Abstract
Meningiomas are the most common primary nervous system tumor. The tumor suppressor NF2 is disrupted in approximately half of meningiomas1 but the complete spectrum of genetic changes remains undefined. We performed whole-genome or whole-exome sequencing on 17 meningiomas and focused sequencing on an additional 48 tumors to identify and validate somatic genetic alterations. Most meningiomas exhibited simple genomes, with fewer mutations, rearrangements, and copy-number alterations than reported in other adult tumors. However, several meningiomas harbored more complex patterns of copy-number changes and rearrangements including one tumor with chromothripsis. We confirmed focal NF2 inactivation in 43% of tumors and found alterations in epigenetic modifiers among an additional 8% of tumors. A subset of meningiomas lacking NF2 alterations harbored recurrent oncogenic mutations in AKT1 (E17K) and SMO (W535L) and exhibited immunohistochemical evidence of activation of their pathways. These mutations were present in therapeutically challenging tumors of the skull base and higher grade. These results begin to define the spectrum of genetic alterations in meningiomas and identify potential therapeutic targets.
Meningiomas, tumors that arise from arachnoidal cap cells of the leptomeninges, constitute approximately one-third of primary central nervous system (CNS) tumors1. Most meningiomas (80%) are World Health Organization (WHO) grade I and treated by surgical resection. However, resection of some meningiomas, particularly at the skull base, is associated with high morbidity. Moreover, 18% of these will recur within five years, and patients with grade I tumors have significantly reduced long-term survival related to both tumor recurrence and stroke risk2. Recurrence rates for grade II and III meningiomas can be as high as 40% and 80%, with 5-year overall survival of approximately 76% and 32%, respectively1,3,4. Although there are recent reports of a stepwise progression of a subset of grade I meningiomas to higher grades5,6, these secondary grade II/III meningiomas may differ fundamentally from spontaneously arising grade II/III tumors. Radiation is frequently used as an adjunct to surgery; however, there are no effective chemotherapeutic options when surgery and radiation fail to offer durable long-term disease control7.
We sequenced DNA from a discovery set of grade I meningiomas using whole-genome (n = 11) and whole-exome (n = 6) techniques to identify somatic copy-number alterations (SCNAs), rearrangements, mutations, and insertions/deletions throughout the genome. We then sequenced 645 known cancer-associated genes, including genes altered in this discovery set, in a validation set of 30 additional grade I tumors. We also sequenced these genes in an extrapolation set of 18 grade II-III meningiomas to evaluate whether findings in grade I tumors extended to the tumors of higher grades (Online Methods, Supplementary Tables 1 and 2).
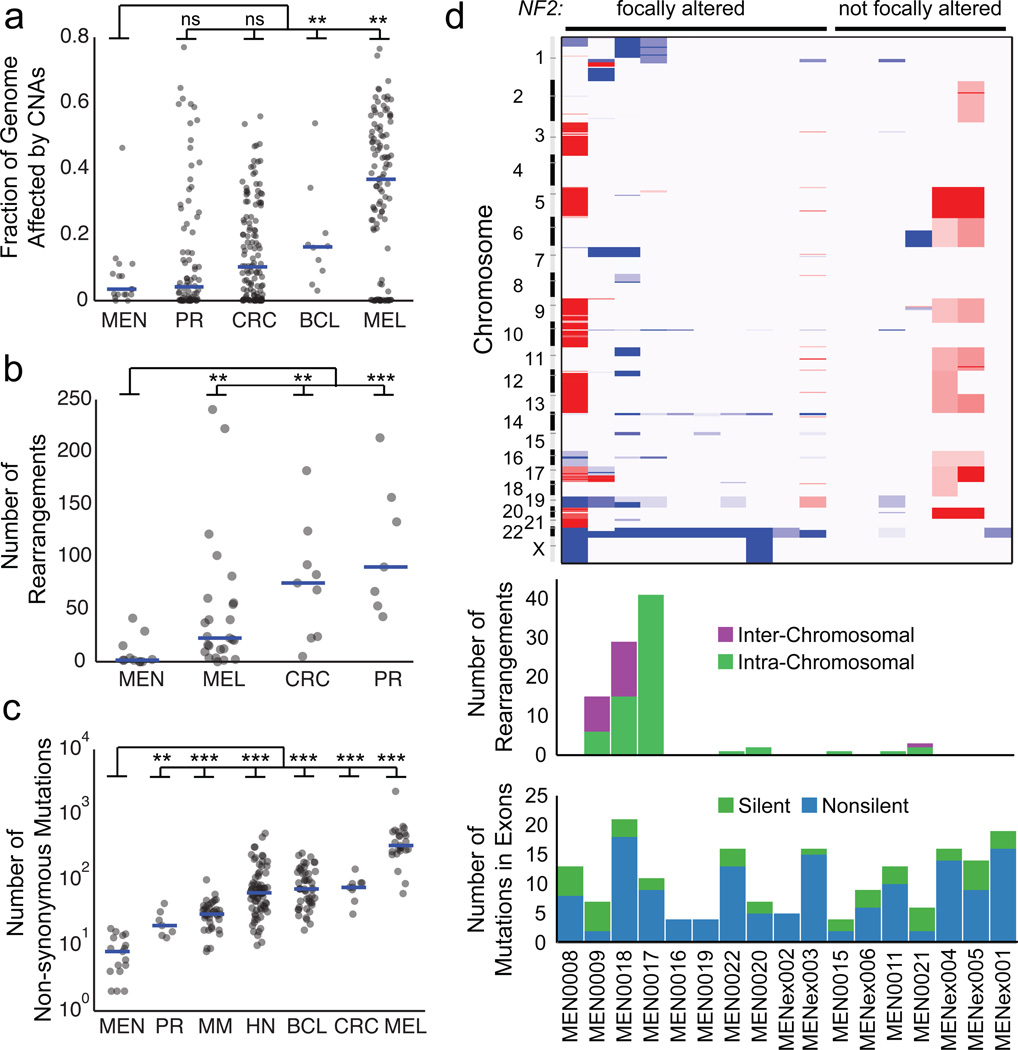
Meningiomas in the discovery set exhibited a small number of somatic genetic events compared to other tumor types8–14. We sequenced to high depth (median 57X genome, 181X exome, and 154X validation/extrapolation coverage) and utilized large-insert libraries in whole-genome samples (500 bp and 800 bp) to optimize our detection of rearrangements and other events. Nevertheless, we found that the median meningioma exhibited SCNAs affecting only 3.3% of the genome, one rearrangement, and 8 non-synonymous mutations. Among mutations, cytosine to thymidine transitions within a CpG context predominated, consistent with spontaneous deamination (Supplementary Fig. 1a)15. The rates of alteration we observed in meningiomas were significantly lower (often by a factor of 10 or more) than previously determined rates for other tumors that have been sequenced to lower depth and with smaller insert sizes (Fig. 1a–c). These findings may reflect differences in mitotic index, prior treatments, or carcinogen exposure between these tumor types.
