Abstract
It has recently been shown that alteration of the methylation pattern of imprinted genes is associated with different types of male infertility. The objective of our study was to investigate the methylation pattern of selected gene promoters in sperm of patients with abnormal protamine replacement. The promoters of OCT4, SOX2, NANOG, HOXC11, miR-17 and CREM were analyzed using bisulfite sequencing and the percentage of DNA methylation was compared between patients with an abnormal protamine 1/protamine 2 (P1/P2) ratio and normozoospermic controls. No significant quantitative differences were found between groups of patients with either an abnormally high or low P1/P2 ratio compared to normal controls. However, two individual samples from infertile subjects (2/20, 10%) showed an altered methylation pattern for the CREM gene promoter that was not found in control samples. These two samples had a significantly higher (P<0.05) promoter methylation (5.58 and 4.23%, respectively) compared to the control group (0.46%). In conclusion, in our pilot study, extreme methylations defects were not seen broadly in severely infertile men. However, two patients exhibited altered methylation of the CREM gene, which may be either causative or a result of abnormal protmaine replacement.
Keywords: Epigenetic, gene promoter, male infertility, methylation, protamine
Introduction
Infertility affects 10–15% of the population and male factor is probably involved in about half of infertility cases. Several classes of male infertility have a well-characterized pathology, but approximately 60% of the cases are classified as idiopathic.1, 2 Genetic variations have long been known to account for a considerable fraction of male infertility, and it is likely that the number of the known genetic risk factors will increase in the future.3 Along with genetic variations, it is now believed that epigenetic alterations may contribute to a significant percentage of male infertility cases.4
Epigenetic marks can affect gene expression without altering the gene coding sequence, and include the methylation of cytosines in the DNA itself. DNA methylation in the promoter region plays an important role in gene expression regulation.5 A causative relationship of abnormal promoter methylation of methylenetetrahydrofolate reductase gene with non-obstructed azoospermia has been reported.6 In addition several recent studies have revealed an increased abnormal methylation of imprinted loci in men with oligozoospermia7, 8 and with idiopathic infertility.9 There is also evidence that the pattern of DNA methylation at imprinted loci is altered in patients with abnormal protamination.10
During spermiogenesis, canonical histones are largely replaced with protamines in a multistep process. This results in a more condensed nucleus that is believed to be important for protecting the sperm chromatin against nucleases and mutagens and enhancing the capability of sperm motility.11, 12 Two types of protamines, protamine 1 (P1) and protamine 2 (P2), are expressed in most mammals, and in humans the ratio of the incorporation of these is strictly regulated.13 Fertile men have a P1/P2 ratio of 1.11, 12, 14, 15 Several studies have reported that diminished sperm quality, increased DNA fragmentation and reduced pregnancy rates are associated with an altered protamine ratio.14, 15, 16, 17, 18, 19, 20, 21 However, the mechanism to explain how abnormal protamination results in altered semen quality and reduced pregnancy rates is not known.
Our laboratory has recently demonstrated that protamine replacement results in an interesting pattern of epigenetic modifications that may poise paternal genes for expression during early embryogenesis.22 During the protamine replacement, retained histones are retained at transcription and signaling factors that guide embryo development, at promoters of embryonic developmental genes and at microRNAs. Furthermore, these regions showed a significant DNA hypomethylation.22 These data have been supported in two recent papers studying both human and mouse sperm.23, 24 These studies may indicate that patients with aberrant protamine replacement might also have an altered histone retention localization that could affect the DNA methylation profile on genes important for early embryonic development.
In this pilot study we have studied methylation patterns in the promoters of six genes including a gene involved in spermiogenesis (CREM), a gene from the developmentally important HOX locus (HOXC11), transcription factors (OCT4, SOX2 and NANOG), and a microRNA (miR-17). These selected genes show diverse promoters methylation characteristics including those with and without distinct CpG islands. Ninety-one CpGs per subject in 10 replicates and a total of 27 300 CpGs were analyzed. The aim of this study was to evaluate sperm from patients with altered P1/P2 ratios (both high and low P1/P2 ratios) for possible changes in the methylation pattern of the promoter regions of these empirically selected genes.
