Abstract
MicroRNAs (miRNAs) are a class of small regulatory noncoding RNAs that modulate the expression of their target genes through either mRNA degradation or inhibition of protein translation. In recent years, miRNAs have been shown to be critical regulators of hematopoiesis and have important roles in the differentiation of specific lineages. Here, we summarize our current understanding of miRNAs involved in hematopoiesis with a focus on the role of miRNAs in regulating erythroid and megakaryocytic differentiation and megakaryocyte–erythroid progenitor lineage commitment.
Keywords: microRNA, erythropoiesis, megakaryocytopoiesis, megakaryocyte–erythroid progenitor
INTRODUCTION
Each day the average human produces over 1011 new erythrocytes and platelets. If this output is not maintained, the individual is at risk for significant pathologic conditions including anemia and thrombocytopenia. Because of this rapid turnover in circulating blood cells, the hematopoietic system is poised to continually replenish these lineages through the proliferation, differentiation and maturation of immature progenitor populations that are the progeny of self-renewing stem cells. In contrast to most other organ systems, the hematopoietic system is unique because the output of these various progenitors can be functionally validated in vivo through the use of transplantation assays.1 Because of the ability to experimentally validate the hierarchy of progenitor cell populations within the hematopoietic system, as well as the ability to prospectively identify and purify these same cell populations, the hematopoietic system has served as a useful paradigm for understanding the process of differentiation of stem and progenitor cells in normal physiology and development.1,2
A great deal has been learned about the molecules necessary for normal differentiation, including identification of extrinsic regulators of hematopoiesis such as lineage-specific cytokines. In addition, there has been an intense focus on the identification of the intrinsic regulators of this process.1 There is significant experimental support for the notion that transcription factors act as critical intrinsic regulators of hematopoiesis and also serve to integrate the distal regulatory output of extrinsic signals.3,4 The importance of transcription factors in normal hematopoiesis is illustrated by their frequent disruption as translocation partners in a variety of leukemias and other hematopoietic malignancies.1 Even less is understood about the regulation of hematopoiesis by other classes of intrinsic regulators. Here, we discuss how microRNAs (miRNAs), a recently described class of~23 nucleotide RNA molecules that bind to sequences in mRNA and mediate their degradation or translational repression,5,6 have a role in the regulation of erythroid and megakaryocytic cells within the hematopoietic hierarchy. An extensive amount of literature has been published on this topic in just the past several years and numerous comprehensive reviews on this topic exist.7–10 Here, we cover vignettes that illustrate the types of regulatory roles that miRNAs have in erythroid and megakaryocytic differentiation and megakaryocyte–erythroid progenitor (MEP) lineage commitment.
A BACKGROUND ON ERYTHROID AND MEGAKARYOCYTIC DIFFERENTIATION AND MEP LINEAGE COMMITMENT
The traditional view of hematopoiesis involves a hierarchical model where long-term self-renewing hematopoietic stem cells, which stably maintain hematopoiesis in the adult, give rise to more rapidly cycling short-term stem and multipotential progenitor cells, some of which may have limited self-renewal capabilities.1,2 There is a split between the lymphoid and myeloid lineages, with the production of a common myeloid progenitor (CMP) and a common lymphoid progenitor. The common lymphoid progenitor is then responsible for the production of committed progenitors that give rise to all of the cells of the lymphoid lineage: T, B, natural killer and a subset of dendritic cells.2,11 In contrast, the CMP has to produce progenitors that can allow for the production of all other hematopoietic lineages, which are classified as myeloid or myeloerythroid cells.2,12,13 This includes erythrocytes, platelets, mast cells, neutrophils, eosinophils, basophils, monocytes, macrophages and a subset of dendritic cells. The CMP is able to differentiate into two prospectively isolatable progenitor populations, which include MEP and the granulocyte–monocyte progenitor (GMP).2,13 The MEP and GMP solely give rise to lineage-committed progenitors that can undergo further differentiation to the particular lineages that they are dedicated to—megakaryocytes and erythroid cells for the MEP and neutrophils, eosinophils, basophils, monocytes and macrophages for the GMP.
