Abstract
Background:
Multiple myeloma (MM) is still a fatal plasma cell cancer. Novel compounds are currently clinically tested as a single agent in relapsing patients, but in best cases with partial response of a fraction of patients, emphasising the need to design tools predicting drug efficacy. Histone deacetylase inhibitors (HDACi) are anticancer agents targeting epigenetic regulation of gene expression and are in clinical development in MM.
Methods:
To create a score predicting HDACi efficacy, five MM cell lines were treated with trichostatin A (TSA) and gene expression profiles were determined.
Results:
The expression of 95 genes was found to be upregulated by TSA, using paired supervised analysis with Significance Analysis of Microarrays software. Thirty-seven of these 95 genes had prognostic value for overall survival in a cohort of 206 newly diagnosed MM patients and their prognostic information was summed up in a histone acetylation score (HA Score); patients with the highest HA Score had the shorter overall survival. It is worth noting that MM cell lines or patients' primary MM cells with a high HA Score had a significant higher sensitivity to TSA, valproic acid, panobinostat or vorinostat.
Conclusion:
In conclusion, the HA Score allows identification of MM patients with poor survival, who could benefit from HDACi treatment.
Keywords: multiple myeloma, HDACi, GEP-based score, prediction of drug sensitivity
The molecular events governing the onset and progression of malignant transformation involve oncogenic activation and inactivation of tumour suppressor genes, which help cancer cells over-riding the normal mechanisms controlling cellular survival and proliferation (Hahn and Weinberg, 2002; Vogelstein and Kinzler, 2004). These molecular events are triggered by DNA alterations (translocations, amplifications or deletions, mutations) and also by epigenetic modifications (Baylin, 2005). Epigenetic modifications include methylation of DNA cytosine residues and histone methylation or acetylation, and are critical in the initiation and progression of many cancers (Kondo, 2009). Acetylation of histone releases condensed chromatin into a more relaxed structure that is associated with greater levels of gene transcription. Histone acetylation and then gene transcription is controlled by histone acetyl transferases and histone deacetylases (HDACs) bringing and removing acetyl groups. Eighteen HDACs have been described and classified into four classes based on cellular localisation and function (Lane and Chabner, 2009). Class I HDACs are located in the nucleus and comprise HDACs 1, 2, 3 and 8. Class II HDACs comprise HDACs 4, 5, 7 and 9 (class IIa), which shuttle back and forth between the nucleus and the cytoplasm, and HDACs 6 and 10 (class IIb), which are expressed in the cytoplasm only. Class III HDACs include the sirtuin family, which does not act primarily on histones, and class IV includes HDAC11 (Lane and Chabner, 2009; Neri et al, 2012). Histone deacetylase inhibitors (HDACi) are now being used in the treatment of some hematologic malignancies (Kelly et al, 2010). Histone deacetylase inhibitors are classified into four classes according to their chemical structure: aliphatic acids (valproic acid (VPA) and sodium phenylbutyrate), hydroxamates (panobinostat, trichostatin A (TSA), vorinostat, belinostat (PXD101), NVP-LAQ824 and givinostat (ITF2357)), cyclic peptide (romidepsin (depsipeptide)) and benzamides (MS-275, MGCD0103) (Neri et al, 2012). Histone deacetylase inhibitors include inhibitors specific to class I HDACs (MGCD0103, romidepsin and MS-275) and pan-HDACi (TSA, panobinostat, vorinostat and belinostat) (Neri et al, 2012). Multiple myeloma (MM) is a plasma cell (PC) neoplasm characterised by the accumulation of malignant PCs, termed MM cells (MMCs), primarily within the bone marrow (BM). Despite the recent introduction of novel agents such as bortezomib or lenalidomide, MM remains an almost incurable disease (Moreau, 2012). Multiple myeloma arises through progressive accumulation of multiple genetic abnormalities that include primarily overexpression of a D-type cyclin gene, CCND1 (cyclin D1) in the case of t(11;14)(q13;q32.3) translocation, CCND3 (cyclin D3) in the case of the rare t(6p23;14q32) translocation or CCND2 (cyclin D2) on the background of a t(14q32;16q23) translocation involving CMAF or t(4;14)(p16.3;q32.3) involving WHSC1/FGFR3. CCND genes are also overexpressed in hyperdiploid MM patients because of gene amplification or downregulation of miRNAs that target CCND genes (Bergsagel and Kuehl, 2005; Rio-Machin et al, 2013). Secondary genetic abnormalities include NRAS or KRAS mutations, TP53 monoallelic deletion and mutations, MYC alterations, mutations of genes coding for NF-κB pathway (Hideshima et al, 2004; Bergsagel and Kuehl, 2005; Morgan et al, 2012). Histone deacetylase inhibitors have already been evaluated in MM including TSA (Lavelle et al, 2001), vorinostat (Mitsiades et al, 2003, 2004), depsipeptide (Khan et al, 2004), KD5170 (Feng et al, 2008), NVP-LAQ824 (Catley et al, 2003), VPA (Kaiser et al, 2006; Neri et al, 2008) and panobinostat (Neri et al, 2012). Histone deacetylase inhibitors induce G1 cell cycle arrest in MMCs through dephosphorylation of retinoblastoma protein and increase expression of p53 and p21 (Lavelle et al, 2001; Mitsiades et al, 2003; Neri et al, 2008). Histone deacetylase inhibitors induce apoptosis by dowregulation of Bcl-2 family members (Mitsiades et al, 2003; Khan et al, 2004) and overcome drug resistance mediated by the BM environment (Mitsiades et al, 2003). Furthermore, glucose-regulated protein 78 (GRP78) was recently identified as a novel non-histone target of HDACi (Rao et al, 2010; Kahali et al, 2011). Glucose-regulated protein 78 has a central role in the unfolded protein response (UPR). Glucose-regulated protein 78 acetylation following HDACi treatment was described to activate UPR and contributes to the antitumour activity of HDACi. Class 1 HDACs binding to GRP78, within the endoplasmic reticulum (ER), represent a novel mode of UPR regulation and an interesting mechanism of HDACi action (Kahali et al, 2012). The ER of normal PCs and MMCs is well developed to accommodate the production and secretion of large amounts of immunoglobulins. That is why association of HDACi with proteasome inhibitors could be promising in MM treatment (Hideshima and Anderson, 2013). When used as a single agent in patients with relapsing/refractory MM, HDACi have shown modest antitumour activity (Richardson et al, 2008; Niesvizky et al, 2011). In combination with other anti-MM treatments, HDACi can induce durable antitumour responses (Badros et al, 2009; Harrison et al, 2011).
To improve the clinical testing of the efficacy of novel agents, a major stake is identify patients who could benefit from treatment by finding biomarkers predictive for sensitivity of MMCs to HDACi. We recently reported the development of a gene expression-based risk score predicting the sensitivity of MMCs to DNA methylation inhibitors (Moreaux et al, 2012). In this study, we used the same strategy to build a histone acetylation (HA) score, based on genes whose expression is deregulated by HDACi in MMCs. Histone acetylation score score makes it possible to identify a subgroup of 42% of patients with short overall survival (OS), whose MMCs are highly sensitive to HDAC inhibition.
Materials and methods
Human myeloma cell lines
Human myeloma cell lines (HMCLs, N=40) were obtained as described previously (Zhang et al, 1994; Rebouissou et al, 1998; Tarte et al, 1999; Gu et al, 2000; Moreaux et al, 2011). Human myeloma cell lines' phenotypic and molecular characteristics have been described previously (Moreaux et al, 2011). Human myeloma cell lines' microarray data have been deposited in the ArrayExpress public database (accession numbers: E-TABM-937 and E-TABM-1088).
Primary MMCs and gene expression profiling
Patients presenting with previously untreated MM (N=206) or monoclonal gammopathy of undetermined significance (N=5) at the university hospitals of Heidelberg and Montpellier as well as seven healthy donors have been included in the study with the approval of the ethics committee of Montpellier and Heidelberg after obtaining written informed consent in accordance with the Declaration of Helsinki. Clinical parameters and treatment regimens of the MM patients included in the Heidelberg–Montpellier cohort were described previously (Moreaux et al, 2012).