Materials and methods
Sperm samples and P1/P2 ratio
Institutional Review Board approval was obtained before the initiation of the study. Sperm DNA was extracted from 10 normozoospermic, fertile controls, 10 patients with a high P1/P2 ratio (1.65–3.72) and 10 patients with low P1/P2 ratio (0.13–0.64). Semen analysis was carried out according to the World Health Organization (2010) standards.25 The P1/P2 ratio was determined using acetic acid–urea gel electrophoresis as described previously.17
DNA extraction and bisulfite conversion
Two microgram of extracted DNA of each sample was treated with sodium bisulfite. Briefly, DNA was denatured with 0.2 mol l−1 NaOH at 70 °C for 15 min, then 30 µl of freshly prepared 10 mmol l−1 hydroquinone was added along with 520 µl of fresh 3 mol l−1 sodium metabisulfite (pH 5.0). Samples were overlaid gently with mineral oil and incubated for 16 h at 50 °C then transferred to a new tube and desalted using PB buffer (Qiagen, Germantown, MD, USA). DNeasy Blood & Tissue Kit (Qiagen) was used for DNA cleanup. Ethanol traces were removed by centrifugation and the DNA was eluted with 200 µl distilled water. Twenty µl of 3 mol l−1 NaOH was added and incubated for 15 min at 37 °C, and then DNA was precipitated with 22 µl 4 mol l−1 NaOAc and 400 µl of ice-cold ethanol using tRNA (10 µg ml−1) as a carrier. The precipitate was centrifuged at 16 000g for 60 min at 4 °C and resuspended in 30 µl of TE (pH 8.0). Two µl of the sample was used for the amplification of each sequence analyzed.
PCR amplification, cloning and sequencing
Amplicons from the promoter region of the genes for OCT4, SOX2, NANOG, HOXC11, miR-17 and CREM were amplified by PCR (Table 1). AccuPrime Taq DNA Polymerase System (Invitrogen, Carlsbad, CA, USA) was used according to manufacturer's instructions with a slight modification. Briefly, 50 µl reactions were prepared for each gene consisting of 1× PCR buffer containing 1.5 mmol l−1 MgCl2 and 0.2 mmol l−1 of each dNTP, 0.4 µmol l−1 of each primer, 1 µl of Taq polymerase and 1× PCRx Enhancer solution (Invitrogen). MgCl2 concentration for each PCR reaction is specified in Table 1. PCR conditions were: initial denaturation 94 °C for 10 min; 35 amplification cycles of denaturation at 94 °C for 30 s, primer annealing at 52–58 °C (Table 1) for 45 s and strand elongation for 1 min at 72 °C followed by a final extension of 10 min at 72 °C. The size of the PCR products and the number of CpGs analyzed for each amplicon is indicated in Table 1.
Table 1. Parameters of PCR amplifications and characteristics of the selected sequences.
| Primers (5′-3′ forward, reverse) | MgCl2 concentration (mmol l−1) | Annealing temperature (°C) | Amplicon size (bp) | Location from transcription start site (+1) | No. of analyzed/total CpGs | |
|---|---|---|---|---|---|---|
| OCT4a | AATAGATTTTGAAGGGGAGTTTAGG | 4.0 | 56 | 181 | −1734 to −1553 | 6/6 |
| TTCCTCCTTCCTCTAAAAAACTCA | ||||||
| SOX2 | GTGGTGTGATTTGTTGTTGYGAG | 3.5 | 58 | 374 | −685 to −312 | 19/20 |
| CCTCTTCTTTCTCTCAATCCTAATCTTAAAA | ||||||
| NANOGa | TTAATTTATTGGGATTATAGGGGTG | 4.0 | 58 | 164 | −549 to −385 | 4/4 |
| AAACCTAAAAACAAACCCAACAAC | ||||||
| HOXC11 | GTTGTGGTATGGAGAGAAGGGG | 3.5 | 52 | 337 | −304 to +33 | 19/19 |
| ACAAAAATTACCCAAATTAACCRAATTAAACA | ||||||
| miR-17 | GAATTATAATGTGAAGAAGGAGGTGTTT | 3.5 | 56 | 461 | −3730 to −3269 | 17/17 |
| CTTCCTAAAAACCCTACTCTCCC | ||||||
| CREM | GGAGGAAAAGGGTTATTAGTTAAGGTATG | 3.5 | 58 | 493 | −492 to +1b | 26/30 |
| CAAAAATAAAACRCCACAATCCAACAATA |
SOX forward primer contains a Y (= C or T) and CREM (G or A) reverse primer contains an R.
