Abstract
White matter hyperintensities (WMHs) of the brain are important markers of aging and small-vessel disease. WMHs are rare in healthy children and, when observed, often occur with comorbid neuroinflammatory or vasculitic processes. Here, we describe a complex 4 kb deletion in 2q36.3 that segregates with early childhood communication disorders and WMH in 15 unrelated families predominantly from Southeast Asia. The premature brain aging phenotype with punctate and multifocal WMHs was observed in ∼70% of young carrier parents who underwent brain MRI. The complex deletion removes the penultimate exon 3 of TM4SF20, a gene encoding a transmembrane protein of unknown function. Minigene analysis showed that the resultant net loss of an exon introduces a premature stop codon, which, in turn, leads to the generation of a stable protein that fails to target to the plasma membrane and accumulates in the cytoplasm. Finally, we report this deletion to be enriched in individuals of Vietnamese Kinh descent, with an allele frequency of about 1%, embedded in an ancestral haplotype. Our data point to a constellation of early language delay and WMH phenotypes, driven by a likely toxic mechanism of TM4SF20 truncation, and highlight the importance of understanding and managing population-specific low-frequency pathogenic alleles.
Introduction
Age-related WMHs, observed as hyperintensities on T2 weighted and fluid-attenuated inversion recovery (FLAIR) brain MRI sequences, are recognized to be neuroimaging expression of brain aging and cerebral small-vessel disease. With a prevalence that ranges from 11% to 21% in individuals ∼64 years of age and increasing to more than 90% by age 82,1 these lesions are known to be associated with an increased risk of cerebrovascular events and cognitive decline.2 Multiple large studies indicate that white matter signal abnormalities are infrequent in healthy individuals in their 20s and 30s3,4 and are infinitesimal (∼1%) in a neurologically healthy pediatric population.5 Conversely, in pathological conditions such as pediatric systemic lupus, causing occlusive vascular disease, WMHs are reported in almost 50% of children, primarily in the subcortical frontal regions.6 Children with unexplained cognitive and/or developmental delay also have white matter signal abnormalities in an estimated 17%–26% of cases.7–9 Although the imaging appearance of white matter pathology in such cases may be nonspecific and suggestive of injury resulting from prematurity, hypoxia, ischemia, or fetal-maternal infection,10 WMHs are never considered innocuous findings when observed in the pediatric age group.11–13
In a genomic study of 15,493 children referred to the Medical Genetics Laboratories (MGL) at Baylor College of Medicine (BCM) (Houston, TX), by using 180,000 oligonucleotide-based whole-genome exon-focused array comparative genomic hybridization (CGH),14 we identified a 4 kb deletion at 2q36.3 that removed the penultimate exon 3 of TM4SF20 in 12 unrelated individuals with language delay and WMHs (penetrance of ∼70%). With two other independent study groups (group 2, Vietnamese ethnicity, Texas Children’s Hospital, n = 76; group 3, Vietnamese ethnicity, Signature Genomics, n = 171), we confirmed the segregation of the deletion allele with familial WMHs and disorders of communication. We show that this highly penetrant 4 kb deletion cosegregates with WMHs and early childhood language delay in multiple families extending from Burma to Micronesia. We present evidence to illustrate that this population-specific deletion with allele frequency of ∼1% causes truncation of the cognate protein and is associated with communication disorders, with near complete penetrance in the Vietnamese, Thai, Burmese, Filipino, Indonesian, and Micronesian pediatric subpopulations. We also demonstrate that the frequency of WMHs related to TM4SF20 is significantly higher than that reported in other large pediatric studies of WMHs with unexplained cognitive and/or developmental delay, suggesting a link between the observed brain imaging abnormalities and the deletion allele.7–9
Subjects and Methods
Subjects
The study was performed in accordance with protocols approved by the appropriate human subjects ethics committees at Baylor College of Medicine and each participating institution. Written informed consents were obtained from the parents of subjects with the TM4SF20 deletion.
Study Group 1: Array CGH at MGL
Between May 2009 and July 2012, 15,493 children of all ethnicities were studied with the 180,000 oligonucleotide-based whole-genome exon-focused array CGH for intellectual disability, epilepsy, facial dysmorphisms, congenital heart defects, multiple congenital anomalies, and/or speech and language delay. The clinical array was performed at the MGL at BCM. Twelve subjects with the TM4SF20 deletion were evaluated by geneticists, developmental pediatricians, and/or neurologists for language delay and/or brain imaging abnormalities. Written informed consents were obtained from the parents of subjects with the TM4SF20 deletion, identified on the clinical array CGH. Many Southeast Asian families in the study were either refugees (Burmese) or young Vietnamese immigrants to the United States. Vietnamese and Burmese language interpreters were employed for recruitment of the non-English-speaking families. The first and second degree relatives of the probands were enrolled whenever available and written informed consents were obtained.
DNA was extracted from whole blood by the Puregene DNA Blood Kit (Gentra) according to the manufacturer’s instructions. The procedures for DNA digestion, labeling, and hybridization for the oligo arrays were performed according to the manufacturers’ instructions. Array CGH was performed with V8 OLIGO custom-designed genome-wide array with approximately 180,000 interrogating oligonucleotides manufactured by Agilent Technologies as previously described.14,15 The clinical array is designed by the MGL at BCM with exon-by-exon coverage for about 1,700 genes and 700 microRNAs to detect intragenic single exon deletions and duplications.
The clinical array includes tiling coverage of mitochondrial genome and provides whole-genome coverage at the resolution of 30 kb. Log2 ratios were analyzed for copy-number change via circular binary segmentation16 to minimize the possibility for false-negative results; we performed postprocessing of the oligo level data to ensure all copy-number variation (CNV) events comprise at least three oligos with consistent median log-ratios.
Long-Range Polymerase Chain Reaction and Sequencing
Long-range PCR (LR-PCR) reactions were performed to amplify the predicted junction fragments in the breakpoint regions in TM4SF20 deletion carriers according to the manufacturer’s specifications (Takara Bio). The 1 kb junction fragment of the deletion was amplified by LR-PCR with primers forward 5′-ACAAGCATAAGCCATTTGAGATCAACTAGTCC-3′ and reverse 5′-CAACAGAACTGGAGTAAGTATGAAGCAGTCG-3′. The PCR products were purified with the PCR Purification Kit (QIAGEN) and sequenced (Lone Star).
Brain Magnetic Resonance Imaging
Brain MRI of all 32 subjects with the deletion (14 unrelated children, 13 carrier parents, and 5 related individuals) was performed at 1.5 T or higher with 5 mm slice thickness. Sequences included sagittal T1-weighted imaging, axial T2-w imaging, and T2-w FLAIR imaging acquired in all subjects, in addition to diffusion-weighted imaging and gradient echo imaging. Each case was evaluated by at least two neuroradiologists, blinded to the other’s radiological interpretation and the genetic data.
Linkage Analysis
Families TM200, TM800, TM900, TM1000, TM1100, and 017-1 were evaluated for linkage. Two-point parametric linkage analysis was performed with MLINK of the FASTLINK package.17 An autosomal-dominant mode of inheritance and a disease allele frequency of 1% were used in the analysis. For the language delay phenotype, a fully penetrant model with a phenocopy rate of 0.05 was used. The phenocopy rate of 5% was selected because this was approximately the prevalence of language delay in the general population. For the WMH phenotype, a reduced penetrance model was used with a penetrance of 0.7. Because of the rarity of this phenotype in healthy children, no phenocopies were incorporated in the model.
Determination of TM4SF20 Deletion Haplotype
To set phase of the haplotype on which the TM4SF20 deletion is segregating, trios (mother, father, child) from five families segregating the TM4SF20 deletion were genotyped with either the Omni 2.5-8 or Human Omni Express arrays from Illumina per manufacturer’s instruction. One family was of Vietnamese origin, one Filipino, and three families were of Burmese ancestry. Genotypes for 139 SNPs typed across all individuals and spanning 500 kb across TM4SF20 were used to determine haplotypes by family inheritance patterns.
