Abstract
The sensitivity of an organism and its cells to hypoxic injury varies widely; yet, few genetic determinants of metazoan hypoxic sensitivity have been identified. We report here the isolation of a profoundly hypoxia resistant mutant and its identification as a reduction-of-function allele of rrt-1, which encodes an arginyl-tRNA synthetase. rrt-1 knockdown before or after the hypoxic injury rescues animals from death. RNAi knockdown of most other aminoacyl-tRNA synthetases also confers hypoxia resistance, the level of which inversely correlates with translation rate. rrt-1(RNAi) blocked hypoxic induction of the unfolded protein response and tunicamycin toxicity. Disruption of the unfolded protein response partially suppressed the hypoxia resistance of rrt-1(lf). The data support a model where translational suppression induces hypoxia resistance, in part by reducing unfolded protein toxicity.
Forward genetic screens offer the possibility of discovering genes not previously known to control hypoxic sensitivity. Such genes are likely to play an important role in emergent organismal traits such as habitat range and ability to hibernate. Additionally, these genes may lead to the development of novel therapies for conditions where cellular hypoxic sensitivity is a pathological determinant such as stroke, myocardial infarction, and cancer. We have previously shown that wild type C. elegans when placed in a severe hypoxic environment ([O2] < 0.3 vol%) become immobile but fully recover when returned to normoxia within 4 hours (1). If the hypoxic incubation is prolonged past 4 hours, permanent behavioral deficits, cellular death, and eventually organismal death ensue. After a 24-hour hypoxic incubation, greater than 99% of wild type animals are dead. We used this easily scored organismal hypoxic death endpoint as the basis for a mutant screen to identify genes that control hypoxic sensitivity of the whole animal and its cells. Specifically, we screened for EMS-derived mutants that survived a 22 hour hypoxic incubation. In a screen of 3884 F1 mutant genomes, we recovered 14 mutants that had a significant hypoxia resistant phenotype (Table S1). These mutants fell into 13 complementation groups. gc47, one of the strongest hypoxia resistant mutants, was chosen for further characterization and mapping.
After outcrossing to the wild type strain N2, the hypoxia resistance of gc47 was quantified and found to be substantial. Immediately after removal from a 20 hour hypoxic incubation both N2 and gc47 were paralyzed, but gc47 recovered the ability to move completely over the next 1 to 2 hours. After a 24 hour recovery, essentially all of the gc47 animals were alive whereas almost all wild type failed to survive (Fig. 1); gc47 prolonged the hypoxic incubation time required for complete killing by more than three fold (Fig. 1). The hypoxia resistant phenotype was fully recessive and segregated as a single locus in a Mendelian fashion (Fig. 1D and Materials and Methods). gc47 was mapped based on its hypoxia resistant phenotype to the left arm of chromosome III (Fig. 2A). A combination of two factor and three factor mapping with visible mutations and single nucleotide polymorphisms placed the mutation in a 106 kb interval containing 32 known or predicted genes. RNAi of 29 of the 32 genes in the interval identified only one gene, rrt-1, whose knockdown resulted in high level hypoxia resistance (Fig. 2B). Simultaneously, five fosmids that together spanned the entire interval were individually injected along with a transformation marker to attempt transformation rescue of gc47; only one fosmid restored normal hypoxia sensitivity to gc47 (Fig. 2C). The rescuing fosmid contained the rrt-1 gene that was implicated by RNAi. Sequencing of the rrt-1 gene in gc47 found a single mutation, a G to A transition at nucleotide 811, resulting in a change of amino acid residue 271 from an aspartate in wild type to an asparagine in gc47 (Fig. 2A). Based on mapping, transformation rescue, phenocopy with RNAi, and the identification of a missense mutation, we assign gc47 as an allele of rrt-1. Given that the hypoxia-resistant phenotype of gc47 is phenocopied by rrt-1(RNAi) and that the mutation is fully recessive, gc47 behaves like a reduction-of-function allele of rrt-1.
