Summary
Fusion of myoblasts is essential for the formation of multi-nucleated muscle fibers. However, the identity of myogenic proteins that directly govern this fusion process has remained elusive. Here, we discovered a muscle-specific membrane protein, named Myomaker, that controls myoblast fusion. Myomaker is expressed on the cell surface of myoblasts during fusion and is down-regulated thereafter. Over-expression of Myomaker in myoblasts dramatically enhances fusion and genetic disruption of Myomaker in mice causes perinatal death due to an absence of multi-nucleated muscle fibers. Remarkably, forced expression of Myomaker in fibroblasts promotes fusion with myoblasts, demonstrating the direct participation of this protein in the fusion process. Pharmacologic perturbation of the actin cytoskeleton abolishes the activity of Myomaker, consistent with prior studies implicating actin dynamics in myoblast fusion. These findings reveal a long-sought myogenic fusion protein both necessary and sufficient for mammalian myoblast fusion and provide new insights into the molecular underpinnings of muscle formation.
Introduction
Myoblast fusion is a complex and tightly controlled process required for the formation of skeletal muscle fibers1. The fusion process must be highly cell type-specific to ensure that fusogenic myoblasts do not form syncytia with non-muscle cell types. While the transcriptional mechanisms governing skeletal muscle development have been elucidated in detail2–5, the mechanisms that coordinate myoblast fusion remain poorly understood, and no muscle-specific protein that directly regulates myoblast fusion has been identified in any organism6,7. In contrast, numerous proteins involved in cell-cell adhesion and actin dynamics have been implicated in myoblast fusion8–12. However, none of these proteins are muscle-specific, necessary and sufficient for mammalian myoblast fusion, suggesting that muscle-specific components of this process remain to be discovered. Here we describe the discovery of a muscle-specific membrane protein called Myomaker that is transiently expressed during myoblast fusion and is both necessary and sufficient to drive merger of plasma membranes in vivo and in vitro.
Discovery and regulation of Myomaker
To search for novel skeletal muscle regulatory genes, we interrogated the NCBI UniGene database for genes with expression profiles similar to those of Myod and Myogenin, which encode important muscle-specific transcription factors13,14. Among the genes identified in this screen, was Transmembrane protein 8c (Tmem8c), which had not been previously studied. Based on the observations described below, we named this gene Myomaker.
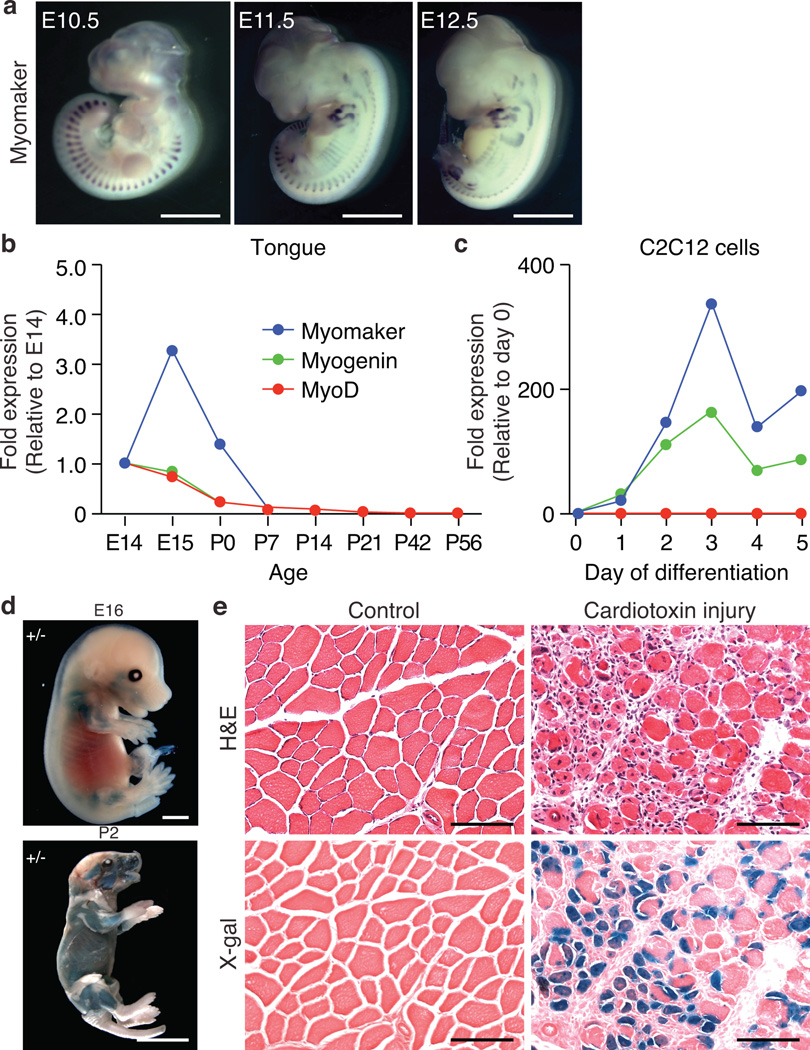
During mouse embryogenesis, Myomaker is robustly expressed in the myotomal compartment of the somites, and later is expressed in limb buds and axial skeletal muscles (Fig. 1a and Supplemental Figure 1a). Expression of Myomaker in the myotomes coincides with expression of other known muscle transcripts, such as Myogenin and M-cadherin (Supplemental Figure 1a). Myomaker mRNA is expressed in skeletal muscle of the tongue and is subsequently down-regulated upon completion of muscle formation, similar to the expression pattern of Myod and Myogenin (Fig. 1b). Myomaker expression was not detected in tissues other than skeletal muscle in E19 embryos (Supplementary Figure 1b and 1c). In the C2C12 skeletal muscle cell line, Myomaker mimics Myogenin expression, increasing sharply during differentiation and fusion (Fig. 1c).
Figure 1. Muscle-specific expression of Myomaker.
a, In situ hybridization for Myomaker in WT embryos illustrates muscle specificity. b, qPCR for Myomaker, Myogenin, and MyoD on tongues at the indicated ages shows down-regulation after myogenesis. c, Gene expression during differentiation of C2C12 myoblasts. d, X-gal staining on E16 and P2 Myomaker+/− (Myomaker-LacZ) mice confirms expression in all skeletal muscles. e, Cardiotoxin-injured and X-gal stained tibialis anterior (TA) muscle from 6 week old Myomaker+/− mice shows the re-activation of Myomaker. Serial H&E stained sections indicate muscle injury. Control represents uninjured contralateral TA. Scale bars: a, d, 2 mm; e 200 µm.
To begin to assess the function of Myomaker in skeletal muscle, we obtained ES cells that contained a LacZ-Neo cassette in intron 1 of the Myomaker locus (Supplementary Figure 2a). In this allele, exon 1 of Myomaker is spliced to lacZ, preventing expression of a functional Myomaker transcript. We refer to mice heterozygous and homozygous for the Myomaker-lacZ allele as Myomaker+/− and Myomaker−/− mice, respectively. X-gal staining of Myomaker+/− mice showed expression of the targeted lacZ allele specifically in skeletal muscle, and not in other muscle tissues or non-muscle tissues (Fig. 1d and Supplementary Figure 2b and 2c). Like the endogenous Myomaker gene, skeletal muscle expression of the Myomaker-lacZ allele declined postnatally (Supplementary Figure 2d).
