Abstract
While chromatin modifications can offer a useful readout for enhancer activities, it is less clear whether these modification marks are a cause or consequence of transcription factor occupancy and enhancer activation. We have examined in details the temporal events of acetyltransferase associations and histone acetylations at different regulatory regions of the Myod1 locus. Our studies demonstrate that the histone acetyltransferase (HAT) p300 is stepwise enriched at distinct Myod1 regulatory regions during myogenic differentiation. This enrichment of p300 is associated with increased histone acetylation in a discrete pattern. Inhibition of p300 HAT activity impedes myogenic differentiation, which is coupled with decreased histone acetylation at specific Myod1 regulatory regions. We show for the first time that p300 is directly involved in the early regulation of Myod1 enhancer, and provide molecular insights into how p300 HAT activity and histone acetylation are related to enhancer activation and, consequently, gene transcription.
Gene expression is governed by an integrated action of many cis-regulatory elements including the long-range enhancers, proximal regulatory elements, and promoters. Dynamic interplay among this constellation of regulatory elements is a key attribute to the control of target gene transcription1. To command the intricate interaction of these regulatory elements, histone acetyltransferases (HAT) or transcriptional coactivators recruited by sequence specific transcription factors play leading roles in activating target enhancers and consequently the transcriptional program.
The transcriptional coactivator p300, containing an intrinsic HAT activity, is capable of interacting with a myriad of regulatory proteins and is therefore essential for many aspects of cellular processes2,3. The basic functional mode of p300 is to serve as a HAT enzyme to coordinate chromatin remodeling. Particularly, p300 is known as the best chromatin signature of enhancers4,5. The enrichments of H3K27 acetylation are often observed at p300 positive enhancers6,7,8,9. Thus, p300 association and histone acetylation are powerful means to identify subclass of putative enhancers. However, the relationship of different histone acetylation marks with chromatin signature of p300 occupancy, and the molecular basis for the histone acetylation-mediated enhancer activation remain to be determined.
Beside histone acetylation, p300 can also act as a scaffold for complex assembly and act as a bridge to connect the transcription factors with the basal transcriptional machinery10. Genetic evidence has revealed a particular role of p300 in the specification and terminal differentiation of skeletal muscle lineage. Mutation in the HAT domain of p300 results in the loss of Myf5 and MyoD gene expression, and ES cells lacking the p300 HAT activity exhibit severe defects in skeletal muscle formation11. Intriguingly, mutations in the HAT activity of CBP, a close functional homologue of p300, do not cause a clear muscle impairment, and the CBP mutant cells are still able to form myotubes11. Thus, p300 HAT activity is specifically required for normal muscle development.
Skeletal myogenesis is a highly ordered process coordinated through a succession of signal cascades which activate interconnected regulatory networks12. Sequential expression of the myogenic regulatory factors such as Myf5, MyoD, and myogenin, is essential for spatiotemporal expression of muscle specific genes, which is controlled by progressive activation of complex cis-regulatory elements13.
Within the Myod1 locus, a highly conserved core enhancer region (CER) controls normal spatiotemporal expression pattern of MyoD, and directs embryonic activation of Myod1 in muscle progenitor cells14,15. The distal regulatory region (DRR) and the proximal regulatory region (PRR) are important for the maintenance of MyoD gene expression16,17. While the DRR and PRR are not sufficient for the early activation of MyoD gene, they contain essential regulatory elements for sustaining MyoD expression in adult muscles18. Therefore, different Myod1 regulatory regions have distinct regulatory function in skeletal muscle development.
It is known that p300 HAT activity is involved in lysine specific histone acetylation, such as H3K27, to regulate Myf5 early enhancer during stem cell differentiation19. Previous studies have also suggested that Myod1 activation is regulated at multiple levels including chromatin structure and histone deposition14,20. However, whether p300 is directly involved in MyoD gene expression and its potential role in specific enhancer activation remain to be determined.
On the other hand, p300/CBP-associated factor (PCAF) has also been linked to myogenic differentiation21,22. The HAT activity of PCAF is essential for MyoD to function as a DNA-binding protein and for cell-cycle arrest in terminal differentiation22,23. Interestingly, inactivation of either PCAF or p300 abolishes the myogenic differentiation program, suggesting that p300 and PCAF have divergent functioning, and thereby cannot compensate for each other22.