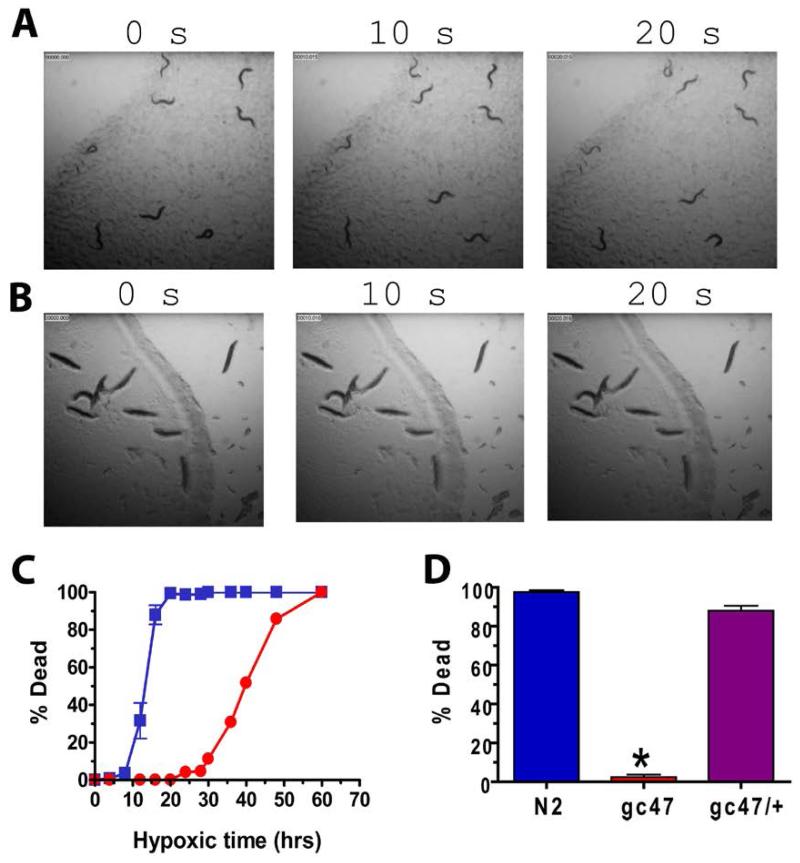
Fig. 1. gc47 is a potent regulator of hypoxic cell death in C. elegans.
(A,B) Time lapse images of (A) gc47 and (B) N2 adult worms following a 24 hr recovery from a 20 hr hypoxic insult. (C) Percent dead animals for N2 (blue squares) and gc47 (red circles) after a 24 hour recovery as a function of length of hypoxic insult. (D) gc47 is recessive. Percent death of homozygous gc47 (red; trials = 13), heterozygous gc47/+ (purple; trials = 9), and N2 (blue; trials = 16). mean ± s.e.m with at least 30 animals/trial.; * p < 0.01 (two-tailed t test).
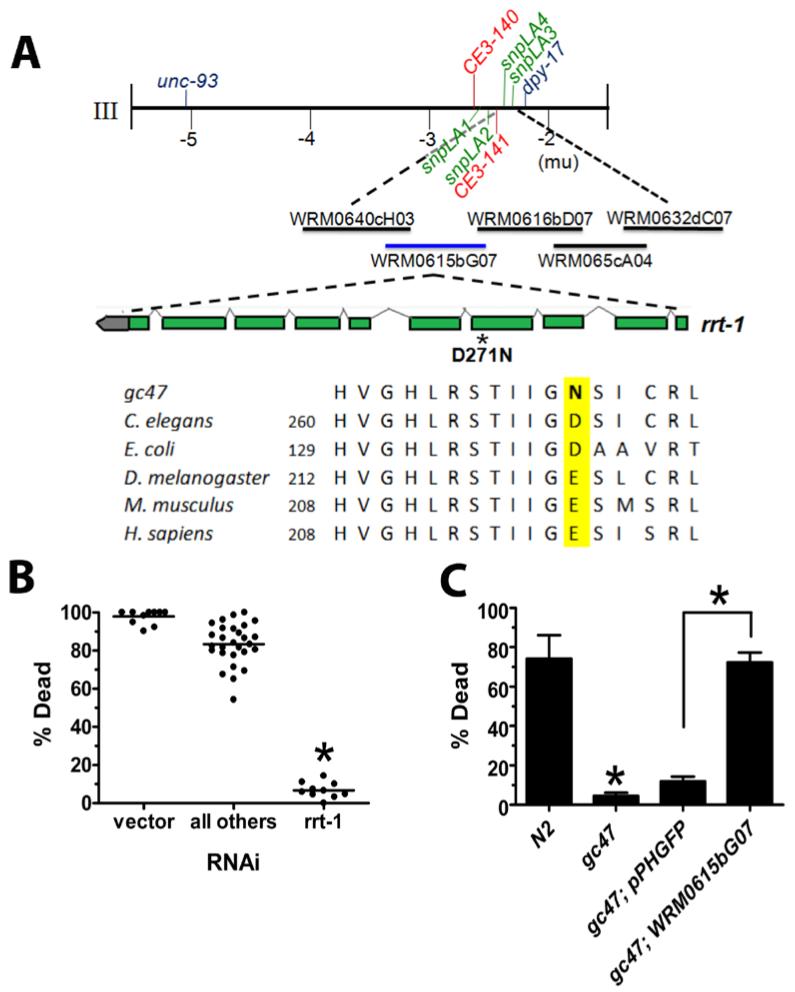
Fig. 2. gc47 is an allele of rrt-1.
