Abstract
To better understand the limits of antigenic reactivity and epitope accessibility of the V3 domain of primary HIV-1 isolates, we evaluated three human anti-V3 monoclonal antibodies (mAbs) and selected guinea pig vaccine sera for neutralization against reference panels of subtype B and C pseudoviruses derived from early stage infections. The mAbs and vaccine sera potently neutralized several prototype viruses, but displayed substantially less neutralization of most reference strains. In the presence of soluble CD4 (sCD4), the breadth of V3-mediated neutralization was increased; up to 80% and 77% of the subtype B and C viruses respectively were sensitive to V3-mediated neutralization. Unlike sCD4, the reaction of CD4-binding site mAbs b12 and F105 with native virus did not lead to full exposure of the V3 domain. These findings confirm that V3 antibodies recognize most primary viral strains, but that the epitope often has limited accessibility in the context of native envelope spike.
Introduction
The trimeric HIV-1 Env spike is composed of the surface unit gp120 and transmembrane gp41 glycoproteins. Viral entry into susceptible cells is mediated by the interaction of gp120 with the cell surface receptor CD4, leading to conformational changes that form and expose the co-receptor binding region of gp120 (Kwong et al., 1998; Salzwedel et al., 2000; Wyatt and Sodroski, 1998; Xiang et al., 2002). The V3 region of gp120 is critical for co-receptor recognition and determines which co-receptor, CCR5 or CXCR4, is used for viral entry (Cormier and Dragic, 2002; Huang et al., 2005; Suphaphiphat et al., 2003). Hence, while relatively variable in linear sequence, the V3 region has some level of functional and structural conservation (Cardozo et al., 2007; Haynes and Montefiori, 2006; Huang et al., 2005; Rosen et al., 2005; Sharon et al., 2003).
During HIV-1 infection, antibodies to the V3 loop are common (Broliden et al., 1992; Gorny et al., 2006; Haynes and Montefiori, 2006; Krachmarov et al., 2001; Kraft et al., 2007; Pantophlet and Burton, 2006; Profy et al., 1990; Schreiber et al., 1994; Spenlehauer et al., 1998; Wu et al., 1995; Zolla-Pazner, 2004). However, the V3 region appears to play a limited role in the neutralization of most primary virus isolates (Binley et al., 2004; Burton et al., 2004; Lusso et al., 2005; Stamatos et al., 1998; Vancott et al., 1995). Early vaccine studies using V3 peptides as immunogens showed a highly type-specific neutralizing antibody (NAb) response to V3 (Javaherian et al., 1990), while more recent studies of V3 mAbs and immune sera suggest that the V3 NAb response can be more broadly reactive (Derby et al., 2007; Haynes et al., 2006; Moore et al., 1995b; Wu et al., 2006; Yang et al., 2004; Zolla-Pazner, 2005). Human anti-V3 mAbs from both subtype B and non-subtype B infected individuals can neutralize a subset of subtype B and non-subtype B primary virus strains. Interestingly, the breadth and potency of this neutralizations is maximized when the V3 region is built into an unmasked V3 sensitive Env such as on virus SF162 (Binley et al., 2004; Gorny et al., 2006; Krachmarov et al., 2006; Li et al., 2005; Moore et al., 1995a; Pantophlet et al., 2007; Patel, Hoffman, and Swanstrom, 2008; Zolla-Pazner et al., 2008). These data, along with recently described atomic level structures of V3 mAb liganded to cognate peptides, confirm that there are conserved motifs within the V3 region (Cardozo et al., 2007; Huang et al., 2005; Sharon et al., 2003; Stanfield et al., 2004; Stanfield et al., 2006).
These findings are consistent with our understanding that the V3 region is displayed to varying degrees in the context of the quaternary structure of the native viral spike of individual strains of HIV-1. However after binding to the CD4 receptor, conformational changes in Env result in exposure of specific regions previously inaccessible to antibody. CD4 binding significantly enhances gp120 binding by mAb 17b, which recognizes the co-receptor binding site (Decker et al., 2005; Hoffman et al., 1999; Salzwedel et al., 2000; Sullivan et al., 1998a; Sullivan et al., 1998b; Xiang et al., 2002). Similarly, the V3 loop appears to be accessible to antibody when gp120 is in a CD4-bound state (Krachmarov et al., 2006; Lusso et al., 2005; Mbah et al., 2001; Potts et al., 1993; Sullivan et al., 1998b). We therefore postulated that the limited breadth of neutralization by recently isolated broadly reactive V3 mAbs, and by V3-directed vaccine sera, may be due to poor epitope accessibility rather than antigenic diversity of the V3 region (Bou-Habib et al., 1994; Krachmarov et al., 2005; Stamatos et al., 1998; Vancott et al., 1995).
To study the breadth and potency of anti-V3 antibody mediated virus neutralization, we used recently established reference panels of 12 acute subtype B and 12 acute subtype C Env-pseudoviruses (Li et al., 2005; Li et al., 2006). We evaluated three well-characterized anti-V3 mAbs and five guinea pig (GP) vaccine-induced immune sera known to contain high levels of anti-V3 antibodies. The mAbs and immune sera were assayed in the presence of varying concentrations of sCD4 to test if the resulting conformational change in Env would lead to exposure and recognition of the V3 loop. One anti-V3 mAb, 447-52D, is known to react with the GPGR sequence at the tip of the V3 loop which is conserved among most subtype B isolates (Gorny et al., 1992; Zolla-Pazner et al., 2004). The other two mAbs, 2219 and 3074, were derived from subtype B and the circulating recombinant form (CRF) 02_AG infected individuals respectively, and were selected for their broad profile of V3 reactivity (Gorny et al., 2002; Gorny et al., 2006; Krachmarov et al., 2005; Krachmarov et al., 2006). The GP vaccine sera were the result of plasmid DNA prime, recombinant adenoviruses serotype-5 (rAd5) boost immunization with Env constructs containing a deletion of the V1V2 loop and two small deletions on both arms of the V3 stem. This immunization strategy produced high V3-directed NAb responses (Chakrabarti et al., 2002; Chakrabarti et al., 2005; Wu et al., 2006; Yang et al., 2004). Two GPs were immunized with a subtype B immunogen and three were immunized with a subtype C immunogen.
Results
SCD4 enhances neutralization by V3-directed mAbs
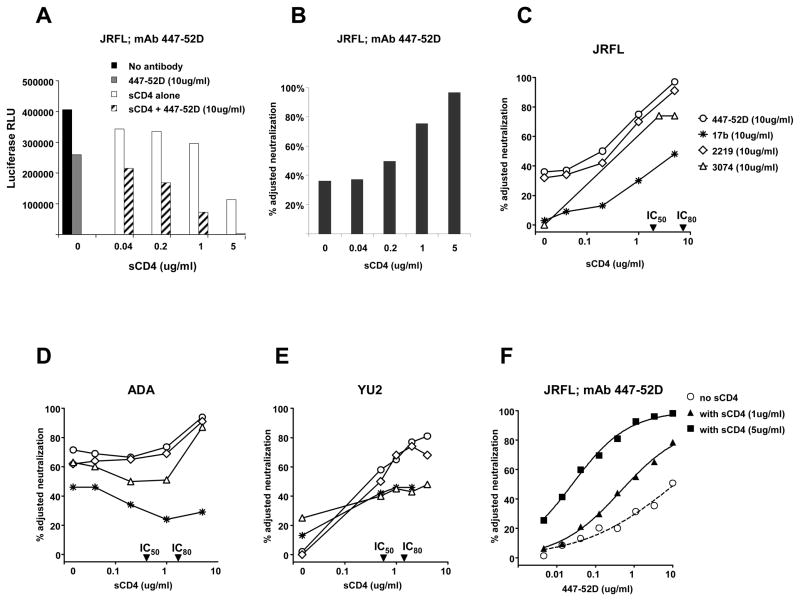
We tested the subtype B-derived anti-V3 mAbs 447-52D and 2219 and the CRF02_AG-derived mAb 3074 against subtype B viruses SF162, BaL.26, ADA, JRFL and YU2. All three mAbs neutralized SF162 and BaL.26 by > 90% at 10 ug/ml (data not shown), but displayed more variable activity against ADA, JRFL and YU2 (Table 1). Of interest, all three mAbs displayed less than 50% neutralizing activity against JRFL even though this virus contain an identical V3 amino acid sequence to BaL.26 (Supplementary Fig. 1). However, pre-incubation with sCD4 substantially increased the sensitivity of JRFL, ADA and YU2 to neutralization by each of the V3 mAbs. In our luciferase-based pseudovirus neutralization assay, viral entry is quantified by relative light units (RLU) and neutralization is calculated as the percent reduction in virus entry caused by antibody compared to baseline viral entry with no antibody present (Fig. 1A). To account for the effect on virus entry by sCD4, antibody-mediated neutralization was calculated using a baseline of viral entry that occurred with the relevant concentration of sCD4 present. Adjusted neutralization (Fig. 1B) depicts the V3 mAb-mediated neutralization that occurred with various concentrations of sCD4 present. As sCD4 concentration increased from zero to 5 ug/ml, 447-52D adjusted neutralization of JRFL increased from 38% to 95%. In these experiments, the mAb was used at a fixed concentration of 10 ug/ml. A similar pattern of JRFL neutralization was observed for mAbs 2219 and 3074 (Fig. 1C). Hence, the weak JRFL neutralization by these V3 mAbs was not due to poor antigenic recognition of the JRFL V3 region, but rather due to limited accessibility of the epitope. SCD4 also enhanced the V3 mAb neutralization of ADA and YU2 (Fig. 1D and 1E). Note that the CRF02_AG-derived mAb 3074 was able to neutralize subtype B viruses JRFL and ADA when sCD4 was present, though not as potently as the two subtype B-derived mAbs. Interestingly, mAb 17b neutralization was only modestly increased in the presence of sCD4, and was never above 50% against JRFL, ADA and YU2 (Fig. 1C, 1D and 1E). This suggests that while the level of sCD4 used in these experiments was enough to inhibit viral entry by 50% – 80%, and was enough to expose the V3 region, it was insufficient to result in full conformational exposure of the co-receptor binding site. The experiments described above were performed with varying concentrations of sCD4 and a fixed concentration of the V3 mAbs. We also determined the effect of a fixed concentration of sCD4 on the potency of mAb 447-52D. The neutralization IC50 for 447-52D decreased from >10 ug/ml in the absence of sCD4 to 0.67 ug/ml and 0.025 ug/ml in the presence of 1 and 5 ug/ml of sCD4 respectively, a greater than 400-fold increase in mAb neutralization potency (Fig. 1F).