Assessment of Deletion Frequency in Worldwide Populations
Population-based data were obtained from genotyping 2,089 Vietnamese Kinh cord blood samples, initially recruited and utilized as controls in a study of genetic susceptibility to dengue fever.18 The 2,089 cord blood controls were genotyped with the Illumina Human 660W Quad BeadChips according to the manufacturer’s instructions. After exclusions resulting from quality control (relatedness and genome-wide genotyping success rate), SNP genotype data from 2,018 Vietnamese Kinh (1,022 males, 996 females) were available for analysis. Genotypes, log R ratio (LRR), and B allele frequency (BAF) for each probeset were determined with the Illumina BeadStudio software.
The Illumina Human 660W Quad BeadChip array contains one SNP in the deletion of interest, rs10933181. Genotype plus LRR and BAF for this SNP were used to infer the presence or absence of the deletion in each individual, a genotype and a BAF (>0.97 or <0.01) that was consistent with a homozygous call and a logR ratio that was less than two SDs from the mean LRR for rs10933181 in this cohort. Subsequently, each individual suspected of having the deletion based on these criteria (n = 53) plus an additional 72 samples were genotyped by PCR assay, as described above, in order to confirm the TM4SF20 deletion status for each individual. A total of 46 out of 53 individuals typed positive for the deletion by PCR and one of these 53 subjects was equivocal. The 72 samples that were negative for deletion by SNP data were also negative by PCR assay. In summary, the performance of the SNP array-based assay for assessment of the TM4SF20 deletion showed sensitivity = 1, specificity = 0.92, positive predictive value = 0.87, and negative predictive value = 1 in this cohort.
The frequency of this deletion in worldwide populations was based on data from the 1000 Genomes Project variant calls from the callset released 2012-02-14.
Generation of DNA Constructs for In Vitro Studies
A minigene construct (gDNA-ΔEX3) to assay mRNA splicing was generated by PCR amplification from genomic DNA harboring the TM4SF20 deletion with the forward primer beginning at the 5′ end of exon 2 and the reverse primer at the stop codon of exon 4. This fragment was cloned into pCR8/GW-TOPO (Invitrogen), sequence confirmed, and recombineered into pcDNA6.2-N-EmGFP (Invitrogen) with LR clonase II (Invitrogen).
The WT protein expression construct was generated by amplification of the full-length TM4SF20 open reading frame (ORF) from whole human brain cDNA (QUICK-clone, Clontech); the ΔEX3 construct was generated by amplification of the partial TM4SF20 ORF (exons 1 and 2) from whole human brain cDNA with the introduction of a stop codon in the reverse primer. Both WT and ΔEX3 pCR8/GW constructs were recombineered into pcDNA6.2-N-EmGFP (Invitrogen) with LR clonase II (Invitrogen).
Cell Culture and Immunostaining
mRNA Splicing Assay
HEK293-FT cells were seeded in 6-well plates, and at 70% confluency, 1 μg pcDNA6.2-N-EmGFP-gDNA-ΔEX3 was transfected with X-tremeGENE transfection reagent (Roche). Cells were harvested in Trizol (Invitrogen) at 48 hr after transfection subsequent to either 8 hr of treatment with Emetine (100 μg/ml) or no treatment. We generated oligo-dT-primed cDNA with Quantitect reverse transcriptase (QIAGEN), PCR amplified TM4SF20 according to standard procedures, and Sanger sequenced PCR products.
Protein Localization
Neuro-2a cells were seeded on glass coverslips in 6-well plates and were transfected with 1 μg pcDNA6.2-N-EmGFP-WT or 1 μg pcDNA6.2-N-EmGFP-ΔEX3 with Dharmafect1 transfection reagent (Dharmacon). Cells were fixed with cold methanol at 24 hr after transfection and immunolabeled with α-tubulin antibody (Sigma; 1:500), mouse secondary antibody conjugated to Alexa Fluor 568 (Invitrogen; 1:1,000), and nuclei were stained with 10 μg/ml Hoeschst 3342 (1:1,000, Invitrogen). Confocal images were visualized with a Nikon Eclipse 90i microscope and C1 confocal scanner controlled by EZ-C1 v.3.10 software (Nikon). Confocal images were acquired at 60×, 1.4OD objective, and 5 μm z-stack.
Results
Study Group 1: Array CGH at MGL
By using 180,000 oligonucleotide-based whole-genome exon-focused array CGH14 in 15,493 children referred to the MGL at BCM, we identified an ∼4 kb deletion at 2q36.3 that removed the penultimate exon 3 of TM4SF20 (RefSeq accession number NM_024795.3) in 12 unrelated individuals (Figures 1A, 1B, and 2A). Group 1 had diverse ethnic distribution and wide-ranging referral indications, including facial dysmorphisms, congenital cardiovascular malformations, multiple congenital anomalies, intellectual impairment, and/or speech and language delay (Table S1 available online). The index case, individual TM101 (II-1 in Figure 1A) was a 3-year-old Burmese boy, referred for evaluation of language delay and left-sided hemiparesis. His brain MRI showed bilateral periventricular confluent WMHs with a thin corpus callosum. The same deletion was also found in his 35-year-old, apparently healthy, Burmese mother, TM102 (I-2 in Figure 1A) However, her brain MRI subsequently showed similar multifocal T2 hyperintensities in the periventricular and subcortical deep white matter of both cerebral hemispheres, suggestive of cerebral small-vessel disease with mild diffuse cerebral and cerebellar volume loss. This identical deletion was observed in 11 other unrelated pediatric subjects, all of whom shared a diagnosis of communication disorder, ranging from early language delay to autism spectrum disorder. None of them had any significant facial dysmorphisms or other congenital anomalies. Driven by the discovery of this genetic lesion, we performed systematic brain MRI in other children with the deletion and in all, found punctate WMHs in the deep white matter with enlarged perivascular (VR) spaces to multifocal T2 hyperintensities in the periventricular white matter in 8 out of 11 pediatric subjects (72%) imaged (TM101, TM301, TM501, TM601, TM801, TM1101, TM1301, and TM1401) (Figure 3; Table 1). The parents declined brain imaging studies for TM1201. Interestingly, most children with the 4 kb TM4SF20 deletion originated from Southeast Asia or the Far East, including Burma, Vietnam, Philippines, Thailand, Indonesia, and Micronesia (Table 1). TM301 was reported to be Hispanic and TM1301 was of unspecified Asian origin. We noted that the frequency of WMHs in this group was significantly higher than previously reported for healthy children between the ages of 1 month and 18 years (p = 2.589 × 10−11),5 and 3- to 4-fold higher than reported for children with unexplained intellectual disability with IQ < 70 (p = 1.966 × 10−4)7 or those with idiopathic developmental delay (p = 2.922 × 10−3)9 from large studies (Table S2). In addition, the presence of punctate and multifocal T2 hyperintensities in the periventricular and deep white matter in 8/10 unrelated carrier parents similar to those observed in the affected children strongly suggested a link between the observed brain imaging abnormalities and the deletion allele (Table 2). Children exposed to early hypoxic or infectious insult sometimes demonstrate nonspecific T2 hyperintensities on brain imaging, but the extent of the findings and the consistency across unrelated families from a distinct geographical region strongly suggested that these were related to the shared deletion allele. In further support of this, of all the 15,493 cases (all ethnicities) studied for the deletion allele, the variant was observed only in those individuals referred for evaluation of developmental delay, speech/language impairment, and/or brain imaging abnormalities (n = 6,390) and not in those with craniofacial dysmorphisms, congenital cardiovascular malformations, multiple anomalies, musculoskeletal concerns, urogenital abnormalities, and other concerns (n = 9,103) (Table S1). The difference in the frequency of deletion allele in these two classes of phenotypes is highly significant (p = 1.7 × 10−6), suggesting that the deletion is distinct to individuals with communication disorders and brain imaging abnormalities in this group.