Figure 1.
The landscape of somatic alterations in the grade I meningioma genome. (a) Fraction of the genome altered by somatic copy-number alterations (SCNAs), (b) number of somatic intra- and inter-chromosomal rearrangements, and (c) number of non-synonymous mutations in meningiomas and other tumor types. MEN: meningioma; PR: prostate; CRC: colorectal carcinoma; BCL: diffuse large B-cell lymphoma; MEL: melanoma; MM: multiple myeloma; HN: head and neck cancers. (d) Somatic genetic alterations in individual grade I meningiomas of the discovery cohort. Top: Copy-number profiles (red = gain, blue = loss). Middle: Number of somatic rearrangements per tumor. Samples labeled as MENex are exome-sequenced, in which no rearrangements were detected. Bottom: Total non-synonymous and synonymous mutations in exons. ** p<0.01, *** p<0.0001.
In contrast, rates of genomic disruption were higher among the grade II-III meningiomas in the extrapolation set than their grade I counterparts (Supplementary Fig. 2). The mean rate of non-synonymous mutations within the 645 sequenced genes was nearly twice as high in higher-grade tumors (3.0 vs. 1.6; p<0.02), and the proportion of the genome affected by SCNAs was dramatically higher (median 12.8 vs. 0.3%; p<0.001).
Among SCNAs, loss of chromosome 22 (containing NF2) was the most frequent genetic alteration in the discovery cohort, occurring in all 10 tumors with focal NF2 alteration (defined as mutation, insertion/deletion, or rearrangement within the NF2 locus) and one without (Fig. 1d, Supplementary Fig. 3). The association between NF2 mutation and chr22 loss extended to the validation and extrapolation sets (p<0.0001) and confirms prior reports using other methods16. In the grade I tumors, we also found recurrent significant losses on 1p, 7p, 14p, and 19 and gains on chr5 and chr20 (q<0.1), some of which have been described previously17. Higher-grade tumors of the extrapolation set exhibited additional recurrent losses on 10q and 14q, previously reported in atypical and anaplastic meningiomas1. We did not observe previously described losses of DAL-1, a gene related to NF2 on 18p18.
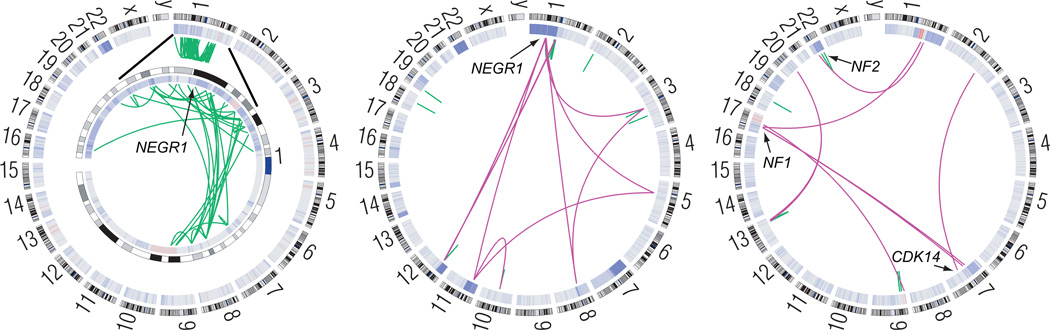
We identified a total of 110 candidate somatic rearrangements in whole-genome samples, of which 93 were confirmed to have reads spanning the putative fusion site (Supplementary Table 3, Supplementary Fig. 4). Most of these were concentrated in three tumors (Fig. 2), one of which exhibited chromothripsis, a simultaneous rearrangement of one or more chromosomes estimated to occur in 2–3% of aggressive malignancies19,20. One of the genes disrupted by chromothripsis was the putative tumor suppressor NEGR1, which was also rearranged in a second tumor. NEGR1 plays a role in CNS development and is recurrently deleted in neuroblastoma21, though its role in tumorigenesis remains to be elucidated. A third sample harbored disruptions in the known and putative tumor suppressors NF1, NF2, and CDK14. A copy-neutral 12.5Mb inversion caused reciprocal fusion of NF2 (before exon 2) and TCF20, representing a novel mechanism of inactivating NF2 in meningiomas. We also identified other translocations in samples that were not subjected to whole-genome analysis, including a truncating translocation in CHEK2 (Supplementary Fig. 4i and Supplementary Table 3). The loss of CHEK2 may lead to impaired DNA repair, as has been described in meningioma cell lines22.