(A) Genetic mapping of gc47. A portion of the genetic map of chromosome III is shown, with genes causing visible phenotypes in blue and relevant single nucleotide polymorphisms between Hawaiian CB4856 and N2 indicated in red (previously reported) or green (reported herein). Three-factor mapping with the visible markers and SNPs placed gc47 in a 106 kb interval between CE3-141 and snpLA3. Fosmids used to attempt transformation rescue are shown below with the rescuing fosmid shown in blue. Alignment is with rrt-1 orthologs. (B) RNAi of genes within the 106 kbp mapping interval. Hypoxia-induced death of animals treated with 29/32 predicted genes within the mapping interval are shown. The empty vector is shown as a non-resistant control. rrt-1 RNAi confers highly significant hypoxia resistance (P < 0.01; two-tailed t test, n = 30 animals/data point). (C) Transformation rescue of gc47. Hypoxia-induced animal death was scored in N2 wild type, gc47 (full genotype: rrt-1(gc47) dpy-17(e164)), and gc47 transformed with the transformation marker pPHGFP alone or in addition, the rescuing fosmid WRM0615bG07. mean ± s.e.m. of > 2 trials of > 20 animals/trial); * p < 0.01 (two-tailed t test).
rrt-1 encodes an arginyl-tRNA synthetase, one of the aminoacyl-tRNA synthetases (aaRS). aaRS catalyze the ATP-dependent acylation of their cognate tRNA(s) with a specific amino acid (2). aaRS fall into two distinct structural classes. RRT-1 is a class I enzyme, which is characterized by HIGH and MRSK domains (3, 4). aaRS can also be grouped according to whether they can form a multisubunit complex; in higher eukaryotes, the RRT-1 ortholog has been isolated in a cytoplasmic complex with six other aaRS and three accessory subunits (5, 6). Finally, some aaRS are specific for mitochondrial tRNAs. Besides their role in translation, a subset of the aaRS has been implicated in non-translation-related functions (7, 8). Thus, a critical question to answer is whether rrt-1 is unique among aaRS in controlling hypoxic sensitivity or if additional aaRS also have this property and if so, whether these fall into a particular structural or functional class. To test the other aaRS, we used feeding RNAi constructs against 23 of the 33 predicted aaRS in the C. elegans genome. All but one of the 23 RNAi constructs conferred significant hypoxia resistance (Table S2). Thus, most, if not all, aaRS control hypoxic sensitivity. However, knockdown of some aaRS produced a strong hypoxia-resistant phenotype and some did not. This variable strength of phenotype was not explained by the class of aaRS or whether the tRNA substrate was cytoplasmic or mitochondrial. One trivial explanation for this range of phenotypes is that the RNAi constructs are variably effective at knocking-down the target gene. Indeed, quantitative real-time PCR to compare transcript abundance before and after RNAi knockdown found that some of the non-hypoxia resistant RNAis produced a small or even undetectable reduction in transcript abundance (Table S2). However, RNAi efficacy did not explain all of the phenotypic variance. For example, the levels of the methionine-tRNA synthetase transcript were knocked down seven-fold, yet the level of hypoxia resistance was modest and much less than that seen with histidine-, tyrosine, or isoleucine-tRNA synthetases with a smaller degree of knockdown observed. To examine whether the variable levels of hypoxia resistance can be explained by differences in the degree of translational suppression, we measured 35S-methionine incorporation in animals treated with aaRS RNAis spanning the range of hypoxia resistant phenotypes as well as in rrt-1(gc47) (Figure S1). The level of hypoxia resistance had a strong inverse correlation with the relative translation rate. However, the absolute amount of translational suppression was relatively modest even in the strongest hypoxia resistant animals where the 35S-methionine incorporation was about half that of vector controls. Thus, hypoxic sensitivity appears to be exquisitely sensitive to even small changes in translation rate. Consistent with translational suppression as the proximate mechanism of hypoxia resistance, treatment with cycloheximide also conferred hypoxia resistance (Fig. S2).