Figure 1.
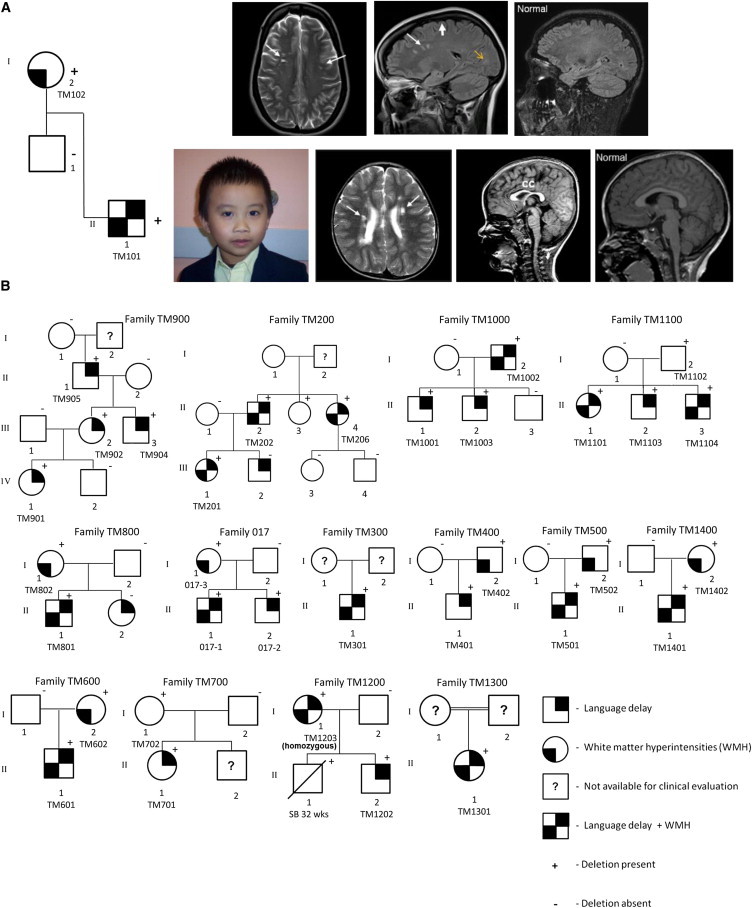
Identification of TM4SF20 Exon 3 Deletion in Multiple Families Predominantly of Southeast Asian Descent
(A) Brain MRI study in family TM100 is highlighted, revealing hyperintensities on axial T2-weighted spin echo and confirmed on axial T2 FLAIR, consistent with foci of gliotic changes, and observed in the periventricular and deep subcortical white matter (marked with long white arrow) in the carrier child (II-1) and parent (I-2). The maternal brain MRI images (TM102) are shown at the top, and the images of the proband are illustrated below. Comparison is made with the age-matched normal corresponding MRI image. Note the thin corpus callosum (cc) in TM101. Generous biparietal extra-axial CSF spaces with mild diffuse volume loss is observed in the carrier parent (marked with short white arrow). Note the prominent Virchow-Robin (VR) spaces highlighted by the yellow arrow.
(B) Fourteen additional pedigrees are shown. Positive sign indicates presence of the deletion and negative sign highlights absence of the deletion.
Figure 2.
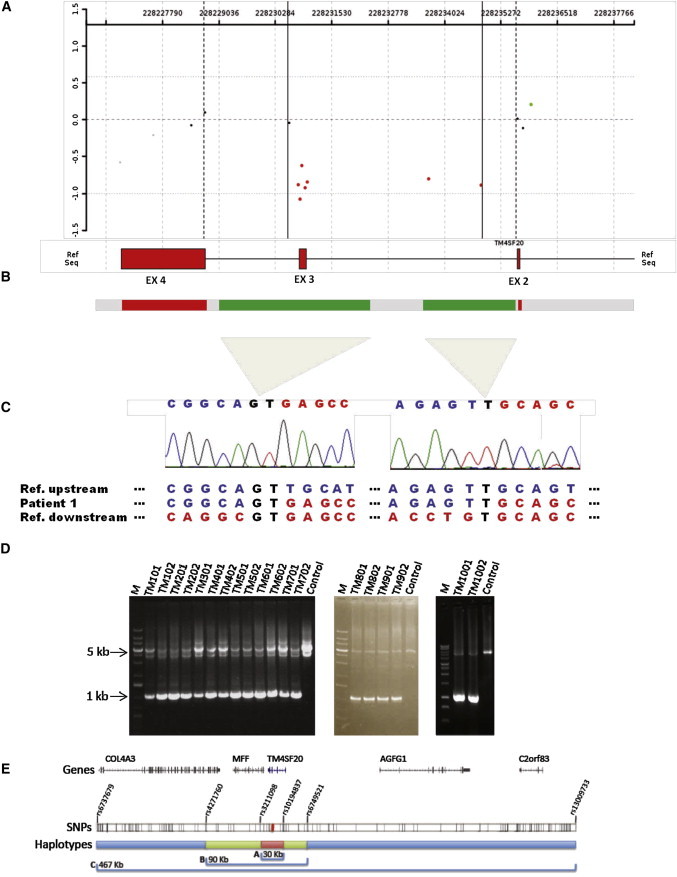
TM4SF20 Heterozygous Deletion and Breakpoint Analysis
(A and B) High-resolution array CGH analysis revealed a 4 kb deletion within 2q36.3 involving exon 3 of TM4SF20 (UCSC Genome Browser build GRCh37/hg19: 228,230,759–228,234,864).
(C and D) Long-range PCR analysis identified a common 1 kb junction fragment in all the affected subjects. Sequence analysis revealed a complex rearrangement with a 2.3 kb deletion with microhomology of GT at the first junction, followed by a neutral copy number region of approximately 100 bp (marked as gray bar-interrupted green), then a 1.7 kb deletion with insertion of a T at the second junction.
(E) The haplotype on which the TM4SF20 deletion segregates was determined for one Vietnamese, one Filipino, and three Burmese families. A haplotype of 30 kb (bracket labeled A, red) was found to be shared across all families. A longer haplotype of 90 kb (bracket labeled B, green) was shared between the Burmese and Filipino families. Burmese families shared a longer 467 kb haplotype (bracket labeled C, blue). All SNPs typed in these individuals and utilized in haplotype analyses are shown as black tick marks and the SNPs delineating haplotypes are labeled. RefSeq genes for this region are shown.
Figure 3.
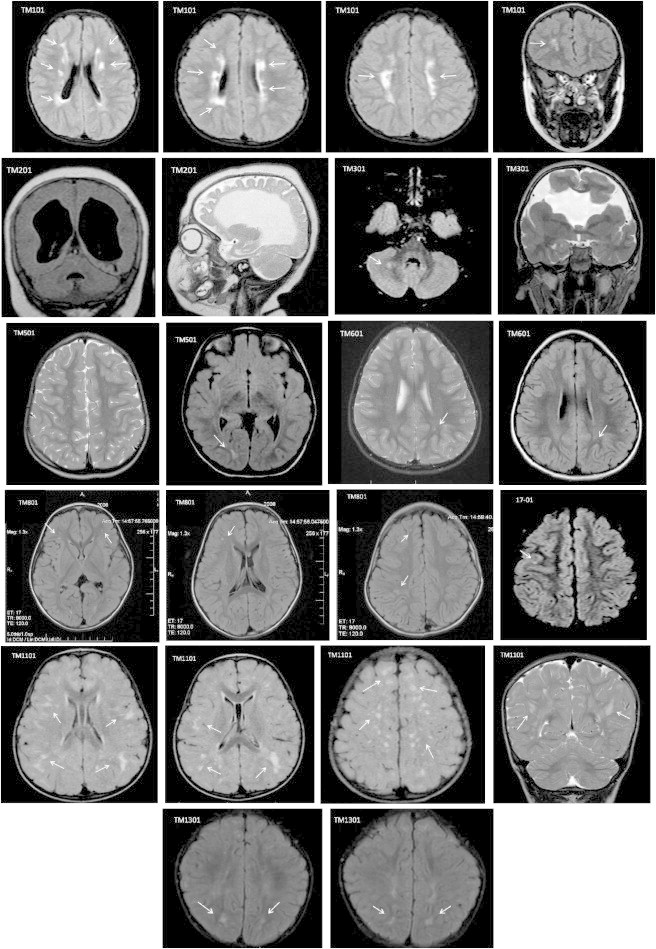
Spectrum of WMHs in the Children with TM4SF20 Deletion
T2 weighted and fluid-attenuated inversion recovery (FLAIR) sequence magnetic resonance imaging (MRI) scans of nine unrelated subjects with TM4SF20 deletion are presented, showing varying degrees of white matter disease observed as punctate T2 hyperintensities in the subcortical white matter and multifocal T2 hyperintensities in the periventricular and deep white matter of both cerebral hemispheres. Subject TM201, born prematurely, has gross enlargement of the third and lateral ventricles with near-complete loss of periventricular white matter. Subject TM301 has bilateral open lip schizencephaly in the frontal lobes. Prominent perivascular (VR) spaces are seen in TM501.
