Abstract
Takayasu arteritis is a rare inflammatory disease of large arteries. The etiology of Takayasu arteritis remains poorly understood, but genetic contribution to the disease pathogenesis is supported by the genetic association with HLA-B∗52. We genotyped ∼200,000 genetic variants in two ethnically divergent Takayasu arteritis cohorts from Turkey and North America by using a custom-designed genotyping platform (Immunochip). Additional genetic variants and the classical HLA alleles were imputed and analyzed. We identified and confirmed two independent susceptibility loci within the HLA region (r2 < 0.2): HLA-B/MICA (rs12524487, OR = 3.29, p = 5.57 × 10−16) and HLA-DQB1/HLA-DRB1 (rs113452171, OR = 2.34, p = 3.74 × 10−9; and rs189754752, OR = 2.47, p = 4.22 × 10−9). In addition, we identified and confirmed a genetic association between Takayasu arteritis and the FCGR2A/FCGR3A locus on chromosome 1 (rs10919543, OR = 1.81, p = 5.89 × 10−12). The risk allele in this locus results in increased mRNA expression of FCGR2A. We also established the genetic association between IL12B and Takayasu arteritis (rs56167332, OR = 1.54, p = 2.18 × 10−8).
Main Text
Takayasu arteritis (MIM 207600) is a rare large-vessel vasculitis characterized by nonspecific inflammation of the aorta and its major branches.1 This inflammatory process leads to blood vessel wall thickening, fibrosis, progressive occlusion, and dilatation, resulting in limb- and organ-threatening ischemia and the characteristic absence of pulses in the extremities. Other nonspecific symptoms may include weight loss, fever, night sweats, fatigue, arthralgia, and myalgia.2 The disease typically presents in the second or third decade, but approximately 20% of cases occur before the age of 20 and 20% after the age of 40.3 The disease is most prevalent in Far East Asia, India, and Mexico, and tends to be less common in European-derived populations. The incidence in North America has been reported as 2.6 cases per million per year.4 Takayasu arteritis is more common in women; however, the female-to-male ratio is dependent upon ethnicity and geographical location.2,5
The etiology and pathogenesis of Takayasu arteritis is largely unknown. The evidence for a genetic contribution to Takayasu arteritis comes from ethnic differences in the prevalence of the disease, familial aggregation, and the confirmed genetic association within the HLA region.5 The association with HLA-B∗52 is the most significant association in the HLA region in Japanese, but other HLA alleles (including HLA-B∗39, HLA-DRB1∗1502, and HLA-DRB1∗0405) have also been associated with the disease in Japanese.1 The association with HLA-B∗52 has been confirmed in cases from Korea,6 India,7 Thailand,8 and Turkey.9 Interestingly, individuals with Takayasu arteritis with HLA-B∗52 tend to have more severe disease with a higher incidence of left ventricular wall abnormalities and aortic regurgitation and an earlier disease onset.9,10 In contrast, the presence of HLA-B∗39 allele is associated with the development of renal artery stenosis in Takayasu arteritis.11 Genetic associations with polymorphisms within cytokine genes such as IL2 (MIM 147680), IL6 (MIM 147620), and IL12B (MIM 161561) have been recently reported in Turkish cases.12
We studied two independent cohorts of Takayasu arteritis cases and controls from Turkey and North America. The Turkish cohort consisted of 339 cases and 516 healthy controls, and the North American cohort consisted of 112 European-American (EA) cases enrolled in the Vasculitis Clinical Research Consortium Longitudinal Study of Takayasu Arteritis and 599 EA controls. All cases fulfilled the 1990 American College of Rheumatology classification criteria for Takayasu arteritis.13 Our study was approved by the Institutional Review Boards and the Ethics Committees at our institutions, and all study participants signed an informed written consent.