A reduction in translation rate might be expected to produce a variety of phenotypes besides hypoxia resistance. In particular, reduced translation has been shown to lengthen lifespan in C. elegans and other organisms (9-11). Additionally, we have previously reported that a known, very long-lived mutant was also highly hypoxia resistant (1). Thus, we hypothesized that rrt-1(gc47) would be long lived. Indeed, rrt-1(gc47) had a small but significantly increased lifespan (Figure S3). While this further links hypoxia resistance and long lifespan, the striking difference in the strength of the two phenotypes suggests that their mechanisms downstream of RRT-1 are distinct or that hypoxic sensitivity is much more responsive than lifespan to alterations in the translation machinery. Other phenotypes seen in gc47 were less remarkable. It had a modest decrease in fecundity, a small but significant level of embryonic lethality, moved about half as fast as wild type animals, and developed to adults about 15% slower than wild type (Table S3). Thus, as expected for mutation of a gene with such an essential function, rrt-1(gc47) has a pleiotropic phenotype but the only strong phenotype was hypoxia resistance.
RRT-1 presumably functions in all cells to mediate translation. However because of their high metabolic activity, germ cells might be particularly vulnerable to hypoxic injury and thereby determine the hypoxic sensitivity of the whole organism. To determine if RRT-1 acts exclusively in the germline to control whole organismal hypoxia sensitivity or if it acts in somatic cells such as neurons and myocytes, we made use of a mutation in the rrf-1 gene. rrf-1 encodes an RNA-directed RNA polymerase required for the somatic but not germline actions of RNAi (12). Thus the hypoxia resistant phenotype of rrt-1(RNAi) in an rrf-1 mutant background would be diminished if RRT-1 acts in somatic cells to control organismal hypoxic sensitivity whereas no effect of the rrf-1 mutant would be seen if RRT-1 acted exclusively in germ cells. Indeed, the hypoxia resistance of rrt-1(RNAi) was greatly reduced in an rrf-1 mutant versus wild type background (Figure S4). We next directly examined neurons and myocytes to determine if rrt-1(gc47) prevents hypoxic injury of these somatic cell types. We have previously observed that a sublethal hypoxic insult produces characteristic pathological changes including axonal beading and myocyte nuclear fragmentation (1, 13). Both axonal beading and myocyte nuclear fragmentation were abated in rrt-1(gc47) (Fig. S5).
When does RRT-1 function to regulate hypoxic sensitivity? In particular, we wanted to know whether rrt-1 acts around the time of the hypoxic insult, either during or after hypoxia. Alternatively, given the fundamental role of RRT-1 in translation, reduction of rrt-1 function might produce developmental defects that indirectly confer hypoxia resistance. In order to answer this question, we applied RNAi to wild type animals early in development, early in adulthood prior to the hypoxic insult, or in adulthood after the hypoxic insult (Fig. 3). RNAi either during early development or during early adulthood protected equally well from subsequent hypoxic death (Fig. 3B). Thus, the hypoxia resistant phenotype of rrt-1 reduction-of-function is not dependent on developmental stage and can be induced after development is complete. Importantly, rrt-1(RNAi) after the hypoxic insult increased survival from delayed hypoxic death (Fig. 3C). This result has significant implications for the mechanism whereby RRT-1 controls hypoxic sensitivity. rrt-1 reduction-of-function almost certainly reduces global translation rate, and as a consequence, should reduce oxygen consumption. Indeed, the rate of paralysis by hypoxia, which should correlate with oxygen consumption, is decreased in rrt-1(gc47) (Figure S6). Reduced oxygen consumption by translational arrest is a logical and established mechanism for reducing cellular injury during hypoxia but not after (14-16). Thus, the mechanism of protection by rrt-1 knockdown, at least that functioning after the hypoxic insult, appears to be more complex than a global reduction in oxygen consumption by translational arrest.