Normal BM PCs and myeloma cells were purified as published previously (Moreaux et al, 2012) and whole genome gene expression profiling assayed with Affymetrix U133 2.0 plus microarrays (Affymetrix, Santa Clara, CA, USA) (ArrayExpress accession number E-MTAB-372). Affymetrix data of an independent cohort of 345 MM patients from the University of Arkansas for Medical Sciences (UAMS, Little Rock, AR, USA) were also used (Gene Expression Omnibus accession number GSE2658. http://www.ncbi.nlm.nih.gov/geo/). T(4;14) translocation was evaluated using MMSET spike expression (Kassambara et al, 2012b) and del17p13 surrogated by TP53 probe set signal (Xiong et al, 2008) for UAMS-TT2 patients.
Change in gene expression profile of myeloma cell lines by HDACi
Five HMCLs (XG-5, XG-6, XG-7, XG-20 and LP1) were treated without or with 0.33 μmol l−1 TSA (Sigma, St Louis, MO, USA) for 24 h in RPMI 1640, 10% fetal bovine serum supplemented with IL-6 for IL-6-dependent HMCLs (Moreaux et al, 2011, 2012). Whole genome gene expression profiling was assayed with Affymetrix U133 2.0 plus microarrays (Affymetrix).
Sensitivity of myeloma cell lines and primary myeloma cells to HDACi
Human myeloma cell lines were cultured with graded TSA, VPA (Sigma), vorinostat (SAHA) (Sigma) or panobinostat (LBH-589) (Sigma) concentrations. Human myeloma cell lines' cell growth was quantified with a Cell Titer Glo Luminescent Assay (Promega, Madison, WI, USA) as described(Moreaux et al, 2012). The half inhibitory concentration (IC50) was determined using GraphPad Prism (http://www.graphpad.com/scientific-software/prism/).
Primary myeloma cells of 13 patients were cultured with or without graded concentrations of TSA and MMC cytotoxicity was evaluated using anti-CD138-PE mAb (Immunotech, Marseille, France) as described (Mahtouk et al, 2004; Moreaux et al, 2012).
Bioinformatics and statistics
Gene expression data were analysed using SAM (Significance Analysis of Microarrays) software (Cui and Churchill, 2003) and our bioinformatics platforms (http://rage.montp.inserm.fr/ and http://amazonia.montp.inserm.fr/) (Reme et al, 2008; Tanguy Le Carrour, 2010) as published (Moreaux et al, 2012). All computations were performed using R 2.15.1 (http://www.r-project.org/) and bioconductor 2.0. Survival analyses were investigated using the Kaplan–Meier method and Cox's proportional hazards model as published (Moreaux et al, 2012).
The HA Score was built using our previously published methodologies to develop prognostic scores using a set of prognostic genes coding for related proteins (Kassambara et al, 2012a, 2012b; Moreaux et al, 2012). Histone acetylation score is the sum of the Cox β-coefficients of each of the 37 TSA-deregulated genes with a prognostic value, weighted by ±1 if the patient MMC signal for a given gene is above or below the probe set Maxstat value of this gene (Moreaux et al, 2012).
Results
Identification of prognostic genes whose expression is upregulated by TSA treatment of MMCs
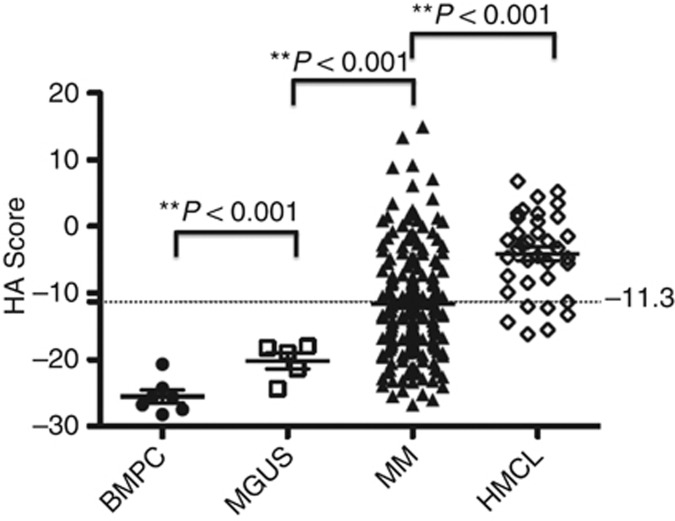
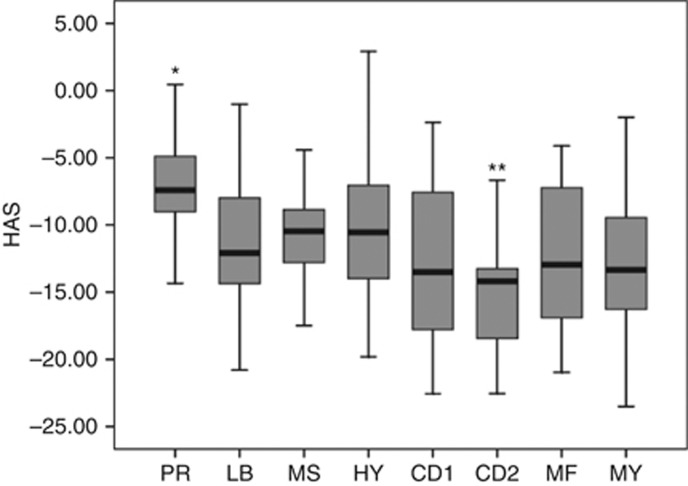
Genes upregulated by TSA treatment of MMCs were identified by treating five HMCLs with 0.33 μM TSA for 24 h, a concentration-inducing histone acetylation in mammalian cells, in particular in myeloma cells in vitro (Yoshida et al, 1990; Heller et al, 2008). In addition, this TSA treatment did not affect myeloma cell viability (Supplementary Table S1) (Heller et al, 2008). Using SAM supervised paired analysis, expression of 95 genes was found significantly upregulated by TSA treatment (FDR <5% Supplementary Table S2). Trichostatin A-regulated genes are significantly enriched in genes related to ‘Immunological disease and Inflammatory disease' pathway (P<0.05; Ingenuity pathway analysis, data not shown). It is worth noting that TSA-deregulated genes were also found to be upregulated by Panobinostat treatment in MMC (Supplementary Figure S1). We next investigated associations of TSA-deregulated genes with OS using Maxstat R function. The aim was to identify genes regulated by HDAC that have potentially important disease-modulating functions. The results of our analysis were corrected for multiple testing using the Benjamini–Hochberg algorithm. Investigating the expression of these 95 TSA-regulated genes in primary MMCs of a cohort of 206 newly diagnosed patients (HM cohort), 16 genes had a bad prognostic value and 21 a good one (Table 1). The prognostic information of HDACi-regulated genes was gathered within an HA Score as indicated in Materials and Methods. The value of HA Score in normal, premalignant or malignant PCs is displayed in Figure 1. Cells from MGUS patients had a significantly higher HA Score than normal BMPCs (P<0.001), MMCs of patients had a significantly higher HA Score than normal BMPCs or PCs from MGUS patients (P<0.001), and HMCLs had the highest score (P<0.001) (Figure 1). Figure 2B shows the contribution of the 16 bad prognostic and the 21 good prognostic genes for HA score. Bad prognostic genes are highly expressed in patients with high HA score and the reverse for good prognostic genes. Investigating the HA Score in the eight groups of the molecular classification of MM (Zhan et al, 2006), HA Score was significantly higher in the proliferation subgroup (P<0.001) associated with a poor prognosis and significantly lower in the CD2 subgroup (P<0.001) (Zhan et al, 2006) (Figure 3). Among the 37 genes of the HA Score, 7 code for proteins that have been described as lysine acetylation target proteins and 18 have been identified as HDACi targets (Choudhary et al, 2009; Niesen and Blanck, 2009; Bantscheff et al, 2011; Iwahashi et al, 2011) (Supplementary Tables S5 and S6). Gene expression profiles of HA Score genes in purified MMC and normal BM subpopulations are listed in Supplementary Figure S2. Supplementary Figure S2 shows a highly variable expression of each of the 37 genes in primary MMCs of the patients, indicating they all contribute to unravel disease heterogeneity.