Genotyping was performed with the Human Immuno DNA Analysis BeadChip Kit (Immunochip), which includes 196,524 genetic variants primarily in genetic susceptibility loci previously shown to confer risk for autoimmune or inflammatory diseases. Rigorous quality-control measures were applied and samples were removed from the analysis based on population stratification by principal components analysis (>4 standard deviations), identity by descent (IBD > 0.4), and autosomal heterozygosity (>2 standard deviation around the mean). Principal components analysis was performed with Eigenstrat.14 IBD and heterozygosity analysis were performed with PLINK.15 Genotyped markers were filtered for minor allele frequency (MAF > 0.01), genotype success rate (GSR > 0.9), and Hardy-Weinberg equilibrium p value (HWPControls > 0.01, HWPCases > 0.0001). In addition, markers with differential missingness between cases and controls (p < 0.05) were also excluded from the analysis. After applying the aforementioned quality-control measures, a total of 124,393 variants in the Turkish cohort and 124,312 in the EA cohort were evaluated. A total of 305 Turkish cases and 483 controls and 106 EA cases and 553 controls were included in the final analysis. A set of 3,120 variants that are not associated with autoimmunity and also included on the Immunochip were filtered for MAF, GSR, HWP, and differential missingness as above, and then used as “null” variants to calculate genomic control lambda (λGC) in our cohorts. We detected no to minimal residual population stratification in our cohorts (λGC in the Turkish cohort = 1.00 and in the EA cohort = 1.06). Meta-analysis was performed with PLINK and haplotype analysis was performed with Haploview v.4.2.16 Adjusted associations between SNPs were performed with logistic regression in PLINK.
The 1000 Genomes Project data were used to impute additional genetic variants in the associated loci that we discovered, from a combined reference panel consisting of 1,092 individuals.17 PLINK-formatted files were converted to the appropriate format with GTOOL and imputation was performed with Impute 2.18 A stringent posterior probability imputation threshold of 0.9 was applied. Variants with MAF > 1%, imputation success rate > 90%, and HWP > 0.0001 in controls were included in the analysis.
Imputation of classical HLA alleles was performed with 9,156 genotyped variants in the HLA region. These variants were first cleaned, aligned, and phased with the HLA∗IMP software package according to established quality-control measures.19 Phased haplotypes consisting of 5,776 variants were used to impute the classical HLA alleles in HLA-A (MIM 142800), HLA-B (MIM 142830), HLA-C (MIM 142840), HLA-DQA1 (MIM 146880), HLA-DQB1 (MIM 604305), and HLA-DRB1 (MIM 142857). An imputation posterior probability of 0.9 was applied to assign allele calls to phased and imputed haplotypes. Genetic association analysis for imputed HLA alleles was performed in PLINK with only samples in which both alleles were successfully imputed in each locus. Adjusted associations between SNPs and classical HLA alleles were calculated with logistic regression in PLINK.
We detected evidence for genetic association in two loci with a genome-wide level of significance (p < 5 × 10−8) in Takayasu arteritis (Figure 1). The most significant genetic association detected was in the HLA region (rs12524487; pmeta = 5.57 × 10−16, pTurkish = 4.42 × 10−7, pEA = 4.05 × 10−12). We also detected a genetic association in the region that includes the genes encoding Fc-gamma receptor IIA (FCGR2A [MIM 146790]) and Fc-gamma receptor IIIA (FCGR3A [MIM 146740]). Two additional association effects that confer risk for Takayasu arteritis but did not pass the threshold for genome-wide significance were detected on chromosome 21 downstream of the gene encoding proteasome (prosome, macropain) assembly chaperone 1 (PSMG1 [MIM 605296]) and on chromosome 5 in the gene encoding for the P40 subunit of the interleukin-12 (IL-12) and IL-23 (IL12B) (Figure 1).
Figure 1.
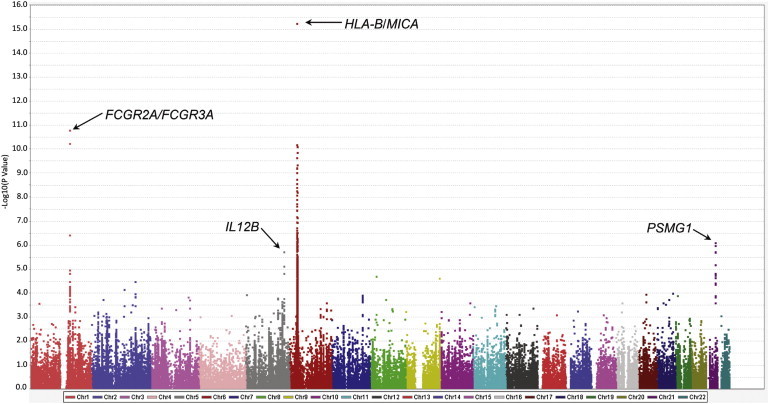
Manhattan Plot Showing the Results of a Meta-analysis between the Two Cohorts of Cases with Takayasu Arteritis and Controls Included in This Study
The −log10 p values for each genetic variant evaluated are plotted against its chromosomal location.