Table 1.
Clinical Characteristics of Children with TM4SF20 Heterozygous Deletion
| No. | Patient ID | Study Group | Ancestry | Inheritance | Current Age | Gender | Gestation | Language and/or Speech Disordera | WMH | MRI Study Results |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | TM101 | 1 | Burmese | maternal | 3 years | male | term | language delay | + | periventricular white matter loss with T2 hyperintensities, not typical of periventricular leukomalacia |
| 2 | TM301 | 1 | Hispanic/unknown | unknown (adopted) | 3 years | male | term | moderate to severe global delay, nonverbal, epilepsy | + | bilateral open-lipped schizencephaly and deformity of the corpus callosum; right-sided peridentate nuclear T2 hyperintensity |
| 3 | TM401 | 1 | Burmese | paternal | 7 years | male | 32 weeks | language delay | − | slight prominence to the T2 hyperintensity returned from the terminal zones in the bilateral peritrigonal regions; one or two prominent VR spaces |
| 4 | TM501 | 1 | Burmese | paternal | 2 years | male | term | language delay, autism spectrum disorder | + | prominent VR spaces with prominence to the terminal zones with increased T2 hyperintensity returned from the periventricular white matter outlining the occipital horns with delays in myelination to the subcortical U-fibers in the high parietal regions |
| 5 | TM601 | 1 | Thai/Hispanic | maternal | 4 years | male | term | nonverbal, autism spectrum disorder | + | two small focal T2 hyperintensities returned from the bilateral deep white matter |
| 6 | TM801 | 1 | Pakistani/Filipino | maternal | 3 years | male | term | autism spectrum disorder | + | multifocal punctate T2 hyperintensities returned from the bilateral fronto-parietal regions |
| 7 | TM901 | 1 | Filipino | maternal | 2 years | female | term | language delay | − | normal study |
| 8 | TM1001 | 1 | Indonesian/European | paternal | 1 year | male | term | language delay, global delay | − | within normal limits appearances to the brain which is incompletely myelinated, but age appropriate, with prominent terminal zones |
| 9 | TM1101 | 1 | Burmese | paternal | 1 year | female | term | language delay | + | multifocal subcortical and deep T2 hyperintensities returned from all lobes of both cerebral hemispheres without mass effect |
| 10 | TM1201 | 1 | Vietnamese/European | maternal | 4 years | male | term | speech and language disorder | NA | MRI not done |
| 11 | TM1301 | 1 | unspecified Asian ancestry | unknown (adopted) | 1 year | female | term | language delay | + | symmetric patchy T2 hyperintense signals in the occipital subcortical white matter |
| 12 | TM1401 | 1 | Micronesian/European | maternal | 13 years | male | term | language delay, developmental delay | + | few scattered nonspecific high T2 FLAIR foci seen throughout the subcortical white matter, most prominently in the left frontal lobe |
| 13 | TM201 | 2 | Vietnamese | paternal | 1 year | female | 33 weeks | severe global delay, epilepsy | + | near complete agenesis of the corpus callosum with evidence for gliotic change returned in a left posterior cerebral artery distribution; ex-vacuo dilation of the supratentorial ventricular system |
| 14 | 017-1 | 2 | Vietnamese | maternal | 7 years | male | term | severe language delay | + | at least one focal T2 hyperintensity returned from the right posterior frontal subcortical white matter |
| 15 | TM701 | 2 | Vietnamese | maternal | 4 years | female | term | language delay, autism spectrum disorder | − | normal study |
Abbreviation is as follows: VR, Virchow-Robin spaces.
Details of deficits are as follows. For TM101: receptive language abilities, 3-year-old level; expressive language, 2-year-old level (Preschool Language Scale, Fourth Edition [PLS-4]). For TM301: general developmental score, 40; communication, 50; cognitive, 50; adaptive behavior, 50 (DP-3). For TM401: deficiencies in LNF (letter-naming fluency), in PSF (phoneme-segmentation fluency), and in NWF (nonsense-word fluency) (mCLASS: Dynamic Indicators of Basic Early Literacy Skills). For TM501: visual problem-solving skills, DQ 68; language, DQ 43 (Cognitive Adaptive Test/Clinical Linguistic Auditory Milestone Scale). For TM601: responses exceed cutoffs on autism spectrum disorder questionnaires (Social Communication Questionnaire - Lifetime [SCQ; 2003] form and a Social Responsiveness Scale [SRS; 2005] parent form). For TM701: adaptive behavior composite, 68; communication, 54; daily living skills, 75; socialization, 63; motor skills, 94 (Vineland Adaptive Behavior Assessment II Survey Interview). For TM801: expressive communication, standard score 56, age equivalent of 6 months at 19 months of chronological age; auditory comprehension, standard score of 61, age equivalent of 9 months; total language score 54, percentile rank 1, age equivalent of 7 months (PLS-4). For TM1101: visual problem-solving skills, DQ 87; language, DQ 67; overall developmental quotient is 77. For TM1201: at age 3.1, informal observation of expressive language skills showed use of up to five single words per sentence to describe, request, and interact communicatively, and the use of simple question formulation. Prepositions were apparent but frequently omitted. At age 4.4, the expressive Speech Profile results showed an improvement from the moderate to the mild to moderate level of articulation proficiency. For TM1301: expressive language at 6 months and receptive language at 10 months, when evaluated at 12 months. For TM1401: 40 point difference between verbal and performance abilities, with verbal scores significantly lower (Peabody Picture Vocabulary Test Fourth Edition [PPVT-4]); Test of Nonverbal Intelligence Second Edition [TONI-2]).
Table 2.