Adult skeletal muscle regenerates in response to damage, due to the activation of satellite cells, which fuse with residual muscle fibers4,5. We tested whether Myomaker expression is re-activated during adult muscle regeneration by inducing muscle injury in adult mice. Expression of the Myomaker-LacZ allele and Myomaker mRNA and was strongly induced in regenerating muscle after cardiotoxin injury (Fig. 1e and Supplementary Figure 2e). We conclude that Myomaker is expressed specifically in skeletal muscle during embryogenesis and adult muscle regeneration.
Genetic loss of Myomaker prevents skeletal muscle formation
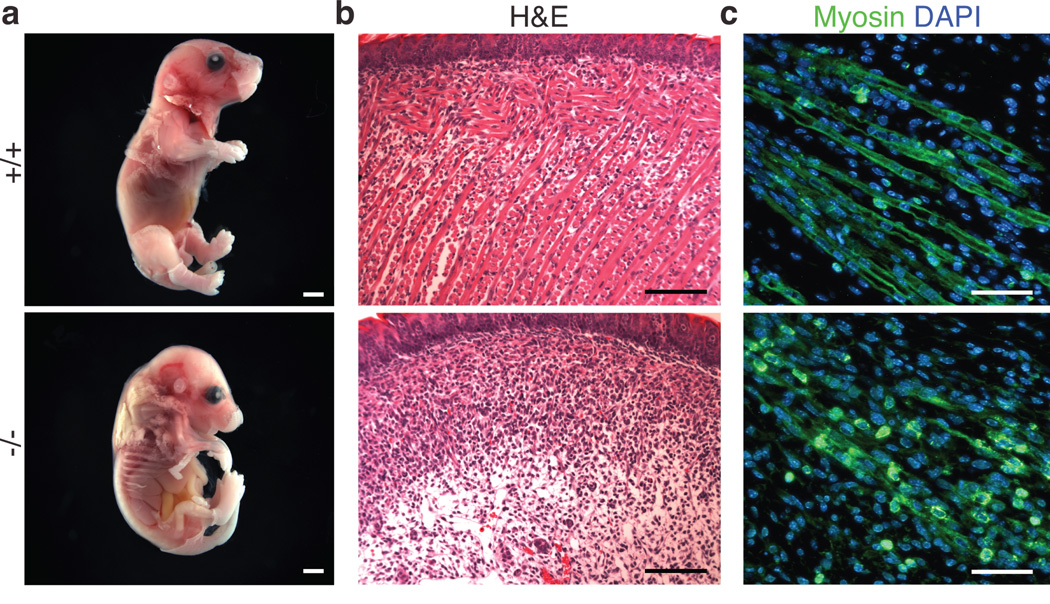
We generated Myomaker−/− mice by interbreeding of heterozygous mice. Myomaker transcripts were absent in skeletal muscle of Myomaker−/− mice, confirming that the targeting strategy created a null allelle (Supplementary Figure 2f). Myomaker−/− mice were observed at normal Mendelian ratios at E15 and E17.5, however we failed to detect any live Myomaker−/− mice at P7, suggesting earlier lethality due to muscle dysfunction (Supplementary Figure 2g). Full-term Myomaker−/− embryos were alive, as their hearts were beating, but were paralyzed and kyphotic with flaccid limbs, hallmarks of skeletal muscle deficiency (Fig. 2a). Strikingly, no semblance of differentiated muscle tissue was present in the trunk, limbs, or head of Myomaker−/− animals (Fig. 2b and Supplementary Figure 3a and 3b).
Figure 2. Myomaker is essential for skeletal muscle development.
a, Full term WT (+/+) and Myomaker−/− embryos were dissected and skinned to illustrate the lack of muscle surrounding Myomaker−/− limbs. b, Paraffin sectioning and H&E staining on tongues reveal a lack of muscle fibers in E17.5 Myomaker−/− embryos. c, Longitudinal sections of E14 hindlimb muscles stained with a myosin antibody to determine the multi-nucleation of the muscle cells. WT limbs exhibit myofibers containing multiple nuclei, which are absent in Myomaker−/− sections. Scale bars: a, 2 mm; b, 100 µm; c, 40 µm.
Muscle formation requires myoblast specification, migration, differentiation, and fusion2–5. In principle, dysfunction of one or more of these processes could contribute to lethality and lack of muscle formation in Myomaker−/− embryos. To begin to define the mechanistic actions of Myomaker, we tested the functionality of these processes. The muscle-specific transcription factors, MyoD and Myogenin, were expressed normally in Myomaker−/− embryos (Supplementary Figure 3c and 3d), suggesting that specification of the skeletal muscle lineage occurred normally in the absence of Myomaker.
Muscle tissues were present in Myomaker−/− embryos, indicating that muscle precursor cells were organized appropriately in the absence of Myomaker (Supplementary Figure 3e). Desmin, a marker of muscle cells, was expressed comparably in Myomaker−/− and wild-type (WT or +/+) forelimbs, confirming that myoblast migration was unaltered (Supplementary Figure 3f). These findings suggested Myomaker functions after myoblast specification and migration. Longitudinal sections through hindlimb muscles of Myomaker−/− embryos at E14 revealed the expression of myosin, a muscle differentiation marker, but an absence of multi-nucleated myofibers (Fig. 2c). These findings imply that Myomaker−/− myoblasts can activate muscle-specific gene expression and differentiate, but lack the ability to fuse.
Myomaker−/− muscle tissues contained only mononucleated cells, however, the cell number was clearly reduced in each muscle analyzed. One possible explanation for this decrease is cell death, which has previously been associated with a failure to fuse15,16. Indeed, TUNEL staining revealed increased apoptotic nuclei in muscle forming regions of Myomaker−/− mice, suggesting that fusion defective myoblasts are non-viable (Supplementary Figure 3g).