To further fine-map and localize the genetic association detected in the HLA region, we first imputed additional genetic variants up to the density of the 1000 Genomes Project in the region extending from chr6: 28,889,000 to chr6: 33,000,000 (UCSC Genome Browser build hg19) as previously described.20 A meta-analysis of genotyped or imputed variants in the HLA extended region in both cohorts was performed (Figure 2). Two independent genetic associations were detected in the HLA class I and HLA class II regions (r2 < 0.2; Table S1 available online). The most significant association was with a variant in the HLA-B/MICA locus between HLA-B and MICA (rs12524487, p = 5.57 × 10−16). An additional effect was observed in the HLA-DQB1/HLA-DRB1 locus (rs113452171, p = 3.74 × 10−9, and rs189754752, p = 4.22 × 10−9) (Table 1, Figures 2, S1, and S2). Conditional logistic regression confirmed that these two effects in the HLA class I and HLA class II regions are independent genetic susceptibility loci for Takayasu arteritis (Table S2). The most significant variants in the HLA-DQB1/HLA-DRB1 locus, however, are in tight linkage disequilibrium (r2 = 0.907 and 1, in the Turkish and EA cohorts, respectively). Therefore, the genetic susceptibility locus in HLA class II probably represents one genetic effect and could not be further localized to an individual genetic variant.
Figure 2.
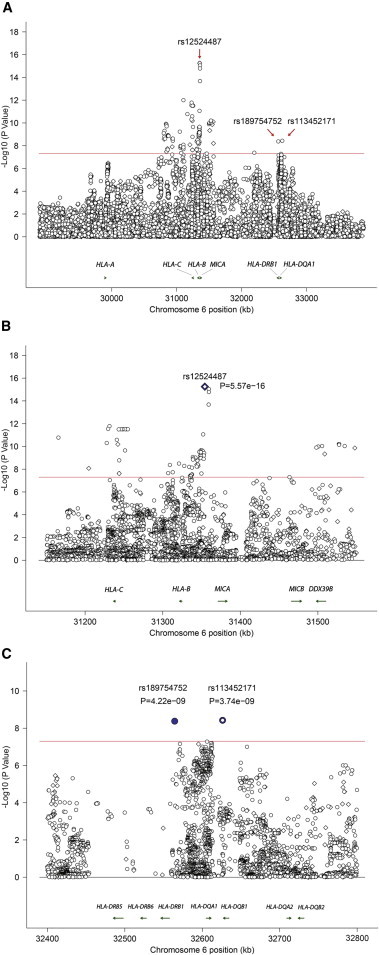
Results of the Meta-analysis Showing Genotyped and Imputed Variants in the HLA Region
Two independent genetic effects are detected (r2 < 0.2).
(A) The entire HLA region evaluated in this study.
(B and C) Regional plots for the meta-analysis results in the HLA-B/MICA locus (B) and the HLA-DQB1/HLA-DRB1 locus (C).
The red line represents the genome-wide level of significance (p = 5 × 10−8). Genotyped and imputed variants are shown in diamonds and circles, respectively.
Table 1.
Genetic Association Results Showing the Most Significant Variant in Each of the Two Independent Genetic Effects Observed in the HLA Region in Cases with Takayasu Arteritis
| SNP (Minor Allele) |
Turkish |
European-American |
Meta-analysis |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAFCase | MAFControl | OR | 95% CI | p | MAFCase | MAFControl | OR | 95% CI | p | OR | p | Q Statistic p | |
| HLA-B/MICA | |||||||||||||
| rs12524487 (T) | 0.105 | 0.04 | 2.79 | (1.85–4.21) | 4.42 × 10−7 | 0.217 | 0.067 | 3.85 | (2.57–5.76) | 4.05 × 10−12 | 3.29 | 5.57 × 10−16 | 0.27 |
| HLA-DQB1/HLA-DRB1 | |||||||||||||
| rs113452171 (T) | 0.185 | 0.093 | 2.21 | (1.64–2.99) | 1.42 × 10−7 | 0.048 | 0.015 | 3.43 | (1.53–7.66) | 0.0015 | 2.34 | 3.74 × 10−9 | 0.32 |
| rs189754752 (G) | 0.163 | 0.0795 | 2.26 | (1.63–3.13) | 5.46 × 10−7 | 0.0529 | 0.013 | 4.24 | (1.90–9.47) | 0.00014 | 2.47 | 4.22 × 10−9 | 0.16 |
Abbreviations are as follows: MAF, minor allele frequency; OR, odds ratio; CI, confidence interval.