Table 1.
Neutralization by the indicated anti-V3 mAbs and GP vaccine sera against subtype B viruses with and without sCD4 present
| sCD4 IC80 ug/ml | 447-52D 10 ug/ml | 2219 10 ug/ml | 3074 10 ug/ml | 82-2 1:10 | 82-4 1:10 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| −sCD4 | +sCD4 | −sCD4 | +sCD4 | −sCD4 | +sCD4 | −sCD4 | +sCD4 | −sCD4 | +sCD4 | ||
| JRFL | 7.2 | 36% | 99% | 32% | 98% | 0% | 94% | 29% | 100% | 31% | 97% |
| ADA | 1.7 | 72% | 82% | 62% | 78% | 63% | 69% | 55% | 73% | 58% | 67% |
| YU2 | 1.4 | 2% | 73% | 0% | 64% | 25% | 41% | 9% | 97% | 29% | 40% |
| 6535.3 | 8.6 | 87% | 93% | 93% | 93% | 24% | 74% | 17% | 85% | 4% | 16% |
| QH0692.42 | 6.4 | 27% | 51% | 28% | 45% | 68% | 73% | 46% | 66% | 12% | 47% |
| SC422661.8 | 125.3 | 0% | 16% | 18% | 25% | 18% | 18% | 41% | 0% | 37% | 22% |
| PVO.4 | 46.0 | 4% | 43% | 35% | 63% | 39% | 57% | 56% | 90% | 44% | 71% |
| TRO.11 | >200 | 50% | 48% | 31% | 37% | 29% | 13% | 70% | 75% | 66% | 67% |
| AC10.0.29 | 80.9 | 8% | 39% | 47% | 57% | 13% | 12% | 59% | 80% | 13% | 55% |
| RHPA4259.7 | 21.6 | 36% | 34% | 29% | 37% | 34% | 45% | 15% | 12% | 9% | 11% |
| THRO4156.18 | 3.1 | 25% | 0% | 14% | 12% | 10% | 8% | 38% | 39% | 23% | 21% |
| REJO4541.67 | 22.5 | 31% | 21% | 47% | 74% | 29% | 55% | 34% | 78% | 6% | 41% |
| TRJO4551.58 | 17.7 | 0% | 86% | 0% | 83% | 5% | 65% | 0% | 87% | 14% | 73% |
| WITO4160.33 | 50.0 | 40% | 76% | 15% | 67% | 12% | 41% | 31% | 68% | 34% | 55% |
| CAAN5342.A2 | 95.2 | 30% | 26% | 39% | 26% | 32% | 17% | 46% | 67% | 34% | 59% |
| Breadth | 2(13%) | 7(47%) | 2(13%) | 9(60%) | 2(13%) | 7(47%) | 4(27%) | 12(80%) | 2(13%) | 8(53%) | |
| P-value | 0.0625 | 0.016 | 0.0625 | 0.008 | 0.031 | ||||||
Values indicate percent neutralization when no sCD4 was present in the assay (−sCD4) and with sCD4 present (+sCD4) at a concentration sufficient to inhibit viral entry by 80%. The later value is the percent adjusted neutralization and indicates the effect of the mAb or GP sera calculated based on baseline viral entry with sCD4 present. The second column shows the concentration of sCD4 required to inhibit viral entry by 80%. Note that the IC80 for sCD4 was derived separately for each virus and the percent adjusted neutralization mediated by mAb or GP sera was calculated using viral entry values at this level of sCD4 (e.g., Fig. 3). Values greater than 50% neutralization are shown in bold and were used to calculate the breadth of neutralization. P-values were computed using the two-sided McNemar’s test.
Fig. 1.
Effect of sCD4 on neutralization of JRFL, ADA and YU2 by anti-V3 mAbs 447-52D, 2219, 3074 and anti-co-receptor mAb 17b. (A) Viral entry expressed as luciferase (RLU) of JRFL is shown with varying concentrations of sCD4. Black bar shows viral entry with no mAb and no sCD4 present. Grey bar shows viral entry with 447-52D. White bars show the effect of sCD4 alone on viral entry. Hatched bars show the effect of each concentration of sCD4 with a fixed amount (10 ug/ml) of mAb 447-52D. (B) The percent adjusted neutralization was calculated from (A) using RLU values for each concentration of sCD4 as the baseline for viral entry. The additional effect on viral entry by 10 ug/ml of mAb 447-52D is the percent adjusted neutralization. (C), (D) and (E) show the percent adjusted neutralization by several mAbs against viruses JRFL, ADA and YU2. Legend in (C) applies to (D) and (E). (F) Serial dilutions of mAb 447-52D alone, or with a fixed concentration of sCD4 were tested to calculate the impact of sCD4 on the potency of mAb 447-52D neutralization. Note that for graphs C, D and E, the IC50 and IC80 effect of sCD4 against each virus are indicated on the X-axis.
SCD4 enhances and broadens neutralization by GP vaccine sera
We previously demonstrated that DNA prime, rAd5 boost immunization with Env constructs containing a deletion of the V1V2 loop and two small deletions on both arms of the V3 stem, produced high V3-directed NAb responses (Chakrabarti et al., 2002; Chakrabarti et al., 2005; Wu et al., 2006; Yang et al., 2004). In the present study, we examined two such GP sera from animals immunized with an HIV-1 subtype B-based Env immunogen. GP sera 82-2 and 82-4 had high and moderate NAb levels respectively (Supplementary Fig. 2). Virus neutralization by both sera was largely mediated by antibodies to the V3 loop, as shown by peptide competition studies (Wu et al., 2006).
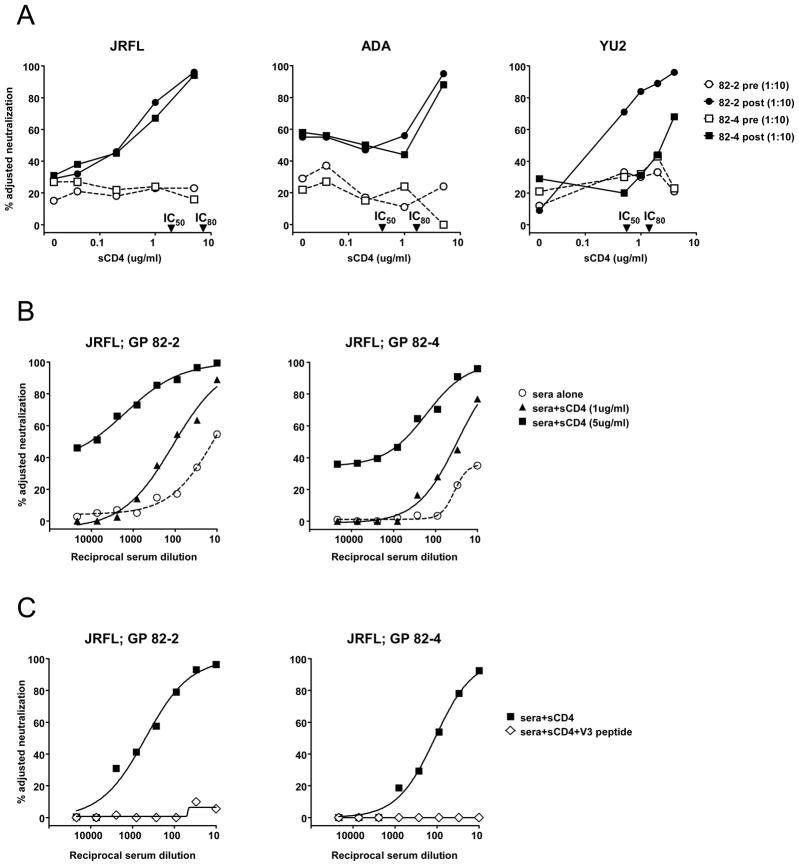
As we previously described, these sera displayed little neutralization of viruses JRFL, ADA and YU2. However, in the presence of sCD4, a 1:10 dilution of the sera neutralized all three viruses (Fig. 2A). To assess the increase in potency of the sera caused by sCD4, the sera were serially diluted in the presence of 1 ug/ml or 5 ug/ml of sCD4 (Fig. 2B). The neutralization IC50 of serum 82-2 against JRFL improved from 1:12 in the absence of sCD4 to 1:100 and >1:1,000 in the presence of 1 and 5 ug/ml of sCD4, respectively. Similarly, the neutralization IC50 of 82-4 increased from < 1:10 in the absence of sCD4 to 1:30 and 1:625 in the presence of 1 and 5 ug/ml of sCD4, respectively. The V3 specificity of these neutralization activities was confirmed by peptide competition assays using a V3 peptide that matched the immunogen V3 sequence (Fig. 2C).