Primers are published by Deb-Rinker et al..38
Has multiple transcription start sites; coordinate is in reference to the first exon.
Amplicons were cloned using a TOPO TA Cloning kit (Invitrogen) according to manufacturer's recommendations. Ten alleles from positive colonies were sequenced at the University of Utah Core Research Facilities using an ABI 3730xl instrument. For additional technical replicates, five samples from the gene NANOG and five samples from the gene CREM were reamplified, cloned and sequenced.
Data analysis
The conversion efficiency was checked internally and only sequences with a conversion rate of 95% or higher were included in data analysis. On the amplicons of SOX2 and CREM, not every CpG was sequenced because the bisulfite conversion produced long poly-T sequences that could not be read. All the statistical analysis was carried out using Stata 9.2 (Stat Corp., College Station, TX, USA). Differences in semen analysis parameters between groups were assessed using analysis of variance. The percentage of methylated CpGs at each promoter was compared with Kruskal–Wallis test between the three groups. For the comparison of fertile and infertile groups (high and low P1/P2 groups combined), the Mann–Whitney test was used. When comparing a single sample to the control group, the percent methyaltion for each allele was transformed by arc sine transformation and analyzed by one-way analysis of variance. Differences with P<0.05 were considered as significant.
Results
The P1/P2 ratio was different in all three groups (P<0.0001). The control group had a significantly higher mean P1/P2 ratio (0.90±0.43; P<0.0001) than the low P1/P2 group (0.55±0.48) and a significantly lower P1/P2 ratio than the high P1/P2 group (2.08±0.20; P<0.0001). In agreement with previously published data, sperm concentration was the highest in the control group (132.50±14.81; P<0.05). Sperm concentration was not different between the high and low P1/P2 groups. Sperm motility was similar across the different groups. Normal head morphology was reduced in the high and low P1/P2 groups compared to the control groups (P<0.001; Table 2).
Table 2. Sperm parameters in the study groups.
| Control | High P1/P2 | Low P1/P2 | P value | |
|---|---|---|---|---|
| P1/P2 | 0.90±0.43 | 2.08±0.20 | 0.55±0.48 | 0.0001a |
| Concentration (×106 ml−1) | 132.50±14.81b | 60.37±9.56 | 69.31±24.45 | 0.0167 |
| Motility (%) | 50.00±2.41 | 56.20±6.86 | 40.80±6.59 | 0.1817 |
| Normal head (%) | 68.67±4.35c | 24.20±3.05 | 26.56±5.42 | 0.0001 |
Abbreviation: P1/P2, protamine 1/protamine 2.
All three groups were significantly different (P<0.001).
Significantly different from high (P=0.0011) and low (P=0.0435) P1/P2 groups.
Significantly different from high and low P1/P2 groups (P<0.001).
Our technical replicates were in good accordance with the initial methylation data with exception of one sample that was therefore removed from further analysis. We did not find any significant difference in the frequency of DNA methylation between the three groups for any of the genes analyzed. Also, no difference was observed between the fertile control group and the combined infertile group in any of the gene promoters investigated (Table 3).