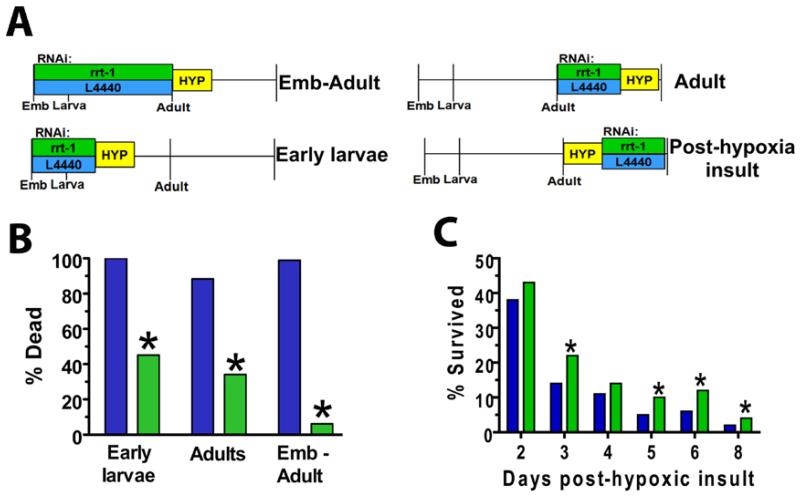
Fig. 3. rrt-1 acutely controls hypoxic sensitivity, during and after the insult.
(A) Schematic of the experimental protocol. Wild-type animals were exposed to rrt-1 or L4440 empty vector RNAi at the times indicated and treated with a 20 hour (B) or 16 hour (C) hypoxic incubation (HYP, yellow boxes). “Emb” represents embryo. (B) Hypoxia resistance by RNAi knockdown of rrt-1 is not developmental stage dependent. n = 40 animals/condition. *p < 0.05 versus L4440 (Fisher’s exact test, two-sided). (C) Inhibition of rrt-1 is effective after hypoxic insult. % Survived = 100(#animals alive at day of interest/# animals alive initially after 24 hr recovery). *p < 0.05 vs L4440 (Fisher’s exact test, two-sided). n > 300 initially alive worms per RNAi over 3 independent trials.
Hypoxia has been shown to produce intracellular misfolded proteins and thereby induce the unfolded protein response (UPR)(17). One effect of UPR induction is phosphorylation of the translation initiation factor eIF2-α, thereby suppressing translation (18). Translational suppression by the UPR has been proposed as an adaptive mechanism to reduce the load of newly synthesized and unfolded proteins, particularly in the context of cancer cell biology. Thus, we considered the hypothesis that the hypoxia resistant phenotype of rrt-1(rf) may be, at least in part, due to a reduction in unfolded proteins. To examine this question, we utilized strains carrying a transgene consisting of a fusion between the hsp-4 promoter and GFP, Phsp-4::GFP. Phsp-4::GFP expression has been shown to be a reliable indicator of the level of unfolded proteins and of activation of the UPR(19-21). Hypoxia induced a significant increase in expression of Phsp-4::GFP that peaked four hours after recovery from hypoxia and was dependent on the length of hypoxic incubation (Fig. 4A,B,C). As would be expected, the glycosylation-inhbitor tunicamycin, which increases the level of unfolded proteins, also induced Phsp-4::GFP expression. Induction of Phsp-4::GFP expression by either hypoxia or tunicamycin was blocked virtually completely by a loss-of-function mutation in ire-1, which encodes an ER transluminal kinase essential for the UPR (21, 22) (Fig. 4A,B). Consistent with a reduction in unfolded protein load, rrt-1(RNAi) completely blocked hypoxic induction of Phsp-4::GFP (Fig. 4A,B); however, it did not diminish induction by tunicamycin. Thus importantly, the level of translational suppression by rrt-1(RNAi) does not preclude synthesis of the GFP marker under strong inducing conditions. Further supporting the hypothesis that rrt-1(rf) reduces the load of unfolded proteins, rrt-1(gc47) was highly resistant to tunicamycin-induced developmental arrest (Fig. 4D). Finally, we found that ire-1(lf) and xbp-1(lf), a downstream target of ire-1(21), significantly suppressed the hypoxia resistance produced by rrt-1(RNAi), but neither ire-1(lf) nor xbp-1(lf) induced hypersensitivity in rrt-1(+) animals (Fig. 4E). Rather, the ire-1(lf) and xbp-1(lf) animals were weakly resistant. These data indicate that inhibition of translation by rrt-1(lf) and the UPR interact synergistically to reduce hypoxic sensitivity, but that in the absence of translational suppression by rrt-1(lf), an intact UPR weakly promotes death after a severe hypoxic insult.