Fig. 2.
Neutralization by GP vaccine sera. (A) The effect of sCD4 on neutralization by GP sera 82-2 and 82-4. Pre-immune and vaccine sera are shown by open and filled symbols respectively. The IC50 and IC80 of sCD4 against each virus are indicated on the X-axis. (B) Serial dilutions of GP vaccine sera alone, or with a fixed concentration of sCD4 were tested to calculate the impact of sCD4 on the potency of serum neutralization. (C) With 5 ug/ml of sCD4 present, GP vaccine sera neutralize JRFL predominantly via anti-V3 antibodies. A V3 peptide (30 ug/ml) matched to the vaccine immunogen was used to inhibit the V3 mediated neutralization by GP vaccine sera. A scrambled V3 peptide had no inhibitory effect (not shown).
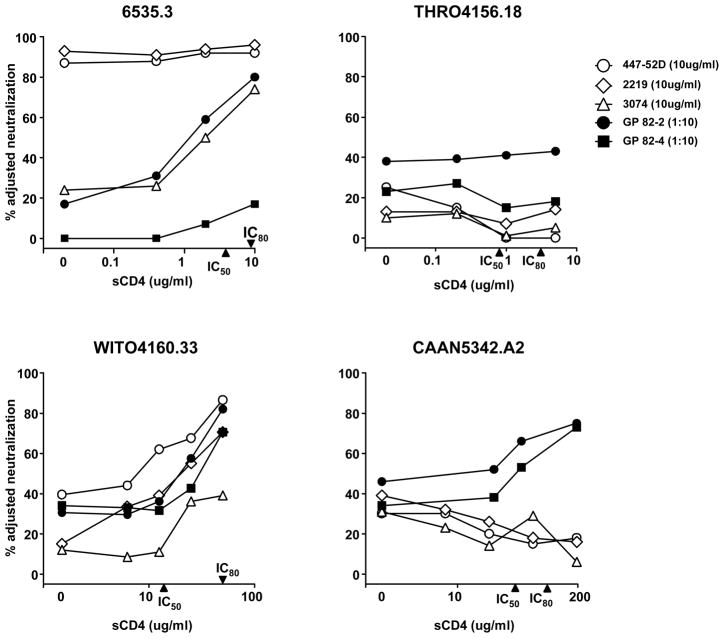
To understand the potential breadth of reactivity of these V3 mAbs and V3-directed GP vaccine sera, we extended this analysis to a recently described reference panel of 12 acute subtype B viruses (Li et al., 2005). The neutralization profiles of three mAbs and two GP sera against four selected viruses are graphically shown in Fig. 3. These four viruses demonstrate the spectrum of V3 neutralization observed among the subtype B reference viruses. Virus 6535.3 was highly sensitive to V3-mediated neutralization by two mAbs without sCD4 present, but displayed increased sensitivity to mAb 3074 and GP 82-2 serum after sCD4 was added; virus WITO4160.33 became generally sensitive to mAb and serum V3-mediated neutralization after sCD4 was added to the assay; virus CAAN5342.A2 became sensitive to the GP sera, but not the mAbs and virus THRO4156.18 was resistant to both anti-V3 mAbs and GP sera, even after sCD4 was added. Note that the concentration of sCD4 required to inhibit 50% or 80% viral entry, and hence to expose the V3 loop to antibody, varied among viruses (Fig. 3 and Table 1). As summarized in Table 1, the anti-V3 mAbs at 10 ug/ml, and the GP vaccine sera at a 1:10 dilution, displayed limited neutralization breadth among the 15 viruses tested. It should be noted that this panel of 15 pseudoviruses, which includes JRFL, ADA, YU2 and 12 reference strains, was chosen to exclude viruses such as BaL.26 and SF162 that are highly sensitive to V3-mediated neutralization. We used a value of 50% virus neutralization by 10 ug/ml of mAb, or a 1:10 serum dilution, as measure of positive virus neutralization. Our prior published data suggests that a GP vaccine serum neutralization value of 50% or greater is a readily reproducible measure of antibody-mediated virus neutralization (Shu et al., 2007; Wu et al., 2006; Yang et al., 2004). By this measure, the three mAbs and two GP sera neutralized two to four of the 15 viruses (13–27%). In the presence of sCD4, the mAbs and GP sera both displayed increased neutralization breadth. For example, mAb 2219 neutralized two of 15 viruses without sCD4 and nine of 15 viruses in the presence of sCD4, and serum 82-2 neutralization increased from four to 12 of 15 viruses. Hence, up to 80% of the subtype B viruses tested were sensitive to V3-mediated neutralization when sCD4 was present. This increase in breadth was statistically significant for mAb 2219 and for both GP sera (Table 1). Statistical analysis comparing the percent neutralization by each mAb and serum against all 15 viruses, with and without sCD4, was also performed. The inclusion of sCD4 in the assay resulted in a statistically significant increase in the median neutralization values for all three anti-V3 mAbs and both GP vaccine sera, but not for mAb 17b (Supplementary Fig. 3).
Fig. 3.
The effect of sCD4 on V3-mediated neutralization of four selected subtype B reference viruses. Data for anti-V3 mAbs and subtype B GP vaccine sera are shown against each virus. The IC50 and IC80 of sCD4 against each virus are indicated on the X-axes. Note that the scale of the X-axes differs among viruses.
SCD4 broadens neutralization of V3-directed mAbs and subtype C GP vaccine sera against a reference panel of subtype C viruses
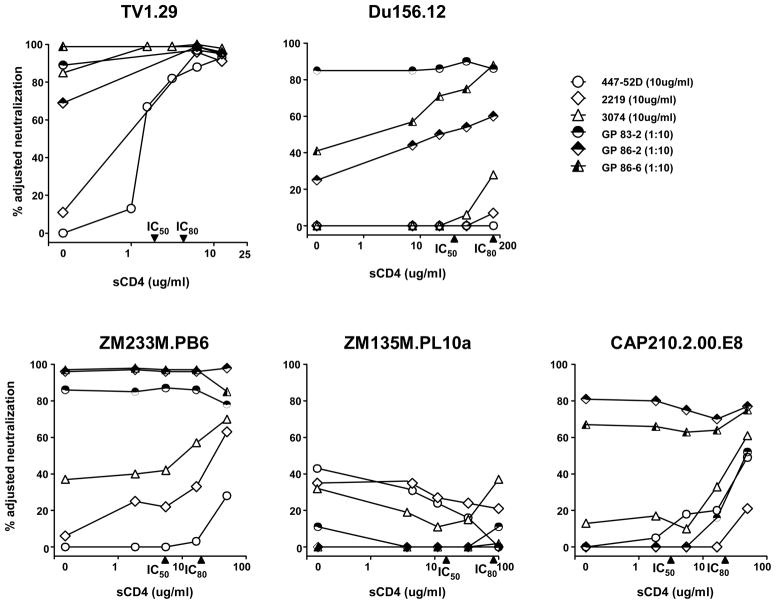
We further evaluated the effect of sCD4 on neutralization of a standardized panel of 12 subtype C reference viruses (Li et al., 2006). We used the same three mAbs, including the CRF02_AG-derived 3074, and included sera from three selected GPs that received a subtype C Env immunogen. The subtype C Env immunogen was similar to the subtype B immunogen except that the V3 region was replaced with a subtype C V3 sequence (Wu et al., 2006). All three GP sera neutralized V3 sensitive subtype C viruses (Supplementary Fig. 2) and had modest to high levels of anti-V3 NAbs (Wu et al., 2006). The neutralization profiles of five selected subtype C viruses are graphically shown in Figure 4. TV1.29 was chosen because it was previously shown to be sensitive to V3 neutralization by subtype C GP vaccine sera (Wu et al., 2006); the other four viruses demonstrate the spectrum of V3 neutralization observed among the subtype C reference viruses. The anti-V3 mAbs displayed little neutralization of the subtype C reference viruses, but the presence of sCD4 resulted in mAb neutralization of some viruses such as ZM233M.PB6 and CAP210.2.00.E8 (Fig. 4). The GP vaccine sera displayed neutralization of some subtype C viruses without sCD4 present, but against other viruses such as Du156.12, neutralization was increased by adding sCD4 to the assay (Fig. 4). Using the same 50% value to denote positive virus neutralization, mAb 3074 neutralization increased from two to five of 13 viruses when sCD4 was present and GP serum 86-6 neutralization increased from five to ten of 13 viruses (Table 2). While these increases in neutralization breadth were not statistically significant (Table 2), comparisons of median percent neutralization values with and without sCD4 indicate significant increases in potency for mAbs 2219, 3074 and GP serum 86-6 (Supplementary Fig. 4). We verified the specificity of the neutralization by GP sera 83-2, 86-2 and 86-6 against virus TV1.29 by V3 peptide competition (Fig. 5). The presence of 5 ug/ml of sCD4 markedly increased the neutralization titers of all three immune sera against TV1.29, and this was inhibited by 30 ug/ml of a V3 peptide that matched the V3 immunogen sequence. These data indicate that similar to subtype B viruses, the subtype C V3 region is recognized by known V3 mAbs and by immune sera, but is poorly accessible to antibody on the functional virion.
Fig. 4.
The effect of sCD4 on V3-mediated neutralization of TV1.29 and four selected subtype C reference viruses. Data for anti-V3 mAbs and subtype C GP vaccine sera are shown against each virus. The IC50 and IC80 of sCD4 against each virus are indicated on the X-axes. Note that the scale of the X-axes differs among viruses.
Table 2.