Table 3. Overall abnormal promoter methylation in different groups.
| Normal P1/P2 (n=10) | High P1/P2 (n=10) | Low P1/P2 (n=10) | Combined infertile group | P value | ||
|---|---|---|---|---|---|---|
| Overall abnormal methylation (%) | Between all groups | Fertile versus infertile groups | ||||
| OCT4 | 4.77 | 3.73 | 5.97 | 4.85 | 0.1039 | 0.5804 |
| SOX2 | 0.50 | 0.28 | 0.17 | 0.22 | 0.4304 | 0.2371 |
| NANOG | 13.89a | 10.50 | 14.00 | 12.25 | 0.9904 | 0.9431 |
| HOXC11 | 0.58 | 0.58 | 0.37 | 0.47 | 0.4802 | 0.5003 |
| miR-17 | 0.12 | 0.06 | 0.29 | 0.18 | 0.1174 | 0.5659 |
| CREM | 0.46 | 1.21 | 0.73 | 1.11 | 0.3702 | 0.5561 |
Abbreviation: P1/P2, protamine 1/protamine 2.
Only nine donor samples were included in the analysis.
Although groups did not differ, we found an interesting methylation pattern on the promoter of CREM in two infertile patients (Figure 1). Samples from patient #4 and patient #19 were significantly different from the control group (P=0.0005 and P=0.0412, respectively). Sample #4 is from a subject that had a P1/P2 ratio of 2.04 and low motility but normal sperm count. Sample #19 was a patient that had a P1/P2 ratio of 0.59 and severe oligoasthenozoospermia.
Figure 1.
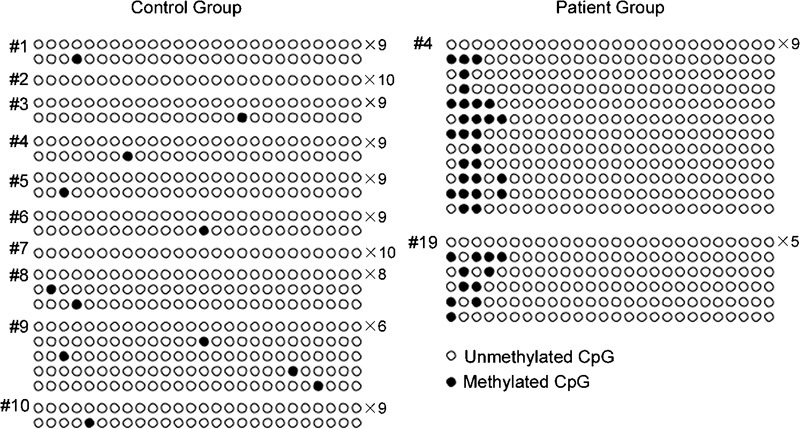
The promoter methylation profile of the CREM in the control group and in patients #4 and #19. The number to the right of each profile indicates the number of alleles with the same methylation pattern.
Conclusions
Our study focused on a population of infertile men with aberrant protamine replacement of histones during spermiogenesis. It has previously been shown that generally these patients have lower sperm concentrations and motility than fertile men, and a higher frequency of abnormal sperm morphology and DNA damage than fertile controls.(14–17,19–21) Furthermore, lower pregnancy rates have also been reported in patients with abnormal protamine replacement, even when intracytoplasmic sperm injection is performed.15, 17, 18, 21 Whether aberrant protamination is the cause of infertility or simply an indication of other abnormalities during spermatogenesis is not known. Additionally, the possible causes of abnormal embryogenesis resulting in lower in vitro fertilization pregnancy rates have not been evaluated beyond the possible effects of elevated DNA damage. The purpose of this study was to evaluate one possible cause of altered embryogenesis, epigenetic marking via DNA methylation of gene promoter regions, in men with altered protamination of the sperm chromatin.