Figure 2.
Somatic rearrangements disrupt tumor suppressors in several meningiomas. Circos plots show the SCNAs (inner ring heat map) and intra- and inter-chromosomal rearrangements (green and purple arcs, respectively) in three whole-genome sequenced meningiomas. Left: Chromothripsis of chromosome 1 (enlarged in inset) in an NF2-mutated sample (MEN0017) disrupts the putative tumor suppressor NEGR1. Middle: A second NF2-mutated sample (MEN0018) harbors 29 rearrangements including disruption of NEGR1. Right: An inversion on chromosome 22 disrupts NF2, and other rearrangements affect NF1 and CDK14 (MEN0009). Data from the remaining tumors are shown in Supplementary Figure 4.
The most frequently mutated gene was NF2, which exhibited a total of nine nonsense mutations, nine splice site mutations, nine frame-shift insertions-deletions, and the above-mentioned translocation (Supplementary Fig. 5). The majority of these events overlap with previously reported events, and our rates of NF2 inactivation are consistent with published rates1. The non-synonymous mutation rates of tumors with and without focal NF2 alterations did not differ significantly, nor did the spectra of their mutation subtypes (Supplementary Fig. 1b–d).
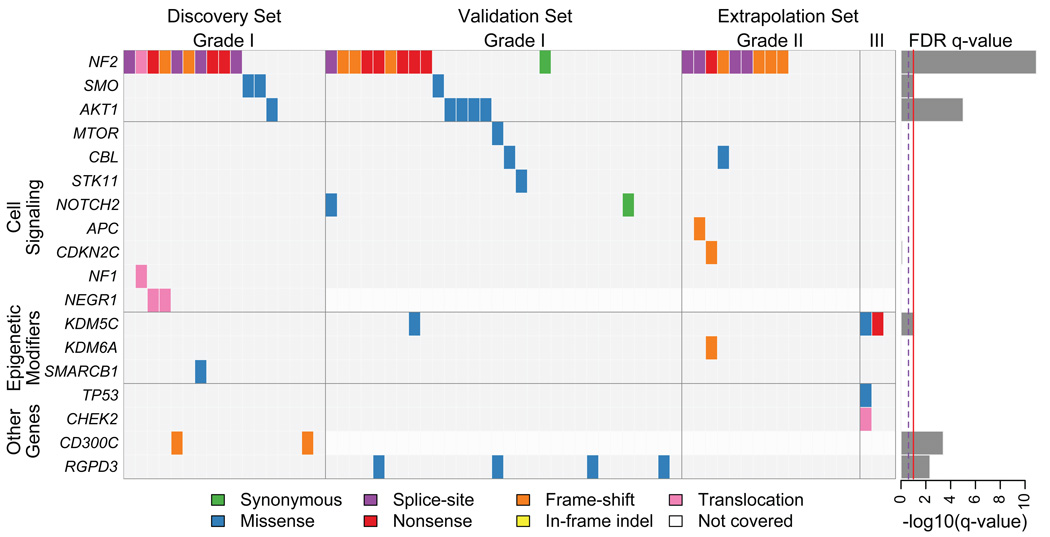
We also identified non-synonymous mutations in 190 other genes (Supplementary Tables 4 and 5). We determined the significance of mutation rates in individual genes relative to genome-wide background rates. Six genes reached statistical significance (Fig. 3, Supplementary Fig. 5, Supplementary Table 4). Four of these (NF2, KDM5C, SMO, and AKT1) have previously been implicated in cancer, and the other two (RGPD3 and CD300C) have not. Isolated or rare mutations that did not reach significance across the entire cohort were observed in 23 genes with known involvement in cancer, including TP53, APC, CBL, STK11 and NOTCH2.
Figure 3.
Significant and selected cancer-related somatic mutations, insertion-deletions, and translocations in meningiomas. Mutation subtypes are denoted by the indicated colors. If multiple mutations were found in a gene in a single sample, only one is shown. Discovery set tumors are in the same order as in Figure 1d. Right: Significance of mutations in each gene, as false discovery rate q-values. The full list of mutated genes is shown in Supplementary Table 4.