Clinical Characteristics of Parents with TM4SF20 Deletion
| No. | Parent ID | Study Group | Carrier Parent | Ancestry | Age | Comorbidity | WMH | MRI Study Results |
|---|---|---|---|---|---|---|---|---|
| 1 | TM102 | 1 | mother | Burmese | 35 years | none | + | T2 multifocal punctate subcentimeter hyperintensities returned from the bifronto-parietal subcortical white matter with dilated VR spaces |
| 2 | TM402 | 1 | father | Burmese | 52 years | end-stage liver disease (alcoholic cirrhosis, hepatitis C) | + | cerebellar greater than cerebral atrophy with mild ex-vacuo dilatation of the supraventricular system in association with one or two subcortical white matter focal T2 hyperintensities |
| 3 | TM502 | 1 | father | Burmese | 39 years | none | + | punctate focal 2–3 mm subcortical white matter T2 hyperintensities returned from the high bilateral posterior fronto-parietal subcortical white matter |
| 4 | TM602 | 1 | mother | Thai | 35 years | none | + | tiny foci of T2 hyperintensities returned from predominately the left posterior parietal subcortical white matter |
| 5 | TM802 | 1 | mother | Filipino | 36 years | none | + | bilateral fronto-parietal, less than 5 mm subcortical T2 bright white matter hyperintensities |
| 6 | TM902 | 1 | mother | Filipino | 32 years | none | − | normal study |
| 7 | TM1002 | 1 | father | Indonesian | 30 years | none | + | multifocal punctate T2 hyperintensities returned from the bifronto-parietal subcortical white matter, slightly more marked on the right when compared to the left |
| 8 | TM1102 | 1 | father | Burmese | 42 years | none | − | normal study |
| 9 | TM1203 | 1 | mother | Vietnamese | 36 years | none | + | there are at least 1–2, 3 mm focus of abnormal T2 hyperintensity returned from the subcortical white matter in the right insular region |
| 10 | TM1402 | 1 | mother | Micronesia | 50 years | hypertension | + | multiple scattered foci of increased T2 signal throughout the periventricular white matter |
| 11 | TM202 | 2 | father | Vietnamese | 37 years | none | + | multifocal punctate T2 hyperintensities returned from the bilateral fronto-parietal subcortical white matter |
| 12 | 017-3 | 2 | mother | Vietnamese | 37 years | none | + | multifocal punctate, less than 5 mm, T2 hyperintensities returned from the bifronto-parietal subcortical white matter |
| 13 | TM702 | 2 | mother | Vietnamese | 29 years | none | − | normal study |
Abbreviation is as follows: VR, Virchow-Robin spaces.
Detection of a Complex Genomic Rearrangement
LR-PCR in children and their carrier parents’ genomic DNA yielded an ∼1 kb deletion-specific junction fragment (Figures 2B–2D). Subsequent sequencing revealed a complex rearrangement consisting of a 2.3 kb deletion with a GT microhomology at the first junction and a 1.7 kb deletion with insertion of a T at the second junction; both deletions are separated by a neutral copy-number genomic segment of approximately 100 bp that is oriented directly with respect to the reference human genome sequence (UCSC Genome Browser GRCh37/hg19) (Figure 2C). The identical complex rearrangement in these unrelated pediatric subjects suggested that the deletion occurred on a single ancestral haplotype and was not due to recurrent de novo events secondary to susceptibility to genomic instability rendered by the innate genomic architecture.
Study Group 2: Brain MRI Study of 76 Children of Vietnamese Origin from Texas Children’s Hospital
To investigate further the potential role of this gene in language delay and WMHs, we reviewed the brain-imaging studies in an ethnicity-specific independent group of 76 children of Vietnamese descent (group 2), evaluated at Texas Children’s Hospital in Houston, for speech and language delay, unexplained developmental delay, and/or autism spectrum disorder (Figure S1). The two independent neuroradiologists blinded to the TM4SF20 deletion status observed diverse brain imaging abnormalities in multiple cases, including distinct white matter abnormalities in 19/76 (25%) children. The TM4SF20 deletion was confirmed by PCR in 4/76 (∼5%) cases, three of whom had distinct white matter signal abnormalities (TM201, 017-1, 18-01) (Table S3). One child (TM701) without the WMHs was also found to have the deletion. The frequency of WMHs in this study was noted to be different in deletion versus nondeletion cases (p = 0.0461, Fisher’s exact test), indicating that the deletion carriers were more likely to have WMHs than the nondeletion carriers. TM201, 017-1, and TM701 were then followed further for family studies. Parents declined further evaluation of 18-01, who harbored the deletion with WMHs.
The individual TM201 (III-1 in Figure 1B), one of the severely affected individuals with the deletion, was born prematurely at 33 weeks gestation with extensive loss of periventricular white matter and evidence of stroke in the posterior cerebral artery (PCA) distribution. Pedigree analysis identified periventricular subcortical T2 hyperintensities in her otherwise healthy carrier father, TM202 (II-2), and her paternal aunt, TM206 (II-4), both of whom were <40 years of age with no cerebrovascular risk factors (Figure S2). Subject 017-1 (II-1 in Figure 1B) presented at 7 years of age with significant language delay. We subsequently found his 5-year-old sibling (II-2) to also harbor the deletion and exhibit severe language impairment, but his brain imaging study was within normal limits. Their 37-year-old asymptomatic mother (I-1), who also carried the deletion, showed multifocal periventricular and subcortical T2 hyperintensities (Figure S3). Subject TM701 (II-1 in Figure 1B) with normal brain imaging had early language delay and was diagnosed with autism spectrum disorder. Her mother (I-1) was also scanned and had normal brain imaging, confirmed by two neuroradiologists.
Combining groups 1 and 2, a total of 15 unrelated pediatric subjects with the TM4SF20 deletion were studied (Table 1). Of the 14 individuals imaged, 10 had WMHs (∼71%). Further, with the exception of two individuals (TM201 and TM301) with moderate to severe disease, the phenotype of most children with the deletion was consistent with language delay without significant craniofacial or other congenital anomalies and with preserved motor skills. Few hyperpigmented macules were observed variably within this group. Formal speech/language and development assessment in the deletion carriers showed significant discrepancies between verbal and nonverbal skills (Table 1).
Study Group 3: Vietnamese Group from Signature Genomics
The study of the 15 ascertained families indicated near-complete penetrance for early language delay in the pediatric population, with high penetrance of WMHs in carriers (∼70%). To exclude the possibility that this low-frequency, population-specific deletion allele was a benign polymorphism in the Vietnamese population, we obtained unidentified DNA samples from a third independent set, comprised of 171 pediatric cases of Vietnamese origin referred to Signature Genomic Laboratories, LLC for array CGH analysis. Of these, 79 Vietnamese children were referred for evaluation of communication disorders, developmental delay, or brain imaging abnormalities. Via LR-PCR, we determined the TM4SF20 deletion allele in 4 children, all from within this subgroup of 79 (4/79 = 0.050), and none in those referred for other concerns including multiple congenital anomalies, facial dysmorphisms, congenital heart defects, or growth failure (0/92).
Thus, combining all Vietnamese cases from groups 1 (n = 168), 2 (n = 76), and 3 (n = 171), there were 415 individuals with a diverse range of phenotypes studied for the TM4SF20 deletion. In aggregate, we found the deletion in 10/189 = 0.053 (95% CI 0.026–0.095) cases evaluated for early language delay, developmental delay, autism spectrum disorder, and brain MRI abnormalities, whereas no deletions were detected in children with congenital heart defects, multiple congenital anomalies, facial dysmorphisms, urogenital or skeletal abnormalities, or other concerns (0/207 = 0.0 [95% CI 0.0–0.018]) (Table S4). The difference in the frequency of the deletion in these two groups is statistically significant (p = 5.4 × 10−4), demonstrating that the deletion is unique to Vietnamese individuals with communication disorders and brain imaging abnormalities.
Pedigree and Linkage Analysis
These data suggested a causal link between the deletion in TM4SF20 and WMHs with associated language delay of variable expressivity. We determined that the early language and/or speech delay phenotype was self-reported by parents in pedigrees TM200, TM900, TM1000, TM1200, and TM1400 and in siblings of TM1100 and 17-01 even before the deletion was identified in the probands. We performed further genetic and phenotypic family studies by using standardized criteria for early language delay, defined as fewer than 50 words or no word combinations between 20 and 34 months of age,19 as well as by brain imaging studies. In six families (TM200, TM900, TM1000, TM1100, TM1200, and 017; Figure 1B), we found the deletion allele to fully segregate with early childhood language delay. In family TM900, the proband’s mother (III-2 in Figure 1B), maternal uncle (III-3), and maternal grandfather (II-1) reported late expressive language development, between 3 and 4 years of age prior to the deletion testing in the proband; all three were found to have the deletion (Figure S4). These individuals demonstrated high educational achievement, holding doctorate degrees. The deletion was not present in the proband’s brother (IV-2), father (III-1), maternal grandmother (II-2), or maternal great-grandmother (I-1). The paternal great-grandfather (I-2) (aged 92 years) declined genetic testing. Brain MRI studies were carried out in the proband and her 32-year-old mother, which were essentially normal.