Abnormal methylation of imprinted loci has been reported in infertile men. Kobayashi et al. analyzed seven imprinted loci in sperm of oligozoospermic patients and found severely altered abnormal methylation patterns in both paternal and maternal imprints.7 Hypomethylation of H19 and hypermethylation of MEST were also observed at a higher frequency in oligozoospermic men than in normozoospermic controls.8 Our laboratory has reported alterations in the methylation pattern of imprinted loci of not only oligozoopermic patients, but also in the sperm of patients with abnormal P1/P2 ratios.10 Poplinsky et al. have also reported abnormal methylation of imprinted genes in the sperm of men with idiopathic infertility.9 Lastly, abnormal promoter methylation of non-imprinted genes has also been associated with impaired spermatogenesis.6 These studies have consistently found very low levels of methylation abnormalities in the sperm of fertile controls, but have demonstrated that methylation defects are commonly seen in various types of genes analyzed in a broad spectrum of infertility phenotypes.
It has been shown that sperm promoter methylation pattern resembles to those found in pluripotent cell types such as embryonic stem and embryonic germ cells.26 Additionally, a bivalent histone modification pattern, similar to that found in embryonic stem cells, has also been reported in sperm at promoters of microRNAs, and at imprinted gene clusters, and transcription and signaling factors that are important for embryonic development.22 It is unknown whether the protamine and histone bound regions are the same in patients with normal and abnormal protamination as in fertile men and whether the establishment of epigenetic marks on DNA is somehow regulated by the nucleioprotein pattern. Our hypothesis was that histone retention on promoter regions is crucial for the proper establishment of DNA methylation, and that abnormal protamination might be accompanied, by or causative of, epigenetic changes on non-imprinted genes in these patients, which might explain their cause of infertility or affect development.
With one exception, we selected genes for this study that have important roles in embryo development. OCT4, SOX2 and NANOG are transcription factors that are crucial for the efficient maintenance of the pluripotent state of cells.27 miR-17 (also known as miR-17-5p) is a noncoding microRNA belongs to the miR-17–92 cluster.28 This cluster has important roles in tissue development and has been implicated in several types of cancer.29 HOXC11 has multiple roles including the formation of the central nervous system, the formation of the vertebrae, hindlimb development and the formation of the urogenital system.30 HOXC11 mRNA can be found in eight-cell stage embryos and is thought to be the earliest HOX gene in the developing embryo expressed.31
These genes promoters show different methylation characteristics. SOX2 and miR-17 are hypomethylated and both have distinct CpG islands over 100 bp in size with observed/expected CpG ratio >0.6 and CG content >50%. HOXC11 is also hypomethylated, and its promoter region has no CpG island but has several CpGs. OCT4 and NANOG are both methylated in sperm and their promoters 1× PCRx Enhancer solution (Invitrogen) do not contain CpG islands.
The paternal genome undergoes active demethylation following fertilization that erases most of the epigenetic marks that are established in spermatogenesis.32, 33 The purpose of this methylation reprogramming is not understood, but it is possible that it is required for the embryo to achieve totipotency.34 It has been recently proposed that promoters in sperm do not undergo extensive genome-wide methylation reprogramming at fertilization. Demethylation likely occurs on transposons and inter and intragenic regions.26
There is a subset of important genes in sperm whose promoters do not show this epigenetic modification resemblance to pluripotent cells. Promoters of NANOG and OCT4 are hypermethylated in sperm and hypometylated in embryonic stem and embryonic germ cells. These key regulators of pluripotency are normally silenced during spermatogenesis, possibly preventing the formation of teratomas. After fertilization these marks needs to be removed in order to achieve pluripotency in the embryo. NANOG has been shown to be demethylated rapidly after fertilization as a result of both active and passive processes.26 Thus, abnormalities in the methylation patterns of these genes likely reflect aberrant re-establishment of these epigenetic marks during spermatogenesis rather than a deficiency that will be transmitted to the next generation.
We did not find any significant difference in promoter methylation of the selected developmental genes between the patient group with abnormal protamination and fertile subjects. Our comparison was made based on the number of methylated CpGs, assuming that all the CpGs are of equal importance. It has to be taken account that smaller changes in methylation still might be biologically relevant. Animal models might be useful in understanding the effect of small but possibly meaningful differences in the methylation profile of gene promoters in the future. In this pilot study, we did not find extreme differences between samples from patients with abnormal protamination and known fertile subjects at least in the patients and on the genes we evaluated.