In this study, we examined the association of p300 to the Myod1 locus and its coupling with histone acetylation and MyoD gene expression. Our studies provide molecular insights into the mechanisms by which the early expression of MyoD gene is regulated and the molecular basis for the requirement of p300 and histone acetylation at the Myod1 locus.
Results
C2C12 myogenic differentiation
C2C12 cells are myoblasts that have already committed to the skeletal muscle lineage. They proliferate when cultured with growth factors, but differentiate under low mitogen conditions24. By day 3 of differentiation, majority of cells developed into myocytes and formed multinucleated myotubes (Fig. 1A). The Myf5 protein was expressed in proliferating myoblasts, but not in differentiating cells which subsequently express myogenin protein (Fig. 1B). The up-regulation of MyoD transcripts was evident by day 1 of differentiation and peaked by day 2 (Fig. 1C). Since Myf5 is absent in the differentiating C2C12 cells, we employed this model to study a potential direct connection of p300 with the early regulation of MyoD gene expression during myogenic differentiation.
Figure 1. Association of p300 to the Myod1 locus.
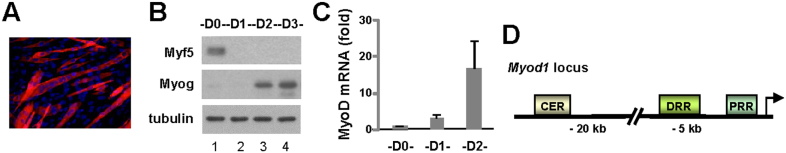
(A) C2C12 cells were cultured in differentiation medium and stained for myosin heavy chain (red) and the nuclei (blue) on day 3 of differentiation. Shown is the representative microscopy image. (B) The protein levels of Myf5 and myogenin were analyzed on day 1–3 of differentiation (d1, d2 or d3) by Western blotting. Proliferating myoblasts were included as a comparison and β-tubulin was used as a loading control. Shown are the cropped blot images representing indicated proteins. (C) The levels of MyoD transcripts on days 1 and 2 of differentiation were determined by real-time RT-PCR and presented as the fold variances relative to the proliferating myoblasts after being normalized to GAPDH. Error bars are the standard deviations of three independent experiments. (D) Schematic diagram of the Myod1 locus with presentation of the core enhancer region (CER), the distal regulatory region (DRR), and the proximal regulatory region (PRR).
The core enhancer region (CER) of Myod1 is important for the early activation of MyoD gene transcription whereas the distal regulatory region (DRR) and proximal regulatory region (PRR) play critical roles in maintaining Myod1 transcription at a late stage (Fig. 1D). Since the transcriptional coactivator p300 is essential for MyoD gene expression11, we sought to examine the association of p300 to the Myod1 locus by a real-time PCR based ChIP assay to examine the role of p300 in the regulation of MyoD gene expression.