There were six families (TM200, TM800, TM900, TM1000, TM1100, and 017) who were sufficiently large for linkage analysis for language delay. Of these families, only four were informative for WMHs. When the deletion was tested for linkage at theta = 0, the total LOD score for these six families for the language delay phenotype was 3.1, whereas the combined LOD score at theta = 0 for the four families for WMHs (TM200, TM800, TM1000, and 017) was 0.75. Although the penetrance for language delay appears to be high (in pediatric cases) with an autosomal-dominant mode of inheritance, there is reduced penetrance for the WMH phenotype.
Overall, we performed brain MRI study in the following deletion carriers: 14 unrelated children, 13 parents, and 5 related individuals (TM205, TM206, 017-02, TM1103, and TM1104) from 15 families (Table S5). Abnormalities consistent with WMHs were observed in 22/32 (∼69%) deletion carriers, indicating a high (but not complete) penetrance probably due to the deletion allele (Figures S5 and 3). Each case was evaluated by at least two neuroradiologists who were blinded to the radiological interpretation and the genetic data. An individual was designated to have WMHs only if the diagnosis was concurred by two neuroradiologists. Across all individuals studied, the radiological interpretation for WMHs was discordant for only two subjects (TM401 and 17-02); these were both scored as normal to err on the side of conservative interpretation. The pattern of distribution of the WMHs and the apparently fixed abnormalities in the young carrier adult individuals are more suggestive of ischemic injury resulting from small brain vessel disease as opposed to demyelination. No overt neurological deficits were observed in the young adult carriers. Neuropsychological testing was undertaken for a subset of these young apparently healthy carrier parents to assess the impact of WMHs on memory, information processing, and executive functioning. The results suggested mild impairments in visuospatial functioning, verbal learning, and problem solving (Table S6).
Diverse Human Populations and 4 kb TM4SF20 Deletion
Taken together, our segregation and linkage data indicated that the deletion allele is a likely driver of WMHs and language delay in the families investigated. We next asked what the frequency of this allele might be in diverse human populations, for which we turned to the 1000 Genomes Project20 data set. The individuals (n = 1,092) in this released call set represent African, European, Indigenous American, and Asian populations. The TM4SF20 deletion was not observed in this sampling. However, we noted that no Southeast Asians were included in this data set and the sample size of each individual population conferred limited power to detect variants of very low frequency (∼1%). Therefore, we employed array-based SNP genotyping from 2,018 umbilical cord blood samples from Vietnamese Kinh infants.18 Kinh is the most common ethnic group of Vietnam, comprising ∼86% of the population. Strikingly, we found that 46/2,018 (2.3%) individuals were carriers of the deletion, a finding confirmed in each case by PCR of the junction fragment, indicating an allele frequency of 1.1% in the Vietnamese Kinh (46/4,036, 95% CI = 0.0076–0.0143).
Haplotype Analysis
The enrichment of the deletion in the subjects suggested a possible founder effect. To determine the haplotype background on which the TM4SF20 deletion is segregating, child-parent trios from five families (one Vietnamese [TM200], three Burmese [TM100, TM400, and TM500], and one Filipino [TM900] ancestry) were genotyped. Haplotype phase was established via 150 SNPs spanning 500 kb centered on the deletion. We observed a core haplotype of ∼30.4 kb that was shared among all individuals with the deletion (Figure 2E). A longer 90 kb haplotype was shared among Burmese and Filipino individuals with the deletion and a haplotype of 467 kb was shared among the Burmese families studied. This finding of a short shared haplotype across Southeast Asian populations along with the unique and complex structure of the deletion suggests that the TM4SF20 deletion may represent a founder mutation that occurred prior to the dispersal of these sub populations (Figure S6).
Functional Studies
Finally, we turned to the question of a possible mechanism of disease. To test the potential consequences of the TM4SF20 deletion on mRNA splicing, and given that we could not detect TM4SF20 message in either human lymphocytes or fibroblasts, we deployed a surrogate in vitro system. We generated a minigene encompassing the 3.2 kb genomic fragment spanning exon 2 through exon 4 by amplifying genomic DNA from an individual harboring the deletion (Figure 4A). Subsequent to transient transfection of the deletion construct into HEK293-FT cells, we cultured cells in the presence or absence of Emetine (to block nonsense-mediated decay). TM4SF20 RT-PCR studies of cell lysates from either treatment group yielded a single mRNA splicing product; sequence confirmation revealed splicing of the exon 2 splice donor site with the exon 4 splice acceptor (Figure 4A). This in-frame deletion of exon 3 introduced a premature stop codon (p.Met84∗) in the terminal exon, resulting in a stable truncated message (Figures 4A and 4B).
Figure 4.
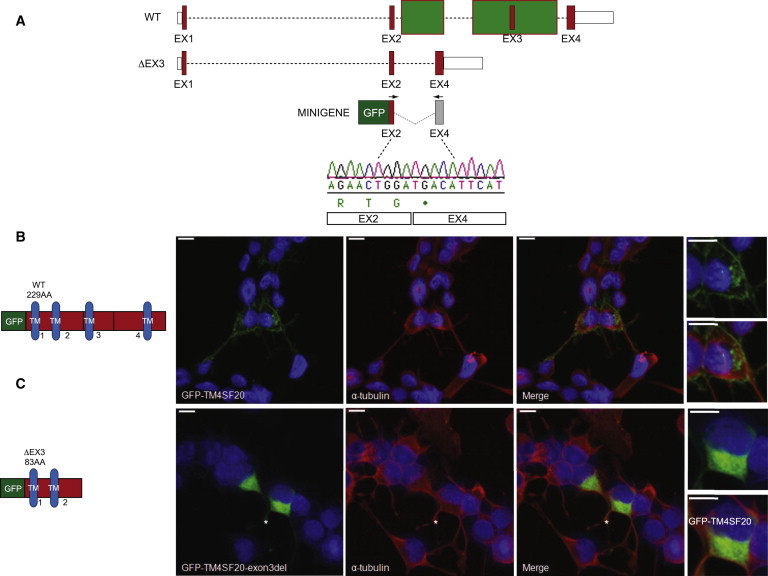
Molecular and Cellular Consequences of TM4SF20 Exon 3 Deletion
(A) Schematic of the wild-type (WT) and mutant (ΔEX3) TM4SF20 loci on human chromosome 2; red boxes, coding exons; white boxes, untranslated regions; dashed lines, intronic sequence; green boxes, deleted regions. Minigene construct to assay the splicing effect of ΔEx3; N-terminal GFP was cloned in-frame to a 3.2 kb region spanning exons 2 through 4. Chromatogram of the minigene RT-PCR product; black arrows indicate position of primers. Exons 2 and 4 are spliced together, introducing a stop codon at the end of exon 2.
(B) Schematic of WT and ΔEX3 TM4SF20 GFP fusion proteins. Blue, transmembrane domains (TM); black numbers indicate corresponding exons; amino acids abbreviated as AA.
(C) Confocal images of Neuro-2a cells transfected with N-terminal GFP TM4SF20-WT or TM4SF20-ΔEX3 constructs (green; left panels) and costained with α-tubulin (red; center panels) 24 hr after transfection. Blue, Hoeschst staining of nuclei; white asterisks, cells depicted in the insets (far right panels). Scale bars represent 10 μm.
TM4SF20 is a member of the 4-transmembrane L6 superfamily, encoding a surface protein with four transmembrane domains. These proteins interact with integrins21 to perform roles in cell adhesion, proliferation, and motility22 and promote angiogenic activities in endothelial cells through VEGF induction.21–23 To test the cellular effect of the TM4SF20 exon 3 deletion, which encodes only two of the transmembrane domains, we transfected Neuro-2a mouse neuroblastoma cells with N-terminal GFP-TM4SF20 expression constructs and asked whether the mutant has either a different half-life or localization compared to WT. We saw no differences in the former (data not shown). However, in contrast to the full-length GFP-TM4SF20 that gave the expected localization to the cell membrane in essentially all transfected cells, truncated GFP-TM4SF20-ΔEX3 protein mislocalized consistently to the cytoplasm (Figures 4B and 4C; minimum 50 transfected cells scored in duplicate experiments).