Table 1. Prognostic value of TSA-deregulated genes in primary MMC of newly diagnosed patients.
| Probe set | Name | Adjusted P-value (Benjamini–Hochberg multiple testing correction) | Hazard ratio |
|---|---|---|---|
|
Bad prognostic genes | |||
| 204563_at | SELL | 0.04 | 1.94 |
| 203567_s_at | TRIM38 | 0.04 | 1.96 |
| 201012_at | ANXA1 | 0.02 | 2.01 |
| 205352_at | SERPINI1 | 0.04 | 2.03 |
| 204944_at | PTPRG | 0.01 | 2.12 |
| 222651_s_at | TRPS1 | 0.03 | 2.17 |
| 214875_x_at | APLP2 | 0.01 | 2.19 |
| 203854_at | IF | 0.03 | 2.34 |
| 209958_s_at | PTHB1 | 0.01 | 2.35 |
| 209969_s_at | STAT1 | 0.009 | 2.37 |
| 205552_s_at | OAS1 | 0.01 | 2.50 |
| 226269_at | GDAP1 | 0.008 | 2.69 |
| 210432_s_at | SCN3A | 0.007 | 2.71 |
| 224701_at | PARP14 | 0.01 | 2.94 |
| 214079_at | DHRS2 | 4.76e−05 | 3.11 |
| 226158_at |
KLHL24 |
0.01 |
3.44 |
|
Good prognostic genes | |||
| 34408_at | RTN2 | 2.42e−05 | 0.28 |
| 225842_at | — | 9.96e−05 | 0.32 |
| 208894_at | HLA-DRA | 0.01 | 0.36 |
| 212464_s_at | FN1 | 0.01 | 0.37 |
| 202391_at | BASP1 | 7.01e−05 | 0.37 |
| 228726_at | SERPINB1 | 0.009 | 0.38 |
| 235301_at | KIAA1324L | 0.01 | 0.39 |
| 206385_s_at | ANK3 | 0.007 | 0.40 |
| 230233_at | RASGEF1B | 0.04 | 0.42 |
| 215193_x_at | HLA-DRB1 | 0.01 | 0.43 |
| 212636_at | QKI | 0.02 | 0.44 |
| 212998_x_at | HLA-DQB1 | 0.01 | 0.47 |
| 223218_s_at | NFKBIZ | 0.03 | 0.47 |
| 209198_s_at | SYT11 | 0.03 | 0.48 |
| 211990_at | HLA-DPA1 | 0.02 | 0.49 |
| 218918_at | MAN1C1 | 0.04 | 0.49 |
| 215388_s_at | CFH | 0.04 | 0.52 |
| 228152_s_at | FLJ31033 | 0.03 | 0.53 |
| 216834_at | RGS1 | 0.04 | 0.54 |
| 203695_s_at | DFNA5 | 0.04 | 0.54 |
| 219833_s_at | EFHC1 | 0.04 | 0.55 |
Abbreviations: TSA=trichostatin A; MMC=multiple myeloma cell.
Figure 1.
Histone acetylation score in normal and malignant PCs. Histone acetylation score in normal BMPCs (N=7), in premalignant PCs of patients with monoclonal gammopathy of undetermined significance (MGUS, N=5), in MM cells of patients with intramedullary MM (N=206) and in HMCLs (N=40). ** Indicate that the score value is significantly different with a P-value <0.01.
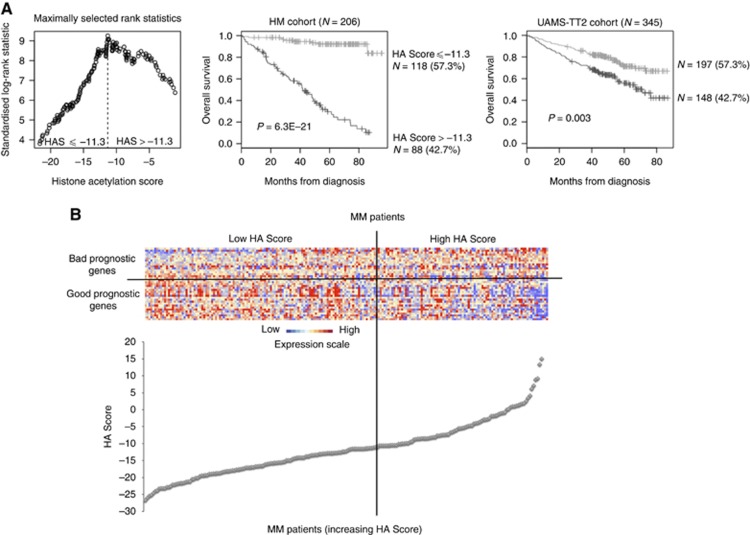
Figure 2.
Prognostic value of HA Score in MM. (A) Patients of the HM cohort were ranked according to increased HA Score and a maximum difference in OS was obtained with HA Score=−11.3 splitting patients into high-risk (42.7%) and low-risk (57.3%) groups. Histone acetylation score also had a prognostic value of an independent cohort of 345 patients from University of Arkansas for Medical Science (UAMS) treated with TT2 therapy (UAMS-TT2 cohort). The parameters to compute the HA Score of patients of UAMS-TT2 cohort and the proportions delineating the two prognostic groups were those defined with HM cohort. (B) Clustergram of HA Score genes ordered from best to worst prognosis. The level of the probe set signal is displayed from low (deep blue) to high (deep red) expression. MM patients (N=206) were ordered by increasing GE-based risk score.
Figure 3.
Histone acetylation score in MM molecular subgroups. The HA Score was computed for MMCs of patients belonging to the eight groups of the University of Arkansas for Medical Science (UAMS) molecular classification of multiple myeloma, using UAMS-TT2 cohort. *Score value is significantly higher in the group compared with all the patients of the cohort (P<0.05). **Score value is significantly lower in the group compared with all the patients of the cohort (P<0.05). Abbreviations: CD1, cyclin D1; CD2, cyclin D2; HY, hyperdiploid; LB, low bone disease; MF, MAF; MS, MMSET; MY, myeloid; PR, proliferation.
Prognostic value of HA score in two independent cohorts of patients
When used as a continuous variable, HA Score had prognostic value (P⩽10−4, results not shown). Using Maxstat R function, a maximum difference in OS was obtained with an HA Score=−11.3, splitting patients into a high-risk group of 42.7% patients (HA Score >−11.3) with a 43.5 months median OS and a low-risk group of 57.3% patients (HA Score ⩽−11.3) with not reached median survival (Figure 2A). High-risk patients are characterised by a higher expression of the 16 bad prognostic genes and a lower expression of the 21 good ones used for HA Score building (Figure 2B). Using univariate Cox analysis, HA Score, UAMS-HRS, IFM-score and GPI had prognostic value as well as t(4;14), del17p, β2m, albumin and ISS using the HM patient cohort (Supplementary Table S3). When these parameters were compared two by two, HA Score tested with β2m and t(4;14) remained significant. When tested together, HA Score, β2m, t(4;14) and GPI kept prognostic value. The HA Score, computed using HM cohort parameters, is also prognostic in an independent cohort of 345 patients from UAMS (UAMS-TT2 cohort). The median OS of patients with high HA Score was 71.4 months and not reached for patients with low HA Score (P<0.0001) (Figure 2A). Using Cox univariate analysis, UAMS-HRS, IFM and GPI scores as well as t(4;14) and del17p had prognostic value. Serum concentrations of β2m or albumin are not publicly available for this cohort. When analysed two by two, HA Score remained significant compared with UAMS-HRS, IFM, GPI, t(4;14) and del17p in the UAMS-TT2 cohort (Supplementary Table S3). When these parameters were tested together, HA Score, UAMS-HRS, t(4;14) and del17p kept prognostic value in UAMS-TT2 cohort.