Among genes mutated in our cohort and previously associated with cancer, three (KDM5C, KDM6A, and SMARCB1) are epigenetic modifiers. Mutations involving these genes affected 8% of the cohort. Both KDM5C (also known as JARID1C) and KDM6A are histone demethylases23; SMARCB1 (also named SNF5 and INI1) is a member of the SWI/SNF chromatin-remodeling complex. SMARCB1 is located 6 Mb from NF2, and a “four hit” model of biallelic inactivation of both genes has been described in familial schwannomas24. The mutation we identified (R374Q) is near a mutational hotspot (R377H) described in meningiomas25 and germline mutations in SMARCB1, including R374Q, have been reported in the congenital disorder, Coffin-Siris syndrome26.
We observed mutations of SMO, a member of the Hedgehog (Hh) signaling pathway, in three tumors (5%). None of these tumors exhibited focal alterations of NF2. Two samples harbored SMO W535L, a known oncogenic mutation in basal cell carcinomas27, and a third harbored an L412F mutation, previously reported in desmoplastic medulloblastoma28. The allelic fractions of these mutations (45%, 55% and 47%) were among the highest in these tumors, suggesting clonality. Moreover, dysregulation of the Hh pathway has been previously described in meningiomas29,30. Gorlin syndrome (nevoid basal-cell carcinoma syndrome) is caused by a germline PTCH1 mutation (upstream of SMO), and is characterized by predisposition to multiple cancers, including desmoplastic medulloblastomas and meningiomas31,32. Although we did not observe somatic mutations in PTCH1, two samples harbored germline alterations that were also present in the tumor (D436N, E44G/P1282L). However, these patients did not exhibit manifestations of Gorlin syndrome, so the functional significance of these specific events is unclear.
Six samples exhibited mutations of the PI3K/AKT/mTOR pathway. None of these had mutations of NF2 or SMO (p=0.03). Five samples harbored identical AKT1 mutations (E17K) known to be oncogenic in breast, colorectal and lung cancers33. The E to K substitution results in constitutive AKT1 activation, which stimulates downstream mTOR signaling. The sixth sample exhibited a novel MTOR mutation (D1279V).
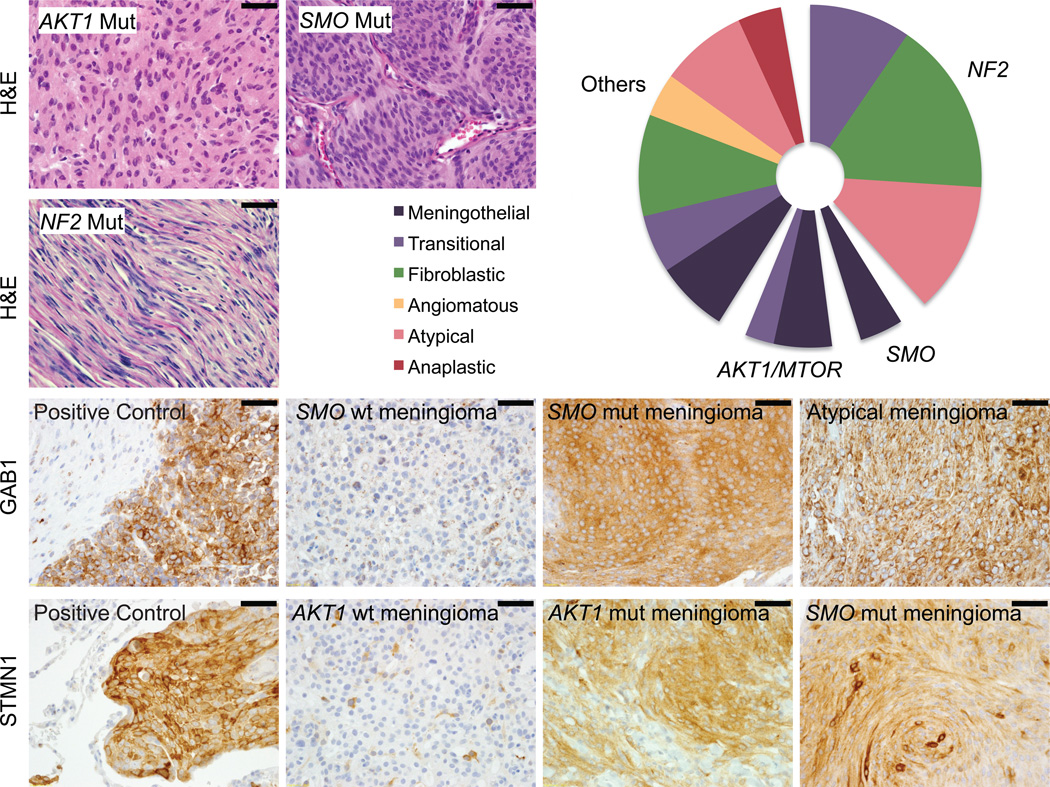
The Hh and PI3K pathway mutations were also associated with particular histopathologic subtypes (Fig. 4). SMO- and AKT1/MTOR- mutated meningiomas were predominantly of the meningothelial subtype (p=0.009, p=0.005, Fisher’s exact test). This is in contrast to the grade I NF2-altered tumors, which were predominantly fibroblastic and/or transitional (p=0.013), consistent with prior reports34. We did not find statistically significant correlations with other clinically relevant variables including age, location or prior resection. However, one AKT1- and two SMO-mutated tumors were resected from the skull base, a region that offers particular challenges to resection and higher rate of recurrence35 (Supplementary Table 5).
Figure 4.