One can argue that the empirical approach we used for the selection of the genes for our study is inefficient and genome-wide studies help for a better selection of target genes. It is, however, expensive and cumbersome to screen genome-wide for genes that are differentially methylated in patients and in controls. Based on the data generated in this study, a genome-wide study is underway in our laboratory to investigate whether differentially methylated gene promoters are randomly localized in the genome or whether they are gene specific. We are also interested in finding possible target genes that are characteristic for abnormal protamination.
Our study population was selected based on the P1/P2 ratio. Although, abnormal P1/P2 ratio is clearly associated with diminished semen quality,17 our study groups showed lower but still normal sperm concentrations and normal motility. Most of the studies that have investigated the methylation status of imprinted control regions reported abnormalities in oligozoospermic men.7, 8 It is possible that the inclusion of a study group with low sperm count would have given us more information. Future studies will address these questions.
Little is known about the methylation status of spermiogenesis regulatory genes. Also, the expression patterns of these genes in human sperm are not well characterized. For obvious reasons it is very difficult to obtain testicular tissue that could allow us to gain more functional information. CREM is a key transcription factor during spermiogenesis that targets several important genes including P1 and transition protein 1 among others in the process of structuring the spermatozoon.35, 36 We found no difference in CREM promoter methylation between groups with different P1/P2 ratios, although we did find two samples with an interesting methylation pattern on the promoter of CREM. It is not known whether these changes could be the underlying reason for their abnormal P1/P2 ratio and if so, whether these changes are accompanied by other disturbances in the establishment of epigenetic marks. It is possible that abnormal P1/P2 ratio could be the result of several different types of misregulation during spermiogenesis, and our finding is intriguing with a 10% frequency in this phenotype, albeit using a small number of patients. However, the analysis of additional samples will be necessary to validate our finding. A follow-up study is underway in our laboratory analyzing more patients with an extended target region.
In our study, we analyzed a heterogeneous sperm population. Most of the earlier studies used samples that were fractioned with either density centrifugation8, 37 or with swim-up.7, 9 There is no information regarding the methylation status of the whole sample compared to a fractioned sample. However, there is a possibility that the unselected sample can mask differences that might exist between the better quality sperm populations in different samples. We found a very low level of hypermethylation and a moderate level of hypomethylation even in samples with low quality, which contradicts to this theory. Also, comparison of unselected samples resulted in detectable differences analyzing imprinted genes.10
In conclusion, while no statistically significant changes in promoter methylation of selected genes were found using bisulfite sequencing comparing patients with protamination abnormalities with controls, we did find a distinct methylation pattern on the promoter of CREM in two patients that need to be followed up in a larger study. It is clear, however, that more robust techniques such as pyrosequencing or promoter methylation array must be employed to be able to detect marginal but possibly meaningful differences in methylation.
Author contributions
The work presented here was carried out in collaboration between both authors. L.N. and D.T.C. were involved in study design and data analysis. L.N. prepared samples and performed all of the experiments.
The authors declare no competing financial interests.