Association of coactivators to the Myod1 locus
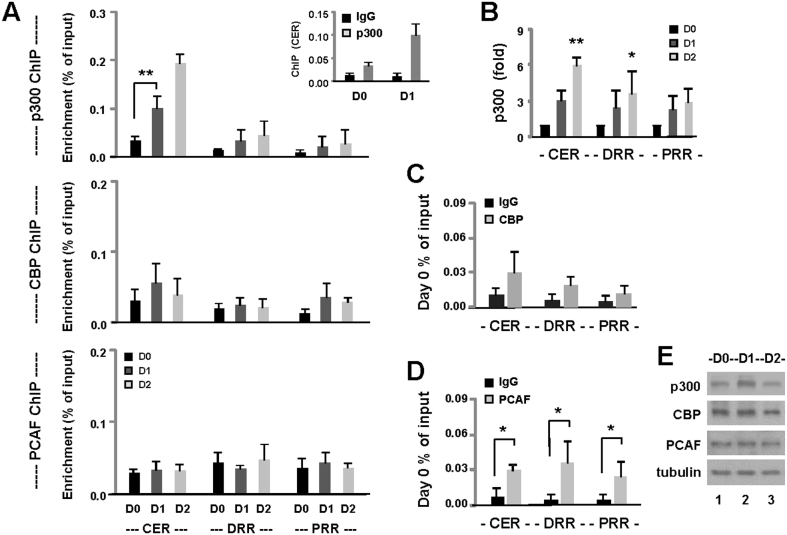
The C2C12 myoblasts were conditioned to differentiate for 1 and 2 days and processed for ChIP with a p300 specific antibody to examine in detail the profiles of p300 association to the Myod1 locus. Proliferating myoblasts were used as controls. As shown in figure 2A, p300 was mostly enriched at the CER comparing to the DRR and PRR. The enrichment of p300 at the CER was over 10-fold by day 1 when compared with the IgG ChIP control in the differentiating myoblasts, whereas it was only about 3-fold in the proliferating cells (Fig. 2A inset). More importantly, the association of p300 to the CER increased about 3-fold by day 1 of differentiation and about 6-fold by day 2, when compared to the proliferating cells (Fig. 2B). On the other hand, the enrichments of p300 occupancy at the DRR and PRR, only increased about 3-fold by day 2 of differentiation (Fig. 2B).
Figure 2. Association of coactivators to the Myod1 locus.
(A) Myoblasts were differentiated for 1 and 2 days, and then processed for the ChIP analysis using antibody against p300, CBP and PCAF. Proliferating myoblasts (D0) were used as controls. Quantification is presented as the percentage of enrichment in relation to the input chromatin DNA (** p < 0.01 in relation to the day 0 control). Error bars represent the standard deviations of three independent experiments. (B) The enrichments of p300 association were also plotted as the fold variations comparing to proliferating myoblasts (* p < 0.05 and ** p < 0.01 in relation to the day 0 control). (C) The enrichments of CBP at the locus were presented as a comparison to the normal IgG ChIP. (D) The enrichments of PCAF were presented as a comparison to the normal IgG ChIP (* p < 0.05 in relation to the day 0 control). (E) Western analysis of the protein levels of p300, CBP and PCAF, with β-tubulin as a loading control. Shown are the cropped blot images representing the indicated proteins.
We also examined the association of other nuclear HATs to the Myod1 locus by the ChIP analysis. As shown in figure 2A, CBP was somewhat enriched at the CER and PRR on day 1 of differentiation, but not significantly. It was only about 2–3 fold, when compared with the IgG ChIP control (Fig. 2C). While PCAF was also detected at the CER, DRR and PRR, the degree of enrichment was relatively low in comparison to p300 (Fig. 2A). Nonetheless, it was about 4–7 fold, when compared with the IgG ChIP control (Fig. 2D). Importantly, the levels of PCAF occupancy were similar in both proliferating and differentiating myoblasts (Fig. 2A), suggesting that PCAF is not involved in the activation of Myod1 enhancer during C2C12 differentiation. The protein levels of these HATs remained steady (Fig. 2E). Taken together, our data suggest that p300, but not CBP, is directly involved in the early regulation of the MyoD gene and we next sought to examine the molecular basis for the requirement of p300 in myogenic differentiation.
Effects of curcumin on myogenic differentiation
To determine the effects of p300 HAT activities on myogenic differentiation, we employed curcumin, a cell permeable compound from curcuma longa rhizome, which inhibits the HAT activity of p300 and CBP, but not that of PCAF25,26,27. Myoblasts were conditioned to differentiate in the presence of increasing concentrations of curcumin for 1–3 days. The cells were then stained for myosin heavy chain and subjected to quantitative immunofluorescence microscopy.