Associations between mutations in Hh and Akt/mTOR pathways and histologic findings. (a) AKT/MTOR and SMO-mutated samples are predominantly of the meningothelial subtype (p=0.009, p=0.005). NF2-mutated samples are predominantly fibroblastic and transitional (p=0.013). (b) Immunohistochemistry indicates activation of Hh and Akt/mTOR pathways in tumors harboring SMO and AKT1 mutations, respectively (p=0.0008, p=3×10−6). GAB1 staining was used as a marker of Hh pathway activation, and STMN1 staining was used as a marker of PI3K/Akt/mTOR pathway activation. Scale bars denote 50 micrometers.
To validate these findings, we also genotyped an additional 46 grade I and 49 grade II/III meningiomas for AKT1 (E17K) and SMO (W535L and L412F) mutations using a mass spectrometric based method (Sequenom hME). We found one SMO (L412F) mutation and three additional AKT1 (E17K) mutations (Supplementary Table 6). One of these was in a grade III tumor, and the remaining three were in skull base tumors, of which two were of meningothelial histology.
Mutations in SMO, AKT1, and MTOR also correlated with markers of pathway activation. GAB1 immunoreactivity is used in medulloblastomas to characterize tumors with Hh pathway activation36. Among the 65 meningiomas, seven exhibited strong immunoreactivity for GAB1, including the three tumors harboring SMO mutations (p=0.0008). The other four GAB1-positive tumors were grade II meningiomas. Likewise, STMN1 expression is a marker of PI3K/Akt pathway activation37. Ten meningiomas exhibited strong STMN1 expression, including all six AKT1 and MTOR-mutated meningiomas (p=3×10−6). Notably, all three SMO-mutated meningiomas also exhibited strong (n=2) or moderate (1) STMN1 staining. This is consistent with studies that have suggested that Hh and PI3K/AKT/mTOR pathways may interact38.
We observed recurrent mutations in signaling pathways and epigenetic modifiers, but our data also highlight the heterogeneity of mutations in these tumors. Among the 17 tumors we comprehensively characterized, three (18%) did not exhibit mutations in any of the significantly mutated genes. Genomic characterization of additional tumors, particularly of higher grades, is likely to reveal additional oncogenic mechanisms.
These observations also have the potential to guide new therapeutic strategies. We observed SMO and AKT1 mutations in tumors that pose special therapeutic challenges, including a high-grade and six skull-base tumors. Inhibitors of SMO have generated high response rates in patients with basal cell carcinoma, many of which are driven by Hh pathway mutations39. Likewise, inhibitors of the PI3K/AKT/mTOR pathway have shown promise in preclinical and clinical trials in multiple cancer types40. The paucity of additional genetic events in meningiomas harboring mutations of PI3K and Hh signaling pathways suggests that patients with these meningiomas may benefit from such targeted therapies already in development or use.
ONLINE METHODS
Sample Selection and Preparation
This study was reviewed and approved by the human subjects institutional review boards of the Dana-Farber Cancer Institute, Brigham and Women’s Hospital, and the Broad Institute of Harvard and MIT. Written informed consent was obtained from all participants. We chose to initially perform whole-exome sequencing in 6 meningiomas as a pilot study to determine the optimal depth of whole-genome sequencing needed to provide adequate tumor coverage in spite of stromal contamination. These six tumors and 11 additional whole-genome sequenced meningiomas comprised the discovery cohort. To validate clinically actionable mutations identified in the discovery set, we performed focused sequencing on a validation cohort consisting of an additional 30 grade I meningiomas. We also assembled an extrapolation cohort of 15 grade II and 3 grade III meningiomas (Supplementary Table 1) in order to compare genetic aberrations of low- and high-grade tumors. Our approach of deep, broad sequencing of a small representative cohort followed by validation in an independent larger group is similar to the approach used by others to discover IDH141 and PIK3CA42 mutations. Three study pathologists (SS/DL/ASR) reviewed the histopathologic diagnosis, grade and purity of each tumor.
Representative fresh-frozen blocks with estimated purity of ≥90% were selected. DNA was extracted from tissue shavings and buffy coat preparations of paired blood using standard techniques (QIAGEN, Valencia CA) then quantified using PicoGreen® dye (InVitrogen, Carlsbad CA). Identities of tumor-normal pairs were confirmed by mass spectrometric genotyping using a well-established 48-SNP panel (Sequenom, San Diego CA)43.
Data, including sequence data and analyses, are available for download via dbGaP (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000552.v1.p1).
Sequencing
For whole-genome sequencing (n=11), DNA was randomly fragmented and libraries of two insert sizes were prepared (500 and 800bp) for paired-end sequencing on Illumina HiSeq 200010,11. Target depth (60X-tumor/30X-normal) was achieved in all but one sample (MEN0015) that failed 800bp library preparation; however, adequate coverage of the 500bp library was achieved.
For whole-exome sequencing (n=6), 250bp libraries were prepared by Covaris sonication (Covaris, Woburn MA), followed by double size-selection (Agencourt AMPure XP beads) and ligation to specific barcoded adaptors (Illumina TruSeq) for multiplexed analysis. Exome hybrid capture was performed with NimbleGen SeqCap EZ Exome Library SR v2.2, Agilent Sure Select All Exon v2.0 hybrid capture kit, or both approaches. All samples but one achieved >150X coverage in exon regions (MENex004, 79X-tumor/90X-normal).