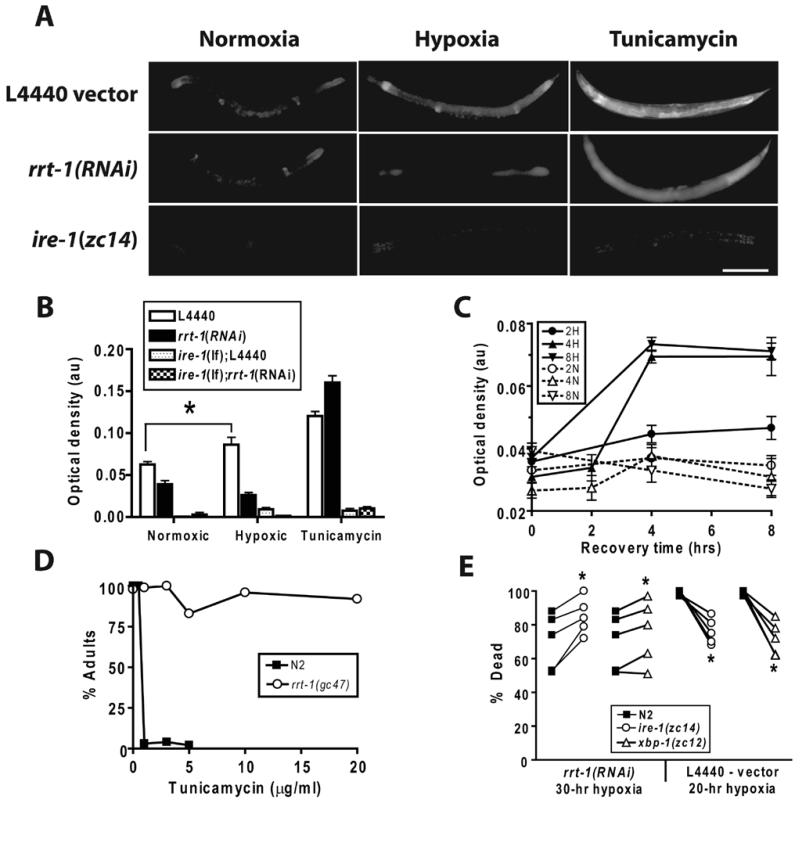
Fig. 4. The unfolded protein response is induced by hypoxia and is required for high level hypoxia resistance of rrt-1(RNAi).
(A) Phsp-4::GFP expression in age matched young adult C. elegans after incubation for six hours in M9 buffer in a normoxic or hypoxic environment or with 25 μg/ml tunicamycin. zcIs4[Phsp-4::GFP] animals were raised on empty vector or rrt-1(RNAi) bacteria; ire-1(zc14);zcIs4 animals were raised on empty vector. scale bar = 200 μm (B) Quantification of Phsp-4::GFP expression under the various conditions and genetic backgrounds. * - p < 0.01, unpaired 2-sided t-test. (C) Time course of Phsp-4::GFP induction after hypoxic or normoxic incubations of 2, 4, or 8 hours. (D) Sensitivity to developmental arrest by tunicamycin in wild type and rrt-1(gc47) animals. Freshly laid eggs were allowed to develop on agar plates containing the indicated concentrations of tunicamycin. The percent of animals reaching adulthood after seven days of development was scored. (E) Hypoxic sensitivity of wild type or UPR pathway mutant animals exposed to rrt-1(RNAi) (30 hour hypoxic incubation) or empty vector control (20 hour hypoxic incubation). * - p < 0.05, paired t-test.
Translational repression is well-established as a mechanism of survival for hibernating animals in a prolonged hypoxic environment (23). Translational mechanisms are important in the tumorigenicity of cancer cells (24) and are increasingly implicated in the sensitivity of normal cell types to hypoxic and ischemic injury (14, 25). In these diverse scenarios, translational repression results in several secondary changes in the biology of the cell, including a decrease in both ATP consumption and protein aggregates, and an alteration of relative protein abundance. Our data show that a modest suppression of translation that still allows relatively normal growth and physiology can produce a profound hypoxia resistance that requires the UPR for its full phenotypic expression. An important point to reiterate in this regard is that one effect of UPR activation is translational suppression by a mechanism distinct from limiting aminoacylated-tRNA levels. A logical model derived from our data is that a reduction in translation rate by a decrement in aaRS activity reduces the unfolded protein load to a level that is manageable by the UPR and may synergize with the translational suppression produced by the UPR itself. However, without translational inhibition, the activity of the UPR is maladaptive in the context of hypoxic injury. Understanding fully this interaction between translational activity and the UPR is critical if these cellular processes are to be exploited to regulate hypoxic cell death.