To localize the genetic effect tagged by the variant rs12524487 in the HLA-B/MICA locus, we identified all genotyped and imputed variants that are in LD (r2 > 0.7) with rs12524487 in the two cohorts included in this study (Tables S3 and S4). In the EA cohort, the genetic effect in rs12524487 remained significant when adjusted for the effect in any of these additional variants, with the exception of variants with very strong LD (≥0.9) in which case conditional analysis was less informative. However, in the Turkish cohort, there were only five additional genetic variants in LD (r2 > 0.7) with rs12524487. Conditional analysis suggests that the effect in two of these variants is dependent on rs12524487, although very high LD (r2 ≥ 0.99) in the other three variants made conditional analysis impossible. Taken together, transancestral mapping and conditional regression analysis fine maps the effect tagged by rs12524487 in the HLA class I region to four genetic variants (rs12524487, chr6: 31,359,160:D, chr6: 31,359,428:D, and rs141706050). All these variants are located in the intergenic region between HLA-B and MICA (Tables S3 and S4).
Next, we used the genotyped variants in the HLA region to impute the HLA classical alleles in both cohorts and performed genetic association analysis to determine the association between classical HLA alleles and Takayasu arteritis. We detected genetic association with two classical alleles with a genome-wide level of significance (HLA-B∗5201, p = 8.98 × 10−10, and HLA-Cw∗1202, p = 4.01 × 10−10) (Table S5). HLA-B∗5201 and HLA-Cw∗1202 are in high linkage disequilibrium (r2 = 0.827 and 0.873 in the Turkish and the EA cohorts, respectively) and therefore probably represent the same genetic effect. The imputed allele frequency of HLA-B∗5201 in normal Turkish controls in our study (3.4%) is very similar to the reported genotyped allele frequency of HLA-B∗52 in Turkey (3.5%).21
We performed conditional regression analysis to determine the relationship between the genetic effect in the HLA-B/MICA locus tagged by rs12524487 and the classical HLA alleles HLA-B∗5201/HLA-Cw∗1202 (Table S6). In the EA cohort, the effect in HLA-B∗5201 and HLA-Cw∗1202 remained significant after adjusting for rs12524487 and the effect in rs12524487 remained significant after adjusting for HLA-B∗5201 and HLA-Cw∗1202. On the other hand, both effects are significantly attenuated when adjusted for one another in the Turkish cohort (Table S6). These findings are also supported by a relatively high LD between rs12524487 and the HLA-B∗5201 in the Turkish cohort (r2 = 0.674) and low LD between them in the EA cohort (r2 = 0.154). These data suggest that both rs12524487 and the HLA classical alleles in the HLA class I region contribute to the genetic effect in this locus.
Other suggestive associations (p < 5 × 10−5) between classical HLA alleles and Takayasu arteritis in the meta-analysis included HLA-B∗1302 and HLA-Cw∗0701. The suggested genetic association with HLA-B∗1302 confers risk for Takayasu arteritis, whereas HLA-Cw∗0701 seems to exert a protective effect against the disease in both the Turkish and the EA cohorts. These effects, however, do not pass the threshold for genome-wide significance and will require independent confirmation.
To fine map and localize the genetic association between Takayasu arteritis and FCGR2A/FCGR3A, we imputed additional genetic variants in this locus up to the 1000 Genomes Project density. The most significant association in this locus was detected with the SNP rs10919543 (pmeta = 5.89 × 10−12, pTurkish = 1.49 × 10−8, pEA = 6.71 × 10−5) (Figure 3). We identified two and three additional associated genetic variants that are in LD (r2 > 0.7) with the index SNP rs10919543 in this locus in the Turkish and the EA cohorts, respectively (Table 2). Very high linkage disequilibrium precluded conditional analysis among these variants and therefore this genetic effect could not be further localized to a single variant. The index SNP rs10919543 is not included in available expression quantitative trait loci data sets. However, rs2099684, which is in high linkage disequilibrium with rs10919543 (r2 = 0.977 and 0.98 in the Turkish and EA cohorts, respectively) (Table 2), is included in the GENe Expression VARiation (Genevar) expression quantitative trait loci database.22 The disease-risk variant in rs2099684 (allele G) is associated with increased mRNA expression of FCGR2A in lymphoblastoid cells (p = 3 × 10−4) (Figure S3).
Figure 3.