Neutralization by the indicated anti-V3 mAbs and GP vaccine sera against subtype C viruses with and without sCD4 present
| sCD4 IC80 ug/ml | 447-52D 10 ug/ml | 2219 10 ug/ml | 3074 10 ug/ml | 83-2 1:10 | 86-2 1:10 | 86-6 1:10 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| −sCD4 | +sCD4 | −sCD4 | +sCD4 | −sCD4 | +sCD4 | −sCD4 | +sCD4 | −sCD4 | +sCD4 | −sCD4 | +sCD4 | ||
| TV1.29 | 4.3 | 0% | 100% | 11% | 87% | 85% | 96% | 89% | 95% | 69% | 95% | 99% | 99% |
| Du156.12 | 200.0 | 0% | 4% | 0% | 8% | 0% | 28% | 85% | 88% | 25% | 62% | 41% | 85% |
| Du172.17 | 10.2 | 24% | 31% | 20% | 24% | 25% | 66% | 0% | 6% | 69% | 85% | 66% | 79% |
| Du422.1 | 100.0 | 16% | 10% | 17% | 28% | 1% | 40% | 1% | 16% | 8% | 4% | 0% | 58% |
| ZM197M.PB7 | 58.1 | 1% | 0% | 2% | 11% | 0% | 8% | 0% | 0% | 0% | 16% | 0% | 59% |
| ZM214M.PL15 | 160.0 | 24% | 25% | 9% | 3% | 0% | 16% | 0% | 31% | 0% | 34% | 0% | 59% |
| ZM233M.PB6 | 19.9 | 0% | 6% | 7% | 43% | 37% | 58% | 86% | 87% | 96% | 97% | 97% | 97% |
| ZM249M.PL1 | 43.9 | 4% | 15% | 10% | 28% | 0% | 6% | 0% | 0% | 0% | 0% | 0% | 1% |
| ZM53M.PB12 | 45.3 | 23% | 10% | 10% | 9% | 12% | 54% | 0% | 0% | 5% | 26% | 0% | 89% |
| ZM109F.PB4 | 2.5 | 17% | 31% | 45% | 46% | 75% | 70% | 56% | 47% | 84% | 81% | 87% | 67% |
| ZM135M.PL10a | 83.5 | 44% | 10% | 36% | 22% | 33% | 31% | 1% | 0% | 11% | 0% | 0% | 0% |
| CAP45.2.00.G3 | 93.3 | 2% | 12% | 6% | 25% | 4% | 28% | 0% | 0% | 0% | 0% | 7% | 43% |
| CAP210.2.00.E8 | 22.7 | 0% | 31% | 0% | 0% | 13% | 41% | 0% | 22% | 82% | 76% | 67% | 69% |
| Breadth | 0(0%) | 1(8%) | 0(0%) | 1(8%) | 2(15%) | 5(38%) | 4(31%) | 3(23%) | 5(38%) | 6(46%) | 5(38%) | 10(77%) | |
| P-value | 1 | 1 | 0.25 | 1 | 1 | 0.0625 | |||||||
Values indicate percent neutralization when no sCD4 was present in the assay (−sCD4) and with sCD4 present (+sCD4) at a concentration sufficient to inhibit viral entry by 80%. The later value is the percent adjusted neutralization and indicates the effect of the mAb or GP sera calculated based on baseline viral entry with sCD4 present. The second column shows the concentration of sCD4 required to inhibit viral entry by 80%. Note that the IC80 for sCD4 was derived separately for each virus and the percent adjusted neutralization mediated by mAb or GP sera was calculated using viral entry values at this level of sCD4 (e.g., Fig. 4). Values greater than 50% neutralization are shown in bold and were used to calculate the breadth of neutralization. P-values were computed using the two-sided McNemar’s test.
Fig. 5.
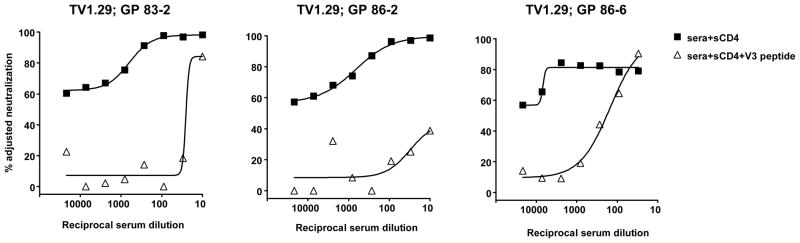
In the presence of 5 ug/ml of sCD4, GP vaccine sera neutralized subtype C virus TV1.29 predominantly via anti-V3 antibodies. A V3 peptide (30 ug/ml) matched to the subtype C vaccine immunogen was used to inhibit the V3-mediated neutralization by the sera. A scrambled V3 peptide had no inhibitory effect (not shown).
Approximately 25% of pseudoviruses in the subtype B and C panels we tested remained resistant to V3-antibody neutralization, even in the presence of sCD4 at IC80 or higher concentrations. The explanation of this observation could be that sCD4 did not trigger exposure of V3 loop or that antigenic variation resulted in lack of recognition by any of the antibody reagents tested. This could be partially addressed by testing antibody reactivity to the soluble monomeric g120 captured on an ELISA plate. Of the three subtype B viruses that were resistant to V3-mediated neutralization, the gp120 of SC422661.8 and RHPA4259.7 was strongly bound by mAb 447-52D (data not shown). This suggests that this neutralization resistance was due to a lack of exposure of the V3 loop on the native virus. In contrast, the gp120 from virus THRO4156.18, which has an unusual GPGG sequence at the tip of the V3 loop, did not react with 447-52D. Hence, both mechanisms may be occurring; the masked V3 loop of some viruses are not readily exposed, even when high concentrations of sCD4 are present and amino acid sequence variation may account for the neutralization resistance of other viruses. Although V1V2 length and positioning of glycosylation have been suggested as major factors in masking the V3 epitope (Krachmarov et al., 2006), we did not find significant differences in the length of V1V2 or number of potential N-linked glycosylation sites between viruses that were sensitive to anti-V3 antibodies and those that were resistant.
Anti-CD4-binding site (CD4bs) mAbs b12 and F105 did not affect V3-mediated neutralization
In the hope of finding an antibody that would bind to the CD4bs of the HIV-1 Env and induce conformational changes similar to those induced by sCD4, we tested the anti-CD4bs mAbs b12 and F105 for their effect on neutralization of V3-directed mAb and GP immune serum. Although the presence of sCD4 increased neutralization of JRFL by 447-52D and immune serum 82-4, the presence of b12 and F105 at concentrations sufficient to inhibit JRFL entry by greater than 50% did not induce V3 mediated neutralization (Fig. 6). Similar results were obtained with virus YU2 (data not shown). These data suggest that mAbs b12 and F105 do not induce a conformation alteration in Env that exposes the V3 loop to NAbs.
Fig. 6.
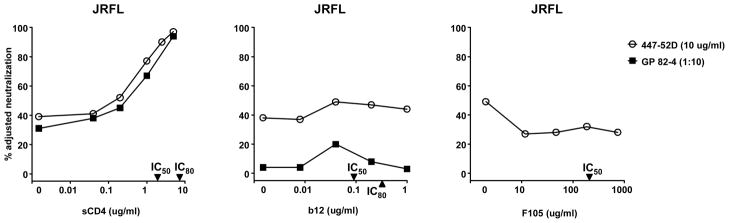
Lack of effect of anti-CD4bs mAbs b12 and F105 on neutralization of JRFL by mAb 447-52D and GP vaccine serum 82-4. Unlike sCD4, which increases neutralization sensitivity of JRFL to anti-V3 antibodies, b12 and F105 did not affect neutralization sensitivity of JRFL to anti-V3 antibodies. The IC50 and IC80 of sCD4, b12 and F105 against JRFL are indicated on the X-axes.
Discussion
The V3 region of HIV-1 has been considered a potential vaccine target because of its immuno-dominance in eliciting antibody responses. Anti-V3 antibodies are common during natural infection and are readily elicited by numerous peptide- or Env-based vaccine immunogens (Broliden et al., 1992; Javaherian et al., 1990; Krachmarov et al., 2005; Krachmarov et al., 2001; Kraft et al., 2007; Letvin et al., 2001; Profy et al., 1990; Vancott et al., 1995; Wu et al., 2006; Yang et al., 2004; Zolla-Pazner, 2004; Zolla-Pazner et al., 2008). Early studies examined vaccine sera against T-cell line adapted HIV-1 strains that were highly sensitive to V3-mediated neutralization. Vaccine sera displayed potent neutralization against prototype viruses such as MN, RF and IIIB, but usually failed to neutralize primary virus stains that were not adapted to growth in cell lines (Javaherian et al., 1990; Mascola et al., 1996; Montefiori et al., 1993; Spenlehauer et al., 1998; Wrin et al., 1995; Wrin and Nunberg, 1994). In addition, V3-elicited antibodies were often found to be highly strain specific, suggesting that the antigenic diversity of the V3 loop posed a major obstacle for NAbs to this variable region of Env (Lu, Putney, and Robinson, 1992; Nara et al., 1990; Palker et al., 1988; Park et al., 1998; Zwart et al., 1992). However, our current understanding of the V3 domain suggests a significant degree of functional conservation, likely due to the interaction of V3 with the CCR5 or CXCR4 co-receptor prior to gp41-mediated viral fusion with the target cell membrane (Cormier and Dragic, 2002; Huang et al., 2005; Stanfield et al., 2004; Stanfield et al., 2006). This functional conservation is consistent with some level of antigenic conservation and the observation that human anti-V3 mAbs can be reactive with the V3 loop of numerous strains of HIV-1 (Gorny et al., 2002; Gorny et al., 2006; Gorny et al., 1993; Krachmarov et al., 2005; Krachmarov et al., 2006; Moore et al., 1995b). These more broadly reactive human anti-V3 mAbs are able to neutralize a subset of primary strains of HIV-1, but are ineffective against many other HIV-1 isolates. This likely results because the V3 loop is partially or completely inaccessible to antibody in the context of the native structure of the trimeric gp120-gp41 complex (Binley et al., 2004; Krachmarov et al., 2005; Lusso et al., 2005; Moore et al., 1995a; Pantophlet et al., 2007; Pantophlet and Burton, 2006; Spenlehauer et al., 1998).