References
- Dohle GR, Halley DJ, van Hemel JO, van den Ouwel AM, Pieters MH, et al. Genetic risk factors in infertile men with severe oligozoospermia and azoospermia. Hum Reprod. 2002;17:13–6. doi: 10.1093/humrep/17.1.13. [DOI] [PubMed] [Google Scholar]
- Erenpreiss J, Spano M, Erenpreisa J, Bungum M, Giwercman A. Sperm chromatin structure and male fertility: biological and clinical aspects. Asian J Androl. 2006;8:11–29. doi: 10.1111/j.1745-7262.2006.00112.x. [DOI] [PubMed] [Google Scholar]
- O'Flynn O'Brien KL, Varghese AC, Agarwal A. The genetic causes of male factor infertility: a review. Fertil Steril. 2010;93:1–12. doi: 10.1016/j.fertnstert.2009.10.045. [DOI] [PubMed] [Google Scholar]
- Carrell DT, Hammoud SS. The human sperm epigenome and its potential role in embryonic development. Mol Hum Reprod. 2010;16:37–47. doi: 10.1093/molehr/gap090. [DOI] [PubMed] [Google Scholar]
- Hatada I, Fukasawa M, Kimura M, Morita S, Yamada K, et al. Genome-wide profiling of promoter methylation in human. Oncogene. 2006;25:3059–64. doi: 10.1038/sj.onc.1209331. [DOI] [PubMed] [Google Scholar]
- Khazamipour N, Noruzinia M, Fatehmanesh P, Keyhanee M, Pujol P. MTHFR promoter hypermethylation in testicular biopsies of patients with non-obstructive azoospermia: the role of epigenetics in male infertility. Hum Reprod. 2009;24:2361–4. doi: 10.1093/humrep/dep194. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Sato A, Otsu E, Hiura H, Tomatsu C, et al. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum Mol Genet. 2007;16:2542–51. doi: 10.1093/hmg/ddm187. [DOI] [PubMed] [Google Scholar]
- Marques CJ, Costa P, Vaz B, Carvalho F, Fernandes S, et al. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol Hum Reprod. 2008;14:67–74. doi: 10.1093/molehr/gam093. [DOI] [PubMed] [Google Scholar]
- Poplinski A, Tuttelmann F, Kanber D, Horsthemke B, Gromoll J. Idiopathic male infertility is strongly associated with aberrant methylation of MEST and IGF2/H19 ICR1. Int J Androl. 2010;33:642–9. doi: 10.1111/j.1365-2605.2009.01000.x. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Purwar J, Pflueger C, Cairns BR, Carrell DT. Alterations in sperm DNA methylation patterns at imprinted loci in two classes of infertility. Fertil Steril. 2010;94:1728–33. doi: 10.1016/j.fertnstert.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Oliva R. Protamines and male infertility. Hum Reprod Update. 2006;12:417–35. doi: 10.1093/humupd/dml009. [DOI] [PubMed] [Google Scholar]
- Carrell DT, Emery BR, Hammoud S. Altered protamine expression and diminished spermatogenesis: what is the link. Hum Reprod Update. 2007;13:313–27. doi: 10.1093/humupd/dml057. [DOI] [PubMed] [Google Scholar]
- Corzett M, Mazrimas J, Balhorn R. Protamine 1: protamine 2 stoichiometry in the sperm of eutherian mammals. Mol Reprod Dev. 2002;61:519–27. doi: 10.1002/mrd.10105. [DOI] [PubMed] [Google Scholar]
- Balhorn R, Reed S, Tanphaichitr N. Aberrant protamine 1/protamine 2 ratios in sperm of infertile human males. Experientia. 1988;44:52–5. doi: 10.1007/BF01960243. [DOI] [PubMed] [Google Scholar]
- Carrell DT, Liu L. Altered protamine 2 expression is uncommon in donors of known fertility, but common among men with poor fertilizing capacity, and may reflect other abnormalities of spermiogenesis. J Androl. 2001;22:604–10. [PubMed] [Google Scholar]
- Belokopytova IA, Kostyleva EI, Tomilin AN, Vorob'ev VI. Human male infertility may be due to a decrease of the protamine P2 content in sperm chromatin. Mol Reprod Dev. 1993;34:53–7. doi: 10.1002/mrd.1080340109. [DOI] [PubMed] [Google Scholar]
- Aoki VW, Liu L, Carrell DT. Identification and evaluation of a novel sperm protamine abnormality in a population of infertile males. Hum Reprod. 2005;20:1298–306. doi: 10.1093/humrep/deh798. [DOI] [PubMed] [Google Scholar]