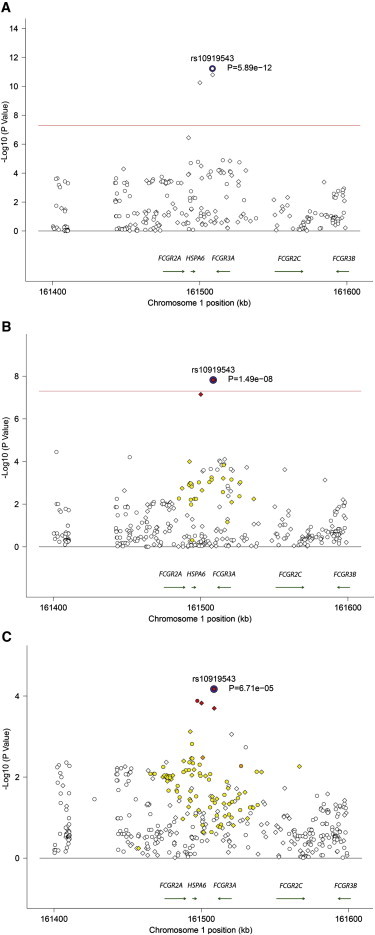
Genetic Association Results in the FCGR2A/FCGR3A Region in Takayasu Arteritis
(A) Association results from the meta-analysis.
(B and C) Association results in the Turkish (B) and the European-American (C) cohorts.
The red line represents the genome-wide level of significance (p = 5 × 10−8). Genotyped and imputed variants are shown in diamonds and circles, respectively. The colors represent r2 values with the index SNP in each locus (white, r2 < 0.2; yellow, r2 ≥ 0.2–0.5; orange, r2 ≥ 0.5–0.8; red, r2 ≥ 0.8).
Table 2.
Genetic Association Results of the Most Significant SNP, rs10919543, in the FCGR2A/FCGR3A Locus and All Genetic Variants in High LD with This SNP
| SNP (Minor Allele) |
Turkish |
European-American |
Meta-analysis |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAFCase | MAFControl | OR | 95% CI | p | MAFCase | MAFControl | OR | 95% CI | p | OR | pmeta | Q Statistic p | |
| rs10919543 (G) | 0.559 | 0.413 | 1.80 | (1.47–2.22) | 1.49 × 10−8 | 0.462 | 0.320 | 1.82 | (1.35–2.46) | 6.71 × 10−5 | 1.81 | 5.89 × 10−12 | 0.952 |
| rs10919544 (C) | 0.560 | 0.413 | 1.81 | (1.47–2.22) | 1.50 × 10−8 | 0.458 | 0.325 | 1.75 | (1.30–2.36) | 0.0002012 | 1.79 | 1.61 × 10−11 | 0.874 |
| rs2099684 (G) | 0.554 | 0.415 | 1.75 | (1.43–2.15) | 7.17 × 10−8 | 0.458 | 0.323 | 1.77 | (1.32–2.39) | 0.0001497 | 1.76 | 5.70 × 10−11 | 0.951 |
| rs141730227 (T) | NA | NA | NA | NA | NA | 0.462 | 0.324 | 1.79 | (1.33–2.42) | 0.000131 | NA | NA | NA |
NA: The SNP rs141730227 was not evaluated in the Turkish cohort because the genotype success rate (GSR) of 0.898 was below our GSR threshold for SNP inclusion. However, when analyzed, it did show evidence for association (OR = 1.735 [1.397–2.155], p = 5.754 × 10−7).
The effect detected on chromosome 21q22 did not pass the threshold for genome-wide significance and is primarily derived from the EA cohort. The most significantly associated variant in this locus (p = 4.39 × 10−7) is an imputed indel (chr21: 40,462,378:D) located downstream of PSMG1 (Figure S4). Genetic association results in all genotyped or imputed markers in this locus that are in linkage disequilibrium (r2 > 0.7) with the index indel variant are presented in Table S7.
The most significant variant associated with Takayasu arteritis on chromosome 5 in our study is rs56167332 (p = 1.88 × 10−6), located ∼70 kb upstream of IL12B (Figure S5). This effect is primarily detected in the Turkish cohort, and six additional variants in LD (r2 > 0.7) with rs56167332 also show evidence for suggestive association (Table S8). Conditional analysis suggests that rs56167332 or rs6871626 (r2 = 0.98) explain the genetic effect in this locus. The genetic association in all the other variants in LD (r2 > 0.7) with rs56167332 became nonsignificant upon conditioning on rs56167332 (Table S8). There were 30 variants in LD (r2 > 0.7) with rs56167332 in the EA cohort, but only three variants (including rs56167332) showed evidence for a suggestive association (p < 0.05). Meta-analysis results for these three variants in both cohorts are presented in Table S9. To further determine whether the genetic association between IL12B and Takayasu arteritis can be established with a genome-wide level of significance, we imputed the variant rs56167332 into an additional set of 1,278 healthy controls from Turkey (dbGaP Study Accession: phs000272.v1.p1). Genetic association analysis between rs56167332 and Takayasu arteritis in our Turkish cohort combined with the additional Turkish controls was performed (OR = 1.60, 95% CI = 1.34–1.91, p = 1.34 × 10−7). A meta-analysis with the EA cohort established this genetic association in IL12B with a genome-wide level of significance (p = 2.18 × 10−8, OR = 1.54, Q statistic p = 0.40).