Env binding to CD4 induces a conformational stabilization of Env that results in the formation of the co-receptor binding surface, as well as movement of the V1V2 domain and exposure of the V3 loop. Studies with soluble gp120 and Env expressed on the surface of transfected cells have shown increased binding of co-receptor binding site mAbs such as 17b and 48d, and anti-V3 mAbs after Env binds to sCD4 (Krachmarov et al., 2006; Lusso et al., 2005; Mbah et al., 2001; Salzwedel et al., 2000; Sullivan et al., 1998b; Wyatt and Sodroski, 1998; Xiang et al., 2002). These data suggested that sCD4-induced exposure of the V3 loop could maximize V3-mediated neutralization of free virus and would allow us to test whether the lack of anti-V3 neutralization of most viruses was due to poor recognition of the V3 loop or to limited accessibility of the epitope on native virus. To address this question with representative viruses, we used panels of well-characterized subtype B and C Env-pseudoviruses that were derived during the acute phase of HIV-1 infection. We selected three human anti-V3 mAbs, two derived from subtype B and one from a CFR02-AG infected patient, and GP vaccine sera elicited by subtype B or C Env immunogens that were known to contain moderate to high levels of anti-V3 antibodies. Consistent with previous reports, the anti-V3 mAbs and GP vaccine sera strongly neutralized several prototype V3 sensitive viruses, but displayed more variable neutralization against the subtype B and C reference viruses (Binley et al., 2004; Li et al., 2005; Li et al., 2006; Wu et al., 2006; Yang et al., 2004). However, upon pre-incubation with sCD4, 12 of 15 (80%) subtype B viruses and 10 of 13 (77%) subtype C viruses were sensitive to V3-mediated neutralization. These findings suggest that the V3 loop of most HIV-1 isolates is recognized by anti-V3 mAbs and immune sera, and that the low level of V3-mediated neutralization of many primary virus strains is largely a result of inaccessibility of the V3 loop. The level of accessibility of the V3 loop is in part dependent on the antibody tested. For example, we found mAb 447-52D to have weak neutralization activity against virus JRFL, but mAb B4e8, not tested here, is reported to neutralize JRFL with an IC50 of 2.4 ug/ml. (Pantophlet et al., 2007).
Of note, the amount of sCD4 required to trigger exposure of the V3 loop varied among viruses. Generally, concentrations of sCD4 that inhibited viral entry by 50% to 80% were sufficient to trigger exposure of the V3 loop. For some viruses this required as little as 1 ug/ml of sCD4 (e.g., JRFL), but the sCD4 IC80 value for other viruses was as high as 200 ug/ml. As has been previously described, the CRF02_AG-derived anti-V3 mAb 3074 displayed greater cross-subtype neutralizing activity than the subtype B-derived mAbs 447-52D and 2219. Even in the presence of sCD4, 447-52D and 2219 displayed little or no cross neutralization against the subtype C viruses while 3074 neutralized seven of 15 subtype B viruses. Therefore, a subtype A V3 sequence or a consensus V3 sequence that is representative of multi-subtypes may be a better immunogen than a subtype B V3 sequence. This is borne out by our experience with subtype B and C Env immunogens. The subtype C Env generated anti-V3 antibodies that cross-neutralized sensitive subtype B viruses, whereas the anti-V3 response elicited by the subtype B Env was restricted to subtype B viruses (Wu et al., 2006). Also, priming of immune responses with DNA from a subtype C Env elicited a broader response than did priming with a subtype B Env (Zolla-Pazner et al., 2008).
Overall, these data have implications for V3-based immunogen design. In contrast to epitopes such as the membrane proximal external region and the CD4bs of gp120, which appear to be poorly immunogenic, anti-V3 antibodies can be elicited by current Env-based vaccines. Such anti-V3 Abs display cross-subtype V3 reactivity, but do not neutralize the majority of primary viruses and Env-pseudoviruses against which they have been tested. If it were possible for an antibody to induce conformation changes similar to those of sCD4, anti-V3 antibodies could play a major role in virus neutralization. To evaluate this directly, we assayed two well-characterized anti-CD4bs mAbs to determine if their binding to native virus resulted in exposure of the V3 loop. MAb b12 is the only CD4bs-directed mAb that can neutralize most primary virus strains; mAb F105 neutralization is restricted to a smaller subset of neutralization sensitive viruses. We chose virus JRFL because its V3 loop was highly exposed to mAb 447-52D neutralization by incubation with relatively low concentrations of sCD4 (Fig. 1). Also, JRFL is neutralized by both b12 and F105, although the former is much more potent than the latter. Concentrations of either mAb sufficient to inhibit JRFL entry by 50% - 80% had no effect on 447-52D neutralization of JRFL. This was perhaps expected for b12, which binds to the outer domain of gp120 without causing significant conformational rearrangements (Zhou et al., 2007). In contrast, thermodynamic and structural data show that effective binding of sCD4 requires conformational changes in gp120 (Kwong et al., 2002; Zhou et al., 2007). F105 also induces moderate conformational rearrangement during binding to gp120, but our data suggest that these do not result in full exposure of the V3 loop to antibody. Whether the characteristic of sCD4 binding that results in exposure of the V3 loop is unique to sCD4 remains to be determined. Of note, numerous studies have suggested additive or synergistic interactions between V3 antibodies and antibodies to other regions of Env (Buchbinder et al., 1992; Cavacini et al., 1993; Laal et al., 1994; Li et al., 1997; McKeating et al., 1992; Montefiori et al., 1993; Moore and Sodroski, 1996; Pinter, Honnen, and Tilley, 1993; Potts et al., 1993; Thali et al., 1992; Tilley et al., 1992; Vijh-Warrier et al., 1996). It therefore remains possible that an antibody-mediated rearrangement of Env conformation could lead to increased sensitivity to V3 loop NAbs.
In summary, selected human anti-V3 mAbs and vaccine immune sera can recognize the antigenic structure of V3 domain of the most circulating subtype B and C virus strains, but the V3 loop is often masked, or partially masked, in the context of the native viral spike. Vaccine elicited anti-V3 antibodies may have enough potency to neutralize a subset of V3 sensitive primary viruses, but this antibody specificity will likely need to be augmented by NAbs against other viral epitopes for an antibody-based vaccine to be effective.
Materials and methods
Antibodies, cells and virus stocks
The anti-V3 mAbs 447-52D, 2219 and 3074 were described previously (Conley et al., 1994; Gorny et al., 1992; Gorny et al., 2002; Gorny et al., 2006; Nyambi et al., 1998). The co-receptor binding site antibody 17b was obtained from James Robinson (Tulane University). The CD4bs mAbs b12 was provided by Dennis Burton and Ralph Pantophlet (Scripps Research Institute); F105 was from the laboratory of Marshall Posner (Dana-Farber Cancer Institute). Two domain sCD4 (sCD4-183) was obtained through the NIH AIDS Research and Reference Reagent Program and from Pharmacia. Human embryonic kidney cell line 293T was purchased from the American Type Culture Collection and maintained in DMEM (Gibco, Invitrogen, Carlsbad, CA) containing 10% heat-inactivated fetal bovine serum and 100 ug/ml of penicillin/streptomycin. The TZM-bl cells were obtained from the NIH AIDS Research and Reference Reagent Program, as contributed by John Kappes and Xiaoyun Wu. This cell line is a genetically engineered HeLa cell clone that expresses CD4, CXCR4, and CCR5, and contains HIV Tat-responsive reporter genes for firefly luciferase and Escherichia coli β-galactosidase under regulatory control of an HIV-1 long terminal repeat.
Pseudovirus was prepared by transfecting 293T cells (6 × 106 cells in 50 ml growth medium in a T-175 culture flask) with 10 μg of rev/env expression plasmid and 30 μg of an env-deficient HIV-1 backbone vector (pSG3ΔEnvelope), using Fugene 6 transfection reagents (Invitrogen). Pseudovirus-containing culture supernatants were harvested two days after transfection, filtered (0.45 μm), and stored at −80°C or in the vapor phase of liquid nitrogen. The virus 50% tissue culture infectious dose (TCID50) was determined on TZM-bl cells by serial 5-fold dilutions of the pseudovirus stock in quadruplicate wells. At approximately 48 hours, the infection levels were determined by a luciferase assay (Bright Glo, Promega, Madison, WI) using the cell lysate. Infections producing the RLU three times above the mock infection background were scored as positive.
Constructions of plasmid DNA, recombinant adenoviruses and vaccination
The construction of plasmid DNA and rAd5 were described previously (Wu et al., 2006). Selective modifications of the HIV-1 Env included ΔCFI, indicating deletions in the cleavage site, fusogenic domain, and spacing of heptad repeat 1 and 2, and ΔV1V2(V3-1AB), indicating deletions of the V1V2 loop and two small deletions on both arms of the V3 stem (Chakrabarti et al., 2002; Chakrabarti et al., 2005; Wu et al., 2006; Yang et al., 2004). HIV-1 env genes encoding BaL gp145ΔCFI, and Hx/BaL gp145ΔCFI were synthesized using human-preferred codons as previously described. In gp145ΔCFIΔV1V2 (ZA012V3-1AB), amino acids 202-388 of BaL V3 were replaced by amino acids 195–382 of subtype C ZA012 V3 sequence.