Histone acetylation score allows identification of HMCLs or patients' primary MMCs sensitive to TSA in vitro
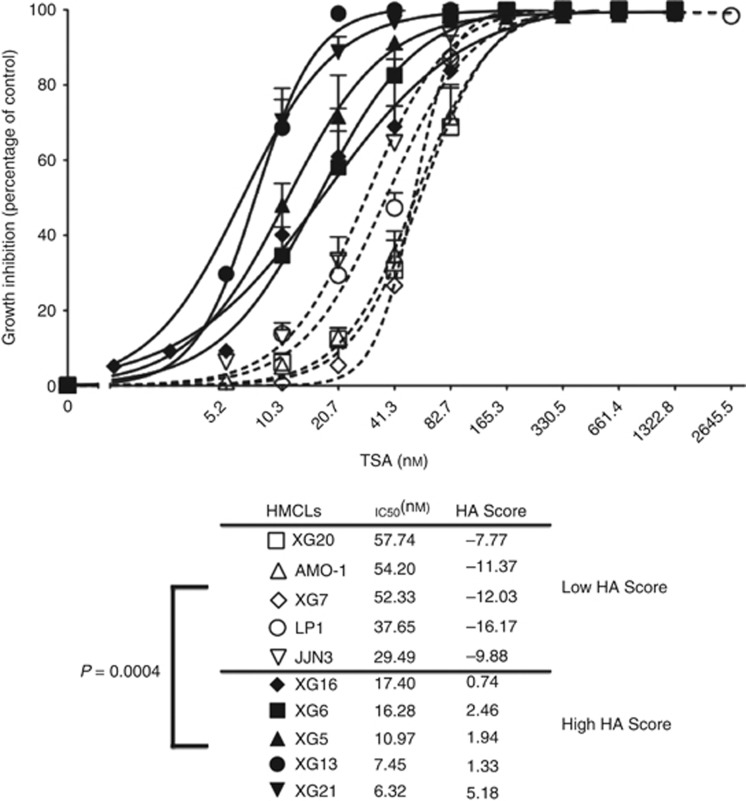
We investigated whether HA Score could predict for the sensitivity of HMCLs to HDAC inhibitors. Ten out of 40 HMCLs (Moreaux et al, 2011) with the highest or lowest HA Score were selected to assay for TSA sensitivity. The five HMCLs with the highest HA Score displayed a significant (P=0.0004) fivefold higher sensitivity to TSA (median IC50=10.97 nM; range: 6.32–17.4 nM) than the five HMCLs with low HA Score (median IC50=52.33 nM; range: 29.49–57.74 nM) (Figure 4). No difference in recurrent genetic abnormalities was found between HMCLs with the highest or lowest HA Score (Table 2). Histone acetylation score could also predict for sensitivity of patients' primary MMCs, cocultured with BM environment, to TSA. The TSA concentrations used to treat primary MM samples were chosen to cover the range of TSA concentrations yielding 50% inhibition of the growth of the 10 HMCLs displaying high and low HA Scores (Figure 4).
Figure 4.
Histone acetylation score predicts for sensitivity of HMCLs to TSA. HMCLs with high HA Score (N=5) exhibit significant higher TSA sensitivity compared with HMCLs with low HA Score (N=5). Human myeloma cell lines were cultured for 4 days in 96-well flat-bottom microtitre plates in RPMI 1640 medium, 10% fetal calf serum, 2 ng ml−1 IL-6 culture medium (control) and graded TSA concentrations. Data are mean values ±standard deviation (s.d.) of five experiments determined on sextuplet culture wells.
Table 2. Characteristics of HDACi-resistant or -sensitive HMCLs.
| HMCL name | IL-6 dependencea | Originb | Diseasec | Patient sampled | Gender | Isotype | t(14q32 or 22q11) | Target genes | Ras | TP53 | CD45 | HMCL classification |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
TSA-resistant HMCLs | ||||||||||||
| XG7 | + | MN | MM | PB | F | Ak | t(4;14) | MMSET | mut | wt | +/− | MS |
| XG20 | ++ | MN | PCL | PB | M | l | t(4;14) | MMSET | wt | abn | − | MS |
| AMO1 | − | CO | PCT | AF | F | Ak | t(12;14) | Unknown | wt | wt | + | CD-2L |
| JJN3 | − | CO | MM | PE | F | Ak | t(14;16) | c-Maf | mut | abn | +/− | MF |
| LP1 |
− |
CO |
MM |
PB |
F |
Gl |
t(4;14) |
MMSET/FGFR3 |
wt |
abn |
− |
MS |
|
TSA-sensitive HMCLs | ||||||||||||
| XG5 | ++ | MN | MM | PB | F | l | t(11;14) | CCND1 | wt | abn | − | CD-1 |
| XG6 | ++ | MN | MM | PB | F | Gl | t(16;22) | c-Maf | wt | wt | + | CTA/MF |
| XG13 | ++ | MN | PCL | PB | M | Gl | t(14;16) | c-Maf | wt | abn | + | MF |
| XG16 | ++ | MN | PCL | PB | M | k | None | None | mut | abn | + | CTA/FRZB |
| XG21 | ++ | MN | MM | PE | M | l | t(11;14) | CCND1 | wt | wt | + | CD-1 |
Abbreviations: AF=ascitic fluid; BM=bone marrow; CO=collected; F=female; HDACi=histone deacetylase inhibitor; HMCL=human myeloma cell line; IL=interleukin; M=male; MM=multiple myeloma; MN=Montpellier or Nantes; PB=peripheral blood; PCL=plasma cell leukemia; PCT=plasmacytoma; PE=pleural effusion; TSA=trichostatin A.
++, if growth is strictly dependent on adding exogenous IL-6; +, if dependent on adding exogenous IL-6; −, if not.
Origin of the HMCL: MN, CO.
Disease at diagnosis: MM, PCL, PCT.
Origin of the sample: AF, BM, PE, PB.
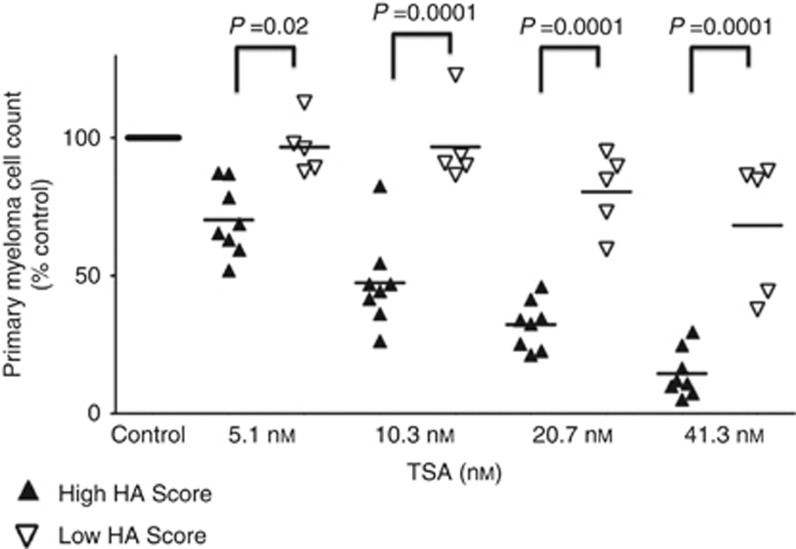
Primary MMCs of eight patients with an HA Score above the Maxstat cutoff (>−11.3; Figures 1 and 2) exhibited significant (P<0.05) 2.4-fold higher TSA sensitivity than MMCs of five patients with HA Score ⩽−11.3 (Figure 5).
Figure 5.
Histone acetylation score predicts for TSA sensitivity of primary myeloma cells of patients. Mononuclear cells from tumour samples of 13 patients with MM were cultured for 4 days in the presence of IL-6 (2 ng ml−1) with or without graded TSA concentrations. At day 4 of culture, the count of viable MMCs was determined using CD138 staining by flow cytometry. Black color represents patients with high HA Score (N=8) and white represents patients with low HA Score values (N=5).