Supplementary Material
Acknowledgments
We thank Meghann Mabon for early efforts on screen design and Laura Metz for help with gonadal injections and the velocity measurements. We thank Malene Hansen for helpful advice on the translation rate measurements and for unpublished strains. This work was funded by the National Institute of Neurological Disorders and Stroke, a Neuroscience of Brain Disorders Award from the McKnight Endowment Fund for Neuroscience, and an American Heart Association Established Investigator Award. Some of the strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by NIH NCRR.
References and Notes
- 1.Scott BA, Avidan MS, Crowder CM. Science. 2002;296:2388. doi: 10.1126/science.1072302. [DOI] [PubMed] [Google Scholar]
- 2.Ibba M, Soll D. Annu Rev Biochem. 2000;69:617. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 3.Landes C, et al. Biochimie. 1995;77:194. doi: 10.1016/0300-9084(96)88125-9. [DOI] [PubMed] [Google Scholar]
- 4.Delagoutte B, Moras D, Cavarelli J. Embo J. 2000;19:5599. doi: 10.1093/emboj/19.21.5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kyriacou SV, Deutscher MP. Molecular Cell. 2008;29:419. doi: 10.1016/j.molcel.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han JM, Kim JY, Kim S. Biochem Biophys Res Commun. 2003;303:985. doi: 10.1016/s0006-291x(03)00485-6. [DOI] [PubMed] [Google Scholar]
- 7.Sampath P, et al. Cell. 2004;119:195. doi: 10.1016/j.cell.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 8.Park SG, Ewalt KL, Kim S. Trends Biochem Sci. 2005;30:569. doi: 10.1016/j.tibs.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Syntichaki P, Troulinaki K, Tavernarakis N. Nature. 2007;445:922. doi: 10.1038/nature05603. [DOI] [PubMed] [Google Scholar]
- 10.Pan KZ, et al. Aging Cell. 2007;6:111. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen M, et al. Aging Cell. 2007;6:95. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 12.Sijen T, et al. Cell. 2001;107:465. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 13.Dasgupta N, Patel AM, Scott BA, Crowder CM. Curr Biol. 2007;17:1954. doi: 10.1016/j.cub.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paschen W, Proud CG, Mies G. Curr Pharm Des. 2007;13:1887. doi: 10.2174/138161207780858401. [DOI] [PubMed] [Google Scholar]
- 15.Wouters BG, et al. Semin Cell Dev Biol. 2005;16:487. doi: 10.1016/j.semcdb.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 16.DeGracia DJ, Kumar R, Owen CR, Krause GS, White BC. J Cereb Blood Flow Metab. 2002;22:127. doi: 10.1097/00004647-200202000-00001. [DOI] [PubMed] [Google Scholar]
- 17.Koumenis C, et al. Methods in Enzymology. Vol. 435. Academic Press; 2007. pp. 275–293. [DOI] [PubMed] [Google Scholar]
- 18.Koumenis C, et al. Mol Cell Biol. 2002;22:7405. doi: 10.1128/MCB.22.21.7405-7416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoneda T, et al. J Cell Sci. 2004;117:4055. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- 20.Urano F, et al. J. Cell Biol. 2002;158:639. doi: 10.1083/jcb.200203086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calfon M, et al. Nature. 2002;415:92. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 22.Shen X, et al. Cell. 2001;107:893. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- 23.Storey KB, Storey JM. Biol Rev Camb Philos Soc. 2004;79:207. doi: 10.1017/s1464793103006195. [DOI] [PubMed] [Google Scholar]
- 24.van den Beucken T, Koritzinsky M, Wouters BG. Cancer Biol Ther. 2006;5:749. doi: 10.4161/cbt.5.7.2972. [DOI] [PubMed] [Google Scholar]
- 25.DeGracia DJ, Hu BR. J Cereb Blood Flow Metab. 2007;27:875. doi: 10.1038/sj.jcbfm.9600388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.