GP immunizations were performed as previously descried (Wu et al., 2006). Briefly, animals were immunized intramuscularly with 500 ug (in 400 ul PBS) of the gp145 version of plasmid DNA at weeks 0, 2 and 6. At week 14, GPs were boosted with 1011 particles (in 400 ul PBS) of rAd5 expressing the corresponding gp140 version of the HIV-1 Env protein. Serum tested in this study was collected two weeks after the rAd5 boost immunization. GP group 82 received a subtype B V3 in a subtype B Env backbone (BaL gp145 ΔCFI ΔV1V2 (V3-1AB)); groups 83 and 86 received a subtype C V3 (ZA012V3-1AB) in a subtype B Env backbone. This backbone was BaL gp145 ΔCFI ΔV1V2 for group 83 and Hx/BaL gp145 ΔCFI ΔV1V2 for group 86.
Neutralization and V3 peptide competition assays
Neutralization was measured using HIV-1 Env-pseudoviruses to infect TZM-bl cells as described previously (Li et al., 2005; Shu et al., 2007). Briefly, 40 ul of virus was incubated for 30 minutes at 37°C with 10 ul of serial dilutions of test antibody or serum samples in duplicate wells of a 96-well flat bottom culture plate. For neutralization assays with sCD4, 5 ul of sCD4 was added to the virus for 30 minutes followed by addition of 5 ul of test mAb or serum sample. To keep assay conditions constant, sham media was used in place of sCD4 or antibody in specified control wells. The mAb and sCD4 concentrations, and serum dilutions, were defined at the point of incubation with virus supernatant. 10,000 TZM-bl cells were then added to each well in a volume of 20 ul, and plates were incubated overnight at 37°C in a 5% CO2 incubator. One set of eight wells received mock antibody followed by virus and cells as controls for virus entry, and a set of eight wells received cells with mock virus to control for luciferase background. After overnight incubation, 150 ul of fresh medium was added to each well. Infection levels were determined the next day using a quantitative luciferase assay measuring luciferase activity present in cell lysate (Promega). The virus input was set at a multiplicity of infection (Ortiz et al.) of approximately 0.1, which generally results in 100,000 to 400,000 RLU in the luciferase assay. Neutralization curves were fit by non-linear regression using a 4-parameter hill slope equation programmed into JMP statistical software (JMP 5.1, SAS Institute Inc., Cary, NC). The 50% or 80% inhibitory concentrations (IC50 and IC80) are reported as the reciprocal serum dilutions or mAb concentrations required to inhibit infection by 50% or 80%.
The V3 peptide competition assays were done in the same assay format as the neutralization assay, except that the control or test peptide was added to antibody or serum 30 min prior to the addition of virus. The concentration of peptide used was 30 ug/ml, and represents the concentration that was present when peptide, serum or antibody and virus were incubated together. The V3 peptide sequences used in this study were based either on BaL.26 (TRPNNNTRKSIHIGPGRAFYTTG) or ZA012 (TRPNNNTRKSMRIGPGQTFYATG), each matching the representative vaccine strain. A scrambled V3 peptide (IGPGRATRPNNNFYTTGTRKSIH) was used as a negative control. These peptides were synthesized by SynPep (Dublin, CA).
Statistical analysis
Comparisons of neutralization breadth in the absence and presence of sCD4 were performed by McNemar’s test using the statistical package within R software. Comparisons of neutralization values in the absence and presence of sCD4 were performed by paired Wilcoxon signed-rank test using the statistical package within GraphPad Prism (V5.0) software. P-values ≤ 0.05 are reported as significant.
Supplementary Material
Acknowledgments
We thank Peter Kwong, Tongqing Zhou and Richard Wyatt for helpful discussions of the data, and Stephen Schmidt for review of this manuscript. This research was supported by the Intramural Research Program of the Office of Clinical Research and the Vaccine Research Center, NIAID, NIH and by NIH grants AI 36085, and research funds from the Department of Veterans Affairs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos CJ, Burton DR. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78(23):13232–52. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou-Habib DC, Roderiquez G, Oravecz T, Berman PW, Lusso P, Norcross MA. Cryptic nature of envelope V3 region epitopes protects primary monocytotropic human immunodeficiency virus type 1 from antibody neutralization. J Virol. 1994;68(9):6006–13. doi: 10.1128/jvi.68.9.6006-6013.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broliden PA, von Gegerfelt A, Clapham P, Rosen J, Fenyo EM, Wahren B, Broliden K. Identification of human neutralization-inducing regions of the human immunodeficiency virus type 1 envelope glycoproteins. Proc Natl Acad Sci U S A. 1992;89(2):461–5. doi: 10.1073/pnas.89.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchbinder A, Karwowska S, Gorny MK, Burda ST, Zolla-Pazner S. Synergy between human monoclonal antibodies to HIV extends their effective biologic activity against homologous and divergent strains. AIDS Res Hum Retroviruses. 1992;8(4):425–7. doi: 10.1089/aid.1992.8.425. [DOI] [PubMed] [Google Scholar]
- Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5(3):233–6. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- Cardozo T, Kimura T, Philpott S, Weiser B, Burger H, Zolla-Pazner S. Structural basis for coreceptor selectivity by the HIV type 1 V3 loop. AIDS Res Hum Retroviruses. 2007;23(3):415–26. doi: 10.1089/aid.2006.0130. [DOI] [PubMed] [Google Scholar]
- Cavacini LA, Emes CL, Power J, Buchbinder A, Zolla-Pazner S, Posner MR. Human monoclonal antibodies to the V3 loop of HIV-1 gp120 mediate variable and distinct effects on binding and viral neutralization by a human monoclonal antibody to the CD4 binding site. J Acquir Immune Defic Syndr. 1993;6(4):353–8. [PubMed] [Google Scholar]
- Chakrabarti BK, Kong WP, Wu BY, Yang ZY, Friborg J, Ling X, King SR, Montefiori DC, Nabel GJ. Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J Virol. 2002;76(11):5357–68. doi: 10.1128/JVI.76.11.5357-5368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti BK, Ling X, Yang ZY, Montefiori DC, Panet A, Kong WP, Welcher B, Louder MK, Mascola JR, Nabel GJ. Expanded breadth of virus neutralization after immunization with a multiclade envelope HIV vaccine candidate. Vaccine. 2005;23(26):3434–45. doi: 10.1016/j.vaccine.2005.01.099. [DOI] [PubMed] [Google Scholar]
- Conley AJ, Gorny MK, Kessler JA, 2nd, Boots LJ, Ossorio-Castro M, Koenig S, Lineberger DW, Emini EA, Williams C, Zolla-Pazner S. Neutralization of primary human immunodeficiency virus type 1 isolates by the broadly reactive anti-V3 monoclonal antibody, 447-52D. J Virol. 1994;68(11):6994–7000. doi: 10.1128/jvi.68.11.6994-7000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier EG, Dragic T. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J Virol. 2002;76(17):8953–7. doi: 10.1128/JVI.76.17.8953-8957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med. 2005;201(9):1407–19. doi: 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derby NR, Gray S, Wayner E, Campogan D, Vlahogiannis G, Kraft Z, Barnett SW, Srivastava IK, Stamatatos L. Isolation and characterization of monoclonal antibodies elicited by trimeric HIV-1 Env gp140 protein immunogens. Virology. 2007;366(2):433–45. doi: 10.1016/j.virol.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Conley AJ, Karwowska S, Buchbinder A, Xu JY, Emini EA, Koenig S, Zolla-Pazner S. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J Virol. 1992;66(12):7538–42. doi: 10.1128/jvi.66.12.7538-7542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Williams C, Volsky B, Revesz K, Cohen S, Polonis VR, Honnen WJ, Kayman SC, Krachmarov C, Pinter A, Zolla-Pazner S. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J Virol. 2002;76(18):9035–45. doi: 10.1128/JVI.76.18.9035-9045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Williams C, Volsky B, Revesz K, Wang XH, Burda S, Kimura T, Konings FA, Nadas A, Anyangwe CA, Nyambi P, Krachmarov C, Pinter A, Zolla-Pazner S. Cross-clade neutralizing activity of human anti-V3 monoclonal antibodies derived from the cells of individuals infected with non-B clades of human immunodeficiency virus type 1. J Virol. 2006;80(14):6865–72. doi: 10.1128/JVI.02202-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Xu JY, Karwowska S, Buchbinder A, Zolla-Pazner S. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J Immunol. 1993;150(2):635–43. [PubMed] [Google Scholar]
- Haynes BF, Ma B, Montefiori DC, Wrin T, Petropoulos CJ, Sutherland LL, Scearce RM, Denton C, Xia SM, Korber BT, Liao HX. Analysis of HIV-1 subtype B third variable region peptide motifs for induction of neutralizing antibodies against HIV-1 primary isolates. Virology. 2006;345(1):44–55. doi: 10.1016/j.virol.2005.08.042. [DOI] [PubMed] [Google Scholar]
- Haynes BF, Montefiori DC. Aiming to induce broadly reactive neutralizing antibody responses with HIV-1 vaccine candidates. Expert Rev Vaccines. 2006;5(4):579–95. doi: 10.1586/14760584.5.4.579. [DOI] [PubMed] [Google Scholar]