Our data reveal multiple genetic susceptibility loci for Takayasu arteritis, a rare and poorly understood inflammatory disease of the large arteries. This extensive genotyping effort in Takayasu arteritis provides evidence for at least two independent genetic susceptibility loci in the HLA class I and class II regions and identifies and establishes a genetic association in the FCGR2A/FCGR3A and IL12B loci. Further analysis with imputed HLA classical alleles validates and confirms the genetic association with HLA-B∗5201/HLA-Cw∗1202 with a genome-wide level of significance. The genetic association within the HLA class I region remains the most robust genetic susceptibility locus for Takayasu arteritis.
Genetic associations between FCGR2A/FCGR3A and other autoimmune and inflammatory diseases have been well described. For example, the genetic association between this locus and giant cell arteritis has been previously reported in a small cohort from Spain.23 This suggests that these two large-vessel vasculitides can share common genetic susceptibility loci. Indeed, there are notable similarities between Takayasu arteritis and giant cell arteritis in some clinical manifestations, angiographic findings, and histopathological features of arterial lesions.24,25
The association between the FCGR2A/FCGR3A locus and ulcerative colitis is robust and replicated.26,27 Interestingly, retrospective analysis suggests that individuals with Takayasu arteritis might have a higher prevalence of other inflammatory and autoimmune diseases including inflammatory bowel disease.28,29 The genetic susceptibility conferred by variants in FCGR2A/FCGR3A could influence the susceptibility to Takayasu arteritis through altering the immune response against infectious agents that might be involved in the pathogenesis of this disease. Indeed, we show that the risk variant in this locus alters FCGR2A expression. Previous reports suggest enhanced humoral immune response to Mycobacterium tuberculosis antigens, particularly the 65 kD heat shock protein, in Takayasu arteritis compared to controls.30 More recently, genetic sequences of Mycobacterium tuberculosis were detected in aortic tissue from the majority of individuals with Takayasu arteritis, supporting a role for Mycobacterium tuberculosis in the pathogenesis of this disease.31
A genetic association between IL12B and Takayasu arteritis has been previously suggested in a subset of our Turkish cohort consisting of 94 cases and 108 controls.12 Our data established the genetic association in this locus with a genome-wide level of significance.
The genetic effect we detected in chromosome 21q22 requires further validation, because it did not pass the threshold for genome-wide significance in our study. It should be noted that this same locus is also associated with increased susceptibility to ulcerative colitis32 and ankylosing spondylitis,33 two other inflammatory diseases.
In summary, our data indicate that genetic predisposition contributes to the susceptibility for Takayasu arteritis. Genetic susceptibility loci in HLA and non-HLA loci have been revealed with a genome-wide level of significance. Indeed, the putative functional disease-associated variant we identified in FCGR2A/FCGR3A increases FCGR2A expression and might provide a mechanistic link between a suspected role for infectious triggers and genetic predisposition in Takayasu arteritis. The genetic association we established in the IL12B locus (which encodes for the common P40 subunit of IL-12 and IL-23) might have therapeutic implications in Takayasu arteritis. Future studies to further explore the functional consequences of the disease-associated variants upon disease susceptibility and to explore additional genetic susceptibility loci in Takayasu arteritis and other large-vessel vasculitides are warranted.