Myomaker controls myoblast fusion
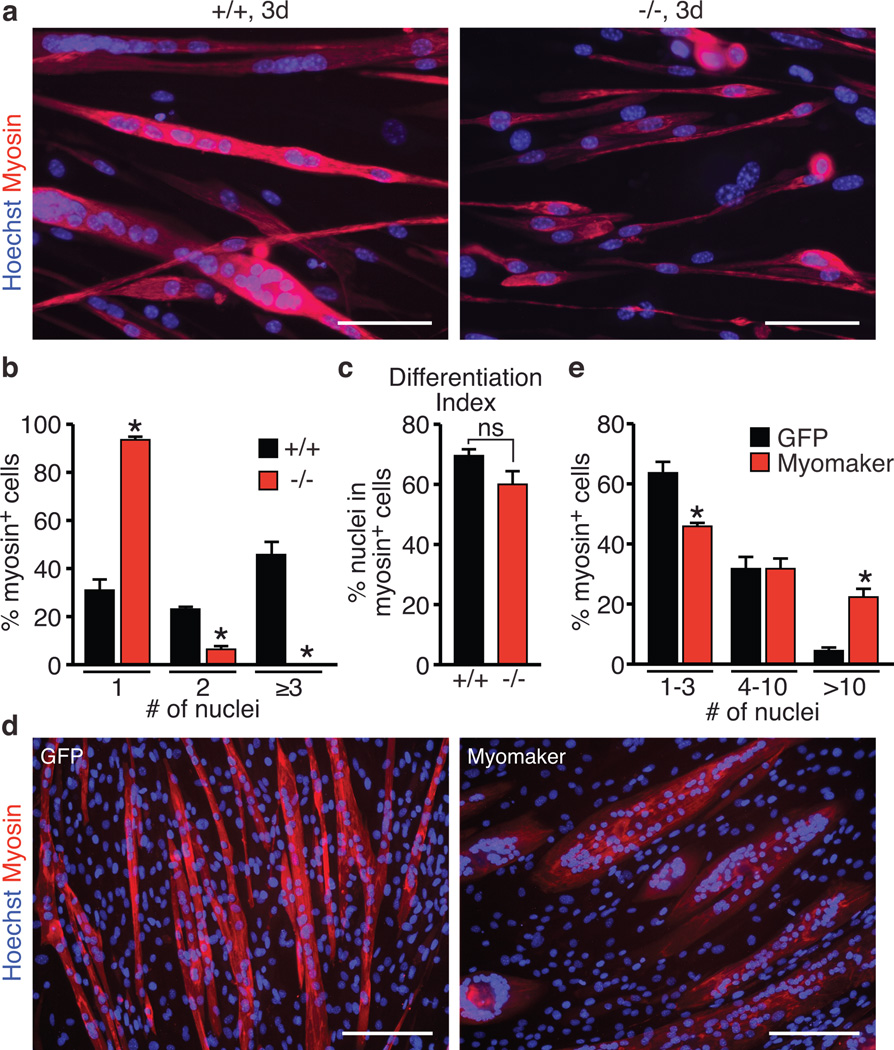
To definitively confirm that Myomaker functions in myoblast fusion, we employed multiple in vitro differentiation assays using primary myoblasts and the C2C12 muscle cell line. First, we isolated myoblasts from WT and Myomaker−/− embryos and after 3 days of differentiation, WT myoblasts formed extensive myotubes containing many nuclei (Fig. 3a). In contrast, the vast majority of Myomaker−/− myoblasts remained mono-nucleated, with only a small percentage forming bi-nucleated myosin positive cells (Fig. 3a and 3b and Supplementary Figure 4a). Quantification of the differentation index revealed no differences in the ability of Myomaker−/− myoblasts to express myosin, however the fusion index was dramatically reduced compared to WT myoblasts (Fig. 3c and Supplementary Figure 4b), even when plated for prolonged periods at higher density than WT myoblasts, indicating that fusion was blocked rather than simply delayed (Supplementary Figure 4c). We conclude that the lack of muscle formation in Myomaker−/− embryos is due to a block of myoblast fusion, representing the cellular mechanism of Myomaker function.
Figure 3. Control of myoblast fusion by Myomaker.
a, Myoblasts from WT (+/+) and Myomaker−/− E17 embryos were differentiated for 3 days, and stained for myosin and a nuclear stain (Hoechst). Myomaker−/− myoblasts failed to fuse. b, Quantitation of the number of nuclei present in a myosin+ cell indicates Myomaker−/− myoblasts cannot form myotubes with three or more nuclei. c, Differentiation index, calculated as the percentage of nuclei in myosin+ cells, indicated null myoblasts can activate the myogenic program. d, C2C12 cells infected with a retrovirus encoding GFP or Myomaker were induced to differentiate for 4 days then stained with a myosin antibody and Hoechst (nuclei). e, Quantitation of the percentage of myosin+ cells that contained the indicated number of nuclei. Quantification was performed after 3 days of differentiation in (b), (c), and after 4 days in (e). Scale bars: a, 100 µm e, 200 µm. Data are presented as mean ± SEM from three independent experiments. * P < 0.05 compared to +/+ in b, c or GFP-infected cells in e. ns in c is not statistically significant.
To test whether Myomaker was a limiting factor in myoblast fusion, we infected C2C12 cells with a Myomaker retrovirus one day prior to differentiation and assessed the consequences on myoblast fusion. Myomaker over-expression caused a dramatic increase in fusion after 4 days of differentiation (Fig. 3d). The kinetics of induction of myogenin and myosin, and maximal levels of expression of the terminal differentiation genes (Myogenin, Ckm, and Myh4) were comparable in Myomaker-infected cells and cells infected with a GFP control virus (Supplementary Figure 4d and 4e). Despite no differences in expression of muscle differentiation factors, we observed a robust increase in the appearance of myotubes with multiple nuclei in the cultures infected with Myomaker, further indicating that Myomaker functions specifically in myoblast fusion and does not regulate differentiation per se (Supplementary Figure 4f). Quantitation of the fusion index and the number of nuclei per myotube indicated a robust activity of Myomaker to increase the fusion capability of these cells (Fig. 3e and Supplementary Figure 3g and 3h). Furthermore, through live cell imaging, we visualized myotube-myotube fusion in Myomaker-infected cells (Supplementary Movie 1). These data demonstrate that Myomaker is sufficient to enhance C2C12 myoblast fusion.
Myomaker is 221 amino acids in length and is highly conserved across vertebrate organisms, ranging from fish to humans (Supplementary Figure 5a). Analysis of the hydrophobicity of Myomaker using a Kyte-Doolittle Plot revealed extensive regions of hydrophobic character, suggesting this protein may localize to a cellular membrane (Supplementary Figure 5b). Myomaker does not contain predicted N-glycosylation sites. At the C-terminus, Myomaker possesses a C-A-A-X motif, the consensus for isoprenylation, which mediates membrane association17. Myomaker shares limited homology to a family of putative transmembrane hydrolases, named the CREST family18, but it lacks a potentially critical histidine residue thought to be important for catalytic activity of hydrolases. The closest relative, Tmem8b, shares homology with Myomaker/Tmem8c in three hydrophobic domains, however Tmem8b is not muscle-specific and its forced expression in C2C12 cells did not promote fusion (data not shown). There is also a related protein in Drosophila, but it is more similar to Tmem8a and Tmem8b than to Myomaker/Tmem8c.