- Hoffman TL, LaBranche CC, Zhang W, Canziani G, Robinson J, Chaiken I, Hoxie JA, Doms RW. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc Natl Acad Sci U S A. 1999;96(11):6359–64. doi: 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310(5750):1025–8. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javaherian K, Langlois AJ, LaRosa GJ, Profy AT, Bolognesi DP, Herlihy WC, Putney SD, Matthews TJ. Broadly neutralizing antibodies elicited by the hypervariable neutralizing determinant of HIV-1. Science. 1990;250(4987):1590–3. doi: 10.1126/science.1703322. [DOI] [PubMed] [Google Scholar]
- Krachmarov C, Pinter A, Honnen WJ, Gorny MK, Nyambi PN, Zolla-Pazner S, Kayman SC. Antibodies that are cross-reactive for human immunodeficiency virus type 1 clade a and clade B v3 domains are common in patient sera from Cameroon, but their neutralization activity is usually restricted by epitope masking. J Virol. 2005;79(2):780–90. doi: 10.1128/JVI.79.2.780-790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krachmarov CP, Honnen WJ, Kayman SC, Gorny MK, Zolla-Pazner S, Pinter A. Factors determining the breadth and potency of neutralization by V3-specific human monoclonal antibodies derived from subjects infected with clade A or clade B strains of human immunodeficiency virus type 1. J Virol. 2006;80(14):7127–35. doi: 10.1128/JVI.02619-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krachmarov CP, Kayman SC, Honnen WJ, Trochev O, Pinter A. V3-specific polyclonal antibodies affinity purified from sera of infected humans effectively neutralize primary isolates of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2001;17(18):1737–48. doi: 10.1089/08892220152741432. [DOI] [PubMed] [Google Scholar]
- Kraft Z, Derby NR, McCaffrey RA, Niec R, Blay WM, Haigwood NL, Moysi E, Saunders CJ, Wrin T, Petropoulos CJ, McElrath MJ, Stamatatos L. Macaques infected with a CCR5-tropic simian/human immunodeficiency virus (SHIV) develop broadly reactive anti-HIV neutralizing antibodies. J Virol. 2007;81(12):6402–11. doi: 10.1128/JVI.00424-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PW, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420(6916):678–82. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–59. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laal S, Burda S, Gorny MK, Karwowska S, Buchbinder A, Zolla-Pazner S. Synergistic neutralization of human immunodeficiency virus type 1 by combinations of human monoclonal antibodies. J Virol. 1994;68(6):4001–8. doi: 10.1128/jvi.68.6.4001-4008.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letvin NL, Robinson S, Rohne D, Axthelm MK, Fanton JW, Bilska M, Palker TJ, Liao HX, Haynes BF, Montefiori DC. Vaccine-elicited V3 loop-specific antibodies in rhesus monkeys and control of a simian-human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate envelope. J Virol. 2001;75(9):4165–75. doi: 10.1128/JVI.75.9.4165-4175.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Baba TW, Sodroski J, Zolla-Pazner S, Gorny MK, Robinson J, Posner MR, Katinger H, Barbas CF, Burton DR, Chou TC, Ruprecht RM. Synergistic neutralization of a chimeric SIV/HIV type 1 virus with combinations of human anti-HIV type 1 envelope monoclonal antibodies or hyperimmune globulins. AIDS Res Hum Retroviruses. 1997;13(8):647–56. doi: 10.1089/aid.1997.13.647. [DOI] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human Immunodeficiency Virus Type 1 env Clones from Acute and Early Subtype B Infections for Standardized Assessments of Vaccine-Elicited Neutralizing Antibodies. J Virol. 2005;79(16):10108–25. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, Decker JM, Li Y, Salazar MG, Polonis VR, Mlisana K, Karim SA, Hong K, Greene KM, Bilska M, Zhou J, Allen S, Chomba E, Mulenga J, Vwalika C, Gao F, Zhang M, Korber BT, Hunter E, Hahn BH, Montefiori DC. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J Virol. 2006;80(23):11776–90. doi: 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Putney SD, Robinson HL. Human immunodeficiency virus type 1 entry into T cells: more-rapid escape from an anti-V3 loop than from an antireceptor antibody. J Virol. 1992;66(4):2547–50. doi: 10.1128/jvi.66.4.2547-2550.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusso P, Earl PL, Sironi F, Santoro F, Ripamonti C, Scarlatti G, Longhi R, Berger EA, Burastero SE. Cryptic nature of a conserved, CD4-inducible V3 loop neutralization epitope in the native envelope glycoprotein oligomer of CCR5-restricted, but not CXCR4-using, primary human immunodeficiency virus type 1 strains. J Virol. 2005;79(11):6957–68. doi: 10.1128/JVI.79.11.6957-6968.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Snyder SW, Weislow OS, Belay SM, Belshe RB, Schwartz DH, Clements ML, Dolin R, Graham BS, Gorse GJ, Keefer MC, McElrath MJ, Walker MC, Wagner KF, McNeil JG, McCutchan FE, Burke DS. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. The National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. J Infect Dis. 1996;173(2):340–8. doi: 10.1093/infdis/173.2.340. [DOI] [PubMed] [Google Scholar]
- Mbah HA, Burda S, Gorny MK, Williams C, Revesz K, Zolla-Pazner S, Nyambi PN. Effect of soluble CD4 on exposure of epitopes on primary, intact, native human immunodeficiency virus type 1 virions of different genetic clades. J Virol. 2001;75(16):7785–8. doi: 10.1128/JVI.75.16.7785-7788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeating JA, Cordell J, Dean CJ, Balfe P. Synergistic interaction between ligands binding to the CD4 binding site and V3 domain of human immunodeficiency virus type I gp120. Virology. 1992;191(2):732–42. doi: 10.1016/0042-6822(92)90249-o. [DOI] [PubMed] [Google Scholar]
- Montefiori DC, Graham BS, Zhou J, Bucco RA, Schwartz DH, Cavacini LA, Posner MR. V3-specific neutralizing antibodies in sera from HIV-1 gp160-immunized volunteers block virus fusion and act synergistically with human monoclonal antibody to the conformation-dependent CD4 binding site of gp120. NIH-NIAID AIDS Vaccine Clinical Trials Network. J Clin Invest. 1993;92(2):840–7. doi: 10.1172/JCI116658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, Cao Y, Qing L, Sattentau QJ, Pyati J, Koduri R, Robinson J, Barbas CF, Burton DR, Ho DD. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J Virol. 1995a;69(1):101–9. doi: 10.1128/jvi.69.1.101-109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J Virol. 1996;70(3):1863–72. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, Trkola A, Korber B, Boots LJ, Kessler JA, 2nd, McCutchan FE, Mascola J, Ho DD, Robinson J, Conley AJ. A human monoclonal antibody to a complex epitope in the V3 region of gp120 of human immunodeficiency virus type 1 has broad reactivity within and outside clade B. J Virol. 1995b;69(1):122–30. doi: 10.1128/jvi.69.1.122-130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nara PL, Smit L, Dunlop N, Hatch W, Merges M, Waters D, Kelliher J, Gallo RC, Fischinger PJ, Goudsmit J. Emergence of viruses resistant to neutralization by V3-specific antibodies in experimental human immunodeficiency virus type 1 IIIB infection of chimpanzees. J Virol. 1990;64(8):3779–91. doi: 10.1128/jvi.64.8.3779-3791.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyambi PN, Gorny MK, Bastiani L, van der Groen G, Williams C, Zolla-Pazner S. Mapping of epitopes exposed on intact human immunodeficiency virus type 1 (HIV-1) virions: a new strategy for studying the immunologic relatedness of HIV-1. J Virol. 1998;72(11):9384–91. doi: 10.1128/jvi.72.11.9384-9391.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz GM, Nixon DF, Trkola A, Binley J, Jin X, Bonhoeffer S, Kuebler PJ, Donahoe SM, Demoitie MA, Kakimoto WM, Ketas T, Clas B, Heymann JJ, Zhang L, Cao Y, Hurley A, Moore JP, Ho DD, Markowitz M. HIV-1-specific immune responses in subjects who temporarily contain virus replication after discontinuation of highly active antiretroviral therapy. J Clin Invest. 1999;104(6):R13–8. doi: 10.1172/JCI7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palker TJ, Clark ME, Langlois AJ, Matthews TJ, Weinhold KJ, Randall RR, Bolognesi DP, Haynes BF. Type-specific neutralization of the human immunodeficiency virus with antibodies to env-encoded synthetic peptides. Proc Natl Acad Sci U S A. 1988;85(6):1932–6. doi: 10.1073/pnas.85.6.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantophlet R, Aguilar-Sino RO, Wrin T, Cavacini LA, Burton DR. Analysis of the neutralization breadth of the anti-V3 antibody F425-B4e8 and reassessment of its epitope fine specificity by scanning mutagenesis. Virology. 2007;364(2):441–53. doi: 10.1016/j.virol.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantophlet R, Burton DR. GP120: target for neutralizing HIV-1 antibodies. Annu Rev Immunol. 2006;24:739–69. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- Park EJ, Vujcic LK, Anand R, Theodore TS, Quinnan GV., Jr Mutations in both gp120 and gp41 are responsible for the broad neutralization resistance of variant human immunodeficiency virus type 1 MN to antibodies directed at V3 and non-V3 epitopes. J Virol. 1998;72(9):7099–107. doi: 10.1128/jvi.72.9.7099-7107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MB, Hoffman NG, Swanstrom R. Subtype-specific conformational differences within the V3 region of subtype B and subtype C human immunodeficiency virus type 1 Env proteins. J Virol. 2008;82(2):903–16. doi: 10.1128/JVI.01444-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter A, Honnen WJ, Tilley SA. Conformational changes affecting the V3 and CD4-binding domains of human immunodeficiency virus type 1 gp120 associated with env processing and with binding of ligands to these sites. J Virol. 1993;67(9):5692–7. doi: 10.1128/jvi.67.9.5692-5697.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts BJ, Field KG, Wu Y, Posner M, Cavacini L, White-Scharf M. Synergistic inhibition of HIV-1 by CD4 binding domain reagents and V3-directed monoclonal antibodies. Virology. 1993;197(1):415–9. doi: 10.1006/viro.1993.1604. [DOI] [PubMed] [Google Scholar]
