Abstract
The isolation and sorting of cells has become an increasingly important step in chemical and biological analyses. As a unit operation in more complex analyses, isolating a phenotypically pure cell population from a heterogeneous sample presents unique challenges. Microfluidic systems are ideal platforms for performing cell separations, enabling integration with other techniques and enhancing traditional separation modalities. In recent years there have been several techniques that use surface antigen affinity, physical interactions, or a combination of the two to achieve high separation purity and efficiency. This review discusses methods including magnetophoretic, acoustophoretic, sedimentation, electric, and hydrodynamic methods for physical separations. We also discuss affinity methods, including magnetic sorting, flow sorting, and affinity capture.
1. Introduction
Separating and sorting cells from a heterogeneous mixture is a fundamental step in basic biological, chemical, and clinical studies.1 Cell separations ideally enrich a target cell of interest while minimizing both the presence and effect of unwanted, background cells. Enriching a target cell type simplifies subsequent analyses and reduces experimental error. For example, testing of an anti-cancer compound on cells aspirated from a biopsy requires isolation of the cancer cell of interest from the normal cells in the sample. In the analyses of leukocytes, isolation of a particular cell type plays a major role in AIDS research, immune function, cancer, and a host of other biomedical problems. An extreme example of cell separations, the isolation of circulating tumor cells (CTCs), represents one of the great challenges in the field.2, 3 The enrichment of rare cells presents obstacles that differ from isolation of more abundant cell types. In both cases, however, the goal of any method is to achieve both high cell purity and cell capture/isolation efficiency.
While the field of cell separations is diverse in application and approach, there is an increasing reliance on microfluidic methods to achieve cell isolation. Cell separations are amenable to lab-on-a-chip devices,4, 5 and present the possibility of point of care analyses. In addition to miniaturization, microfluidic cell separation methods provide control over fluids on the cellular scale,6 introducing new separation modalities that have not been realized in larger-scale devices. Microfluidic methods have been applied to both physical- and affinity-based separations. Physical separation methods exploit differences in size, density, morphology, mass, and electric capacitance or resistance.7 The key benefit of physical-based separations is that labeling of target or background cells is usually not required. However, many techniques require large differences in a particular physical property to separate cells. When physically similar cells interfere with isolation of a target cell type, either multiple physical parameters or affinity-based separations must be used. Affinity approaches include Fluorescence Activated Cell Sorting (FACS), Magnetic Activated Cell Sorting (MACS), and cell affinity separations.8–10 While affinity methods often achieve high separation purity and efficiency, a selective affinity ligand is required for cell capture.
The field of cell separations continues to expand to new applications and methodologies. Microfluidic methods, capable of integrating multiple separation steps or interfacing to other analytical techniques, now rival many traditional separation methods. In this review, we will discuss recent developments in microfluidic methods, current challenges, as well as applications of broad interest.
2. Separation Modalities and Figures of Merit
Cell separations isolate or enrich a target cell type of interest from a mixture containing target and background cells. Like chemical separation methods, cell separations require some degree of selectivity for a particular cell type from a complex matrix. Unlike chemical separations, cell separations often must preserve cell viability, limiting the experimental conditions for separation. Additional constraints, such as maintaining sterile conditions, may be placed on the experiment, further limiting separation options in some cases.
The initial composition of the sample affects the separation. The initial concentration of cells in the sample (total cells/unit volume) must not be too low to require overly long separation times. If the concentration is too large, the sample can saturate the separation system and degrade performance. There is therefore an application-specific concentration window for optimal separations. For example, for affinity separations one target cell type is typically isolated from background cells. When the input cell concentration is too large, cell-cell interactions interfere with cell-surface interactions. In fluorescence activated cell sorting, multiple cells can be sorted if they arrive at the same time and space in the sorting mechanism, potentially yielding erroneous sorting events.
At the same time, the ratio of target cells to the total cell concentration in the sample is of great importance. Generally, separation of an abundant target cell in a mixture has less stringent performance requirements than the selection of a “rare” target cell. The term “rare cell” is used broadly in the literature, and rarity can typically represent target cell ratios from 1:109 to 1:104.11,12 This broad range of target cell ratios is due to the wide range of applications for cell isolation. For example, the isolation of fetal blood cells from maternal blood (1 fetal red blood cell per 105–107 maternal red blood cells)13 presents a major challenge to the field of cell separations. Given that nonspecific binding in most cell separations is larger than the fetal red blood cell concentration, isolating pure samples of fetal red blood cells requires multiple purification steps. However, even isolation of more abundant cells, on the order of 0.001% abundance, is still challenging by most cell separation methods.