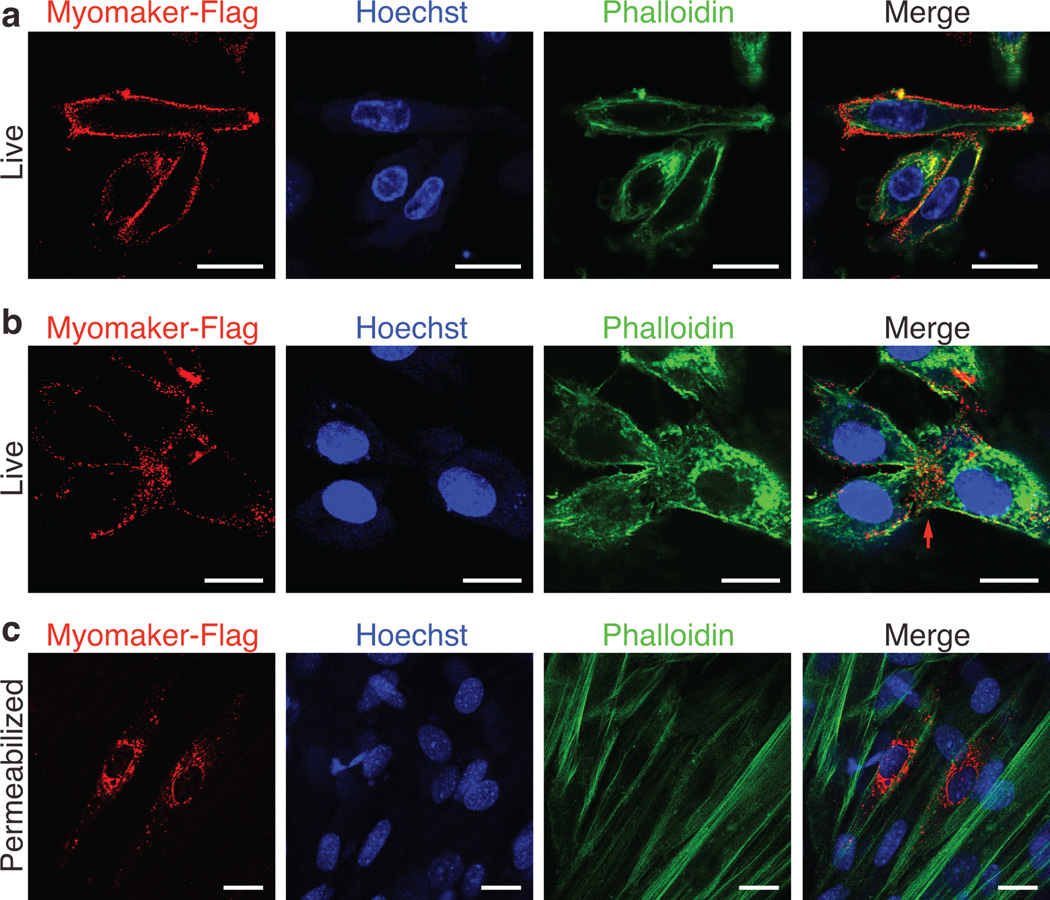
To analyze the cellular distribution of Myomaker, we engineered a Flag epitope after amino acid 61, in a region of the protein that would not be predicted to perturb the hydrophobic domains (Supplementary Figure 5b). The Flag-tagged Myomaker protein, referred to as Myomaker-Flag, was detected in whole cell lysates, by Flag western blots (Supplementary Figure 6a). Retroviral expression of Myomaker-Flag in C2C12 cells confirmed that insertion of the Flag epitope did not alter the function of Myomaker as assayed by its ability to robustly enhance myoblast fusion (Supplementary Figure 6b). Fractionation of C2C12 cells infected with Myomaker-Flag into membrane and cytosolic fractions, showed exclusive localization to the membrane fraction (Supplementary Figure 6c). Myomaker-Flag was readily detected on the surface of myoblasts, by staining live cells with a Flag antibody, a common method used to detect plasma membrane proteins19 (Fig. 4a). Moreover, in myoblast cultures undergoing fusion, Myomaker-Flag was detected at sites of cell-cell interaction (Fig. 4b). Immunocytochemistry of fixed and permeabilized C2C12 cells expressing Myomaker-Flag revealed intracellular vesicle localization of Myomaker-Flag, as expected for a membrane protein (Fig. 4c). Co-staining with intracellular organelle markers revealed some co-localization with endosomes and ER (Supplementary Figure 6d), suggesting that Myomaker transits through one or more intracellular membrane compartments.
Figure 4. Myomaker is expressed on the cell membrane of myoblasts.
a, C2C12 cells were infected with Myomaker-Flag and live cells were stained 2 days after differentiation with Flag antibody on ice. After Flag staining, cells were then fixed and permeabilized and stained with Phalloidin (F-actin) and Hoechst (nuclei) to illustrate cell membrane localization of Myomaker-Flag. b, Cells were stained as in (a) to visualize Myomaker-Flag in fusing cultures. The red arrow depicts sites of cell-cell interaction. f, Myomaker-Flag infected C2C12 cells were fixed, permeabilized, and stained with Flag antibody, Phalloidin, and Hoechst showing the vesicle localization of the intracellular protein. Scale bars: 20 µm.
Myoblast fusion requires actin-cytoskeletal reorganization15,16,20–23. Treating C2C12 cells with cytochalasin D and lantrunculin B, which perturb the cytoskeleton, completely blocked fusion in cells infected with GFP or Myomaker virus suggesting that actin nucleation is required for the fusogenic function of Myomaker (Supplementary Figure 7a). After cytochalasin D treatment, Myomaker-Flag was properly localized to the membrane, indicating that actin dynamics do not regulate transport of the protein to the cell surface (Supplementary Figure 7b).
Investigation of the fusogenic functions of Myomaker
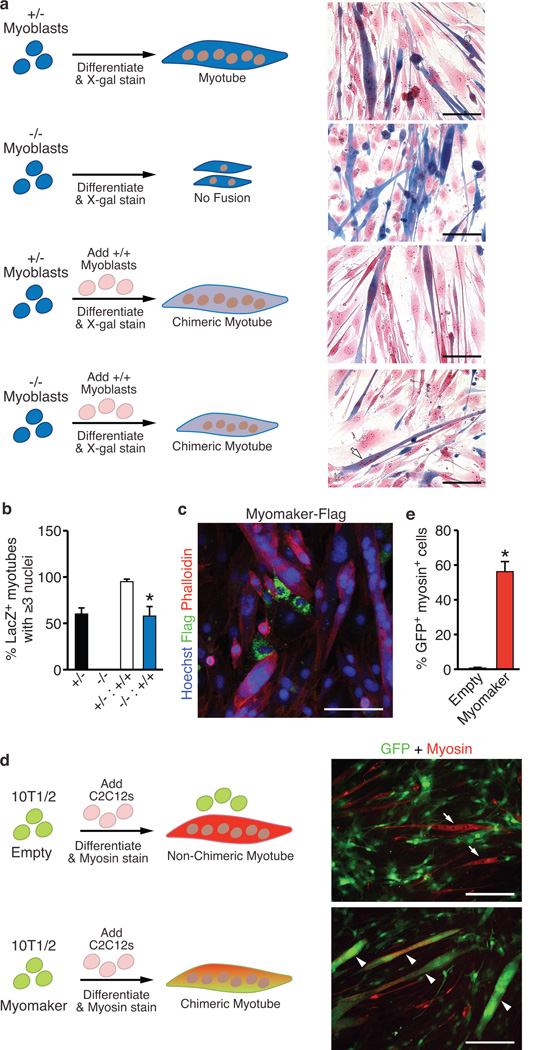
To further understand the mechanism of action of Myomaker, we performed cell-mixing experiments using primary myoblasts from WT, Myomaker+/− and Myomaker−/− embryos (Fig. 5a). After differentiation for 4 days, we visualized beta-galactosidase expression from the lacZ allele in Myomaker+/− and Myomaker−/− myoblasts to monitor fusion between different myoblast populations. As a co-stain, we used nuclear fast red, which stains a nucleus red and confers a pink appearance in the cytoplasm of cells. Myomaker+/− myoblasts formed multi-nucleated myotubes alone, without WT myoblasts, while Myomaker−/− myoblasts failed to fuse (Fig. 5a). Chimeric myotubes (blue/pink) were apparent in cultures containing WT and Myomaker+/− myoblasts, indicating fusion between these two myoblast populations (Fig. 5a). In cultures containing both WT and Myomaker−/− myoblasts, we observed myotubes containing LacZ staining eminating from Myomaker−/− myoblasts (Fig. 5a). Quantification of the percent of LacZ+ myotubes with 3 or more nuclei revealed that Myomaker−/− myoblasts could only form these structures in the presence of WT myoblasts (Fig. 5b). We conclude that a cell with a functional copy of Myomaker can fuse with a Myomaker−/− myoblast, suggesting that Myomaker is absolutely required on the surface of only one of the fusing muscle cells. We further investigated this possibility by analyzing expression of Myomaker-Flag in C2C12 cells and detected Myomaker-Flag in mononuclear C2C12 cells but not in previously fused multi-nucleated myotubes (Fig. 5c).