Histone acetylation score is predictive for sensitivity of human myeloma cells to other clinical grade HDACi in vitro
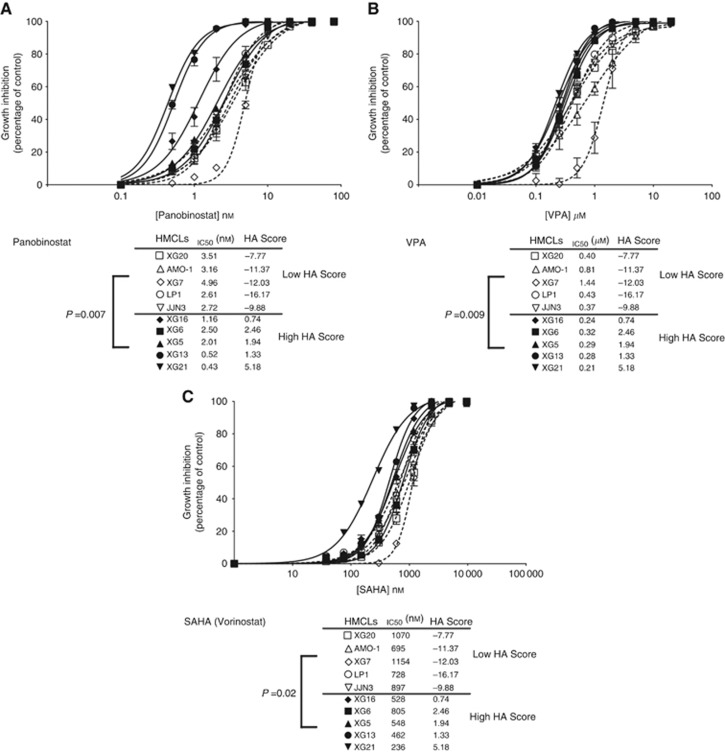
We sought to determine whether HA Score could predict for the sensitivity of myeloma cells to other clinical grade HDAC inhibitors (Neri et al, 2012). The five HMCLs with the highest HA Score exhibited a significant higher sensitivity to panobinostat, VPA or vorinostat (median IC50=1.16 nM, 0.28 μM and 528 nM, respectively) than the five HMCLs with lowest HA Score (P=0.007, P=0.009 and P=0.02; median IC50=3.16 nM, 0.43 μM and 897 nM, respectively) (Figure 6A–C).
Figure 6.
Histone acetylation score predicts for sensitivity of HMCLs to panobinostat, valproic acid (VPA) or SAHA HDACi. Human myeloma cell lines with a high HA Score (N=5) exhibit significant higher sensitivity to panobinostat, VPA or SAHA compared with HMCLs with a low HA Score (N=5). Human myeloma cell lines were cultured for 4 days in 96-well flat-bottom microtitre plates in RPMI 1640 medium, 10% fetal calf serum, 2 ng ml−1 IL-6 culture medium (control) and graded concentration of panobinostat (A), VPA (B) or SAHA (vorinostat) (C). Data are mean values± standard deviation (s.d.) of five experiments determined on sextuplet culture wells.
Discussion
In this study, we have identified a gene expression-based HA Score, which is predictive for patients' survival and for the in vitro sensitivity of HMCLs or patients' primary myeloma cells to the pan-HDACi, TSA and also to other three clinical-grade HDACi. Histone deacetylase inhibitors have been investigated for treating patients with MM, either as a single agent (Richardson et al, 2008; Niesvizky et al, 2011; Neri et al, 2012) or in combination with current drugs used in MM, such as dexamethasone, lenalidomide or bortezomib (Badros et al, 2009; Harrison et al, 2011; Neri et al, 2012). Panobinostat, used as a single agent, demonstrated limited activity in patients with MM, refractory to at least two lines of treatment (Wolf et al, 2012). In association with melphalan, panobinostat yielded a 33% overall response rate in a phase I study, including 12 patients with relapsed/refractory MM (Offidani et al, 2012). Association of panobinostat and bortezomib could be promising as proteasome inhibition affects the UPR pathway, leading to increased HDAC-mediated aggregosome formation (Hideshima et al, 2011). Phase IB and II studies have shown that association of panobinostat to bortezomib and dexamethasone could yield to objective response in relapsing patients refractory to bortezomib therapy (Siegel et al, 2008; San-Miguel et al, 2009; Richardson et al, 2011). Given the encouraging in vitro and in vivo data, association of panobinostat, bortezomib and dexamethasone is now being evaluated in a large phase III randomised trial (San-Miguel et al, 2011). Combination of vorinostat and bortezomib was also investigated in a phase II trial, including patients with MM refractory or ineligible to bortezomib or IMiD therapy (Siegel et al, 2011) and showed a 17% overall response rate and 6 months median response duration. These data also prompt investigating the interest of a combination of vorinostat and bortezomib in phase III trial, including 637 patients with relapsed/refractory MM (Dimopoulos et al, 2011). Final results of the trial remain awaited, but interim results did not demonstrate a marked therapeutic benefit of vorinostat (Dimopoulos et al, 2011). These trials suggest that HDACi could have some benefit for MM. However, their efficacy could be underestimated because it was limited to a subgroup of patients. The current HA Score could be promising to investigate whether the best response to HDACi is found in patients with MMCs displaying a high HA Score.
Only upregulated genes were identified in the HMCLs treated with TSA compared with untreated HMCLs. This may be explained by the 24-h treatment of cell lines with HDACi according to usual protocols. This short treatment makes it possible to release the transcriptional suppressor activity of HDAC yielding to overexpressed genes. However, it is likely too short to get an indirect repression of genes because of the HDACi-induced overexpression of an inhibitor of these genes. Among the current 95 genes deregulated by TSA treatment in HMCLs, 24 genes were commonly identified by Heller et al (2008) (Supplementary Table S4). Seven out of 37 HA Score genes code for proteins that have been described as lysine acetylation target proteins and 18 out of 37 HA Score genes have been identified as HDACi targets (Choudhary et al, 2009; Niesen and Blanck, 2009; Bantscheff et al, 2011; Iwahashi et al, 2011) (Supplementary Tables S5 and S6).
Why HA Score which is built using 37 HDACi-upregulated and prognostic genes could predict for the sensitivity of MMCs to HDACi? Patients with high HA score, and poor survival, are characterised by a higher expression of the 16 bad prognostic genes and a lower expression of the 21 good ones in MMCs (Figure 2B). Thus, one can speculate that primary MMCs of patients with high HA score have a high tumour metabolism and growth, which can be efficiently targeted by the upregulation of gene products encoded by genes upregulated by HDACi, in particular the 21 good prognostic genes. At the opposite, MMCs of patients with a low HA score could be in a more quiescent state and less sensitive to HDCAi. However, a full understanding of the reason why HA score could predict for HDACi sensitivity will be provided by an extensive study of the function of the products encoded by HDACi-regulated genes in promoting MMC survival and/or proliferation. Some genes could highlight pathways involved in MM and we comment below the putative roles of NFKBIZ (nuclear factor of kappa light polypeptide gene enhancer in B-cell inhibitor zeta), BASP1 (brain acid-soluble protein 1) or QKI (Quaking), whose expression in MMCs is induced by HDACi treatment and is associated with good prognosis. NFKBIZ is a member of IκB family (Totzke et al, 2006), localised in the nucleus, where it interacts with and regulates nuclear NF-κB activity. Suppression of endogenous NFKBIZ renders cells more resistant to apoptosis, whereas its overexpression induces cell death (Yamazaki et al, 2001; Totzke et al, 2006). This is of interest because NF-κB pathway is frequently activated through various gene mutations in MM (Annunziata et al, 2007; Keats et al, 2007). More recently, it was demonstrated that NFKBIZ inhibits the transcriptional activity of STAT3, leading to cell growth inhibition and apoptosis induction mediated by the downregulation of a known STAT3 target, Mcl-1 (Wu et al, 2009). Mcl-1 is the major antiapoptotic protein for MMCs, involved in IL-6-mediated survival of MMCs (Derenne et al, 2002; Jourdan et al, 2003). Brain acid-soluble protein 1 is repressed in Myc-transformed cells, and conversely has a strong potential to inhibit cell transformation induced by Myc (Hartl et al, 2009). The inhibition of Myc-induced fibroblast cell transformation by BASP1 also prevents the transcriptional activation or repression of known Myc target genes. Brain acid-soluble protein 1 appears to be a potential tumour suppressor in cancer (Hartl et al, 2009). Myc protein is frequently highly expressed in primary MMCs (Skopelitou et al, 1993) and a vicious circle involving IRF4 and Myc was identified yielding to deregulation of MMC growth (Shaffer et al, 2008). Histone deacetylase inhibitors could be useful to target NF-κB or Myc activation in MMCs through the upregulation of NFKBIZ and BASP1 expression. The RNA-binding protein QKI belongs to the evolutionarily conserved signal-transduction and activator of RNA family. It has been demonstrated that overexpression of QKI induced the G1 cell cycle arrest in oligodendrocyte progenitor cells (Larocque et al, 2005). Furthermore, QKI inhibits colon cancer cell growth, acting as a tumour suppressor (Yang et al, 2010). It was demonstrated that QKI protein is directly transcribed by E2F1, which in turn negatively regulates the cell cycle by targeting multiple cell cycle regulators including p27, cyclin D1 and c-fos (Yang et al, 2011). These results demonstrated that a better understanding of the cellular response to epigenetic-targeted treatments will increase our knowledge of MM development and progression and will provide potential therapeutic advances. Epigenetic therapies could be combined with conventional therapies to develop personalised treatments in MM and render resistant tumours responsive to treatment. These advances may limit the side effects of treatment, improving compliance with dosing regimens and overall quality of life. Our methodology could be extended to other anti-MM treatments.