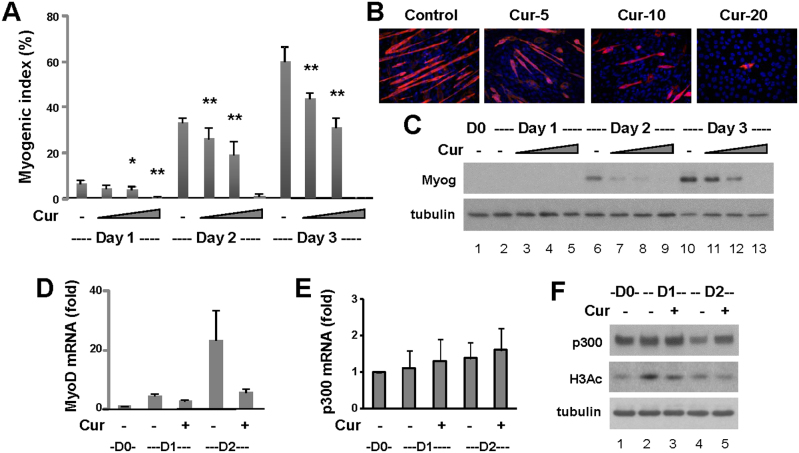
As shown in figure 3A, curcumin significantly inhibited the development of skeletal myocytes, which depends on the concentration and duration of the treatments. A high concentration of curcumin (20 μM) blocked skeletal myocyte development from day 1 of differentiation (Fig. 3A and B). Moderate concentration of curcumin (10 μM) allowed myogenic differentiation at a significantly lower efficacy, about 50–60% of respective untreated controls, and impaired the fusion events, the formation of multinucleated myotubes (Fig. 3A and B).
Figure 3. Effects of curcumin on C2C12 myogenic differentiation.
(A) C2C12 cells were differentiated in the presence of increasing concentrations of curcumin (5, 10, 20 μM) for 1–3 days, and then stained for myosin heavy chain. Hoechst was used to stain the nuclei. Quantitative microscopic analysis was presented as myogenic index, the percentage of myocyte nuclei in relation to the total number of nuclei (* p < 0.05 and ** p < 0.01 in relation to respective untreated controls). Error bars represent the standard deviations of four independent experiments. (B) Shown are the representative images stained for myosin heavy chain (red) and the nuclei (blue) on day 3 of differentiation. (C) Myogenin protein expression was examined by Western analysis following day 1–3 of differentiation, with β-tubulin as a loading control. Shown are the cropped blot images representing indicated protein. Full-length blots are presented in Supplementary Figure S1. (D) The levels of MyoD transcripts were determined by real-time RT-PCR analysis. Quantification is presented as fold variance relative to the proliferating myoblasts control after being normalized to GAPDH (n = 3). (E) The levels of p300 transcripts were also determined by real-time RT-PCR analysis. (F) The levels of p300 protein and global H3 acetylation were analyzed by Western blotting. Shown are the cropped blot images representing indicated proteins.
Western analysis showed that curcumin treatments also resulted in reduced myogenin expression in a concentration-dependent manner, which correlates with the quantitative microscopic analysis (Fig. 3A and C). The inhibitory effects of curcumin on MyoD mRNA were most pronounced on day 2, whereas the levels of p300 transcripts were not affected by curcumin (Fig. 3D and E). In addition, while p300 protein levels remained steady, the levels of global H3 acetylation were decreased by curcumin treatments (Fig. 3F). Thus, curcumin negatively affects myogenic differentiation through inhibiting the expression of myogenic regulators and muscle specific markers. As p300 HAT activity is required for MyoD gene expression in vivo11, upstream of myogenin, this negative effect likely stem from its property as an inhibitor of p300 HAT activity.