We proceeded with sequencing 645 cancer-associated genes in our larger validation and extrapolation sample set (n=48) because this pre-selected gene set (OncoPanel) included all genes of interest that were also clinically actionable (including AKT and SMO) that arose in the discovery set. In addition, a wide breadth of other genes in the same families (such as MTOR in the AKT pathway) and other interesting cancer-related genes in other families were included (Supplementary Table 2). Briefly, DNA was sonicated to achieve an average fragment size of 150bp, then size-selected and barcoded as above. Each multiplexed pool was hybridized with biotinylated baits (Agilent SureSelect, Agilent) designed to capture exonic sequences. Streptavidin-coated beads were used to pull down the complexes, which were loaded on a HiSeq2000 for paired-end sequencing.
Sequencing Analysis Pipeline
For exome- and OncoPanel-sequenced data, pooled sample reads were de-multiplexed using Picard tools (http://picard.sourceforge.net). Read pairs were aligned to the hg19 reference sequence (b37) (ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/technical/reference/) using the Burrows-Wheeler Aligner44 (http://bio-bwa.sourceforge.net/bwa.shtml) with options: -q 5 -l 32 -k 2 -o 1. Data were sorted and duplicate-marked using Samtools (http://samtools.sourceforge.net) and Picard. Bias in base quality score assignments due to flowcell, lane, dinucleotide context, and machine cycle were analyzed and recalibrated, and local realignment around insertions/deletions was achieved using the Genome Analysis Toolkit (GATK) (http://www.broadinstitute.org/gatk/)45,46.
Variant and germline calling were performed within the Firehose environment11 using GenePattern47. All algorithms described are available publicly (https://confluence.broadinstitute.org/display/CGATools). All sample pairs passed a QC pipeline to test for any tumor/normal and inter-individual mix-ups by comparing insert-size distribution and copy-number profile as described previously10. The Mann-Whitney test was used to determine significance of differences between rates of somatic genetic alteration between meningiomas and the other datasets.
Somatic copy-number alterations (SCNAs) and rearrangements
SCNAs were evaluated using SegSeq48, which uses a combination of local change-point analysis and subsequent merging of adjacent chromosomal segments with similar copy-number. For arm-level significance analysis, pseudo-markers were added every 10,000 bases to segmented copy-number data, and these were analyzed by GISTIC 2.0 to determine which events occurred more often than expected by chance14.
Rearrangements and their exact breakpoints were identified using the combination of dRanger and BreakPointer algorithms8,10. Briefly, dRanger identifies potential rearrangements from read pairs mapping to different chromosomes or unexpected positions / orientations on the same chromosome, and BreakPointer locates reads spanning the fusion and maps the exact breakpoint using modified Smith-Waterman alignment10. Rearrangements are assigned a score reflecting the number of tumor reads supporting a breakpoint, the fraction of nearby reads with MAPQ0, the prevalence of other nearby discordant pairs, and the standard deviation of breakpoint starting positions. To minimize false-positive rearrangements, only breakpoints with score ≥8 passing BreakPointer were accepted. The specificity of this type of approach has previously been shown to be >90%10. Copy-number and rearrangement results were displayed using the Circos program (http://mkweb.bcgsc.ca/circos)49.
Identification of base pair substitutions and short insertion-deletions
Mutations were only considered at covered positions (≥8X-normal and 14X-tumor sites). Somatic mutations and short insertions/deletions were called and post-filtered using MuTect and IndelLocator10,11. These were annotated to genes and compared to events in the Catalogue of Somatic Mutations in Cancer (COSMIC) using Oncotator, and spurious calls caused by mis-mapping and other previously identified systematic errors were removed using an established list of known problematic sites11–13. MutSig was used to determine the significance of mutated genes, adjusting for cohort- and genome-wide mutation rate, local background mutation rate, and gene length11–13. We used a cohort-wide background mutation rate instead of the more commonly used sample-specific mutation rate because the observed low mutation rate in these tumors leads to poor sampling of individual mutation subcategories. This may lead to overly large apparent variations between background rates of mutation subtypes in different samples. Mutations are shown in the context of UniProt regions and domains (www.uniprot.org). Post-hoc analyses for significant non-overlap of NF2, SMO, and AKT1 mutations were determined by Fisher’s exact test, as were associations between these mutations and chr22 loss or specific categorical demographic characteristics. The Mann-Whitney test was used for comparisons of mutations and continuous demographic variables.
Germline variants were called using UnifiedGenotyper45,46, filtered against the 1000genomes dataset50, and annotated to genes as above. Genes with known syndromic associations to meningiomas (NF1, NF2, SMARCB1 and PTCH1) were manually reviewed for germline non-synonymous mutations. These were visualized in Integrative Genomics Viewer (www.broadinstitute.org/igv/).
Sequenom Validation
We obtained IRB approval for collection of independent archival paraffin-embedded meningioma samples from the Brigham and Women’s Hospital Pathology Department. We identified 95 samples from patients that did not overlap with our cohort of 65 tumors, resected between 2005 and 2012. Forty-nine of these were higher-grade meningiomas. Tissue was de-paraffinized and DNA extracted using standard techniques (QIAGEN, Valencia CA). Variants in SMO (W535L and L412F) and AKT1 (E17K) were assayed using Sequenom’s homogeneous Mass-Extend (hME) Genotyping system (Sequenom, Inc. San Diego, CA). Bidirectional probes and primers were designed using Sequenom MassARRAY Typer software (v 4.0), and hME genotyping was performed using 5ng of unamplified genomic DNA template per assay pool (maximum of 4 assays per pool).