Recent studies challenge this traditional hierarchical model of hematopoiesis.2,14 It has been suggested that the MEP may arise from a multipotent progenitor that then gives rise to the common lymphoid progenitor and GMP populations,14 although subsequent work has suggested that this model alone may be oversimplified.15 The majority of evidence supports the existence of the more traditional model of hematopoietic differentiation with the bifurcation between the myeloid and lymphoid lineages, although these recent studies suggest that some progenitor populations may be more heterogeneous and/or display more plasticity in differentiation than was once appreciated.2,3 An important molecular underpinning of this increasing complexity in hematopoiesis may be attributable to the existence of lineage priming by transcription factors. This concept arises from the finding that stem cells and multipotential progenitors express a number of lineage-specific transcription factors2,3 that are required for the generation of specific mature lineages to which that progenitor is able to give rise. It is thought that this early expression of these transcription factors facilitates chromatin remodeling to maintain an open and permissive chromatin state that allows for differentiation of cells mediated by the underlying transcriptional program for that particular lineage.2,3 The existence of lineage priming may also underlie the ability of variations in transcription factor levels to mediate alterations in lineage choice or even reprogramming within the hematopoietic system.2
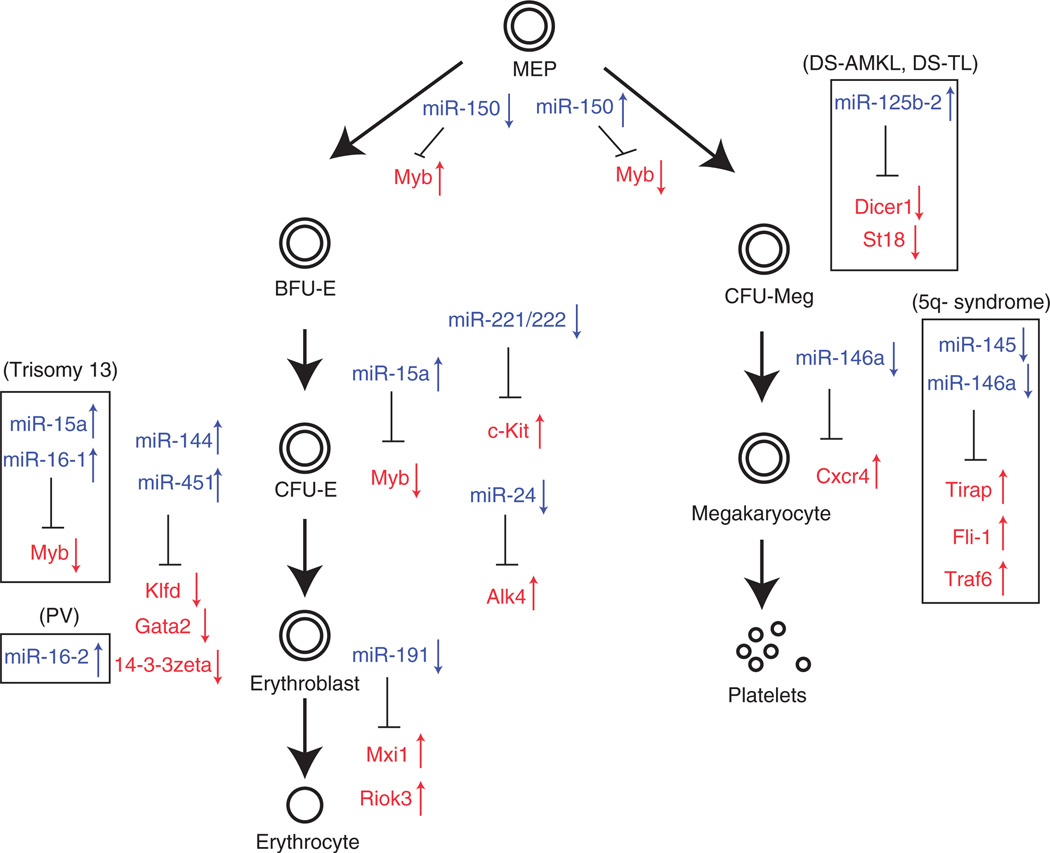
As a result of intensive studies, much is understood about the role of transcription factors in hematopoietic differentiation and particularly in erythropoiesis and megakaryopoiesis.1,2 Far less is understood about the role that miRNAs have in regulating these processes.7 Here, we examine a few well-studied examples of such factors involved in these processes (Figure 1 and Table 1). Recent work suggests that these molecules may be capable of mediating as powerful an effect on lineage choice as transcription factors, as exemplified by the ability of miRNAs to reprogram cells to a pluripotent state.16,17 As further work is done in this area, it is likely that similar types of findings will result for some of the miRNAs necessary during hematopoiesis, particularly those that play roles during erythropoiesis and megakaryopoiesis.
Figure 1.
miRNAs are important regulators of erythroid and megakaryocytic cells production.1,18 CFU-G, colony-forming unit-granulocyte; CFU-GM, colony-forming unit-granulocyte-macrophage; CFU-Eo, colony-forming unit-eosinophil; CFU-M, colony-forming unit-macrophage; CFU-Meg, colony-forming unit-megakaryocyte; MCP, mast cell progenitor. Up arrows represent upregulation of miRNAs or protein-coding genes. Down arrows represent the upregulation of miRNAs or protein-coding genes.
Table 1.
The summary of experimental system, normal developmental function, related human disease, expression pattern, target gene and upstream regulator of microRNAs50
| microRNA | Experimental system | Normal development | Human disease |
Expression pattern | Proposed function | Targets | Upstream regulators |
References |
|---|---|---|---|---|---|---|---|---|
| miR-144 | Zebrafish embryo | Adult α-globin expression | NA | Upregulated during erythroid differentiation | miR-144 and Klfd form a negative feedback loop modulating the expression of an embryonic form of alpha-globin | Klfd | GATA1, Klfd | 22 |
| miR-451 | Zebrafish embryo | Erythropoiesis | NA | Upregulated during erythroid differentiation | miR-451 is required for Zebrafish erythropoiesis | Gata2 | GATA1 | 20,21 |
| Knockout mice | Steady and stress erythropoiesis | NA | Upregulated during erythroid differentiation | miR-451 is required for steady state erythropoiesis and resistance to oxidation during stress erythropoiesis | 14-3-3zeta | GATA1 | 23–25 | |
| miR-221, miR-222 | Human cord blood CD34+ cells, Erythroleukemia cell line TF-1 | Erythroid progenitor proliferation | NA | Downregulated during erythroid differentiation | Downregulation of miR-221 and miR-222 is required for erythroid progenitor proliferation | KIT | NA | 27 |
| miR-24 | Human cord blood CD34+ cells and K562 cell line | Erythroid differentiation | NA | Downregulated during erythroid differentiation | miR-24 inhibits erythropoiesis by targeting ALK4 | ALK4 | NA | 28 |
| miR-15a | Human bone marrow mononuclear cells or CD34+ cells | Erythropoiesis | NA | Upregulated during erythroid differentiation | miR-15a and Myb autoregulatory feedback loop is required for erythropoiesis | MYB | MYB | 30 |
| miR-15a, miR-16-1 | Human adult peripheral blood-derived CD34+ cells | Fetal γ-globin expression | Trisomy 13 | Upregulated in trisomy 13 | Upregulation of miR-15a and miR-16-1 in trisomy 13 resultes in elevated fetal γ-globin expression | MYB | NA | 31 |
| miR-16-2 | CD34+ cells from healthy donor and polycythemia vera patients, mouse model | Erythropoiesis | Polycythemia vera | Upregulated in CD34+ cells from polycythemia vera patients | Dysregulation of miR-16-2 contributes to abnormal expansion of the erythroid lineage in polycythemia vera | NA | NA | 33 |