Acknowledgments
This work was supported by funding from the University of Michigan and the Vasculitis Foundation. Procurement and genotyping of the European-American control samples was supported by the National Institutes of Health grants GM103510, AR053483, AI08714, and AI101914. The Vasculitis Clinical Research Consortium has received support from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (U54AR057319 and U01 AR51874 04), the National Center for Research Resources (U54 RR019497), and the Office of Rare Diseases Research of the National Center for Advancing Translational Sciences.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes, http://browser.1000genomes.org
GTOOL, http://www.well.ox.ac.uk/∼cfreeman/software/gwas/gtool.html
HLA∗IMP, https://oxfordhla.well.ox.ac.uk/hla/
IMPUTE2, http://mathgen.stats.ox.ac.uk/impute/impute_v2.html
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org/
UCSC Genome Browser, http://genome.ucsc.edu
References
- 1.Kobayashi Y., Numano F. 3. Takayasu arteritis. Intern. Med. 2002;41:44–46. doi: 10.2169/internalmedicine.41.44. [DOI] [PubMed] [Google Scholar]
- 2.Johnston S.L., Lock R.J., Gompels M.M. Takayasu arteritis: a review. J. Clin. Pathol. 2002;55:481–486. doi: 10.1136/jcp.55.7.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phillip R., Luqmani R. Mortality in systemic vasculitis: a systematic review. Clin. Exp. Rheumatol. 2008;26(5, Suppl 51):S94–S104. [PubMed] [Google Scholar]
- 4.Hall S., Barr W., Lie J.T., Stanson A.W., Kazmier F.J., Hunder G.G. Takayasu arteritis. A study of 32 North American patients. Medicine (Baltimore) 1985;64:89–99. [PubMed] [Google Scholar]
- 5.Numano F. Hereditary factors of Takayasu arteritis. Heart Vessels Suppl. 1992;7:68–72. doi: 10.1007/BF01744547. [DOI] [PubMed] [Google Scholar]
- 6.Lee S.W., Kwon O.J., Park M.C., Oh H.B., Park Y.B., Lee S.K. HLA alleles in Korean patients with Takayasu arteritis. Clin. Exp. Rheumatol. 2007;25(1, Suppl 44):S18–S22. [PubMed] [Google Scholar]
- 7.Mehra N.K., Jaini R., Balamurugan A., Kanga U., Prabhakaran D., Jain S., Talwar K.K., Sharma B.K. Immunogenetic analysis of Takayasu arteritis in Indian patients. Int. J. Cardiol. 1998;66(Suppl 1):S127–S132. doi: 10.1016/s0167-5273(98)00160-0. [DOI] [PubMed] [Google Scholar]
- 8.Charoenwongse P., Kangwanshiratada O., Boonnam R., Hoomsindhu U. The association between the HLA antigens and Takayasu’s arteritis in Thai patients. Int. J. Cardiol. 1998;66(Suppl 1):S117–S120. doi: 10.1016/s0167-5273(98)00158-2. [DOI] [PubMed] [Google Scholar]
- 9.Sahin Z., Bıcakcıgil M., Aksu K., Kamali S., Akar S., Onen F., Karadag O., Ozbalkan Z., Ates A., Ozer H.T., Turkish Takayasu Study Group Takayasu’s arteritis is associated with HLA-B∗52, but not with HLA-B∗51, in Turkey. Arthritis Res. Ther. 2012;14:R27. doi: 10.1186/ar3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasuya K., Hashimoto Y., Numano F. Left ventricular dysfunction and HLA Bw52 antigen in Takayasu arteritis. Heart Vessels Suppl. 1992;7:116–119. doi: 10.1007/BF01744556. [DOI] [PubMed] [Google Scholar]
- 11.Kitamura H., Kobayashi Y., Kimura A., Numano F. Association of clinical manifestations with HLA-B alleles in Takayasu arteritis. Int. J. Cardiol. 1998;66(Suppl 1):S121–S126. doi: 10.1016/s0167-5273(98)00159-4. [DOI] [PubMed] [Google Scholar]
- 12.Saruhan-Direskeneli G., Biçakçigil M., Yilmaz V., Kamali S., Aksu K., Fresko I., Akkoç N., Kiraz S., Ozer H.T., Tunç E., Rheumatology Education and Research Society Vasculitis Study Group Interleukin (IL)-12, IL-2, and IL-6 gene polymorphisms in Takayasu’s arteritis from Turkey. Hum. Immunol. 2006;67:735–740. doi: 10.1016/j.humimm.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Arend W.P., Michel B.A., Bloch D.A., Hunder G.G., Calabrese L.H., Edworthy S.M., Fauci A.S., Leavitt R.Y., Lie J.T., Lightfoot R.W., Jr. The American College of Rheumatology 1990 criteria for the classification of Takayasu arteritis. Arthritis Rheum. 1990;33:1129–1134. doi: 10.1002/art.1780330811. [DOI] [PubMed] [Google Scholar]