Immunohistochemistry
Tissue sections were deparaffinized and rehydrated using Xylene and graded alcohols. Endogenous peroxidase activity was blocked with 3% H2O2 in 100% ethanol (1:1; 15 minutes). Antigen retrieval was performed with Dako Target Retrieval Solution, pH 6.0 and pressure cooker treatment (120+/−2°C, 15+/−5 psi) for 45 seconds. Sections were incubated with primary antibodies against GAB1 (Abcam ab27439, 1:100×45min), Ki67 (Vector Lab VP-451, 1:2500×45min), or Stathmin (Cell Signaling Technologies #3352, 1:120, 4°C×16hr), followed by secondary antibody (Dako Labeled Polymer-HRP anti-rabbit IgG x30min). Visualization was with 3,3'-diaminobenzidine as a chromogen (Envision+System) and Gill hematoxylin counterstaining. Associations between specific mutations and their differences in histopathological subtypes and immunohistochemical staining were determined post-hoc with Fisher’s exact test.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Mike Lawrence, Nicolas Stransky, Marchin Imielinski, Kristian Cibulskis, Scott Carter, Chip Stewart, Cheng-Zhong Zhang, Yotam Drier, Steven Schumacher and Barbara Tabak for their assistance with genomic analyses; Patrick Wen, Ossama Al-Mefty, Tracy Batchelor, Benjamin Reisler, Mark Johnson, William Richards, and James Kim for assisting with tissue acquisition; and Anna Pisarek-Horowitz, Sophie Adele Horowitz, Guillaume Bergthold and Charilaos H. Brastianos for critical review of the manuscript. This work was supported by the Brain Science Foundation (I.F.D., P.K.B., R.B., S.S.), Pediatric Low Grade Astrocytoma Foundation (R.B., W.C.H.), American Brain Tumor Association (P.K.B.), Conquer Cancer Foundation Young Investigator Award (P.K.B.), NIH grants K08 CA122833 (R.B.), K08 NS064168 (S.S.), and K12 CA090354-11 (P.K.B.), and Department of Defense grant #W81XWH-12-1-0136 (P.H.).
Footnotes
ACCESSION CODES
Data, including sequence data and analyses, are available for download via dbGaP (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000552.v1.p1).
AUTHOR CONTRIBUTIONS
P.K.B, P.M.H., I.F.D., R.B. and W.C.H. conceived the study, designed the experiments, analyzed the data and wrote the paper. P.M.H performed the bio-informatic analyses, and A.M., G.G., R.B. and M.D.D. provided analytical advice. S.S., A.O.S.R., and D.N.L. reviewed the histopathology and S.S. performed immunohistochemistry. K.L.L. managed the tissue repository. R.T.J., E.P., P.V.H., A.R., A.S. and L.E.M. provided technical support and performed sequencing. W.C.H., I.F.D., and R.B. supervised the study. All authors discussed the results and implications and commented on the manuscript.
COMPETING FINANCIAL INTERESTS
WCH is a consultant for Blueprint Medicines and Thermo Fisher. RB is a shareholder for AstraZeneca. WCH and RB are consultants for and receive research support from Novartis.
REFERENCES
- 1.Choy W, et al. The molecular genetics and tumor pathogenesis of meningiomas and the future directions of meningioma treatments. Neurosurg Focus. 2011;30:E6. doi: 10.3171/2011.2.FOCUS1116. [DOI] [PubMed] [Google Scholar]
- 2.van Alkemade H, et al. Impaired survival and long-term neurological problems in benign meningioma. Neuro Oncol. 2012;14:658–666. doi: 10.1093/neuonc/nos013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durand A, et al. WHO grade II and III meningiomas: a study of prognostic factors. J Neurooncol. 2009;95:367–375. doi: 10.1007/s11060-009-9934-0. [DOI] [PubMed] [Google Scholar]
- 4.Perry A, Scheithauer BW, Stafford SL, Lohse CM, Wollan PC. "Malignancy" in meningiomas: a clinicopathologic study of 116 patients, with grading implications. Cancer. 1999;85:2046–2056. doi: 10.1002/(sici)1097-0142(19990501)85:9<2046::aid-cncr23>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 5.Goutagny S, et al. Genomic profiling reveals alternative genetic pathways of meningioma malignant progression dependent on the underlying NF2 status. Clin Cancer Res. 2010;16:4155–4164. doi: 10.1158/1078-0432.CCR-10-0891. [DOI] [PubMed] [Google Scholar]
- 6.Krayenbuhl N, Pravdenkova S, Al-Mefty O. De novo versus transformed atypical and anaplastic meningiomas: comparisons of clinical course, cytogenetics, cytokinetics, and outcome. Neurosurgery. 2007;61:495–503. doi: 10.1227/01.NEU.0000290895.92695.22. discussion 503–4. [DOI] [PubMed] [Google Scholar]
- 7.Wen PY, Quant E, Drappatz J, Beroukhim R, Norden AD. Medical therapies for meningiomas. J Neurooncol. 2010;99:365–378. doi: 10.1007/s11060-010-0349-8. [DOI] [PubMed] [Google Scholar]
- 8.Bass AJ, et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet. 2011;43:964–968. doi: 10.1038/ng.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berger MF, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lohr JG, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mawrin C, Perry A. Pathological classification and molecular genetics of meningiomas. J Neurooncol. 2010;99:379–391. doi: 10.1007/s11060-010-0342-2. [DOI] [PubMed] [Google Scholar]