Acetylation of histones at the Myod1 locus
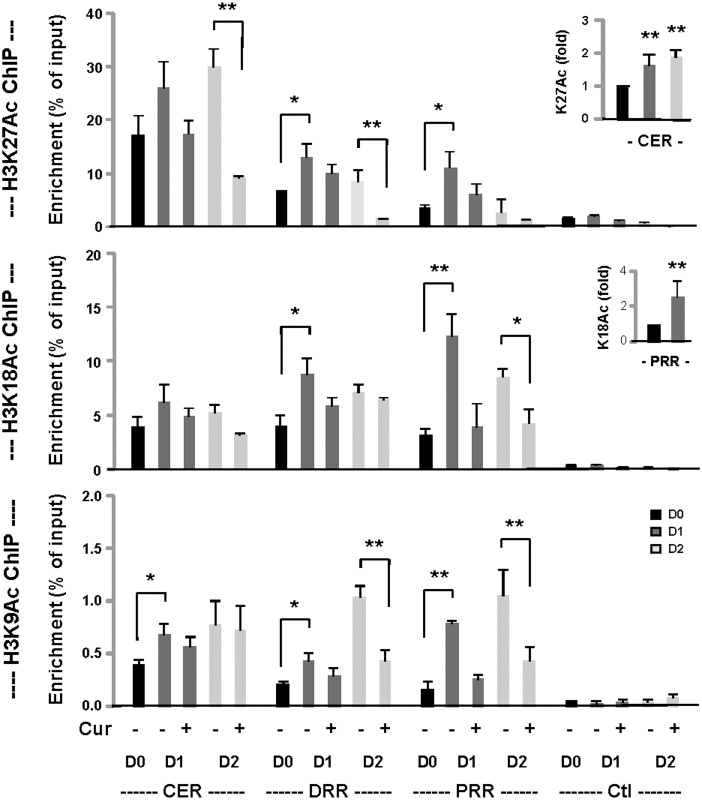
To examine the effects of curcumin on histone acetylation particularly at the Myod1 locus, we sought to profile the acetylation of specific histone H3 lysine residues during the early stage of differentiation. Myoblasts were differentiated for 1–2 days and then subjected to ChIP assay to quantify the levels of H3K27, H3K18 and H3K9 acetylation. As shown in figure 4, the basal level of H3K27Ac was most distinct at the CER and increased significantly in differentiating myoblasts. Interestingly, curcumin treatment at moderate concentration (10 μM) significantly reduced the level of H3K27Ac by day 2 of differentiation (Fig. 4). The increase of H3K27Ac in differentiating myoblasts and its inhibition by curcumin were also evident at the DRR and PRR (Fig. 4). In contrast to H3K27Ac, the basal levels of H3K18Ac were comparable at the CER, DRR and PRR (Fig. 4). However, the augmentation of H3K18Ac in the differentiating myoblasts was most significant at the PRR, which was reduced most significantly following curcumin treatment (Fig. 4).
Figure 4. Histone acetylation at the Myod1 locus.
Myoblasts were differentiated in the presence or absence of curcumin (10 μM) for 1 and 2 days, and then processed for the ChIP analysis using antibody against acetylated H3K27, H3K18 and H3K9 in parallel (* p < 0.05 and ** p < 0.01). Proliferating myoblasts (D0) were used as controls. A negative locus was also included in the ChIP analysis (Ctl). Quantification is presented as the percentage of enrichment in relation to the input chromatin DNA (n = 3). The levels of H3K27 acetylation at the CER on day 1 and 2 of differentiation were also plotted as the fold variations comparing to proliferating myoblasts (inset). Error bars are the standard deviations of four independent experiments (** p < 0.01 in relation to the day 0 control). The level of H3K18Ac at the PRR on day 1 of differentiation was also plotted as the fold variation comparing to proliferating myoblasts (inset). Error bars are the standard deviations of four independent experiments.
As observed for H3K27Ac, the basal level of H3K9Ac was also most distinct at the CER region, consistent with a previous study20. However, similar to H3K18Ac, the increase of H3K9Ac in differentiating myoblasts was most significant at the PRR, about 4-fold by day 1 of differentiation (Fig. 4). Most intriguingly, H3K9Ac was further augmented at the DRR and PRR by day 2 of differentiation, and the inhibition of H3K9Ac by curcumin was most significant at the DRR and PRR (Fig. 4). Thus our data revealed a discrete pattern of histone acetylation at the Myod1 locus and suggests that p300 HAT activity is directly involved in histone acetylation at this locus.
Discussion
In this study, we have examined the temporal events of HAT association and histone modifications at different regulatory regions of the Myod1 locus. Our studies revealed that p300 is temporally enriched at distinct Myod1 regulatory regions during myogenic differentiation. The enrichment of p300 correlates positively with histone acetylation in a region specific manner. We demonstrate for the first time that p300 is directly involved in the early regulation of MyoD gene expression, as well as provide mechanistic mechanistic insights into how p300 HAT activity and histone acetylation are related to enhancer regulation and, consequently, gene transcription.