- Profy AT, Salinas PA, Eckler LI, Dunlop NM, Nara PL, Putney SD. Epitopes recognized by the neutralizing antibodies of an HIV-1-infected individual. J Immunol. 1990;144(12):4641–7. [PubMed] [Google Scholar]
- Rosen O, Chill J, Sharon M, Kessler N, Mester B, Zolla-Pazner S, Anglister J. Induced fit in HIV-neutralizing antibody complexes: evidence for alternative conformations of the gp120 V3 loop and the molecular basis for broad neutralization. Biochemistry. 2005;44(19):7250–8. doi: 10.1021/bi047387t. [DOI] [PubMed] [Google Scholar]
- Salzwedel K, Smith ED, Dey B, Berger EA. Sequential CD4-coreceptor interactions in human immunodeficiency virus type 1 Env function: soluble CD4 activates Env for coreceptor-dependent fusion and reveals blocking activities of antibodies against cryptic conserved epitopes on gp120. J Virol. 2000;74(1):326–33. doi: 10.1128/jvi.74.1.326-333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber M, Petersen H, Wachsmuth C, Muller H, Hufert FT, Schmitz H. Antibodies of symptomatic human immunodeficiency virus type 1-infected individuals are directed to the V3 domain of noninfectious and not of infectious virions present in autologous serum. J Virol. 1994;68(6):3908–16. doi: 10.1128/jvi.68.6.3908-3916.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon M, Kessler N, Levy R, Zolla-Pazner S, Gorlach M, Anglister J. Alternative conformations of HIV-1 V3 loops mimic beta hairpins in chemokines, suggesting a mechanism for coreceptor selectivity. Structure. 2003;11(2):225–36. doi: 10.1016/s0969-2126(03)00011-x. [DOI] [PubMed] [Google Scholar]
- Shu Y, Winfrey S, Yang ZY, Xu L, Rao SS, Srivastava I, Barnett SW, Nabel GJ, Mascola JR. Efficient protein boosting after plasmid DNA or recombinant adenovirus immunization with HIV-1 vaccine constructs. Vaccine. 2007;25(8):1398–408. doi: 10.1016/j.vaccine.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spenlehauer C, Saragosti S, Fleury HJ, Kirn A, Aubertin AM, Moog C. Study of the V3 loop as a target epitope for antibodies involved in the neutralization of primary isolates versus T-cell-line-adapted strains of human immunodeficiency virus type 1. J Virol. 1998;72(12):9855–64. doi: 10.1128/jvi.72.12.9855-9864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatos NM, Mascola JR, Kalyanaraman VS, Louder MK, Frampton LM, Birx DL, VanCott TC. Neutralizing antibodies from the sera of human immunodeficiency virus type 1-infected individuals bind to monomeric gp120 and oligomeric gp140. J Virol. 1998;72(12):9656–67. doi: 10.1128/jvi.72.12.9656-9667.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield RL, Gorny MK, Williams C, Zolla-Pazner S, Wilson IA. Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D. Structure (Camb) 2004;12(2):193–204. doi: 10.1016/j.str.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Stanfield RL, Gorny MK, Zolla-Pazner S, Wilson IA. Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity. J Virol. 2006;80(12):6093–105. doi: 10.1128/JVI.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan N, Sun Y, Binley J, Lee J, Barbas CF, Parren PW, Burton DR, Sodroski J. Determinants of human immunodeficiency virus type 1 envelope glycoprotein activation by soluble CD4 and monoclonal antibodies. J Virol. 1998a;72(8):6332–8. doi: 10.1128/jvi.72.8.6332-6338.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J, Sodroski J. CD4-Induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J Virol. 1998b;72(6):4694–703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suphaphiphat P, Thitithanyanont A, Paca-Uccaralertkun S, Essex M, Lee TH. Effect of amino acid substitution of the V3 and bridging sheet residues in human immunodeficiency virus type 1 subtype C gp120 on CCR5 utilization. J Virol. 2003;77(6):3832–7. doi: 10.1128/JVI.77.6.3832-3837.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thali M, Furman C, Wahren B, Posner M, Ho DD, Robinson J, Sodroski J. Cooperativity of neutralizing antibodies directed against the V3 and CD4 binding regions of the human immunodeficiency virus gp120 envelope glycoprotein. J Acquir Immune Defic Syndr. 1992;5(6):591–9. [PubMed] [Google Scholar]
- Tilley SA, Honnen WJ, Racho ME, Chou TC, Pinter A. Synergistic neutralization of HIV-1 by human monoclonal antibodies against the V3 loop and the CD4-binding site of gp120. AIDS Res Hum Retroviruses. 1992;8(4):461–7. doi: 10.1089/aid.1992.8.461. [DOI] [PubMed] [Google Scholar]
- Vancott TC, Polonis VR, Loomis LD, Michael NL, Nara PL, Birx DL. Differential role of V3-specific antibodies in neutralization assays involving primary and laboratory-adapted isolates of HIV type 1. AIDS Res Hum Retroviruses. 1995;11(11):1379–91. doi: 10.1089/aid.1995.11.1379. [DOI] [PubMed] [Google Scholar]
- Vijh-Warrier S, Pinter A, Honnen WJ, Tilley SA. Synergistic neutralization of human immunodeficiency virus type 1 by a chimpanzee monoclonal antibody against the V2 domain of gp120 in combination with monoclonal antibodies against the V3 loop and the CD4-binding site. J Virol. 1996;70(7):4466–73. doi: 10.1128/jvi.70.7.4466-4473.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrin T, Loh TP, Vennari JC, Schuitemaker H, Nunberg JH. Adaptation to persistent growth in the H9 cell line renders a primary isolate of human immunodeficiency virus type 1 sensitive to neutralization by vaccine sera. J Virol. 1995;69(1):39–48. doi: 10.1128/jvi.69.1.39-48.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrin T, Nunberg JH. HIV-1MN recombinant gp120 vaccine serum, which fails to neutralize primary isolates of HIV-1, does not antagonize neutralization by antibodies from infected individuals. Aids. 1994;8(11):1622–3. doi: 10.1097/00002030-199411000-00017. [DOI] [PubMed] [Google Scholar]
- Wu L, Yang ZY, Xu L, Welcher B, Winfrey S, Shao Y, Mascola JR, Nabel GJ. Cross-clade recognition and neutralization by the V3 region from clade C human immunodeficiency virus-1 envelope. Vaccine. 2006;24(23):4995–5002. doi: 10.1016/j.vaccine.2006.03.083. [DOI] [PubMed] [Google Scholar]
- Wu Z, Kayman SC, Honnen W, Revesz K, Chen H, Vijh-Warrier S, Tilley SA, McKeating J, Shotton C, Pinter A. Characterization of neutralization epitopes in the V2 region of human immunodeficiency virus type 1 gp120: role of glycosylation in the correct folding of the V1/V2 domain. J Virol. 1995;69(4):2271–8. doi: 10.1128/jvi.69.4.2271-2278.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280(5371):1884–8. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- Xiang SH, Doka N, Choudhary RK, Sodroski J, Robinson JE. Characterization of CD4-induced epitopes on the HIV type 1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res Hum Retroviruses. 2002;18(16):1207–17. doi: 10.1089/08892220260387959. [DOI] [PubMed] [Google Scholar]
- Yang ZY, Chakrabarti BK, Xu L, Welcher B, Kong WP, Leung K, Panet A, Mascola JR, Nabel GJ. Selective modifications of variable loops alters tropism and enhances immunogenicity of HIV-1 envelope. J Virol. 2004;78:4029–4036. doi: 10.1128/JVI.78.8.4029-4036.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445(7129):732–7. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S. Identifying epitopes of HIV-1 that induce protective antibodies. Nat Rev Immunol. 2004;4(3):199–210. doi: 10.1038/nri1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S. Improving on nature: focusing the immune response on the V3 loop. Hum Antibodies. 2005;14(3–4):69–72. [PubMed] [Google Scholar]
- Zolla-Pazner S, Cohen SS, Krachmarov C, Wang S, Pinter A, Lu S. Focusing the immune response on the V3 loop, a neutralizing epitope of the HIV-1 gp120 envelope. Virology. 2008;372(2):233–46. doi: 10.1016/j.virol.2007.09.024. [DOI] [PubMed] [Google Scholar]
- Zolla-Pazner S, Zhong P, Revesz K, Volsky B, Williams C, Nyambi P, Gorny MK. The cross-clade neutralizing activity of a human monoclonal antibody is determined by the GPGR V3 motif of HIV type 1. AIDS Res Hum Retroviruses. 2004;20(11):1254–8. doi: 10.1089/aid.2004.20.1254. [DOI] [PubMed] [Google Scholar]
- Zwart G, Wolfs TF, Valk M, Van der Hoek L, Kuiken CL, Goudsmit J. Characterization of the specificity of the human antibody response to the V3 neutralization domain of HIV-1. AIDS Res Hum Retroviruses. 1992;8(11):1897–908. doi: 10.1089/aid.1992.8.1897. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.