- 17.Ketter R, et al. Hyperdiploidy defines a distinct cytogenetic entity of meningiomas. J Neurooncol. 2007;83:213–221. doi: 10.1007/s11060-006-9318-7. [DOI] [PubMed] [Google Scholar]
- 18.Nunes F, et al. Inactivation patterns of NF2 and DAL-1/4.1B (EPB41L3) in sporadic meningioma. Cancer Genet Cytogenet. 2005;162:135–139. doi: 10.1016/j.cancergencyto.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Rausch T, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takita J, et al. Aberrations of NEGR1 on 1p31 and MYEOV on 11q13 in neuroblastoma. Cancer Sci. 2011;102:1645–1650. doi: 10.1111/j.1349-7006.2011.01995.x. [DOI] [PubMed] [Google Scholar]
- 22.Yang HW, et al. Alternative splicing of CHEK2 and codeletion with NF2 promote chromosomal instability in meningioma. Neoplasia. 2012;14:20–28. doi: 10.1593/neo.111574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nature reviews. Molecular cell biology. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 24.Hadfield KD, et al. Molecular characterisation of SMARCB1 and NF2 in familial and sporadic schwannomatosis. J Med Genet. 2008;45:332–339. doi: 10.1136/jmg.2007.056499. [DOI] [PubMed] [Google Scholar]
- 25.Schmitz U, et al. INI1 mutations in meningiomas at a potential hotspot in exon 9. Br J Cancer. 2001;84:199–201. doi: 10.1054/bjoc.2000.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsurusaki Y, et al. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet. 2012;44:376–378. doi: 10.1038/ng.2219. [DOI] [PubMed] [Google Scholar]
- 27.Reifenberger J, et al. Missense mutations in SMOH in sporadic basal cell carcinomas of the skin and primitive neuroectodermal tumors of the central nervous system. Cancer Res. 1998;58:1798–1803. [PubMed] [Google Scholar]
- 28.Jones DT, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laurendeau I, et al. Gene expression profiling of the hedgehog signaling pathway in human meningiomas. Mol Med. 2010;16:262–270. doi: 10.2119/molmed.2010.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aavikko M, et al. Loss of SUFU function in familial multiple meningioma. Am J Hum Genet. 2012;91:520–526. doi: 10.1016/j.ajhg.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorlin RJ. Nevoid basal-cell carcinoma syndrome. Medicine (Baltimore) 1987;66:98–113. doi: 10.1097/00005792-198703000-00002. [DOI] [PubMed] [Google Scholar]
- 32.Santos DC, et al. PTCH1 gene mutations in exon 17 and loss of heterozygosity on D9S180 microsatellite in sporadic and inherited human basal cell carcinomas. Int J Dermatol. 2011;50:838–843. doi: 10.1111/j.1365-4632.2010.04866.x. [DOI] [PubMed] [Google Scholar]
- 33.Bleeker FE, et al. AKT1(E17K) in human solid tumours. Oncogene. 2008;27:5648–5650. doi: 10.1038/onc.2008.170. [DOI] [PubMed] [Google Scholar]
- 34.Wellenreuther R, et al. Analysis of the neurofibromatosis 2 gene reveals molecular variants of meningioma. Am J Pathol. 1995;146:827–832. [PMC free article] [PubMed] [Google Scholar]
- 35.Mathiesen T, Lindquist C, Kihlstrom L, Karlsson B. Recurrence of cranial base meningiomas. Neurosurgery. 1996;39:2–7. doi: 10.1097/00006123-199607000-00002. discussion 8–9. [DOI] [PubMed] [Google Scholar]
- 36.Ellison DW, et al. Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol. 2011;121:381–396. doi: 10.1007/s00401-011-0800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karst AM, et al. Stathmin 1, a marker of PI3K pathway activation and regulator of microtubule dynamics, is expressed in early pelvic serous carcinomas. Gynecol Oncol. 2011;123:5–12. doi: 10.1016/j.ygyno.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riobo NA, Lu K, Ai X, Haines GM, Emerson CP., Jr Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. Proc Natl Acad Sci U S A. 2006;103:4505–4510. doi: 10.1073/pnas.0504337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Von Hoff DD, et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N Engl J Med. 2009;361:1164–1172. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- 40.Janku F, et al. PI3K/AKT/mTOR inhibitors in patients with breast and gynecologic malignancies harboring PIK3CA mutations. J Clin Oncol. 2012;30:777–782. doi: 10.1200/JCO.2011.36.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samuels Y, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 43.Demichelis F, et al. SNP panel identification assay (SPIA): a genetic-based assay for the identification of cell lines. Nucleic Acids Res. 2008;36:2446–2456. doi: 10.1093/nar/gkn089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reich M, et al. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 48.Chiang DY, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.