Genetic studies have elegantly established that p300 HAT activity, but not CBP, is specifically required for the expression of myogenic regulatory factors Myf5 and MyoD11. Nonetheless, it was not clear if the p300 HAT activity is directly involved in the Myf5 and Myod1 gene regulation at the enhancer level. We have previously shown that p300 is enriched at the Myf5 early enhancer during the specification of stem cells into muscle lineage which correlates with H3K27 acetylation19. To discern whether p300 HAT activity is also directly involved in Myod1 enhancer regulation, we used C2C12 myoblasts in this study to minimize the influence of Myf5, an upstream factor of MyoD.
We found that in differentiating myoblasts, p300 is predominantly enriched at the CER, but not at the DRR and PRR if using an arbitrary threshold parameter (>10 fold of IgG ChIP). However, since the basal level of p300 occupancy in proliferating myoblats is lower at the DRR and PRR than at the CER, the fold increase of p300 enrichment at the DRR and PRR becomes discernible in differentiating myoblasts (Fig. 2). While H3K27 acetylation is most abundant at the CER in differentiating myoblasts, H3K18 and H3K9 acetylation is more distinct at the DRR and PRR (Fig. 4). Most importantly, the occupancy of CBP and PCAF at the Myod1 locus is relatively low and at similar levels in both proliferating and differentiating myoblasts (Fig. 2). Thus, p300 appears to be directly involved in the regulation of CER, DRR and PRR through specific histone acetylation.
Temporal priming and rate-limiting of chromatin remodeling is essential for specific enhancer activation and temporal-spatial expression of developmental genes. Many lineage specific enhancers are marked with histone modifications in progenitor cells, although these enhancers are not yet engaged in strong regulatory activities at this stage6,7. In the case of Myod1 locus, H3K27 acetylation is distinctly associated with the CER in proliferating myoblasts, which is further enriched and coincides with p300 association, H3K18 and H3K9 acetylation at the DRR and PRR, and MyoD gene expression during myogenic differentiation (Fig. 2 and 4).
In addition, our data show that the CER is not a simple transcriptional switch. Rather, it acts through stepwise p300 enrichment and histone acetylation. While histone acetylation can provide a powerful readout for regulatory element activities, it is still unclear if the acetylation marks are a cause or consequence of enhancer activation. Our data indicate that histone acetylation at the Myod1 enhancer by p300 is required for MyoD gene expression and myogenic differentiation. Thus, histone acetylation seems to be a driving force in the MyoD gene regulation and plays a causative role in the enhancer activation. Our studies shed new light into the molecular mechanisms that govern the intricate interactions between regulatory elements and target gene transcription.
Methods
Cell culture and differentiation
C2C12 myoblasts (American Type Culture Collection) were maintained in Dulbecco's Modified Eagle Medium (D-MEM, Invitrogen) supplemented with 10% fetal bovine serum (HyClone) at 37°C with 5% CO2. Differentiation medium was D-MEM supplemented with 2% of horse serum (Invitrogen). For differentiation, the cells were grown to 80% confluency and then cultured in the differentiation medium for an additional 1–3 days. Curcumin was purchased from Sigma-Aldrich.
Immunofluorescence microscopy
At indicated time points, cells were fixed with methanol, rehydrated in PBS and incubated with an antibody against myosin heavy chain, and then washed with PBS and incubated with Alexa Fluor®594 secondary antibody (Invitrogen). The cells were also incubated with 0.1 μg/ml Hoechst (Molecular Probes) to stain the DNA28. Finally, the coverslips were mounted on slides with 50% glycerol. Microscopic analysis was performed with the Zeiss Axiovert 200 M microscope. Cells were observed through a Zeiss 20× objective and images were captured with the AxioCam HRM monochrome camera (Zeiss). Images captured through different fluorescence filters were processed and merged using the Zeiss AxioVision Rel 4.8 software. Myogenic index was determined as the percentage of myocyte nuclei in relation to the total number of nuclei. Each experiment was repeated at least three times. The specific antibody for myosin heavy chain was produced as previously described19.