Figure 5. Myomaker participates in the myoblast membrane fusion reaction.
a, Myomaker+/− and Myomaker−/− myoblasts express LacZ, and were either plated alone or mixed with WT myoblasts, induced to differentiate for 4 days, and stained with X-gal and nuclear fast red to determine the amount of fusion. Myomaker+/− myoblasts, alone or in the presence of WT myoblasts fused normally, illustrated by myotubes with robust LacZ staining. Myomaker−/− myoblasts alone exhibited an inability to fuse. Addition of WT myoblasts to Myomaker−/− myoblasts resulted in chimeric myotubes (arrow) indicating fusion between the two cell populations. b, Quantitation of the percentage of LacZ+ myotubes containing ≥3 nuclei shows null myoblasts can only form myotubes with three or more nuclei in the presence of WT myoblasts. c, Phalloidin and Flag staining of C2C12 myoblasts after infection with Myomaker-Flag illustrates the lack of Flag staining in myotubes. d, 10T1/2 fibroblasts were infected with GFP-retrovirus and either Empty- or Myomaker-retrovirus and then mixed with C2C12 cells and differentiated. Myotube formation was monitored by myosin staining, and fusion of fibroblasts was determined by visualization of GFP in myosin+ myotubes. Myosin+ GFP+ myotubes (arrowheads) are evident in cultures containing Myomaker-infected fibroblasts, whereas myosin+ GFP− myotubes (arrows) were observed in Empty-infected cultures. e, Quantitation of the percentage of GFP+ fibroblasts, infected with Empty- or Myomaker-retrovirus, that fused to myosin+ myoblasts. Scale bars: a, 100 µm; c, 20 µm; d, 200 µm. Data are presented as mean ± SEM from three independent experiments. * P < 0.05 compared to −/− in b and compared to empty in e.
To determine whether over-expression of Myomaker could permit fusion of fibroblasts, a cell type that lacks fusion capability, we infected 10T1/2 fibroblasts with a GFP virus and either empty virus, as a control, or Myomaker virus and then mixed these fibroblasts with C2C12 cells (Fig. 5d). We did not detect fusion of GFP-empty virus-infected fibroblasts with myosin-positive cells, however GFP-Myomaker-infected fibroblasts robustly fused with C2C12 cells (Fig. 5d and Supplementary Movie 2). Quantitation of the myotubes expressing both GFP and myosin confirmed a striking ability of fibroblasts expressing Myomaker to fuse with myoblasts (Fig. 5e).
To control for the possibility that Myomaker-expressing fibroblasts were leaky and allowed GFP to diffuse into C2C12 myotubes, we designed a complementary cell mixing experiment in which we tracked fibroblast nuclei by labeling with BrdU, followed by mixing with dsRed-infected C2C12 cells (Supplementary Figure 8a). BrdU-positive nuclei from fibroblasts expressing Myomaker were detected within C2C12 myotubes, confirming that Myomaker expression was sufficient to direct the fusion of fibroblasts to myoblasts (Supplementary Figure 8a and 8b). Myomaker was not sufficient to induce fusion of fibroblasts in the absence of myoblasts. The finding that Myomaker can promote fusion of fibroblasts to myoblasts but cannot promote fibroblast-fibroblast fusion suggests that additional myoblast cell surface proteins are required for proper fusogenic engagement of the two membranes.
Discussion
There are multiple types of membrane fusion, including virus-cell fusion, intracellular vesicle fusion, and cell-cell fusion1. Similarities exist between different fusion mechanisms, but relatively little is known about cell-cell fusion compared to other fusion processes, especially with respect to the fusogenic proteins that directly merge intercellular membranes. Our findings identify Myomaker as a muscle-specific plasma membrane protein expressed specifically during times of myoblast fusion, and required for the formation of multinucleated myofibers. While surface glycoproteins, including cadherins, β-1 integrin, MOR23, and Adam128–12, have been shown to influence myoblast fusion, Myomaker is the only muscle-specific protein yet identified that is absolutely essential for myoblast fusion in vivo. The absence of multinucleated myofibers in Myomaker−/− mice demonstrates the requirement of this membrane protein for the formation of all skeletal muscles.
Myoblast fusion is a multistep process requiring intimate cell-cell interaction followed by membrane coalescence accompanied by actin-cytoskeletal dynamics that drive cell merger. Myomaker clearly participates in the membrane fusion reaction, as demonstrated by its ability to stimulate myoblast fusion and the fusion of fibroblasts to myoblasts. The inability of Myomaker alone to induce fusion of fibroblasts suggests it may require activation or additional myoblast proteins to exert its fusogenic activity, likely reflecting a requirement for close membrane apposition to allow membrane merger. Further evidence that additional myoblast proteins are required for fusion is our finding that WT myoblasts can fuse with Myomaker−/− myoblasts. The requirement for interactions between membrane proteins on opposite cells during myoblast fusion has been shown in zebrafish and Drosophila6,24, suggesting the molecular regulation of myoblast fusion differs from that of virus-cell fusion, which mainly requires the expression of a fusogenic protein25. Changes in the actin-cytoskeleton are required for cell-cell fusion26,27. Consistent with this paradigm, the activity of Myomaker is abolished by cytochalasin D and latrunculin B, which disrupt cytoskeletal events required for fusion, indicating that Myomaker depends on the cytoskeleton to exert its function.
The discovery of Myomaker as a potent myoblast fusion protein opens new opportunities to dissect this fundamental cellular process at a molecular level and to understand how myoblast fusion is perturbed during muscle disease. Moreover, the ability of Myomaker to drive fusion of nonmuscle cells with muscle cells represents an interesting strategy for enhancing muscle repair.
Methods
Generation of Myomaker−/− mice
The Myomaker mouse strain used for this research project was created from ES cell Tmem8c clone (EPD0626_5_C12) obtained from KOMP Repository (www.KOMP.org) and generated by the Wellcome Trust Sanger Institute29. This clone was injected into 3.5-day-old C57BL/6 blastocysts by the Transgenic Core Facility at University of Texas Southwestern Medical Center. High-percentage chimeric male mice were bred to C57BL/6 females to achieve germline transmission of the targeted allele. Myomaker+/− mice were intercrossed to generate Myomaker−/− mice. All experimental procedures involving animals in this study were reviewed and approved by the University of Texas Southwestern Medical Center’s Institutional Animal Care and Use Committee.