Acknowledgments
This work was supported by grants from ARC (SL220110603450, Paris France, to BK), ANR emergence (ETTMM, to BK) and FEDER (141 786 and 42667, to BK and JM) from the Hopp-Foundation, Germany, the University of Heidelberg, Germany, the National Centre for Tumour Diseases, Heidelberg, Germany, the Tumorzentrum Heidelberg/Mannheim, Germany, and the Deutsche Krebshilfe, Bonn, Germany, the Deutsche Forschungsgemeinschaft, Bonn, Germany (to HG). We thank the Microarray Core Facility of IRB (http://irb.montp.inserm.fr/en/index.php?page=Plateau&IdEquipe=6) and the cytometry platform of the Institute of Research in Biotherapy (http://irb.montp.inserm.fr/en/index.php?page=Plateau&IdEquipe=3, Montpellier Rio Imaging).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Author contributions
MJ designed the research and wrote the paper; HD and GH collected BM samples and clinical data and participated in the writing of the paper; RT and VJL participated in the bioinformatics studies; LW and RG provided with technical assistance; and KB designed and supervised the research and wrote the paper.
Supplementary Material
References
- Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, Lenz G, Hanamura I, Wright G, Xiao W, Dave S, Hurt EM, Tan B, Zhao H, Stephens O, Santra M, Williams DR, Dang L, Barlogie B, Shaughnessy JD, Jr, Kuehl WM, Staudt LM. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12 (2:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badros A, Burger AM, Philip S, Niesvizky R, Kolla SS, Goloubeva O, Harris C, Zwiebel J, Wright JJ, Espinoza-Delgado I, Baer MR, Holleran JL, Egorin MJ, Grant S. Phase I study of vorinostat in combination with bortezomib for relapsed and refractory multiple myeloma. Clin Cancer Res. 2009;15 (16:5250–5257. doi: 10.1158/1078-0432.CCR-08-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantscheff M, Hopf C, Savitski MM, Dittmann A, Grandi P, Michon AM, Schlegl J, Abraham Y, Becher I, Bergamini G, Boesche M, Delling M, Dumpelfeld B, Eberhard D, Huthmacher C, Mathieson T, Poeckel D, Reader V, Strunk K, Sweetman G, Kruse U, Neubauer G, Ramsden NG, Drewes G. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat Biotechnol. 2011;29 (3:255–265. doi: 10.1038/nbt.1759. [DOI] [PubMed] [Google Scholar]
- Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2 (Suppl 1:S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol. 2005;23 (26:6333–6338. doi: 10.1200/JCO.2005.05.021. [DOI] [PubMed] [Google Scholar]
- Catley L, Weisberg E, Tai YT, Atadja P, Remiszewski S, Hideshima T, Mitsiades N, Shringarpure R, LeBlanc R, Chauhan D, Munshi NC, Schlossman R, Richardson P, Griffin J, Anderson KC. NVP-LAQ824 is a potent novel histone deacetylase inhibitor with significant activity against multiple myeloma. Blood. 2003;102 (7:2615–2622. doi: 10.1182/blood-2003-01-0233. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325 (5942:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Cui X, Churchill GA. Statistical tests for differential expression in cDNA microarray experiments. Genome Biol. 2003;4 (4:210. doi: 10.1186/gb-2003-4-4-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derenne S, Monia B, Dean NM, Taylor JK, Rapp MJ, Harousseau JL, Bataille R, Amiot M. Antisense strategy shows that Mcl-1 rather than Bcl-2 or Bcl-x(L) is an essential survival protein of human myeloma cells. Blood. 2002;100 (1:194–199. doi: 10.1182/blood.v100.1.194. [DOI] [PubMed] [Google Scholar]
- Dimopoulos MA, Sundar J, Yoon SS, Siegel DS, Lonial S, Hajek R, Facon T, Rosiñol L, Blacklock A, Goldschmidt H, Hungria V, Spencer A, Palumbo A, Reece DE, Graef T, Houp J, Sun L, Eid JE, Anderson KC.2011Vantage 088: vorinostat in combination with bortezomib in patients with relapsed/refractory multiple myeloma: results of a global, randomized phase 3 trial Blood (118)368–369.abstract 811; oral presentation.
- Feng R, Ma H, Hassig CA, Payne JE, Smith ND, Mapara MY, Hager JH, Lentzsch S. KD5170, a novel mercaptoketone-based histone deacetylase inhibitor, exerts antimyeloma effects by DNA damage and mitochondrial signaling. Mol Cancer Ther. 2008;7 (6:1494–1505. doi: 10.1158/1535-7163.MCT-08-0183. [DOI] [PubMed] [Google Scholar]
- Gu ZJ, Vos JD, Rebouissou C, Jourdan M, Zhang XG, Rossi JF, Wijdenes J, Klein B. Agonist anti-gp130 transducer monoclonal antibodies are human myeloma cell survival and growth factors. Leukemia. 2000;14 (1:188–197. doi: 10.1038/sj.leu.2401632. [DOI] [PubMed] [Google Scholar]
- Hahn WC, Weinberg RA. Rules for making human tumor cells. N Engl J Med. 2002;347 (20:1593–1603. doi: 10.1056/NEJMra021902. [DOI] [PubMed] [Google Scholar]
- Harrison SJ, Quach H, Link E, Seymour JF, Ritchie DS, Ruell S, Dean J, Januszewicz H, Johnstone R, Neeson P, Dickinson M, Nichols J, Prince HM. A high rate of durable responses with romidepsin, bortezomib, and dexamethasone in relapsed or refractory multiple myeloma. Blood. 2011;118 (24:6274–6283. doi: 10.1182/blood-2011-03-339879. [DOI] [PubMed] [Google Scholar]
- Hartl M, Nist A, Khan MI, Valovka T, Bister K. Inhibition of Myc-induced cell transformation by brain acid-soluble protein 1 (BASP1) Proc Natl Acad Sci USA. 2009;106 (14:5604–5609. doi: 10.1073/pnas.0812101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller G, Schmidt WM, Ziegler B, Holzer S, Mullauer L, Bilban M, Zielinski CC, Drach J, Zochbauer-Muller S. Genome-wide transcriptional response to 5-aza-2'-deoxycytidine and trichostatin a in multiple myeloma cells. Cancer Res. 2008;68 (1:44–54. doi: 10.1158/0008-5472.CAN-07-2531. [DOI] [PubMed] [Google Scholar]
- Hideshima T, Anderson KC. Histone deacetylase inhibitors in the treatment for multiple myeloma. Int J Hematol. 2013;97 (3:324–332. doi: 10.1007/s12185-013-1290-3. [DOI] [PubMed] [Google Scholar]
- Hideshima T, Bergsagel PL, Kuehl WM, Anderson KC. Advances in biology of multiple myeloma: clinical applications. Blood. 2004;104 (3:607–618. doi: 10.1182/blood-2004-01-0037. [DOI] [PubMed] [Google Scholar]
- Hideshima T, Richardson PG, Anderson KC. Mechanism of action of proteasome inhibitors and deacetylase inhibitors and the biological basis of synergy in multiple myeloma. Mol Cancer Ther. 2011;10 (11:2034–2042. doi: 10.1158/1535-7163.MCT-11-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahashi S, Shimada M, Utsunomiya T, Morine Y, Imura S, Ikemoto T, Mori H, Hanaoka J, Saito Y. Histone deacetylase inhibitor enhances the anti-tumor effect of gemcitabine: a special reference to gene-expression microarray analysis. Oncol Rep. 2011;26 (5:1057–1062. doi: 10.3892/or.2011.1407. [DOI] [PubMed] [Google Scholar]
- Jourdan M, Veyrune JL, Vos JD, Redal N, Couderc G, Klein B. A major role for Mcl-1 antiapoptotic protein in the IL-6-induced survival of human myeloma cells. Oncogene. 2003;22 (19:2950–2959. doi: 10.1038/sj.onc.1206423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahali S, Sarcar B, Chinnaiyan P. The emerging role of histone deacetylases (HDACs) in UPR regulation. Methods Enzymol. 2011;490:159–174. doi: 10.1016/B978-0-12-385114-7.00010-6. [DOI] [PubMed] [Google Scholar]
- Kahali S, Sarcar B, Prabhu A, Seto E, Chinnaiyan P. Class I histone deacetylases localize to the endoplasmic reticulum and modulate the unfolded protein response. FASEB J. 2012;26 (6:2437–2445. doi: 10.1096/fj.11-193706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser M, Zavrski I, Sterz J, Jakob C, Fleissner C, Kloetzel PM, Sezer O, Heider U. The effects of the histone deacetylase inhibitor valproic acid on cell cycle, growth suppression and apoptosis in multiple myeloma. Haematologica. 2006;91 (2:248–251. [PubMed] [Google Scholar]