Real-time RT-PCR
Total RNA was isolated using Total RNA kit I (Omega) according to the manufacturer's protocol including DNase I treatment. Reverse transcription to cDNA was performed using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Real-Time PCR was performed with a MX300P platform (Strategene) using the SYBR green method. Each sample was PCR amplified in triplicates. Results were analyzed by the threshold cycle (Ct) comparative method using GAPDH as an internal control. The experiments were repeated three times. Primers for GAPDH have been described previously19. The primers used for MyoD and p300 were as the following:
MyoD fwd-TGCCTTCTACGCACCTGGA. MyoD rev-ATCATGCCATCAGAGCAGTTGG.
p300 fwd-GCTCCTGGACAGTCAAAGAAG. p300 rev-TCAGGGATTCCTAGAAGCTGC.
Chromatin immunoprecipitation (ChIP)
Cells were differentiated for 1–2 days. At indicated time points, cells were fixed, crosslinked, and sonicated as previously described29. Chromatin DNA was purified using the Omega Bio-tek Cycle Pure Kit (Omega) and quantified using the NanoDrop Spectrophotometer (ND-1000) to ensure equal amounts of chromatin were used in the immunoprecipitation of different samples. For each batch of immunoprecipitation, corresponding normal IgG antiserum was used as a negative control. The immunoprecipitates were captured by incubation with dynabeads protein-A for 2 hours at 4°C. The chromatin DNA was then purified and real-time PCR was performed using the SYBR green method on the MX300P platform (Strategene). Each sample was amplified in triplicate PCR reactions with primer as described previously30. Purified input DNA was used to create a standard curve in the PCR amplification for each immunoprecipitate. Quantification was analyzed as the abundance of immunoprecipitated target DNA in percentage of input chromatin DNA (enrichment as the percentage of input). Each ChIP was repeated at least three times. Antibodies specific for p300, CBP and PCAF were obtained from the Santa Cruz Biotechnology. Antibodies for H3K27Ac, H3K18Ac and H3K9Ac were purchased from Abcam.
Western analysis
Cells were lysed by incubation in whole cell extract buffer (10% glycerol, 50 mM Tris-HCl pH 7.6, 400 mM NaCl, 5 mM EDTA, 1 mM DTT, 1 mM PMSF, 1% NP-40) on a rotator for 30 minutes at 4°C, and cell extracts are prepared as previously described31. The concentration of proteins was determined by Bradford assay using a Bio-Rad Protein Assay Dye Reagent (Bio-Rad) and the Multiscan Spectrum Photospectrometer (Thermo). Antibodies specific for Myf5, p300, CBP, and PCAF were obtained from Santa Cruz Biotechnology. Antibodies for myogenin and β-tubulin were generated as previously described32. All experiments were repeated at least three times.
Author Contributions
M.H. executed microscopy and ChIP analysis. S.K. participated in Western and RT-PCR analysis. J.C. and Q.L. interpreted the data and prepared the manuscript. Q.L. designed the research. All authors reviewed the manuscript.
Supplementary Material
Supplementary figure
Acknowledgments
This work was supported by operating grants to Q.L. from the Canadian Institutes of Health Research. M.H. is a recipient of a Graduate Scholarship from the Ministry of Higher Education of Saudi Arabia. We thank members of the Blais Lab for helpful discussion of the protocols and sharing of the equipment. We also thank Dr. Dilworth for reagents.