Quantitative real-time PCR (qPCR)
Total RNA was extracted from either mouse tissue or cultured cells with TRIZOL (Invitrogen) and cDNA synthesized using Superscript III reverse transcriptase with random hexamer primers (Invitrogen). Gene expression was assessed using standard qPCR approaches with either Power Sybr Green or Taqman Master Mix (Applied Biosystems). Analysis was performed on a 7900HT Fast Real-Time PCR Machine (Applied Biosystems) with the following Sybr primers: Myomaker-F: 5’-ATCGCTACCAAGAGGCGTT-3’, Myomaker-R: 5’-CACAGCACAGACAAACCAGG-3’. Taqman probes for Myogenin, MyoD, Ckm, and Myh4 were purchased from Applied Biosystems. Expression levels were normalized to 18S and represented as fold change.
In situ hybridizations
For whole mount in situ hybridization, embryos were fixed overnight in 4% PFA/PBS at 4°C, then dehydrated in increasing concentrations of methanol and bleached with 6% H202/methanol for 1 hour. Embryos were subsequently rehydrated, treated with proteinase K, and fixed in 4% PFA, 0.2% glutaraldehyde for 20 minutes. Pre-hybridization (50% Formamide, 5× SSC pH 4.5, 2%SDS, 2% blocking reagent (Roche), 250 µg/ml tRNA, 100 µg/ml heparin) was achieved at 70°C for 1 hour followed by incubation with digoxigenin-labeled probe overnight. Embryos were first washed with Solution 1 (50% Formamide, 2× SSC pH 4.5, and 1% SDS) three times, 6 times in Solution 2 (100 mM Maleic Acid, 150 mM NaCl, 0.1% Tween-20, pH 7.5), then blocked with consecutive 1 hour incubations with 2% blocking reagent/Solution 2 and 2% blocking reagent/20% heat-inactivated goat serum/Solution 2. To detect bound probe we performed immunohistochemistry with anti-digoxigenin-Alkaline Phosphatase antibody (1:2000, Roche). To develop the AP signal, embryos were washed with Solution 1, then incubated with Solution 4 (100 mM NaCl, 100 mM Tris-Cl, pH 9.5, 50 mM MgCl2, 0.1% Tween-20) with developing reagents (0.25 mg/ml NBT (Nitro blue tetrazolium chloride), and 0.125 mg/ml BCIP (5-Bromo-4-chloro-3-indolyl phosphate, toluidine salt, Roche). Lastly, the embryos were washed with Solution 4, fixed in 4% PFA/PBS at 4°C overnight, and imaged with a Zeiss 11 Stereoscope. Full length coding sequence was used to generate probes for both MyoD and Myomaker by using the digoxigenin labeling kit (Roche) followed by purification with MicroSpin™ G-25 columns (Amersham).
Radioisotopic in situ hybridization was performed as previously described32. Briefly, sections were deparaffinized, permeabilized, and acetylated prior to hybridization at 55°C with riboprobes diluted in a mixture containing 50% formamide, 0.3M NaCl, 20mM Tris-HCl, pH 8.0, 5mM EDTA, pH 8.0, 10mM NaPO4, pH 8.0, 10% dextran sulfate, 1× Denhardt’s, and 0.5mg/ml tRNA. Following hybridization, the sections were rinsed with increasing stringency washes, subjected to RNAse A (2µg/ml, 30min at 37°C) and dehydrated prior to dipping in K.5 nuclear emulsion gel (Ilford, UK). Autoradiographic exposure ranged from 21 to 28 days. The myogenin probe corresponded to nucleotides 31 through 638 of the coding sequence, whereas nucleotides 181–811 of the coding sequence was used for the M-cadherin probe. The Myomaker probe was full-length coding sequence. 35S-labeled sense and antisense probes were generated by Sp6 and T7 RNA polymerases, respectively, from linearized cDNA templates by in-vitro transcription using the Maxiscript kit (Ambion).
Cardiotoxin injury
Cardiotoxin (CTX) from Naja mossambica mossambica (Sigma) was dissolved in sterile saline to a final concentration of 10 µM and aliquoted and stored at –20°C. Mice were anesthetized by intraperitoneal injection of 2.5% Avertin at (15 µl/g). Mouse legs were shaved and cleaned with alcohol. Tibialis anterior (TA) muscles were injected with 50 µl of CTX with a 26-gauge needle.
X-gal staining
For whole-mount X-gal staining, either embryos or tissues were fixed in 4% PFA/PBS (containing 0.01% deoxycholic acid and 0.02% Igepal) for 45 minutes at 4°C with gentle shaking then rinsed 2 times with cold PBS. Samples were stained overnight in staining solution (5mM K3Fe(CN)6, 5mM K4Fe(CN)6, 2mM MgCl2, 1mg/ml X-gal in PBS) followed by washing twice in PBS and post-fixing with 4% PFA/PBS.
For X-gal staining of cryosections or cells in culture the following procedure was employed: fix with 2% gluraraldehyde/PBS, wash 3 times in 0.1% sodium deoxycholate, 0.2% NP40 Substitute (Fluka), PBS, and incubate in staining solution (4mM K3Fe(CN)6, 4mM K4Fe(CN)6, 0.4mM MgCl2, 1mg/ml X-gal, 0.1% sodium deoxycholate, 0.2% NP40 Substitute in PBS) at 37°C overnight in the dark. The samples were then rinsed in PBS and fixed in 4% PFA/PBS for at least 20 minutes. Tissue sections were co-stained with light eosin, dehydrated, and mounted with Permount (Fisher). Cells were co-stained with nuclear fast red (Sigma).
Northern blot analysis
Total RNA was extracted as previously described. Fifteen micrograms of RNA was extracted, resolved on a 1% agarose/MOPS (0.2M MOPS pH 7.0, 20 mM sodium acetate, 10 mM EDTA pH 8.0) gel, and transferred to Hybond N+ membrane (Amersham). The membrane was then incubated in hybridization buffer (1% crystalline BSA (fraction V), 1mM EDTA, 0.5M NaHPO4, 7% SDS) for at least 2 hours at 68°C followed by overnight incubation with probes labeled with [α-32P]dCTP using the RadPrime DNA Labeling System (Invitrogen). Myomaker probe was generated from full-length coding sequence. The next day the membrane was washed with 1× SSC, 0.1% SDS for 10 minutes at room temperature followed by 3 washes at 68°C with 0.5× SSC, 0.1% SDS. The membrane was exposed to film at −80°C overnight and developed with a SRX101A Tabletop X-Ray Film Processor (Konica Minolta).
Histology and immunohistochemistry
For cryosections, skeletal muscle or limbs were dissected, embedded in gum tragacanth (1% in PBS), and frozen in 2-methylbutane cooled liquid nitrogen. For paraffin sections, tissue was fixed in 10% neutral buffered formalin and processed for routine paraffin histology. Frozen and paraffin sections were cut and stained with H&E using routine procedures. Immunohistochemistry was performed by fixation with 1%PFA/PBS, permeabilization with 0.2% Triton X-100 in PBS, blocking with PBS/1% BSA, 1% heat inactivated goat serum, 0.025% Tween20, incubation with primary antibody for at least 2 hours, incubation with secondary Alexa-Fluor antibodies (Invitrogen) for 1 hour, and mounting with VectaShield containing DAPI (Vector Laboratories). Anti-mouse myosin (my32, Sigma) and desmin (DAKO) antibodies were used at 1:100. The TUNEL (Invitrogen) reaction was performed exactly as described by the manufacturer. Slides were visualized using a Leica DM RXE microscope.