| miR-191 | Mouse primary erythroid progenitors | Erythroblast enucleation | NA | Downregulated during erythropoiesis | The downregulation of miR-191 is required for erythroblast chromatin condensation and enucleation | Riok3 and Mxi1 | NA | 34 |
| miR-150 | Human bone marrow CD34+ cells, mice bone marrow transplantation | MEP lineage commitment | NA | Enriched in MEP and megakaryocytic progenitor relative to erythroid progenitor | miR-150 drives MEP differentiation toward the megakaryocytic lineage at the expense of erythroid lineage cells | MYB | NA | 40 |
| miR-125b-2 | Fetal megakaryocytic progenitors, megakaryoblastic leukemia cell line, DS-TL leukemic blasts | MEP and megakaryocytic progenitor self-renewal and megakaryocytic differentiation | DS-AMKL, DS-TL, non-DS-AMKL | Upregulated in DS-AMKL, DS-TL, non-DS-AMKL | miR-125b-2 is involved in the pathogenesis of megakaryoblastic leukemia | DICER1, ST18 | NA | 41 |
| miR-145 | CD34+ cell culture system and mouse bone marrow transplantation model | Megakaryocytic differentiation | 5q- syndrome | Downregulated in HSPC of 5q- syndrome patients | Loss of miR-145 contributes to the pathogenesis of 5q- syndrome | TIRAP, Fli-1 | NA | 48,49 |
| miR-146a | Human cord blood CD34þ cells, mouse bone marrow transplantation model, and K562 cell line | Megakaryocytic differentiation | 5q- syndrome | Downregulated in HSPC of 5q- syndrome patients; downregulated during megakaryocytopoiesis | Loss of miR-145 contributes to the pathogenesis of 5q- syndrome. The downregulation of miR-146a is required for megakaryocytopoiesis | TRAF6, CXCR4 | PLZF | 48,50 |
Abbreviation: HSPC, hematopoietic stem/progenitor cell.
MIRNAS IN ERYTHROPOIESIS
The earliest lineage-committed erythroid progenitor is the burst-forming unit-erythroid (BFU-E). The proliferation and self-renewal of BFU-Es as well as the differentiation from BFU-E to late erythroid progenitor, the colony-forming unit-erythroid (CFU-E), is regulated by IL-3, IL-6, stem cell factor, glucocorticoids and other extra-cellular signaling molecules. Once the erythroid progenitors have reached the CFU-E stage, they become increasingly dependent on erythropoietin and undergo terminal differentiation, a process that includes three–five more cell divisions, accumulation of important proteins required for hemoglobin synthesis, chromatin condensation and enucleation.18,19 In recent years, a few miRNAs have been linked to the regulation of these different stages of erythropoiesis.
miR-144/451
miR-144 and miR-451 are co-transcribed miRNAs highly induced in the erythroid cell line G1E-ER4 when the important erythroid transcription factor, GATA1, is restored by treatment with estradiol. This is consistent with the observations that miR-144 and miR-451 are upregulated during erythroid differentiation of human CD34+ and murine erythroleukemia cells.20 In both G1EER4 and murine erythroleukemia cells and primary mouse fetal liver cells, chromatin immunoprecipitation experiments demonstrated that GATA1 binds a genomic locus 2.8 kb upstream of miR-144/451, a region containing a predicted GATA1 binding motif. Furthermore, luciferase reporter assays suggested that this motif is indeed a functional erythroid enhancer, implying that miR-144/451 is a direct transcriptional target of GATA1.20
In zebrafish, knockdown of miR-451 by antisense morpholino impairs erythropoiesis, which suggested an important functional role of miR-451 in erythropoiesis.20,21 Erythropoietic defects have also been observed in mnr, a nonanemic zebrafish mutant with reduced expression of miR-144/451. By injection of a green fluorescent protein reporter containing the Gata2 3′UTR into zebrafish embryo, the authors showed that Gata2 expression is downregulated by and is a potential target of miR-144/451. Injection of a Gata2 morpholino partially rescued the erythroid differentiation defects of the mnr mutant, further confirming that Gata2 is a direct target of miR-451.21 In contrast to miR-451 morpholino-injected embryos, zebrafish embryos injected with miR-144 locked nucleic acids to reduce miR-144 expression are morphologically normal. By using in situ hybridization to detect the expression of several hematopoietic- and vascular-specific genes, the authors found that embryonic alpha-globin expression was significantly upregulated in miR-144 locked nucleic acid-injected embryos. In vivo reporter assays suggested that Klfd might be a target of miR-144. Chromatin immunoprecipitation experiments demonstrated that Klfd binds to the promoter region of miR-144 and alpha-globin. Together, this suggested that miR-144 and Klfd form a negative feedback loop modulating the expression of an embryonic form of alpha-globin: Klfd induces miR-144 (and miR-451) expression, and miR-144 downregulates Klfd.22 This model of globin regulation by miR-144 awaits confirmation in other experimental systems.
Recently, several groups created miR-144/451 or miR-451 knockout mice.23–25 miR-144/451 knockout mice displayed moderate erythroid hyperplasia, moderate splenomegaly and mild anemia. When exposed to phenylhydrazine, more than half of the miR-144/451−/− mice died from the resulting hemolytic anemia, whereas all of the wild-type mice fully recovered, which suggests that the knockout mice have an impaired response to oxidative stress.23–25 One direct target gene of miR-451 is 14-3-3ζ, a phospho-serine/threonine-binding protein24,25 that inhibits nuclear accumulation of the transcription factor FoxO3, a positive regulator of erythroid anti-oxidant genes. As a result, the abnormal accumulation of 14-3-3ζ in miR-144/451−/− erythroblasts causes a partial relocalization of FoxO3 from nucleus to cytoplasm and dampens the expression of several FoxO3 target genes that encode important antioxidant proteins, including Cat and Gpx1. shRNA suppression of 14-3-3ζ has been shown to protect miR-144/451−/− erythrocytes against peroxide-induced destruction.24 Together, these results suggest that miR-451 is critical for protecting erythroid cells against oxidant stress. Although the miR-451 knockout mice only display a subtle anemia, the importance of this miRNA in ensuring optimal differentiation of erythrocytes that are capable of mounting a robust antioxidant response is well demonstrated through these elegant in vivo studies. Interestingly, recent work has shown that the maturation of miR-451 does not require Dicer, but instead relies on the enzymatic activity of Ago2. Consistent with the anemic phenotype of miR-144/451 knockout mice, Ago2 knockout mice die shortly after birth from anemia. These mice exhibit dysregulation of expression of miR-451, suggesting that this dysregulation contributes to the anemic phenotype of Ago2 knockout mice.26 However, this work is controversial, given the dramatically different phenotypes seen in the two knockout animals.23–26 The Ago2 knockout animals have a severe fetal anemia, whereas a knockout of miR-451 has very subtle effects. Further work is necessary to better understand the physiological role of these miRNAs in erythropoiesis and is likely to uncover critical reasons for the evolutionary conservation of these miRNAs.
miR-221/222
Originally identified as miRNAs downregulated during erythroid differentiation of human cord blood CD34+ cells, miR-221 and miR-222 are important regulators of early erythroid proliferation.27 The 3′UTR of KIT contains sequences complementary to the similar miR-221 and miR-222 seed sequence and these sequences are responsive to miR-221 and miR-222 in luciferase reporter assays. This suggests that KIT, the stem cell factor receptor and an important regulator of early erythroid progenitor proliferation, may be a direct target downregulated by miR-221 and miR-222. In erythroid cultured human cord blood CD34+ cells, miR-221 and miR-222 are normally downregulated during erythroid differentiation, and overexpression of these two miRNAs by either oligonucleotide transfection or lentiviral infection impairs erythroid progenitor expansion and accelerates erythroid differentiation, accompanied by the decrease of KIT expression. These data suggest that miR-221 and miR-222 together with KIT form a regulatory cascade involved in balancing erythroid progenitor expansion and erythroid differentiation.27 The physiological importance of these miRNAs in in vivo erythropoiesis remains to be established.
miR-24
miR-24 is a negative regulator of activin type I receptor ALK4,28 and the 3′UTR of ALK4 contains sequences complementary to miR-24. Indeed, luciferase reporter assays demonstrated that the 3′UTR of ALK4 is miR-24 responsive, and in HEK293 cells, overexpression of miR-24 downregulated the expression of ALK4. In contrast, overexpression of miR-24 impaired activin-triggered Smad2 phosphorylation and activin-induced luciferase reporter activity, indicators of a defect in activin signaling. In K562 cells, activin A functions as a positive regulator of erythropoiesis and induces the accumulation of hemoglobin. Overexpression of miR-24 partially blocked the accumulation of hemoglobin, whereas knockdown of miR-24 slightly promoted it. This negative effect of miR-24 on erythropoiesis has further been suggested by data from primary human CD34+ hematopoietic progenitor cells where overexpression of miR-24 impaired erythropoiesis and knockdown of miR-24 promoted it, as indicated both by erythroid colony formation assays and a liquid culture erythroid differentiation assay. Together, these data suggested that miR-24 negatively regulates erythropoiesis by downmodulating the activin signaling pathway.28 The in vivo function of miR-24 remains to be tested to better understand the physiological significance of this miRNA.
miR-15a, miR-16-1 and miR-16-2
Initially, miR-15a had been identified as a miRNA that down-regulates MYB expression;29 the 3′UTR of MYB contains sequences complementary to the miR-15a seed sequence. Further, luciferase reporter assays suggested that this DNA fragment is indeed miR-15a responsive. Interestingly, in K562 cells, chromatin immunoprecipitation experiments suggested that MYB binds to the promoter region of miR-15a, and overexpression of miR-15a downregulated MYB expression at the protein level whereas knockdown of miR-15a upregulated MYB expression. The regulation of miR-15a expression by MYB also has been shown in K562 cells where knockdown of MYB impairs miR-15a expression. This data suggested that there exists an autoregulatory loop between miR-15a and MYB, in which MYB promotes expression of miR-15a, and that this miRNA in turn downmodulates MYB expression.29 During erythroid differentiation of cultured human primary CD34+ hematopoietic progenitor cells the expression patterns of MYB and miR-15a were inversely correlated with each other. Overexpression of miR-15a in human bone marrow mononuclear cells or CD34+ cells impaired formation of CFU-E and colony-forming unit-granulocyte-macrophage colonies and had a modest effect on BFU-E colony formation. These data suggested that this autoregulatory feedback loop between miR-15a and MYB is involved in normal hematopoietic regulation.29
Importantly, a recent discovery has linked, via its target MYB, the miR-15a and miR-16-1 miRNA cluster with a certain phenotype of human trisomy 13: an additional chromosome copy of human trisomy 13 results in abnormal upregulation of the human γ-globin (fetal hemoglobin) genes. This segment of chromosome 13 contains the miR-15 and miR-16 genes; the abnormal upregulation of these miRNAs leads to downregulation of their target gene MYB, which in turn elevates fetal hemoglobin expression.30 As increased fetal hemoglobin is important to ameliorate the symptoms of sickle cell disease and beta-thalassemia, alteration of miR-15a/16-1 expression or MYB expression may be useful for developing more targeted therapeutic approaches to treat these conditions.31 Other recently published work has linked miR-16-2 with the regulation of erythropoiesis; the pathological upregulation of miR-16-2 may contribute to the abnormal expansion of erythroid cells in polycythemia vera.32 This finding needs to be confirmed. Furthermore, the in vivo role of these miRNAs in normal erythropoiesis remains to be established.
miR-191
According to a miRNA RNA-seq expression profiling of primary mouse CFU-Es and more mature Ter119+ erythroblasts, the majority of miRNAs are downregulated during terminal erythroid differentiation.33 Among the predominant developmentally downregulated miRNAs, ectopic overexpression of miR-191 in primary mouse fetal liver erythroid progenitors blocked chromatin condensation in late erythroblasts as well as enucleation, but had minor effects on terminal erythroid proliferation or differentiation. Two developmentally upregulated genes, Riok3 and Mxi1, were identified as direct targets of miR-191, and were subsequently shown to be essential for chromatin condensation and enucleation. Both overexpression of miR-191 and knockdown of Riok3 or Mxi1 impaired the normal downregulation of histone acetyltransferase Gcn5. As Mxi1 is a well-known antagonist of Myc34 and as downregulation of c-Myc and its transcriptional target Gcn5 are required for chromatin condensation,35 miR-191, Riok3, Mxi1, c-Myc and Gcn5 may form a cascade that regulates the chromatin condensation process; normal downregulation of miR-191 allows normal upregulation of Riok3 and Mxi1 that in turn promotes downregulation of Gcn5.33 The physiological importance of this miRNA in vivo remains to be tested.
MIRNAS IN MEP LINEAGE COMMITMENT AND MEGAKARYOPOIESIS
Megakaryopoiesis begins with the lineage commitment of a bi-potential MEP to the megakaryocytic lineage. Further differentiation of megakaryocytic progenitors is regulated by thrombopoietin (TPO), the principle hormone that promotes megakaryocytopoiesis. TPO binds to its receptor c-Mpl and triggers several downstream pathways that are important for megakaryopoiesis.36–38 Despite the fact that TPO and some downstream mediators of its effects in megakaryopoiesis have been well characterized, the roles of miRNAs in modulating megakaryopoiesis are less clear. Here, we describe several miRNAs that function as another layer of regulation during megakaryopoiesis.
miR-150
Using miRNA expression profiling of purified human umbilical cord blood hematopoietic progenitors, miR-150 was found to be enriched in MEPs and megakaryocyte progenitors relative to erythroid progenitors.39 In the human CD34+ hematopoietic stem/progenitor cell (HSPC) culture system, in which erythropoietin and TPO promote the differentiation towards erythrocyte and megakaryocyte lineages, respectively, overexpression of miR-150 resulted in an eight-fold increase of megakaryocytic cells. That miR-150 promotes megakaryocyte formation was further supported by data from in vivo HSPC transplantation assays, where overexpression of miR-150 resulted in >15-fold increase of megakaryocytes and a 60% decrease of erythroid cells. In addition, overexpression of miR-150 increases colony-forming unit megakaryocyte (CFU-Mk) formation and decreases CFU-E formation. In contrast, antagomir triggered loss-of-function of miR-150 decreased CFU-Mk formation. Together, these results indicate that miR-150 is important for promoting MEP lineage determination.
Furthermore, the level of miR-150 was dramatically decreased during phenylhydrazine-induced acute anemia, where rapid erythroid reconstitution is required, suggesting that the expression of miR-150 is regulated by the physiological environment.39 MYB is a target gene of miR-150 and MYB downregulation would be one mechanism by which this miRNA promotes formation of megakaryocytes at the expense of erythroid progenitors. The 3′UTR of MYB contains a miR-150 binding site that is responsive to miR-150 in luciferase reporter assays. Moreover, overexpression of miR-150 in the erythroleukemia cell line K562 reduced MYB expression. In addition, shRNA knockdown of MYB in human CD34+ cells promotes megakaryocytic cell production mimicking the miR-150 overexpression phenotype. Together, this work suggested that miR-150 and MYB form a regulatory network involved in the lineage determination of the bi-potential MEP.39
Both miR-150 and miR-15a target MYB, the partial knockdown of which causes thrombocytosis and anemia. Although overexpression of miR-150 blocks the committment of MEP to the erythroid lineage, enforced expression of miR-15a impairs the erythroid differentiation from the BFU-E to the CFU-E stage. This is an interesting example that illustrates how multiple miRNAs regulate the expression of the same gene at sequential developmental stages, contributing to lineage specification and further differentiation. In this scenario, the downregulation of miR-150 from MEP to erythroid progenitor promotes the commitment of MEPs to the erythroid lineage through the upregulation of MYB, and miR-15a fine-tunes the level of MYB in erythroid progenitors and modulates erythroid differentiation. It will be interesting to determine the precise expression patterns of miR-15a in MEP and megakaryocytic progenitors, and to elucidate whether miR-15a is also involved in the MEP lineage specification, in addition to its role in erythroid differentiation.
miR-125b-2
miR-125b-2 is encoded by human chromosome 21. It is upregulated in leukemic blasts from patients with acute megakaryoblastic leukemia with trisomy 21 (Down syndrome) (DSAMKL), transient leukemia with trisomy 21 (DS-TL) and acute megakaryoblastic leukemia without trisomy 21, as compared with those from patients with acute myeloid leukemia FAB M5 and healthy donor CD34+ HSPCs and megakaryocytes. This suggests a correlation between miR-125b-2 upregulation and the pathogenesis of these types of leukemia.40
In mouse megakaryocyte progenitors isolated from embryonic day 12.5 (E12.5) fetal liver, overexpression of miR-125b-2 promoted the formation of CFU-MKs, indicating that overexpression of miR-125b-2 increases proliferation and self-renewal of megakaryocyte progenitors. In a serial replicating assay performed on wild-type fetal liver cells, overexpression of miR-125b-2 resulted in the formation of CFU-MKs that contain hemoglobinized erythroid cells and BFU-Es, whereas empty vector-infected cells mainly formed mast cell colony-forming units. This suggests that overexpression of miR-125b-2 induces the proliferation and self-renewal of MEPs. In addition, when miR-125b-2 is overexpressed in fetal liver cells from Gata1-mutant mice, there is an increase in the number and size of CFU-MKs. In both the megakaryblastic leukemia cell line and the leukemia blasts isolated from DS-TL patients, knockdown of miR-125b-2 impairs their proliferation. The author further identified DICER1 and ST18 as target genes of miR-125b-2, and found that DICER1 and ST18 are significantly downregulated in cells isolated from DS-AMKL and DS-TL patients. Knockdown of these two target genes recapitulates the miR-125b-2 overexpression phenotypes.40
In addition to its role in acute megakaryoblastic leukemia and megakaryocyte formation, miR-125b has also been broadly linked with normal and pathological hematopoiesis, including hematopoietic stem cell self-renewal,41 myelodysplasia and other types of leukemias.42–45
miR-145 and miR-146a
Chromosome 5q deletion syndrome (5q- syndrome) is a subtype of the myelodysplastic syndrome, which is characterized by macrocytic anemia, thrombocytosis, hypolobated megakaryocyte and a common deleted region on chromosome 5q. Although haploinsufficiency of RPS14 contributes to the erythroid differentiation defects of 5q- syndrome,46 the molecular mechanisms underlying the defects related to megakaryocyte lineage are unlikely to be explained by the loss of RPS14.
Among 13 miRNAs encoded in the common deleted region, miR-145 and miR-146a are downregulated in CD34+ HSPC isolated from the bone marrow of 5q- syndrome patients.47 In a mouse bone marrow transplantation model, knockdown of both miRNAs through a ‘miR decoy’ approach caused variable neutropenia, significant thrombocytosis and the formation of hypolobated megakaryocytes, suggesting that the loss of miR-145 and miR-146a contributes to the megakaryocyte lineage defects of 5q- syndrome. Further experiments identified TIRAP (toll-interleukin 1 receptor (TIR) domain containing adaptor protein) and TRAF6 (TNF receptor-associated factor 6), two molecules involved in the Toll-like receptor signaling pathway, as target genes of miR-145 and miR-146a, respectively. In the same mouse bone marrow transplantation model, overexpression of TRAF6 recapitulated the miR-145 and miR-146a loss-of-function phenotype, suggesting that TRAF6 is a functional target of miR-145. Further analysis also suggested a cell non-autonomous effect of TRAF6 on thrombocytosis through induction of IL-6. Taken together, these data indicate that 5q deletion results in the downregulation of miR-145 and miR-146a, which contribute to the pathogenesis of 5q- syndrome.47 Consistent with this report, a recent study shows that miR-145 and its target gene, Fli-1, are dysregulated in 5q- syndrome patients. In both the CD34+ cell culture system and the bone marrow transplantation models, downregualtion of miR-145 and the upregulation of Fli-1 increase the production of megakaryocytic cells relative to erythroid cells. Importantly, it has been shown that miR-145 may functionally cooperate with RPS14 to alter erythroid and megakaryocytic lineage differentiation, illustrating how the loss of protein coding genes and noncoding miRNAs encoded in the same chromosomal region may contribute to a complex phenotype observed in a microdeletion syndrome.48
In addition to its involvement in the pathogenesis of 5q-syndrome, miR-146a has also been suggested to have a negative role in normal megakaryopoiesis.49 In the promyelocytic leukemia zinc-finger (PLZF) overexpressing K562 cell, miR-146a is downregulated and chemokine receptor 4 (CXCR4) is upregulated.49 These expression patterns have been confirmed in megakaryocytic cultures of human cord blood CD34+ -derived cells. Subsequent bioinformatic analysis and luciferase reporter assays suggested that CXCR4 is a direct target of miR-146a, whose 3′UTR contains a miR-146a binding site. Furthermore, chromatin immunoprecipitation assays showed that PLZF is a transcriptional repressor of miR-146a. In megakaryocytic cultures, ectopic overexpression of miR-146a impairs proliferation and differentiation and reduces CXCR4 expression. In contrast, knockdown of miR-146a promotes proliferation and differentiation and increases CXCR4 expression. By using the same megakaryocytic cultures, it was demonstrated that knockdown of either PLZF or CXCR4 impairs megakaryopoiesis. The authors also showed that overexpression of miR-146a disrupts PLZF-induced megakaryocytic differentiation, while overexpression of a 3′UTR-mutated CXCR4, which is no longer responsive to miR-146, blocked the ability of miR-146 to alter megakaryocytic production. Together these data suggest that PLZF, miR-146a and CXCR4 form a positive feedback loop required for megakaryopoiesis: PLZF reduces miR-146a expression, and reduced levels of this miRNA in turn promote expression of CXCR4.49
CONCLUDING REMARKS
From the vignettes described above, it is clear that a great deal has been learned about the role of miRNAs in erythroid and megakaryocytic differentiation and MEP lineage commitment over the past few years. Traditionally, it has been thought that the production of erythroid and megakaryocytic cells are regulated by cytokines, including erythropoietin, IL-3, IL-6, stem cell factor, glucocorticoids and TPO. These extracellular molecules activate several intracellular downstream signaling pathways, including the JAK/STAT, RAS/MAPK and PI3K/AKT pathways, all of which eventually regulate transcription factors, such as GATA1, TAL1/SCL, STAT5 and KLF1, to ultimately modulate the expression of genes required for lineage commitment and differentiation. The discovery that miRNAs are required for erythroid and megakaryocytic cell formation uncovers another layer of this regulatory mechanism.
One type of this miRNA-mediated regulation influences the ‘decision-making’ process through fine tuning of the expression of key regulatory molecules. An intricate example of this type of regulation is visible in the participation of miR-150, miR-15a and their target gene MYB in MEP lineage commitment and further erythroid differentiation. MYB is critical for MEP lineage commitment and erythroid differentiation, and partial loss of MYB expression promotes thrombocytosis and subtle erythroid abnormalities. However, it is not clear how its meticulous expression pattern, expressed higher in MEP and early erythroid progenitors than in megakaryocytic progenitors, and gradually downregulated during further erythroid differentiation, is achieved. The identification of miR-150 and miR-15a as miRNAs that target MYB at sequential stages of MEP lineage commitment and erythroid differentiation, provides one possible explanation. In this case, the higher expression levels of miR-150 in MEP and megakaryocytic progenitors modulate the destiny of such cells. The daughter cell of a MEP that ‘inherits’ an increased level of miR-150 will be more likely to become a megakaryocytic progenitor, and its counterpart with lower levels of miR-150 will be more likely to be an erythroid progenitor. This hypothesis is supported by experimental evidence detailed above, although further studies of such regulatory mechanisms are needed. Following the MEP stage, the upregulation of miR-15a/16-1 may allow for modulation of MYB expression during erythropoiesis.
Another type of miRNA-mediated regulation mainly contributes to the robustness of biological systems that control erythroid and megakaryocytic cell production. A well-studied example is miR-451, where mice with a knockout of this miRNA have a rather subtle phenotype and on its own, this molecule does not appear to contribute in a major way to normal homeostatic erythropoiesis. However, during phenylhydrazine-induced hemolytic anemia, where the demand for erythrocyte production and the resistance to peroxide-induced destruction is greatly elevated, miR-451 becomes indispensible for erythrocyte production.
Despite the fact that much has been learned about the role of miRNAs in erythroid and megakaryocytic cell production, the physiological role of these molecules still remains largely enigmatic, as the vast majority of studies were performed in cell lines rather than primary cells. Although cell lines are easily accessible and can be readily manipulated, there are many instances where phenotypes observed in vitro contrast with those observed when molecules (such as transcription factors) are mutated in vivo. In light of this, it will be critical for the physiological relevance of these miRNAs to be discerned by combining insight from in vitro studies along with in vivo models.
ACKNOWLEDGEMENTS
LZ was supported by a graduate fellowship from the Singapore-Massachusetts Institute of Technology Alliance. VGS received support from NIH grant T32HL007574-30 and funds provided by the Department of Medicine at the Children’s Hospital Boston. This work was supported by SMA grant C-382-641-001-091 (to HFL), and NIH grants DK047618, DK068348 and 5P01 HL066105 (to HFL).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iwasaki H, Akashi K. Myeloid lineage commitment from the hematopoietic stem cell. Immunity. 2007;26:726–740. doi: 10.1016/j.immuni.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Laiosa CV, Stadtfeld M, Graf T. Determinants of lymphoid-myeloid lineage diversification. Ann Rev Immunol. 2006;24:705–738. doi: 10.1146/annurev.immunol.24.021605.090742. [DOI] [PubMed] [Google Scholar]
- 4.Kim SI, Bresnick EH. Transcriptional control of erythropoiesis: emerging mechanisms and principles. Oncogene. 2007;26:6777–6794. doi: 10.1038/sj.onc.1210761. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo H, Ingolia N, Weissman J, Bartel D. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 8.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 9.Zhao G, Yu D, Weiss MJ. MicroRNAs in erythropoiesis. Curr Opin Hematol. 2010;17:155–162. doi: 10.1097/MOH.0b013e328337ba6c. [DOI] [PubMed] [Google Scholar]
- 10.Byon JC, Papayannopoulou T. MicroRNAs: allies or foes in erythropoiesis? J Cell Physiol. 2012;227:7–13. doi: 10.1002/jcp.22729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 12.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 13.Na Nakorn T, Traver D, Weissman IL, Akashi K. Myeloerythroid-restricted progenitors are sufficient to confer radioprotection and provide the majority of day 8 CFU-S. J Clin Invest. 2002;109:1579–1585. doi: 10.1172/JCI15272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Forsberg EC, Serwold T, Kogan S, Weissman IL, Passegue E. New evidence supporting megakaryocyte-erythrocyte potential of flk2/flt3+ multipotent hematopoietic progenitors. Cell. 2006;126:415–426. doi: 10.1016/j.cell.2006.06.037. [DOI] [PubMed] [Google Scholar]
- 16.Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011;29:443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell stem cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lodish H, Flygare J, Chou S. From stem cell to erythroblast: Regulation of red cell production at multiple levels by multiple hormones. IUBMB Life. 2010;62:492–496. doi: 10.1002/iub.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richmond T, Chohan M, Barber D. Turning cells red: signal transduction mediated by erythropoietin. Trends Cell Biol. 2005;15:146–155. doi: 10.1016/j.tcb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 20.Dore LC, Amigo JD, Dos Santos CO, Zhang Z, Gai X, Tobias JW, et al. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc Natl Acad Sci USA. 2008;105:3333–3338. doi: 10.1073/pnas.0712312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pase L, Layton JE, Kloosterman WP, Carradice D, Waterhouse PM, Lieschke GJ. miR-451 regulates zebrafish erythroid maturation in vivo via its target gata2. Blood. 2009;113:1794–1804. doi: 10.1182/blood-2008-05-155812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu YF, Du TT, Dong M, Zhu KY, Jing CB, Zhang Y, et al. Mir-144 selectively regulates embryonic alpha-hemoglobin synthesis during primitive erythropoiesis. Blood. 2009;113:1340–1349. doi: 10.1182/blood-2008-08-174854. [DOI] [PubMed] [Google Scholar]
- 23.Rasmussen KD, Simmini S, Abreu-Goodger C, Bartonicek N, Di Giacomo M, Bilbao-Cortes D, et al. The miR-144/451 locus is required for erythroid homeostasis. J Exp Med. 2010;207:1351–1358. doi: 10.1084/jem.20100458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu D, dos Santos CO, Zhao G, Jiang J, Amigo JD, Khandros E, et al. miR-451 protects against erythroid oxidant stress by repressing 14-3-3zeta. Genes Dev. 2010;24:1620–1633. doi: 10.1101/gad.1942110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patrick DM, Zhang CC, Tao Y, Yao H, Qi X, Schwartz RJ, et al. Defective erythroid differentiation in miR-451 mutant mice mediated by 14-3-3zeta. Genes Dev. 2010;24:1614–1619. doi: 10.1101/gad.1942810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci USA. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q, Huang Z, Xue H, Jin C, Ju XL, Han JD, et al. MicroRNA miR-24 inhibits erythropoiesis by targeting activin type I receptor ALK4. Blood. 2008;111:588–595. doi: 10.1182/blood-2007-05-092718. [DOI] [PubMed] [Google Scholar]
- 29.Zhao H, Kalota A, Jin S, Gewirtz AM. The c-myb proto-oncogene and microRNA-15a comprise an active autoregulatory feedback loop in human hematopoietic cells. Blood. 2009;113:505–516. doi: 10.1182/blood-2008-01-136218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sankaran VG, Menne TF, Šćepanović D, Vergilio JA, Ji P, Kim J, et al. MicroRNA-15a and-16-1 act via MYB to elevate fetal hemoglobin expression in human trisomy 13. Proc Natl Acad Sci USA. 2011;108:1519–1524. doi: 10.1073/pnas.1018384108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sankaran VG, Xu J, Orkin SH. Advances in the understanding of haemoglobin switching. Br J Haematol. 2010;149:181–194. doi: 10.1111/j.1365-2141.2010.08105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guglielmelli P, Tozzi L, Bogani C, Iacobucci I, Ponziani V, Martinelli G, et al. Overexpression of microRNA-16-2 contributes to the abnormal erythropoiesis in polycythemia vera. Blood. 2011;117:6923–6927. doi: 10.1182/blood-2010-09-306506. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L, Flygare J, Wong P, Lim B, Lodish HF. miR-191 regulates mouse erythroblast enucleation by down-regulating Riok3 and Mxi1. Genes Dev. 2011;25:119–124. doi: 10.1101/gad.1998711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schreiber-Agus N, DePinho R. Repression by the Mad(Mxi1)-Sin3 complex. Bioessays. 1998;20:808–818. doi: 10.1002/(SICI)1521-1878(199810)20:10<808::AID-BIES6>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 35.Jayapal S, Lee K, Ji P, Kaldis P, Lim B, Lodish H. Downregulation of MYC is essential for terminal erythroid maturation. J Biol Chem. 2010;285:40252–40265. doi: 10.1074/jbc.M110.181073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deutsch VR, Tomer A. Megakaryocyte development and platelet production. Br J Haematol. 2006;134:453–466. doi: 10.1111/j.1365-2141.2006.06215.x. [DOI] [PubMed] [Google Scholar]
- 37.Kaushansky K. The molecular mechanisms that control thrombopoiesis. J Clin Invest. 2005;115:3339–3347. doi: 10.1172/JCI26674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirito K, Kaushansky K. Transcriptional regulation of megakaryopoiesis: thrombopoietin signaling and nuclear factors. Curr Opin Hematol. 2006;13:151–156. doi: 10.1097/01.moh.0000219660.03657.4b. [DOI] [PubMed] [Google Scholar]
- 39.Lu J, Guo S, Ebert BL, Zhang H, Peng X, Bosco J, et al. MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev Cell. 2008;14:843–853. doi: 10.1016/j.devcel.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klusmann JH, Li Z, Böhmer K, Maroz A, Koch ML, Emmrich S, et al. miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes Dev. 2010;24:478–490. doi: 10.1101/gad.1856210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ooi AG, Sahoo D, Adorno M, Wang Y, Weissman IL, Park CY. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc Natl Acad Sci USA. 2010;107:21505–21510. doi: 10.1073/pnas.1016218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bousquet M, Quelen C, Rosati R, Mansat-De Mas V, La Starza R, Bastard C, et al. Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J Exp Med. 2008;205:2499–2506. doi: 10.1084/jem.20080285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bousquet M, Harris MH, Zhou B, Lodish HF. MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci USA. 2010;107:21558–21563. doi: 10.1073/pnas.1016611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chapiro E, Russell LJ, Struski S, Cav H, Radford-Weiss I, Valle VD, et al. A new recurrent translocation t(11;14)(q24;q32) involving IGH@ and miR-125b-1 in B-cell progenitor acute lymphoblastic leukemia. Leukemia. 2010;24:1362–1364. doi: 10.1038/leu.2010.93. [DOI] [PubMed] [Google Scholar]
- 45.Sonoki T, Iwanaga E, Mitsuya H, Asou N. Insertion of microRNA-125b-1, a human homologue of lin-4, into a rearranged immunoglobulin heavy chain gene locus in a patient with precursor B-cell acute lymphoblastic leukemia. Leukemia. 2005;19:2009–2010. doi: 10.1038/sj.leu.2403938. [DOI] [PubMed] [Google Scholar]
- 46.Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Starczynowski DT, Kuchenbauer F, Argiropoulos B, Sung S, Morin R, Muranyi A, et al. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat Med. 2010;16:49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- 48.Kumar MS, Narla A, Nonami A, Mullally A, Dimitrova N, Ball B, et al. Coordinate loss of a microRNA and protein-coding gene cooperate in the pathogenesis of 5qsyndrome. Blood. 2011;118:4666–4673. doi: 10.1182/blood-2010-12-324715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Labbaye C, Spinello I, Quaranta MT, Pelosi E, Pasquini L, Petrucci E, et al. A threestep pathway comprising PLZF/miR-146a/CXCR4 controls megakaryopoiesis. Nat Cell Biol. 2008;10:788–801. doi: 10.1038/ncb1741. [DOI] [PubMed] [Google Scholar]
- 50.Hattangadi SM, Wong P, Zhang L, Flygare J, Lodish HF. From stem cell to red cell: regulation of erythropoiesis at multiple levels by multiple proteins, RNAs, and chromatin modifications. Blood. 2011;118:6258–6268. doi: 10.1182/blood-2011-07-356006. [DOI] [PMC free article] [PubMed] [Google Scholar]