- de Mateo S, Gazquez C, Guimera M, Balasch J, Meistrich ML, et al. Protamine 2 precursors (Pre-P2), protamine 1 to protamine 2 ratio (P1/P2), and assisted reproduction outcome. Fertil Steril. 2009;91:715–22. doi: 10.1016/j.fertnstert.2007.12.047. [DOI] [PubMed] [Google Scholar]
- Carrell DT, Emery BR, Liu L. Characterization of aneuploidy rates, protamine levels, ultrastructure, and functional ability of round-headed sperm from two siblings and implications for intracytoplasmic sperm injection. Fertil Steril. 1999;71:511–6. doi: 10.1016/s0015-0282(98)00498-1. [DOI] [PubMed] [Google Scholar]
- Aoki VW, Moskovtsev SI, Willis J, Liu L, Mullen JB, et al. DNA integrity is compromised in protamine-deficient human sperm. J Androl. 2005;26:741–8. doi: 10.2164/jandrol.05063. [DOI] [PubMed] [Google Scholar]
- Aoki VW, Liu L, Jones KP, Hatasaka HH, Gibson M, et al. Sperm protamine 1/protamine 2 ratios are related to in vitro fertilization pregnancy rates and predictive of fertilization ability. Fertil Steril. 2006;86:1408–15. doi: 10.1016/j.fertnstert.2006.04.024. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–8. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpanahi A, Brinkworth M, Iles D, Krawetz SA, Paradowska A, et al. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res. 2009;19:1338–49. doi: 10.1101/gr.094953.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, et al. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 2010;17:679–87. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- World Health Organization . WHO Laboratory Manual for the Examination and Processing of Human Semen. 5th ed. Geneva: World Health Organization; 2010. [Google Scholar]
- Farthing CR, Ficz G, Ng RK, Chan CF, Andrews S, et al. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4:e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Smith A. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene. 2004;23:7150–60. doi: 10.1038/sj.onc.1207930. [DOI] [PubMed] [Google Scholar]
- Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. J Mol Biol. 2004;339:327–35. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- Shan SW, Lee DY, Deng Z, Shatseva T, Jeyapalan Z, et al. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat Cell Biol. 2009;11:1031–8. doi: 10.1038/ncb1917. [DOI] [PubMed] [Google Scholar]
- Hostikka SL, Capecchi MR. The mouse Hoxc11 gene: genomic structure and expression pattern. Mech Dev. 1998;70:133–45. doi: 10.1016/s0925-4773(97)00182-2. [DOI] [PubMed] [Google Scholar]
- Li SS, Liu YH, Tseng CN, Singh S. Analysis of gene expression in single human oocytes and preimplantation embryos. Biochem Biophys Res Commun. 2006;340:48–53. doi: 10.1016/j.bbrc.2005.11.149. [DOI] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–2. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, et al. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–8. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–93. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Mali P, Kaipia A, Kangasniemi M, Toppari J, Sandberg M, et al. Stage-specific expression of nucleoprotein mRNAs during rat and mouse spermiogenesis. Reprod Fertil Dev. 1989;1:369–82. doi: 10.1071/rd9890369. [DOI] [PubMed] [Google Scholar]
- Nantel F, Monaco L, Foulkes NS, Masquilier D, LeMeur M, et al. Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant mice. Nature. 1996;380:159–62. doi: 10.1038/380159a0. [DOI] [PubMed] [Google Scholar]
- Boissonnas CC, Abdalaoui HE, Haelewyn V, Fauque P, Dupont JM, et al. Specific epigenetic alterations of IGF2-H19 locus in spermatozoa from infertile men. Eur J Hum Genet. 2010;18:73–80. doi: 10.1038/ejhg.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb-Rinker P, Ly D, Jezierski A, Sikorska M, Walker PR. Sequential DNA methylation of the Nanog and Oct-4 upstream regions in human NT2 cells during neuronal differentiation. J Biol Chem. 2005;280:6257–60. doi: 10.1074/jbc.C400479200. [DOI] [PubMed] [Google Scholar]