Isolation of primary myoblasts and immunocytochemistry
Limbs were dissected from E15 to E17.5 embryos and dissociated in 0.05% Collagenase D (Roche) in PBS at 37°C for 2–3 hrs. Ten milliliters of culture media (20% FBS/Ham F10) was added to the suspension and triturated followed by centrifugation at 1500 × g for 10 minutes at 4°C. The pellet was resuspended in 10 ml of growth media (20% FBS/Ham F10 + 2.5 ng/ml bFGF (Promega)), filtered through a 100 µm cell strainer, and plated on a 10 cm laminin coated culture dish. To enrich for myoblasts, cultures were incubated in a small volume of PBS, and the myoblasts were dislodged by knocking the plate lightly. To induce myogenesis, the cultures were placed in differentiation media (2% horse serum, DMEM) for 3–5 days. Immunocytochemistry was performed by fixing with 4% PFA/PBS, permabilization with 0.2% Triton X-100 in PBS, blocking with 3% BSA/PBS, incubation with primary antibody for at least 2 hours, then incubation with Alexa-Fluor secondary antibodies for 1 hour. Myosin antibody, used as described above, M2 Flag antibody (Sigma) at 1:500, BrdU (Roche) at 1:100, EEA1 (generous gift of Schmid Lab, University of Texas-Southwestern) at 1:500, GM130 (BD Pharmingen) at 1:300, cyclophilin D (Abcam) at 1:200, PDI (Cell Signaling) at 1:500. Cultures were co-stained with Phalloidin-rhodamine (Invitrogen) at 1:200 and nuclei were stained with Hoechst (Invitrogen). For staining of live cells, we first washed the cells with PBS and incubated in blocking buffer (3% BSA/PBS) for 15 minutes. Primary antibody incubation was then performed on ice, followed by fixation with 4% PFA/PBS, and incubation with secondary antibody. These cultures were visualized on a Zeiss LSM 780 Confocal Microscope or a Nikon Eclipse Ti Fluorescent Microscope.
Cloning, generation of retroviruses, and C2C12 infection
We cloned Myomaker coding sequence from P0 WT tongue cDNA using the following primers: Myomaker-F: 5’-ATGGGGACAGTTGTAGCCAA-3’, Myomaker-R: 5’-TCAGACACAAGTGCAGCAGA-3’. Myomaker-Flag was generated by independently cloning the regions immediately upstream (5’ PCR product) and downstream (3’ PCR product) of the site of Flag insertion. These products were used as templates, and Myomaker-F and Myomaker-R as primers, in a standard PCR sewing reaction to generate full-length Myomaker-Flag.
Retroviral plasmid DNA was generated by subcloning Myomaker and Flag-tagged Myomaker cDNA into the retroviral vector pBabe-X31. GFP and dsRed retrovirus have been described previously32. Ten micrograms of retroviral plasmid DNA was transfected using FuGENE 6 (Roche) into Platinum E cells (Cell Biolabs) which were plated on a 10 cm culture dish at a density of 3 × 106 cells per dish, 24 hours before transfection. Forty-eight hours after transfection, viral media was collected, filtered through a 0.45 µm cellulose syringe filter, and mixed with polybrene (Sigma) at a final concentration of 6 µg/ml. C2C12 myoblasts (obtained from ATCC) were plated on 35 mm culture dishes at a density of 3 × 105 cells/dish 24 hours prior to infection with viral media. Eighteen hours after infection, virus was removed, cells were washed with PBS, and replaced with differentiation media. These cultures were assayed between 1 and 5 days of differentiation. The actin inhibitors Cytochalasin D (Sigma) and lantrunculin B (Sigma) were used at a concentration of 0.3 µM and 0.1 µM, respectively.
Subcellular fractionation and western blot analysis
To fractionate C2C12 cells into cytosol and membrane fractions, we first washed a 10 cm dish with cold PBS and lysed the cells by dounce homogenation in hypotonic buffer (10 mM Tris pH 8.0, 1 mM EDTA). The homgenate was centrifuged at 500 × g for 5 min. to pellet nuclei and cell debris. The supernatant was centrifuged at 100,000 × g for 20 min to pellet membrane structures. The supernatant from this step was the cytosol fraction and the membrane fraction was solubilized in an equal volume of hypotonic buffer + 1% n-Dodecyl β-D-maltoside (DDM, Sigma) for further analyses by immunoblotting. For analysis of whole cell extracts, DDM solubilization was used (20 mM HEPES, 150 mM NaCl, 2 mM EDTA, 10% glycerol, 1% DDM). For immunoblotting, equal protein amounts were separated on a 12% SDS-PAGE, transferred to a PVDF membrane (Millipore), blocked in 5% milk in TBS-tween and incubated with primary antibodies. The following antibodies were used: M2 Flag (Sigma, 1:1000), Gapdh (Millipore, 1:10000), VDAC (Santa Cruz, 1:1000), α-tubulin (Sigma, 1:1000), myosin (my32, Sigma, 1:1000), and myogenin (Developmental Studies Hybridoma Bank, 1:1000).
Cell mixing
WT myoblasts were mixed with either Myomaker+/− or Myomaker−/− myoblasts at equal ratios (approximately 1 × 105 cells per genotype), plated on a well of a laminin coated 12-well plate, and induced to differentiate the next day. 10T1/2 fibroblasts were infected with either GFP- and empty-retrovirus or GFP- and Myomaker-retrovirus for 18h. After infection, cells were washed multiple times and then trypsinized, and mixed with C2C12 myoblasts at a 1:1 ratio (1 × 105 of each cell type) and plated on one well of a 6 well plate in differentiation media. GFP and myosin expression was analyzed 4 days after differentiation. A similar protocol was performed to assess incorporation of BrdU-labeled fibroblasts into myotubes with minor modifications. 10T1/2 fibroblasts were incubated with BrdU (Roche) at a final concentration of 10 µM for 18 hours. They were then infected with either empty-retrovirus or Myomaker-retrovirus and mixed with C2C12 myoblasts that had been infected with dsRed-retrovirus.
Time-lapse microscopy
In Movie 1, C2C12 myoblasts were infected with GFP and Myomaker retrovirus. For Movie 2, C2C12 myoblasts were infected with dsRed retrovirus and fibroblasts were infected with GFP and Myomaker retrovirus. GFP and dsRED was visualized using a Perkin Elmer Ultraview Spinning Disk Confocal Microscope with a chamber for control of temperature and CO2. Images were captured every 15 minutes using Volocity 5.4.0 software. Images were analyzed and movies assembled using ImageJ.
Quantitation and statistics
Each histological analysis of embryonic skeletal muscle was performed on four samples per genotype. The differentiation index was calculated as the percentage of nuclei in myosin-positive cells. The fusion index was calculated as the percentage of nuclei contained in myosin-positive myotubes. Structures must contain at least 2 nuclei to be considered a myotube. To quantitate fusion between WT myoblasts and either Myomaker+/− or Myomaker−/− myoblasts, we calculated the percentage of LacZ+ myotubes containing ≥3 nuclei. To quantitate fusion between fibroblasts and myoblasts we calculated the percentage of GFP+ myosin+ cells or the percentage of BrdU+ myotube nuclei. For each quantitation, at least 3 independent experiments were performed in duplicate and at least 6 random fields were imaged per sample. Data are presented as mean ± SEM. Differences between groups were tested for statistical significance using the unpaired two-tailed Student’s t test. P < 0.05 was considered significant.