References
- Heintzman N. D. & Ren B. Finding distal regulatory elements in the human genome. Curr Opin Genet Dev 19, 541–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko V. V., Schiltz R. L., Russanova V., Howard B. H. & Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87, 953–9 (1996). [DOI] [PubMed] [Google Scholar]
- Bedford D. C. & Brindle P. K. Is histone acetylation the most important physiological function for CBP and p300? Aging (Albany NY) 4, 247–55 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N. D. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39, 311–8 (2007). [DOI] [PubMed] [Google Scholar]
- Blow M. J. et al. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet 42, 806–10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton M. P. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 107, 21931–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A. et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–83 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N. D. et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–12 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y. B. & Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet 8, 9–22 (2007). [DOI] [PubMed] [Google Scholar]
- Chen J. & Li Q. Life and death of transcriptional co-activator p300. Epigenetics 6, 957–61 (2011). [DOI] [PubMed] [Google Scholar]
- Roth J. F. et al. Differential role of p300 and CBP acetyltransferase during myogenesis: p300 acts upstream of MyoD and Myf5. Embo J 22, 5186–96 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott S. J. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 132, 2685–95 (2005). [DOI] [PubMed] [Google Scholar]
- Francetic T. & Li Q. Skeletal myogenesis and Myf5 activation. Transcription 2, 109–14 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldhamer D. J. et al. Embryonic activation of the myoD gene is regulated by a highly conserved distal control element. Development 121, 637–49 (1995). [DOI] [PubMed] [Google Scholar]
- Kablar B. et al. Myogenic determination occurs independently in somites and limb buds. Dev Biol 206, 219–31 (1999). [DOI] [PubMed] [Google Scholar]
- Tapscott S. J., Lassar A. B. & Weintraub H. A novel myoblast enhancer element mediates MyoD transcription. Mol Cell Biol 12, 4994–5003 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura A., Lyons G. E. & Tapscott S. J. The regulation of MyoD gene expression: conserved elements mediate expression in embryonic axial muscle. Dev Biol 171, 386–98 (1995). [DOI] [PubMed] [Google Scholar]
- Charge S. B., Brack A. S., Bayol S. A. & Hughes S. M. MyoD- and nerve-dependent maintenance of MyoD expression in mature muscle fibres acts through the DRR/PRR element. BMC Dev Biol 8, 5 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francetic T. et al. Regulation of Myf5 early enhancer by histone acetyltransferase p300 during stem cell differentiation. Mol Biol 1, 103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. H. et al. Myogenic transcriptional activation of MyoD mediated by replication-independent histone deposition. Proc Natl Acad Sci U S A 108, 85–90 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. J., Ogryzko V. V., Nishikawa J., Howard B. H. & Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382, 319–324 (1996). [DOI] [PubMed] [Google Scholar]
- Puri P. L. et al. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell 1, 35–45 (1997). [DOI] [PubMed] [Google Scholar]
- Sartorelli V. et al. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol Cell 4, 725–34 (1999). [DOI] [PubMed] [Google Scholar]
- Silberstein L., Webster S. G., Travis M. & Blau H. M. Developmental progression of myosin gene expression in cultured muscle cells. Cell 46, 1075–81 (1986). [DOI] [PubMed] [Google Scholar]
- Balasubramanyam K., Swaminathan V., Ranganathan A. & Kundu T. K. Small molecule modulators of histone acetyltransferase p300. J Biol Chem 278, 19134–40 (2003). [DOI] [PubMed] [Google Scholar]
- Balasubramanyam K. et al. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J Biol Chem 279, 51163–71 (2004). [DOI] [PubMed] [Google Scholar]
- Morimoto T. et al. The dietary compound curcumin inhibits p300 histone acetyltransferase activity and prevents heart failure in rats. J Clin Invest 118, 868–78 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Germain J. R., Chen J. & Li Q. Involvement of PML nuclear bodies in CBP degradation through the ubiquitin-proteasome pathway. Epigenetics 3, 342–9 (2008). [DOI] [PubMed] [Google Scholar]
- Higazi A., Abed M., Chen J. & Li Q. Promoter context determines the role of proteasome in ligand-dependent occupancy of retinoic acid responsive elements. Epigenetics 6, 202–211 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Chu A., Chakroun I., Islam U. & Blais A. Cooperation between myogenic regulatory factors and SIX family transcription factors is important for myoblast differentiation. Nucleic Acids Res 38, 6857–71 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., St-Germain J. R. & Li Q. B56 Regulatory Subunit of Protein Phosphatase 2A Mediates Valproic Acid-Induced p300 Degradation. Mol Cell Biol 25, 525–32 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le May M. et al. Contribution of Retinoid X Receptor Signaling to the Specification of Skeletal Muscle Lineage. J Biol Chem 286, 26806–12 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure