with HLA in the susceptibility to TAK.
Acknowledgments
We’d like to thank all the individuals with TAK who gave their blood samples and medical staffs to help us for this study; Miki Kokubo for her excellent technique to extract DNA; Kayo Umemoto for coordination of meetings to obtain blood samples; and Masashi Akizuki for his help to collect DNA samples. This study is supported by Kyoto University Step-up grant, Grants-in-aid from Research on rare and intractable diseases, the Ministry of Health, Labor, and Welfare of Japan and from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, grants from SENSHIN Medical Research Foundation (to T. Matsumura), and the Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Program) from the Japan Society for the Promotion of Science (to R.N.).
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Gene Expression Omnibus (GEO), http://www.ncbi.nlm.nih.gov/geo/
Genevar (Gene Expression Variation), http://www.sanger.ac.uk/resources/software/genevar/
International HapMap Project, http://hapmap.ncbi.nlm.nih.gov/
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org/
R statistical software, http://www.r-project.org/
References
- 1.Isobe M. Takayasu arteritis revisited: Current diagnosis and treatment. Int. J. Cardiol. 2013 doi: 10.1016/j.ijcard.2013.01.022. Published online February 13, 2013. [DOI] [PubMed] [Google Scholar]
- 2.Dreyer L., Faurschou M., Baslund B. A population-based study of Takayasu's arteritis in eastern Denmark. Clin. Exp. Rheumatol. 2011;29(1, Suppl 64):S40–S42. [PubMed] [Google Scholar]
- 3.Jennette J.C., Falk R.J., Bacon P.A., Basu N., Cid M.C., Ferrario F., Flores-Suarez L.F., Gross W.L., Guillevin L., Hagen E.C. 2012 revised International Chapel Hill Consensus Conference Nomenclature of Vasculitides. Arthritis Rheum. 2013;65:1–11. doi: 10.1002/art.37715. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida M., Kimura A., Katsuragi K., Numano F., Sasazuki T. DNA typing of HLA-B gene in Takayasu’s arteritis. Tissue Antigens. 1993;42:87–90. doi: 10.1111/j.1399-0039.1993.tb02242.x. [DOI] [PubMed] [Google Scholar]
- 5.Yajima M., Numano F., Park Y.B., Sagar S. Comparative studies of patients with Takayasu arteritis in Japan, Korea and India—comparison of clinical manifestations, angiography and HLA-B antigen. Jpn. Circ. J. 1994;58:9–14. doi: 10.1253/jcj.58.9. [DOI] [PubMed] [Google Scholar]
- 6.Sahin Z., Bıcakcıgil M., Aksu K., Kamali S., Akar S., Onen F., Karadag O., Ozbalkan Z., Ates A., Ozer H.T., Turkish Takayasu Study Group Takayasu’s arteritis is associated with HLA-B∗52, but not with HLA-B∗51, in Turkey. Arthritis Res. Ther. 2012;14:R27. doi: 10.1186/ar3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vargas-Alarcón G., Flores-Domínguez C., Hernández-Pacheco G., Zuñiga J., Gamboa R., Soto M.E., Granados J., Reyes P.A. Immunogenetics and clinical aspects of Takayasu’s arteritis patients in a Mexican Mestizo population. Clin. Exp. Rheumatol. 2001;19:439–443. [PubMed] [Google Scholar]
- 8.Arend W.P., Michel B.A., Bloch D.A., Hunder G.G., Calabrese L.H., Edworthy S.M., Fauci A.S., Leavitt R.Y., Lie J.T., Lightfoot R.W., Jr. The American College of Rheumatology 1990 criteria for the classification of Takayasu arteritis. Arthritis Rheum. 1990;33:1129–1134. doi: 10.1002/art.1780330811. [DOI] [PubMed] [Google Scholar]
- 9.JCS Joint Working Group. Japanese Circulation Society Guideline for management of vasculitis syndrome (JCS 2008) Circ. J. 2011;75:474–503. doi: 10.1253/circj.cj-88-0007. [DOI] [PubMed] [Google Scholar]
- 10.Yoshimura K., Nakayama T., Sekine A., Matsuda F., Kosugi S., Sugino Y., Yoshimura K., Ogawa O., Nagahama Cohort Research Group Prevalence of postmicturition urinary incontinence in Japanese men: Comparison with other types of incontinence. Int. J. Urol. 2013 doi: 10.1111/iju.12074. Published online January 10, 2013. [DOI] [PubMed] [Google Scholar]
- 11.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersson T., Alfredsson L., Källberg H., Zdravkovic S., Ahlbom A. Calculating measures of biological interaction. Eur. J. Epidemiol. 2005;20:575–579. doi: 10.1007/s10654-005-7835-x. [DOI] [PubMed] [Google Scholar]
- 14.Stranger B.E., Forrest M.S., Dunning M., Ingle C.E., Beazley C., Thorne N., Redon R., Bird C.P., de Grassi A., Lee C. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang T.P., Beazley C., Montgomery S.B., Dimas A.S., Gutierrez-Arcelus M., Stranger B.E., Deloukas P., Dermitzakis E.T. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics. 2010;26:2474–2476. doi: 10.1093/bioinformatics/btq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrews J., Mason J.C. Takayasu’s arteritis—recent advances in imaging offer promise. Rheumatology (Oxford) 2007;46:6–15. doi: 10.1093/rheumatology/kel323. [DOI] [PubMed] [Google Scholar]
- 17.Kitamura H., Kobayashi Y., Kimura A., Numano F. Association of clinical manifestations with HLA-B alleles in Takayasu arteritis. Int. J. Cardiol. 1998;66(Suppl 1):S121–S126. doi: 10.1016/s0167-5273(98)00159-4. [DOI] [PubMed] [Google Scholar]
- 18.Warnes C.A., Williams R.G., Bashore T.M., Child J.S., Connolly H.M., Dearani J.A., del Nido P., Fasules J.W., Graham T.P., Jr., Hijazi Z.M., American College of Cardiology. American Heart Association Task Force on Practice Guidelines (Writing Committee to Develop Guidelines on the Management of Adults With Congenital Heart Disease) American Society of Echocardiography. Heart Rhythm Society. International Society for Adult Congenital Heart Disease. Society for Cardiovascular Angiography and Interventions. Society of Thoracic Surgeons ACC/AHA 2008 guidelines for the management of adults with congenital heart disease: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines (Writing Committee to Develop Guidelines on the Management of Adults With Congenital Heart Disease). Developed in Collaboration With the American Society of Echocardiography, Heart Rhythm Society, International Society for Adult Congenital Heart Disease, Society for Cardiovascular Angiography and Interventions, and Society of Thoracic Surgeons. J. Am. Coll. Cardiol. 2008;52:e143–e263. doi: 10.1016/j.jacc.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Okada Y., Takahashi A., Ohmiya H., Kumasaka N., Kamatani Y., Hosono N., Tsunoda T., Matsuda K., Tanaka T., Kubo M. Genome-wide association study for C-reactive protein levels identified pleiotropic associations in the IL6 locus. Hum. Mol. Genet. 2011;20:1224–1231. doi: 10.1093/hmg/ddq551. [DOI] [PubMed] [Google Scholar]
- 20.Mizuki N., Meguro A., Ota M., Ohno S., Shiota T., Kawagoe T., Ito N., Kera J., Okada E., Yatsu K. Genome-wide association studies identify IL23R-IL12RB2 and IL10 as Behçet’s disease susceptibility loci. Nat. Genet. 2010;42:703–706. doi: 10.1038/ng.624. [DOI] [PubMed] [Google Scholar]
- 21.Tsunemi Y., Saeki H., Nakamura K., Sekiya T., Hirai K., Fujita H., Asano N., Kishimoto M., Tanida Y., Kakinuma T. Interleukin-12 p40 gene (IL12B) 3′-untranslated region polymorphism is associated with susceptibility to atopic dermatitis and psoriasis vulgaris. J. Dermatol. Sci. 2002;30:161–166. doi: 10.1016/s0923-1811(02)00072-5. [DOI] [PubMed] [Google Scholar]
- 22.Johnston A., Xing X., Swindell W.R., Kochkodan J., Riblett M., Nair R.P., Stuart P.E., Ding J., Voorhees J.J., Elder J.T., Gudjonsson J.E. Susceptibility-associated genetic variation at IL12B enhances Th1 polarization in psoriasis. Hum. Mol. Genet. 2013;22:1807–1815. doi: 10.1093/hmg/ddt034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cargill M., Schrodi S.J., Chang M., Garcia V.E., Brandon R., Callis K.P., Matsunami N., Ardlie K.G., Civello D., Catanese J.J. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am. J. Hum. Genet. 2007;80:273–290. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans D.M., Spencer C.C., Pointon J.J., Su Z., Harvey D., Kochan G., Oppermann U., Dilthey A., Pirinen M., Stone M.A., Spondyloarthritis Research Consortium of Canada (SPARCC) Australo-Anglo-American Spondyloarthritis Consortium (TASC) Wellcome Trust Case Control Consortium 2 (WTCCC2) Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat. Genet. 2011;43:761–767. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parkes M., Barrett J.C., Prescott N.J., Tremelling M., Anderson C.A., Fisher S.A., Roberts R.G., Nimmo E.R., Cummings F.R., Soars D., Wellcome Trust Case Control Consortium Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat. Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson C.A., Boucher G., Lees C.W., Franke A., D’Amato M., Taylor K.D., Lee J.C., Goyette P., Imielinski M., Latiano A. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H., Irwanto A., Tian H., Fu X., Yu Y., Yu G., Low H., Chu T., Li Y., Shi B. Identification of IL18RAP/IL18R1 and IL12B as leprosy risk genes demonstrates shared pathogenesis between inflammation and infectious diseases. Am. J. Hum. Genet. 2012;91:935–941. doi: 10.1016/j.ajhg.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saruhan-Direskeneli G., Biçakçigil M., Yilmaz V., Kamali S., Aksu K., Fresko I., Akkoç N., Kiraz S., Ozer H.T., Tunç E., Rheumatology Education and Research Society Vasculitis Study Group Interleukin (IL)-12, IL-2, and IL-6 gene polymorphisms in Takayasu’s arteritis from Turkey. Hum. Immunol. 2006;67:735–740. doi: 10.1016/j.humimm.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Morahan G., Huang D., Wu M., Holt B.J., White G.P., Kendall G.E., Sly P.D., Holt P.G. Association of IL12B promoter polymorphism with severity of atopic and non-atopic asthma in children. Lancet. 2002;360:455–459. doi: 10.1016/S0140-6736(02)09676-9. [DOI] [PubMed] [Google Scholar]
- 30.Stanilova S., Miteva L. Taq-I polymorphism in 3’UTR of the IL-12B and association with IL-12p40 production from human PBMC. Genes Immun. 2005;6:364–366. doi: 10.1038/sj.gene.6364213. [DOI] [PubMed] [Google Scholar]
- 31.Basu M., Das T., Ghosh A., Majumder S., Maji A.K., Kanjilal S.D., Mukhopadhyay I., Roychowdhury S., Banerjee S., Sengupta S. Gene-gene interaction and functional impact of polymorphisms on innate immune genes in controlling Plasmodium falciparum blood infection level. PLoS ONE. 2012;7:e46441. doi: 10.1371/journal.pone.0046441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yanagihori H., Oyama N., Nakamura K., Mizuki N., Oguma K., Kaneko F. Role of IL-12B promoter polymorphism in Adamantiades-Behcet’s disease susceptibility: An involvement of Th1 immunoreactivity against Streptococcus Sanguinis antigen. J. Invest. Dermatol. 2006;126:1534–1540. doi: 10.1038/sj.jid.5700203. [DOI] [PubMed] [Google Scholar]
- 33.Yilmaz V., Yentür S.P., Saruhan-Direskeneli G. IL-12 and IL-10 polymorphisms and their effects on cytokine production. Cytokine. 2005;30:188–194. doi: 10.1016/j.cyto.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 34.Yeilding N., Szapary P., Brodmerkel C., Benson J., Plotnick M., Zhou H., Goyal K., Schenkel B., Giles-Komar J., Mascelli M.A., Guzzo C. Development of the IL-12/23 antagonist ustekinumab in psoriasis: past, present, and future perspectives—an update. Ann. N Y Acad. Sci. 2012;1263:1–12. doi: 10.1111/j.1749-6632.2012.06670.x. [DOI] [PubMed] [Google Scholar]
- 35.Sandborn W.J., Gasink C., Gao L.L., Blank M.A., Johanns J., Guzzo C., Sands B.E., Hanauer S.B., Targan S., Rutgeerts P., CERTIFI Study Group Ustekinumab induction and maintenance therapy in refractory Crohn’s disease. N. Engl. J. Med. 2012;367:1519–1528. doi: 10.1056/NEJMoa1203572. [DOI] [PubMed] [Google Scholar]
- 36.Verma D.K., Tripathy N.K., Verma N.S., Tiwari S. Interleukin 12 in Takayasu’s arteritis: plasma concentrations and relationship with disease activity. J. Rheumatol. 2005;32:2361–2363. [PubMed] [Google Scholar]
- 37.Murphy K.M., Reiner S.L. The lineage decisions of helper T cells. Nat. Rev. Immunol. 2002;2:933–944. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- 38.Bettelli E., Carrier Y., Gao W., Korn T., Strom T.B., Oukka M., Weiner H.L., Kuchroo V.K. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 39.Cairo S., Merla G., Urbinati F., Ballabio A., Reymond A. WBSCR14, a gene mapping to the Williams-Beuren syndrome deleted region, is a new member of the Mlx transcription factor network. Hum. Mol. Genet. 2001;10:617–627. doi: 10.1093/hmg/10.6.617. [DOI] [PubMed] [Google Scholar]
- 40.Tsoi L.C., Spain S.L., Knight J., Ellinghaus E., Stuart P.E., Capon F., Ding J., Li Y., Tejasvi T., Gudjonsson J.E., Collaborative Association Study of Psoriasis (CASP) Genetic Analysis of Psoriasis Consortium. Psoriasis Association Genetics Extension. Wellcome Trust Case Control Consortium 2 Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat. Genet. 2012;44:1341–1348. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franke A., McGovern D.P., Barrett J.C., Wang K., Radford-Smith G.L., Ahmad T., Lees C.W., Balschun T., Lee J., Roberts R. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A., 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