Supplementary Material
Acknowledgements
We thank J. Cabrera for graphics; the Transgenic Technology Center at UT Southwestern for ES cell injections; J.A. Richardson for histology; the UT Southwestern Live Cell Imaging Facility (K. Luby-Phelps and A. Budge) for microscopy and live cell-imaging assistance. We thank D. Rosenbaum, W. Snell, S. Schmid, J. Seemann and members of the Olson laboratory for scientific discussions. D.P.M. was funded by a NIH NRSA Fellowship (F32AR05948403). This work was supported by grants from the NIH (HL-077439, HL-111665, HL093039, and U01-HL-100401) and the Robert A. Welch Foundation (grant 1-0025) (E.N.O.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Author Contributions
D.P.M. and E.N.O. conceived the project and designed the experiments. D.P.M., J.R.O., L.B.S., S.B., and J.M.S. performed experiments. D.P.M. and R.B.-D. wrote the animal protocol. D.P.M. and E.N.O. analyzed the data and prepared the manuscript.
The authors declare no competing financial interests.
References
- 1.Chen EH, Olson EN. Unveiling the mechanisms of cell-cell fusion. Science. 2005;308:369–373. doi: 10.1126/science.1104799. [DOI] [PubMed] [Google Scholar]
- 2.Bentzinger CF, Wang YX, Rudnicki MA. Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Buckingham M. Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr Opin Genet Dev. 2006;16:525–532. doi: 10.1016/j.gde.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 5.Kang JS, Krauss RS. Muscle stem cells in developmental and regenerative myogenesis. Curr Opin Clin Nutr Metab Care. 2010;13:243–248. doi: 10.1097/MCO.0b013e328336ea98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abmayr SM, Pavlath GK. Myoblast fusion: lessons from flies and mice. Development. 2012;139:641–656. doi: 10.1242/dev.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rochlin K, Yu S, Roy S, Baylies MK. Myoblast fusion: when it takes more to make one. Dev Biol. 2010;341:66–83. doi: 10.1016/j.ydbio.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charrasse S, et al. M-cadherin activates Rac1 GTPase through the Rho-GEF trio during myoblast fusion. Mol Biol Cell. 2007;18:1734–1743. doi: 10.1091/mbc.E06-08-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charrasse S, Meriane M, Comunale F, Blangy A, Gauthier-Rouviere C. N-cadherin-dependent cell-cell contact regulates Rho GTPases and beta-catenin localization in mouse C2C12 myoblasts. J Cell Biol. 2002;158:953–965. doi: 10.1083/jcb.200202034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwander M, et al. Beta1 integrins regulate myoblast fusion and sarcomere assembly. Dev Cell. 2003;4:673–685. doi: 10.1016/s1534-5807(03)00118-7. [DOI] [PubMed] [Google Scholar]
- 11.Griffin CA, Kafadar KA, Pavlath GK. MOR23 promotes muscle regeneration and regulates cell adhesion and migration. Dev Cell. 2009;17:649–661. doi: 10.1016/j.devcel.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yagami-Hiromasa T, et al. A metalloprotease-disintegrin participating in myoblast fusion. Nature. 1995;377:652–656. doi: 10.1038/377652a0. [DOI] [PubMed] [Google Scholar]
- 13.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 14.Hasty P, et al. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- 15.Vasyutina E, Martarelli B, Brakebusch C, Wende H, Birchmeier C. The small G-proteins Rac1 and Cdc42 are essential for myoblast fusion in the mouse. Proc Natl Acad Sci U S A. 2009;106:8935–8940. doi: 10.1073/pnas.0902501106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruenbaum-Cohen Y, et al. The actin regulator N-WASp is required for muscle-cell fusion in mice. Proc Natl Acad Sci U S A. 2012;109:11211–11216. doi: 10.1073/pnas.1116065109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright LP, Philips MR. Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J Lipid Res. 2006;47:883–891. doi: 10.1194/jlr.R600004-JLR200. [DOI] [PubMed] [Google Scholar]
- 18.Pei J, Millay DP, Olson EN, Grishin NV. CREST--a large and diverse superfamily of putative transmembrane hydrolases. Biol Direct. 2011;6:37. doi: 10.1186/1745-6150-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corcoran JA, Duncan R. Reptilian reovirus utilizes a small type III protein with an external myristylated amino terminus to mediate cell-cell fusion. J Virol. 2004;78:4342–4351. doi: 10.1128/JVI.78.8.4342-4351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen EH, Olson EN. Antisocial, an intracellular adaptor protein, is required for myoblast fusion in Drosophila. Dev Cell. 2001;1:705–715. doi: 10.1016/s1534-5807(01)00084-3. [DOI] [PubMed] [Google Scholar]
- 21.Chen EH, Pryce BA, Tzeng JA, Gonzalez GA, Olson EN. Control of myoblast fusion by a guanine nucleotide exchange factor, loner, and its effector ARF6. Cell. 2003;114:751–762. doi: 10.1016/s0092-8674(03)00720-7. [DOI] [PubMed] [Google Scholar]
- 22.Nowak SJ, Nahirney PC, Hadjantonakis AK, Baylies MK. Nap1-mediated actin remodeling is essential for mammalian myoblast fusion. J Cell Sci. 2009;122:3282–3293. doi: 10.1242/jcs.047597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laurin M, et al. The atypical Rac activator Dock180 (Dock1) regulates myoblast fusion in vivo. Proc Natl Acad Sci U S A. 2008;105:15446–15451. doi: 10.1073/pnas.0805546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Powell GT, Wright GJ. Jamb and jamc are essential for vertebrate myocyte fusion. PLoS Biol. 2011;9:1001216. doi: 10.1371/journal.pbio.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oren-Suissa M, Podbilewicz B. Cell fusion during development. Trends Cell Biol. 2007;17:537–546. doi: 10.1016/j.tcb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 26.Wilson NF, Snell WJ. Microvilli and cell-cell fusion during fertilization. Trends Cell Biol. 1998;8:93–96. doi: 10.1016/s0962-8924(98)01234-3. [DOI] [PubMed] [Google Scholar]
- 27.Shilagardi K, et al. Actin-propelled invasive membrane protrusions promote fusogenic protein engagement during cell-cell fusion. Science. 2013;340:359–363. doi: 10.1126/science.1234781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hargrave M, Koopman P. In situ hybridization of whole-mount embryos. Methods Mol Biol. 2000;123:279–289. doi: 10.1385/1-59259-677-0:279. [DOI] [PubMed] [Google Scholar]
- 30.Kitamura T, et al. Efficient screening of retroviral cDNA expression libraries. Proc Natl Acad Sci U S A. 1995;92:9146–9150. doi: 10.1073/pnas.92.20.9146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song K, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shelton JM, Lee MH, Richardson JA, Patel SB. Microsomal triglyceride transfer protein expression during mouse development. J Lipid Res. 2000;41:532–537. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.