- Kassambara A, Hose D, Moreaux J, Reme T, Torrent J, Rossi JF, Goldschmidt H, Klein B. Identification of pluripotent and adult stem cell genes unrelated to cell cycle and associated with poor prognosis in multiple myeloma. PLoS One. 2012a;7 (7:e42161. doi: 10.1371/journal.pone.0042161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassambara A, Hose D, Moreaux J, Walker BA, Protopopov A, Reme T, Pellestor F, Pantesco V, Jauch A, Morgan G, Goldschmidt H, Klein B. Genes with a spike expression are clustered in chromosome (sub)bands and spike (sub)bands have a powerful prognostic value in patients with multiple myeloma. Haematologica. 2012b;97 (4:622–630. doi: 10.3324/haematol.2011.046821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, Van Wier S, Tiedemann R, Shi CX, Sebag M, Braggio E, Henry T, Zhu YX, Fogle H, Price-Troska T, Ahmann G, Mancini C, Brents LA, Kumar S, Greipp P, Dispenzieri A, Bryant B, Mulligan G, Bruhn L, Barrett M, Valdez R, Trent J, Stewart AK, Carpten J, Bergsagel PL. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12 (2:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2010;28 (10:1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SB, Maududi T, Barton K, Ayers J, Alkan S. Analysis of histone deacetylase inhibitor, depsipeptide (FR901228), effect on multiple myeloma. Br J Haematol. 2004;125 (2:156–161. doi: 10.1111/j.1365-2141.2004.04882.x. [DOI] [PubMed] [Google Scholar]
- Kondo Y. Epigenetic cross-talk between DNA methylation and histone modifications in human cancers. Yonsei Med J. 2009;50 (4:455–463. doi: 10.3349/ymj.2009.50.4.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol. 2009;27 (32:5459–5468. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- Larocque D, Galarneau A, Liu HN, Scott M, Almazan G, Richard S. Protection of p27(Kip1) mRNA by quaking RNA binding proteins promotes oligodendrocyte differentiation. Nat Neurosci. 2005;8 (1:27–33. doi: 10.1038/nn1359. [DOI] [PubMed] [Google Scholar]
- Lavelle D, Chen YH, Hankewych M, DeSimone J. Histone deacetylase inhibitors increase p21(WAF1) and induce apoptosis of human myeloma cell lines independent of decreased IL-6 receptor expression. Am J Hematol. 2001;68 (3:170–178. doi: 10.1002/ajh.1174. [DOI] [PubMed] [Google Scholar]
- Mahtouk K, Jourdan M, De Vos J, Hertogh C, Fiol G, Jourdan E, Rossi JF, Klein B. An inhibitor of the EGF receptor family blocks myeloma cell growth factor activity of HB-EGF and potentiates dexamethasone or anti-IL-6 antibody-induced apoptosis. Blood. 2004;103 (5:1829–1837. doi: 10.1182/blood-2003-05-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsiades CS, Mitsiades NS, McMullan CJ, Poulaki V, Shringarpure R, Hideshima T, Akiyama M, Chauhan D, Munshi N, Gu X, Bailey C, Joseph M, Libermann TA, Richon VM, Marks PA, Anderson KC. Transcriptional signature of histone deacetylase inhibition in multiple myeloma: biological and clinical implications. Proc Natl Acad Sci USA. 2004;101 (2:540–545. doi: 10.1073/pnas.2536759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsiades N, Mitsiades CS, Richardson PG, McMullan C, Poulaki V, Fanourakis G, Schlossman R, Chauhan D, Munshi NC, Hideshima T, Richon VM, Marks PA, Anderson KC. Molecular sequelae of histone deacetylase inhibition in human malignant B cells. Blood. 2003;101 (10:4055–4062. doi: 10.1182/blood-2002-11-3514. [DOI] [PubMed] [Google Scholar]
- Moreau P. The future of therapy for relapsed/refractory multiple myeloma: emerging agents and novel treatment strategies. Semin Hematol. 2012;49 (Suppl 1:S33–S46. doi: 10.1053/j.seminhematol.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Moreaux J, Klein B, Bataille R, Descamps G, Maiga S, Hose D, Goldschmidt H, Jauch A, Reme T, Jourdan M, Amiot M, Pellat-Deceunynck C. A high-risk signature for patients with multiple myeloma established from the molecular classification of human myeloma cell lines. Haematologica. 2011;96 (4:574–582. doi: 10.3324/haematol.2010.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreaux J, Reme T, Leonard W, Veyrune JL, Requirand G, Goldschmidt H, Hose D, Klein B. Development of gene expression-based score to predict sensitivity of multiple myeloma cells to DNA methylation inhibitors. Mol Cancer Ther. 2012;11 (12:2685–2692. doi: 10.1158/1535-7163.MCT-12-0721. [DOI] [PubMed] [Google Scholar]
- Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12 (5:335–348. doi: 10.1038/nrc3257. [DOI] [PubMed] [Google Scholar]
- Neri P, Bahlis NJ, Lonial S. Panobinostat for the treatment of multiple myeloma. Expert Opin Investig Drugs. 2012;21 (5:733–747. doi: 10.1517/13543784.2012.668883. [DOI] [PubMed] [Google Scholar]
- Neri P, Tagliaferri P, Di Martino MT, Calimeri T, Amodio N, Bulotta A, Ventura M, Eramo PO, Viscomi C, Arbitrio M, Rossi M, Caraglia M, Munshi NC, Anderson KC, Tassone P. In vivo anti-myeloma activity and modulation of gene expression profile induced by valproic acid, a histone deacetylase inhibitor. Br J Haematol. 2008;143 (4:520–531. doi: 10.1111/j.1365-2141.2008.07387.x. [DOI] [PubMed] [Google Scholar]
- Niesen MI, Blanck G. Rescue of major histocompatibility-DR surface expression in retinoblastoma-defective, non-small cell lung carcinoma cells by the MS-275 histone deacetylase inhibitor. Biol Pharm Bull. 2009;32 (3:480–482. doi: 10.1248/bpb.32.480. [DOI] [PubMed] [Google Scholar]
- Niesvizky R, Ely S, Mark T, Aggarwal S, Gabrilove JL, Wright JJ, Chen-Kiang S, Sparano JA. Phase 2 trial of the histone deacetylase inhibitor romidepsin for the treatment of refractory multiple myeloma. Cancer. 2011;117 (2:336–342. doi: 10.1002/cncr.25584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offidani M, Polloni C, Cavallo F, Liberati AM, Ballanti S, Pulini S, Catarini M, Alesiani F, Corvatta L, Gentili S, Caraffa P, Boccadoro M, Leoni P, Palumbo A. Phase II study of melphalan, thalidomide and prednisone combined with oral panobinostat in patients with relapsed/refractory multiple myeloma. Leuk Lymphoma. 2012;53 (9:1722–1727. doi: 10.3109/10428194.2012.664844. [DOI] [PubMed] [Google Scholar]
- Rao R, Nalluri S, Kolhe R, Yang Y, Fiskus W, Chen J, Ha K, Buckley KM, Balusu R, Coothankandaswamy V, Joshi A, Atadja P, Bhalla KN. Treatment with panobinostat induces glucose-regulated protein 78 acetylation and endoplasmic reticulum stress in breast cancer cells. Mol Cancer Ther. 2010;9 (4:942–952. doi: 10.1158/1535-7163.MCT-09-0988. [DOI] [PubMed] [Google Scholar]
- Rebouissou C, Wijdenes J, Autissier P, Tarte K, Costes V, Liautard J, Rossi JF, Brochier J, Klein B. A gp130 interleukin-6 transducer-dependent SCID model of human multiple myeloma. Blood. 1998;91 (12:4727–4737. [PubMed] [Google Scholar]
- Reme T, Hose D, De Vos J, Vassal A, Poulain PO, Pantesco V, Goldschmidt H, Klein B. A new method for class prediction based on signed-rank algorithms applied to Affymetrix microarray experiments. BMC Bioinform. 2008;9:16. doi: 10.1186/1471-2105-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P, Mitsiades C, Colson K, Reilly E, McBride L, Chiao J, Sun L, Ricker J, Rizvi S, Oerth C, Atkins B, Fearen I, Anderson K, Siegel D. Phase I trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) in patients with advanced multiple myeloma. Leuk Lymphoma. 2008;49 (3:502–507. doi: 10.1080/10428190701817258. [DOI] [PubMed] [Google Scholar]
- Richardson PG, Alsina M, Weber DM, Coutre SE, Lonial S, Gasparetto C, Warsi G, Ondovik M, Mukhopadhyay S, Snodgrass S, Schlossman R.2011Phase II study of the pan-deacetylase inhibitor panobinostat in combination with bortezomib and dexamethasone in relapsed and bortezomib-refractory multiple myeloma (PANORAMA 2) Blood (118)abstract 814(oral presentation).