- 14.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 15.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 17.Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A., 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dilthey A.T., Moutsianas L., Leslie S., McVean G. HLA∗IMP—an integrated framework for imputing classical HLA alleles from SNP genotypes. Bioinformatics. 2011;27:968–972. doi: 10.1093/bioinformatics/btr061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes T., Coit P., Adler A., Yilmaz V., Aksu K., Düzgün N., Keser G., Cefle A., Yazici A., Ergen A. Identification of multiple independent susceptibility loci in the HLA region in Behçet’s disease. Nat. Genet. 2013;45:319–324. doi: 10.1038/ng.2551. [DOI] [PubMed] [Google Scholar]
- 21.Uyar F.A., Dorak M.T., Saruhan-Direskeneli G. Human leukocyte antigen-A, -B and -C alleles and human leukocyte antigen haplotypes in Turkey: relationship to other populations. Tissue Antigens. 2004;64:180–187. doi: 10.1111/j.1399-0039.2004.00258.x. [DOI] [PubMed] [Google Scholar]
- 22.Yang T.P., Beazley C., Montgomery S.B., Dimas A.S., Gutierrez-Arcelus M., Stranger B.E., Deloukas P., Dermitzakis E.T. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics. 2010;26:2474–2476. doi: 10.1093/bioinformatics/btq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan A.W., Robinson J.I., Barrett J.H., Martin J., Walker A., Babbage S.J., Ollier W.E., Gonzalez-Gay M.A., Isaacs J.D. Association of FCGR2A and FCGR2A-FCGR3A haplotypes with susceptibility to giant cell arteritis. Arthritis Res. Ther. 2006;8:R109. doi: 10.1186/ar1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maksimowicz-McKinnon K., Clark T.M., Hoffman G.S. Takayasu arteritis and giant cell arteritis: a spectrum within the same disease? Medicine (Baltimore) 2009;88:221–226. doi: 10.1097/MD.0b013e3181af70c1. [DOI] [PubMed] [Google Scholar]
- 25.Grayson P.C., Maksimowicz-McKinnon K., Clark T.M., Tomasson G., Cuthbertson D., Carette S., Khalidi N.A., Langford C.A., Monach P.A., Seo P., Vasculitis Clinical Research Consortium Distribution of arterial lesions in Takayasu’s arteritis and giant cell arteritis. Ann. Rheum. Dis. 2012;71:1329–1334. doi: 10.1136/annrheumdis-2011-200795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asano K., Matsushita T., Umeno J., Hosono N., Takahashi A., Kawaguchi T., Matsumoto T., Matsui T., Kakuta Y., Kinouchi Y. A genome-wide association study identifies three new susceptibility loci for ulcerative colitis in the Japanese population. Nat. Genet. 2009;41:1325–1329. doi: 10.1038/ng.482. [DOI] [PubMed] [Google Scholar]
- 27.McGovern D.P., Gardet A., Törkvist L., Goyette P., Essers J., Taylor K.D., Neale B.M., Ong R.T., Lagacé C., Li C., NIDDK IBD Genetics Consortium Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat. Genet. 2010;42:332–337. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohta Y., Ohya Y., Fujii K., Tsuchihashi T., Sato K., Abe I., Iida M. Inflammatory diseases associated with Takayasu’s arteritis. Angiology. 2003;54:339–344. doi: 10.1177/000331970305400310. [DOI] [PubMed] [Google Scholar]
- 29.Reny J.L., Paul J.F., Lefèbvre C., Champion K., Emmerich J., Blétry O., Piette J.C., Fiessinger J.N. Association of Takayasu’s arteritis and Crohn’s disease. Results of a study on 44 Takayasu patients and review of the literature. Ann. Med. Interne (Paris) 2003;154:85–90. [PubMed] [Google Scholar]
- 30.Aggarwal A., Chag M., Sinha N., Naik S. Takayasu’s arteritis: role of Mycobacterium tuberculosis and its 65 kDa heat shock protein. Int. J. Cardiol. 1996;55:49–55. doi: 10.1016/0167-5273(96)02660-5. [DOI] [PubMed] [Google Scholar]
- 31.Soto M.E., Del Carmen Ávila-Casado M., Huesca-Gómez C., Alarcon G.V., Castrejon V., Soto V., Hernandez S., Espinola-Zavaleta N., Vallejo M., Reyes P.A., Gamboa R. Detection of IS6110 and HupB gene sequences of Mycobacterium tuberculosis and bovis in the aortic tissue of patients with Takayasu’s arteritis. BMC Infect. Dis. 2012;12:194. doi: 10.1186/1471-2334-12-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson C.A., Boucher G., Lees C.W., Franke A., D’Amato M., Taylor K.D., Lee J.C., Goyette P., Imielinski M., Latiano A. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reveille J.D., Sims A.M., Danoy P., Evans D.M., Leo P., Pointon J.J., Jin R., Zhou X., Bradbury L.A., Appleton L.H., Australo-Anglo-American Spondyloarthritis Consortium (TASC) Genome-wide association study of ankylosing spondylitis identifies non-MHC susceptibility loci. Nat. Genet. 2010;42:123–127. doi: 10.1038/ng.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