Discussion
Our findings demonstrate a pediatric syndrome characterized by a highly penetrant neurobehavioral trait (language delay) and neuroanatomical abnormality (WMHs), linked to a low-frequency population-specific single-exon deletion of TM4SF20. We show that the deletion occurs on a common genetic background with shared haplotype in the Thai, Burmese, and Vietnamese subpopulations, implying that this is a founder deletion mutation, present in each of these Southeast Asian subpopulations demarcated by distinct cultural, geographical, and linguistic traits. Most speech and language disorders are complex traits with extensive phenotypic and locus heterogeneity. Despite strong evidence for genetic susceptibility, only a few genetic variants have been linked to nonsyndromic communication deficits in children.24,25 Large epidemiological studies estimate the prevalence of early language delay in monolingual English-speaking children to be between 6% and 8%.26,27 In contrast, about 11.68% of Thai children are reported to have early language delay at 2 years of age.28 Our study implicates this TM4SF20 population-specific variant that accounts for a strong effect on disease susceptibility related to familial language delay in the Southeast Asian population. Although children with unexplained cognitive and/or developmental delay reportedly have white matter signal abnormalities in 17%–26% cases,7–9 the frequency of WMHs in children of Vietnamese, Burmese, Thai, Indonesian, Filipino, and Micronesian origin with the TM4SF20 deletion is significantly higher (∼70%) than any of the reported pediatric WMH studies related to seizures, unexplained intellectual disability, or idiopathic developmental delay (Table S2). These data strongly suggest a link between the ancestral deletion allele and the T2-hyperintense cerebral white matter lesions. Although our study shows high penetrance for WMHs in the 15 families with language delay, it remains to be determined whether other asymptomatic young carrier individuals exhibit T2 hyperintensities on brain imaging with similar high penetrance.
Functional studies indicate that the deletion deletes an internal exon and leads to a truncated form of the protein that is missing two of its four transmembrane domains and, although stable, accumulates in the cytoplasm. The combination of linkage (LOD score 3.1), segregation, and functional data all point to a dominant disorder with high penetrance and variable expressivity. TM4SF20 is expressed in the adult brain in mammals, readily detectable in the parietal lobe, occipital lobe, hippocampus, pons, white matter, corpus callosum, and cerebellum (Figure S7). Tm4sf20-null mice are viable and exhibit a neurobehavioral phenotype with a decreased anxiety-like response during open field testing.29 We speculate that the most likely mechanism of disease is a neurotoxic effect of the truncated protein. Haploinsufficiency is unlikely, as is a dominant-negative mechanism, because we identified a young parent (TM1203) homozygous for the deletion whose neuroradiological phenotype was not much different from the heterozygotes in our study (Figure S5). The observed cytoplasmic accumulation of mutant protein in Neuro2A cells is consistent with the genetic model and argues that a toxic effect of the mutant protein is the most plausible driver of the phenotype.
As in most penetrant genetic disorders, there is variable expressivity seen with TM4SF20 deletion. In the two affected individuals TM201 and TM301 with moderate to severe disease, the cognitive impairment was accompanied by other neurological sequelae including spasticity and epilepsy. Based on the brain imaging studies of 32 individuals, we consider that the significant brain abnormalities observed in TM201 are compounded by prematurity and ischemic injury in the left posterior cerebral artery (PCA) territory. It is unclear at present how the specific white matter hyperintensities relate to the neurodevelopmental profile in the carriers. It also remains to be seen whether the deletion causes microvascular or functional changes in the brain, responsible for language delay in these children. From our study, there is no obvious correlation between the extent of WMHs and severity of communication disorder in children. This is particularly highlighted by the brain MRI findings in TM1101, an 18-month-old Burmese female with the deletion, with normal gross motor development and a vocabulary of 3 words at 18 months of age. Her formal speech and language assessment determined her visual problem-solving skills DQ to be 87, language DQ to be 67, and overall developmental quotient to be 77. Her MRI study showed out-of-proportion abnormalities with multifocal subcortical and deep T2 hyperintensities returned from all lobes of both cerebral hemispheres (Figure 3). This study underscores the variability in the extent of white matter involvement in these children, varying from 1–2 focal punctate lesions (TM601) to multifocal lesions observed in multiple individuals with the deletion.
The white matter findings in the families are suggestive of remote small vascular insults as opposed to demyelination. Although these lesions appear to be nonprogressive in the young parents and probably represent gliosis, the natural history of WMHs in older carrier individuals remains to be determined. Our data point to additional contributors to the phenotype that modulate penetrance and expressivity. It will be interesting to evaluate the remaining wild-type allele in the pediatric subjects, because the deletion could also unmask other deleterious alleles, although additional extended families would be required to obtain sufficient statistical power to examine this possibility. It is also likely that other factors might contribute, including trans genetic modifiers, as well as environmental factors such as prematurity, aging, exposure to toxins (such as alcohol), or infectious agents. Although more studies are needed regarding the natural history of language delay in the carriers, the data from the parents and extended pedigree analyses suggest that language delay convalesces over time in most individuals, potentially reflecting compensatory neuronal plasticity. With a 1% allele frequency of this deletion in the Vietnamese Kinh population, this is a distinct example of a population-specific allele that underlies an apparent common disease trait inherent to a distinct world population. This deletion can be simply identified with the PCR-based amplification assay. For this population, this TM4SF20 allele could be responsible for early childhood language delay in a substantial number of the affected pediatric cases. Our findings provide a framework for the evaluation of larger cohorts of children with disorders of communication, including autism spectrum disorder for this deletion in distinct Southeast Asian and Far East subpopulations.
Acknowledgments
We are indebted to the families who participated in this research study. Support for this work was provided by the Doris Duke Charitable Foundation (DDCF) and Gillson Longenbaugh Foundation to S.R.L., National Institutes of Health (RO1-HL091771) to J.W.B. and S.R.L., NINDS (RO1-NS058529-03) and NHGRI (5U54HG006542) to J.R.L.; and NIMH (P50-MH094268) and a grant from the Simons Foundation (SFARI # 239983) to N.K. W.W. is supported by a Career Development Award K23NS078056 grant from the NINDS and Molecular Medicine Scholars Program at BCM (HL-66991). M.B.R. is grateful for the support of grant 5K08NS062711 from the NIH/NINDS. M.M.-S. was supported by the McKnight Endowment for Science, Dana Foundation, and the NIH Intellectual and the Developmental Disabilities Research Grant (P30HD024064). S.A.B. is partially funded by a Children’s Miracle Network endowed chair in pediatric genetics. N.K. is a Distinguished Jean and George W. Brumley Professor. We thank Emily Hall and Jillian Pearring for technical support. The Department of Molecular and Human Genetics at Baylor College of Medicine offers extensive genetic laboratory testing, including chromosomal microarray analysis, and derives revenue from this activity. J.A.R. is an employee of Signature Genomic Laboratories, a subsidiary of PerkinElmer, Inc.