- Rio-Machin A, Ferreira BI, Henry T, Gomez-Lopez G, Agirre X, Alvarez S, Rodriguez-Perales S, Prosper F, Calasanz MJ, Martinez J, Fonseca R, Cigudosa JC. Downregulation of specific miRNAs in hyperdiploid multiple myeloma mimics the oncogenic effect of IgH translocations occurring in the non-hyperdiploid subtype. Leukemia. 2013;27 (4:925–931. doi: 10.1038/leu.2012.302. [DOI] [PubMed] [Google Scholar]
- San-Miguel JF, Hungria VT, Yoon SS, Wikto-Jedrzejczak W, Elghandour A, Siritanaratkul N, Dimopoulos MA, Corradini P, Nakorn TN, Shelekhova T, Günther A, Yong K, Schlossman R, Wroclawska-Swacha M, Weber HJ, Bourquelot P, Hou J, Einsele H, Lee JH, Moreau P, Lonial S, Richardson PG. Update on a phase III study of panobinostat with bortezomib and dexamethasone in patients with relapsed multiple myeloma: PANORAMA 1. Blood. 2011. p. abstract 3976.
- San-Miguel JF, SO, Siegel DS, Sezer O, Siegel DS, Guenther A, Mateos MV, Prosser I, Cavo M, Blade J, Boccadoro M, Bengoudifa BR, Klebsattel M, Bourquelot PM, Anderson KC.2009A phase IB, multi-center, open-label dose-escalation study of oral panobinostat (LBH589) and I.V. bortezomib in patients with relapsed multiple myeloma Blood (114)1481–1482.abstract 3852.19965646
- Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W, Powell J, Dave S, Yu X, Zhao H, Zeng Y, Chen B, Epstein J, Staudt LM. IRF4 addiction in multiple myeloma. Nature. 2008;454 (7201:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel DS, Dimopoulos MA, Yoon S-S, Laubach JP, Kaufman JL, Goldschmidt H, Reece DE, Leleu X, Durrant S, Offner FC, Cavo M, Nagler A, Jagannath S, Graef T, Houp J, Sun L, Howe J, Wear SM, Anderson KC. Vantage 095: vorinostat in combination with bortezomib in salvage multiple myeloma patients: final study results of a global phase 2b trial. Blood. 2011. p. abstract 480.
- Siegel DS, Sezer O, San-Miguel JF, Mateos MV, Prosser I, Cavo M, Jalaluddin M, Hazel K, Bourquelot PM, Anderson KC. A phase IB, multicenter, open-label, dose-escalation study of oral panobinostat (LBH589) and I.V. bortezomib in patients with relapsed multiple myeloma. Blood. 2008. p. abstract 2781.
- Skopelitou A, Hadjiyannakis M, Tsenga A, Theocharis S, Alexopoulou V, Kittas C, Agnantis N. Expression of C-myc p62 oncoprotein in multiple myeloma: an immunohistochemical study of 180 cases. Anticancer Res. 1993;13 (4:1091–1095. [PubMed] [Google Scholar]
- Tanguy Le Carrour SA, Tondeur Sylvie, Lhermitte Ludovic, Lamb Ned, Reme Thierry, Pantesco Veronique, Hamamah Samir, Klein Bernard, De Vos John. Amazonia!: An Online Resource to Google and Visualize Public Human whole Genome Expression Data. Open Bioinform J. 2010;4:5–10. [Google Scholar]
- Tarte K, Zhang XG, Legouffe E, Hertog C, Mehtali M, Rossi JF, Klein B. Induced expression of B7-1 on myeloma cells following retroviral gene transfer results in tumor-specific recognition by cytotoxic T cells. J Immunol. 1999;163 (1:514–524. [PubMed] [Google Scholar]
- Totzke G, Essmann F, Pohlmann S, Lindenblatt C, Janicke RU, Schulze-Osthoff K. A novel member of the IkappaB family, human IkappaB-zeta, inhibits transactivation of p65 and its DNA binding. J Biol Chem. 2006;281 (18:12645–12654. doi: 10.1074/jbc.M511956200. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10 (8:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- Wolf JL, Siegel D, Goldschmidt H, Hazell K, Bourquelot PM, Bengoudifa BR, Matous J, Vij R, de Magalhaes-Silverman M, Abonour R, Anderson KC, Lonial S. Phase II trial of the pan-deacetylase inhibitor panobinostat as a single agent in advanced relapsed/refractory multiple myeloma. Leuk Lymphoma. 2012;53 (9:1820–1823. doi: 10.3109/10428194.2012.661175. [DOI] [PubMed] [Google Scholar]
- Wu Z, Zhang X, Yang J, Wu G, Zhang Y, Yuan Y, Jin C, Chang Z, Wang J, Yang X, He F. Nuclear protein IkappaB-zeta inhibits the activity of STAT3. Biochem Biophys Res Commun. 2009;387 (2:348–352. doi: 10.1016/j.bbrc.2009.07.023. [DOI] [PubMed] [Google Scholar]
- Xiong W, Wu X, Starnes S, Johnson SK, Haessler J, Wang S, Chen L, Barlogie B, Shaughnessy JD, Jr, Zhan F. An analysis of the clinical and biologic significance of TP53 loss and the identification of potential novel transcriptional targets of TP53 in multiple myeloma. Blood. 2008;112 (10:4235–4246. doi: 10.1182/blood-2007-10-119123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Muta T, Takeshige K. A novel IkappaB protein, IkappaB-zeta, induced by proinflammatory stimuli, negatively regulates nuclear factor-kappaB in the nuclei. J Biol Chem. 2001;276 (29:27657–27662. doi: 10.1074/jbc.M103426200. [DOI] [PubMed] [Google Scholar]
- Yang G, Fu H, Zhang J, Lu X, Yu F, Jin L, Bai L, Huang B, Shen L, Feng Y, Yao L, Lu Z. RNA-binding protein quaking, a critical regulator of colon epithelial differentiation and a suppressor of colon cancer. Gastroenterology. 2010;138 (1:231–240, e1-5. doi: 10.1053/j.gastro.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Lu X, Wang L, Bian Y, Fu H, Wei M, Pu J, Jin L, Yao L, Lu Z. E2F1 and RNA binding protein QKI comprise a negative feedback in the cell cycle regulation. Cell Cycle. 2011;10 (16:2703–2713. doi: 10.4161/cc.10.16.15928. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265 (28:17174–17179. [PubMed] [Google Scholar]
- Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, Epstein J, Yaccoby S, Sawyer J, Burington B, Anaissie E, Hollmig K, Pineda-Roman M, Tricot G, van Rhee F, Walker R, Zangari M, Crowley J, Barlogie B, Shaughnessy JD., Jr The molecular classification of multiple myeloma. Blood. 2006;108 (6:2020–2028. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XG, Gaillard JP, Robillard N, Lu ZY, Gu ZJ, Jourdan M, Boiron JM, Bataille R, Klein B. Reproducible obtaining of human myeloma cell lines as a model for tumor stem cell study in human multiple myeloma. Blood. 1994;83 (12:3654–3663. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.