Supplemental Data
Web Resources
The URLs for the data presented herein are as follows:
1000 Genomes, http://browser.1000genomes.org
Customized array CGH, http://www.bcm.edu/geneticlabs/index.cfm?pmid=19394
RepeatMasker, http://www.repeatmasker.org
UCSC Genome Browser, http://genome.ucsc.edu
References
- 1.Ylikoski A., Erkinjuntti T., Raininko R., Sarna S., Sulkava R., Tilvis R. White matter hyperintensities on MRI in the neurologically nondiseased elderly. Analysis of cohorts of consecutive subjects aged 55 to 85 years living at home. Stroke. 1995;26:1171–1177. doi: 10.1161/01.str.26.7.1171. [DOI] [PubMed] [Google Scholar]
- 2.Debette S., Markus H.S. The clinical importance of white matter hyperintensities on brain magnetic resonance imaging: systematic review and meta-analysis. BMJ. 2010;341:c3666. doi: 10.1136/bmj.c3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hopkins R.O., Beck C.J., Burnett D.L., Weaver L.K., Victoroff J., Bigler E.D. Prevalence of white matter hyperintensities in a young healthy population. J. Neuroimaging. 2006;16:243–251. doi: 10.1111/j.1552-6569.2006.00047.x. [DOI] [PubMed] [Google Scholar]
- 4.Katzman G.L., Dagher A.P., Patronas N.J. Incidental findings on brain magnetic resonance imaging from 1000 asymptomatic volunteers. JAMA. 1999;282:36–39. doi: 10.1001/jama.282.1.36. [DOI] [PubMed] [Google Scholar]
- 5.Kim B.S., Illes J., Kaplan R.T., Reiss A., Atlas S.W. Incidental findings on pediatric MR images of the brain. AJNR Am. J. Neuroradiol. 2002;23:1674–1677. [PMC free article] [PubMed] [Google Scholar]
- 6.Muscal E., Traipe E., de Guzman M.M., Myones B.L., Brey R.L., Hunter J.V. Cerebral and cerebellar volume loss in children and adolescents with systemic lupus erythematosus: a review of clinically acquired brain magnetic resonance imaging. J. Rheumatol. 2010;37:1768–1775. doi: 10.3899/jrheum.090983. [DOI] [PubMed] [Google Scholar]
- 7.Decobert F., Grabar S., Merzoug V., Kalifa G., Ponsot G., Adamsbaum C., des Portes V. Unexplained mental retardation: is brain MRI useful? Pediatr. Radiol. 2005;35:587–596. doi: 10.1007/s00247-005-1406-x. [DOI] [PubMed] [Google Scholar]
- 8.Verbruggen K.T., Meiners L.C., Sijens P.E., Lunsing R.J., van Spronsen F.J., Brouwer O.F. Magnetic resonance imaging and proton magnetic resonance spectroscopy of the brain in the diagnostic evaluation of developmental delay. Eur. J. Paediatr. Neurol. 2009;13:181–190. doi: 10.1016/j.ejpn.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 9.Widjaja E., Nilsson D., Blaser S., Raybaud C. White matter abnormalities in children with idiopathic developmental delay. Acta Radiol. 2008;49:589–595. doi: 10.1080/02841850801950087. [DOI] [PubMed] [Google Scholar]
- 10.Back S.A. Perinatal white matter injury: the changing spectrum of pathology and emerging insights into pathogenetic mechanisms. Ment. Retard. Dev. Disabil. Res. Rev. 2006;12:129–140. doi: 10.1002/mrdd.20107. [DOI] [PubMed] [Google Scholar]
- 11.Gupta S.N., Belay B. Intracranial incidental findings on brain MR images in a pediatric neurology practice: a retrospective study. J. Neurol. Sci. 2008;264:34–37. doi: 10.1016/j.jns.2007.06.055. [DOI] [PubMed] [Google Scholar]
- 12.Kalnin A.J., Fastenau P.S., deGrauw T.J., Musick B.S., Perkins S.M., Johnson C.S., Mathews V.P., Egelhoff J.C., Dunn D.W., Austin J.K. Magnetic resonance imaging findings in children with a first recognized seizure. Pediatr. Neurol. 2008;39:404–414. doi: 10.1016/j.pediatrneurol.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kieslich M., Errázuriz G., Posselt H.G., Moeller-Hartmann W., Zanella F., Boehles H. Brain white-matter lesions in celiac disease: a prospective study of 75 diet-treated patients. Pediatrics. 2001;108:E21. doi: 10.1542/peds.108.2.e21. [DOI] [PubMed] [Google Scholar]
- 14.Boone P.M., Bacino C.A., Shaw C.A., Eng P.A., Hixson P.M., Pursley A.N., Kang S.H., Yang Y., Wiszniewska J., Nowakowska B.A. Detection of clinically relevant exonic copy-number changes by array CGH. Hum. Mutat. 2010;31:1326–1342. doi: 10.1002/humu.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Hattab A.W., Smolarek T.A., Walker M.E., Schorry E.K., Immken L.L., Patel G., Abbott M.A., Lanpher B.C., Ou Z., Kang S.H. Redefined genomic architecture in 15q24 directed by patient deletion/duplication breakpoint mapping. Hum. Genet. 2009;126:589–602. doi: 10.1007/s00439-009-0706-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olshen A.B., Venkatraman E.S., Lucito R., Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 17.Cottingham R.W., Jr., Idury R.M., Schäffer A.A. Faster sequential genetic linkage computations. Am. J. Hum. Genet. 1993;53:252–263. [PMC free article] [PubMed] [Google Scholar]
- 18.Khor C.C., Chau T.N., Pang J., Davila S., Long H.T., Ong R.T., Dunstan S.J., Wills B., Farrar J., Van Tram T. Genome-wide association study identifies susceptibility loci for dengue shock syndrome at MICB and PLCE1. Nat. Genet. 2011;43:1139–1141. doi: 10.1038/ng.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rescorla L., Roberts J., Dahlsgaard K. Late talkers at 2: outcome at age 3. J. Speech Lang. Hear. Res. 1997;40:556–566. doi: 10.1044/jslhr.4003.556. [DOI] [PubMed] [Google Scholar]
- 20.Abecasis G.R., Altshuler D.L., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A., 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shih S.C., Zukauskas A., Li D., Liu G., Ang L.H., Nagy J.A., Brown L.F., Dvorak H.F. The L6 protein TM4SF1 is critical for endothelial cell function and tumor angiogenesis. Cancer Res. 2009;69:3272–3277. doi: 10.1158/0008-5472.CAN-08-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berditchevski F. Complexes of tetraspanins with integrins: more than meets the eye. J. Cell Sci. 2001;114:4143–4151. doi: 10.1242/jcs.114.23.4143. [DOI] [PubMed] [Google Scholar]
- 23.Choi S., Lee S.A., Kwak T.K., Kim H.J., Lee M.J., Ye S.K., Kim S.H., Kim S., Lee J.W. Cooperation between integrin alpha5 and tetraspan TM4SF5 regulates VEGF-mediated angiogenic activity. Blood. 2009;113:1845–1855. doi: 10.1182/blood-2008-05-160671. [DOI] [PubMed] [Google Scholar]
- 24.Lai C.S., Fisher S.E., Hurst J.A., Vargha-Khadem F., Monaco A.P. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- 25.Kang C., Drayna D. Genetics of speech and language disorders. Annu. Rev. Genomics Hum. Genet. 2011;12:145–164. doi: 10.1146/annurev-genom-090810-183119. [DOI] [PubMed] [Google Scholar]
- 26.Tomblin J.B., Records N.L., Buckwalter P., Zhang X., Smith E., O’Brien M. Prevalence of specific language impairment in kindergarten children. J. Speech Lang. Hear. Res. 1997;40:1245–1260. doi: 10.1044/jslhr.4006.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Randall D., Reynell J., Curwen M. A study of language development in a sample of 3 year old children. Br. J. Disord. Commun. 1974;9:3–16. doi: 10.3109/13682827409011603. [DOI] [PubMed] [Google Scholar]
- 28.Prathanee B., Purdy S.C., Thinkhamrop B., Chaimay B., Ruangdaraganon N., Mo-suwan L., Phuphaibul R. Early language delay and predictive factors in children aged 2 years. J. Med. Assoc. Thai. 2009;92:930–938. [PubMed] [Google Scholar]
- 29.Tang T., Li L., Tang J., Li Y., Lin W.Y., Martin F., Grant D., Solloway M., Parker L., Ye W. A mouse knockout library for secreted and transmembrane proteins. Nat. Biotechnol. 2010;28:749–755. doi: 10.1038/nbt.1644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