Cell separations can have positive or negative cell enrichment. In positive cell enrichment, the target cell is retained in the separation medium, while background cells are passed through to a waste collector. Positive cell selection requires an affinity ligand or physical parameter unique to the cell type in question. The main benefit of positive selection is that the target cell is retained in the separation medium or passed through to a collection channel with minimal or no dilution. In affinity-based methods, positive selection can increase the target cell concentration. However, as the target cell ratio in the initial sample decreases, the effect of nonspecific binding/sorting increases. Nonspecific binding or sorting is the erroneous separation of background cells into the target cell population. For example, a nonspecific binding or sorting of 0.1% of the total cell population puts a limit on the minimum target cell ratio that can be separated. If the target cell ratio is 1% of the original sample, then the final sample isolated by positive selection would be comprised of 10 target cells for every 1 background cell (91% purity). If the target cell ratio drops to 0.1%, then there will be a 1:1 ratio of target and background cells in the initial sample (50% purity). While a 1:1 target : background ratio is better than the original 1:1000 ratio in the sample, subsequent analyses would require additional separation or identification of target cells from the background cells in the separated population.
An additional drawback of positive selection is that cells are often retained and labeled from the selection process. Labeling may not impede subsequent analysis in most cases, but retention in the separation medium may require some form of elution to collect the cells for additional use. If isolation and counting are the end point of the experiment, then this limitation is minimal.
An alternative separation approach is to use negative selection to enrich a target cell. In negative selection, a unique affinity label is not required for the target cell, as the separation medium retains and depletes background cells. Target cells pass through the device and are collected for analysis or use. An advantage to negative selection is that target cells exiting the device are not labeled, and do not require elution steps. In affinity separations, negative selection requires a ligand or mixture of ligands that can retain all of the background cells in the mixture. As the types of background cells become more diverse, the affinity capture mixture becomes more complex.
Nonspecific capture affects negative selection differently than positive selection. In negative selection, nonspecific capture results in target cells becoming retained in the separation medium. The target cell throughput decreases, while the ratio of target cells in the final population may not change. In a hypothetical example where 100% of background cells are depleted, but with 0.1% nonspecific capture, the target cell would be decreased over time, even though the cell output would be pure. A larger issue in negative selection is the capture efficiency of the separation medium. A system that collected 95% of background cells would still be passing 5% of the background cell to the output, where they would be mixed with the target cell. In many cases, an efficient capture is more important than nonspecific binding for negative selection.
Separation purity and separation efficiency are two figures of merit that can used to assess separation performance. At first glance, both high efficiency and purity would be desired. However, achieving both high separation purity and efficiency is not always possible, and in some separation methods there is a compromise between the two figures of merit. The separation purity can be expressed as
| (1) |
Where ntarget is the number of target cells and ntotal is the total cell number in the separated population. The cell purity should be as large as possible, although the initial cell concentration and ratio should be considered. For example, in recent results where CTCs were separated with a purity of 47%, the new studies that can be conducted with this enabling technology will have a lager impact on cancer research.14 However, in general it is best to strive for the highest purity possible, as it reduces post-separation sample handling and eliminates the need for additional labels or tests on the cells.
A different figure of merit, related to the purity, is the enrichment ratio. That is, the ratio of input to output target cells. This is a somewhat more ambiguous figure, as the definition varies in different studies. The ratio of input and output target cells with respect to sample volume will yield a different figure than the same ratio normalized to the background cell concentration. To further complicate matters, the background cell count for normalization may be before separation, after separation, or both. The enrichment ratio is still important, but should always be reported along with the separation purity. In work by Smirnov and co-workers, a 10,000-fold enrichment of CTCs was reported using immunomagnetic (MACS) isolation. The large enrichment resulted in a purity of 1–10%, as the CTCs were outnumbered by nonspecifically captured cells. However, for their studies of gene expression profiling, this enrichment factor was sufficient, allowing them to identify cancer-specific genes in CTCs.15
The separation efficiency is a measure of the number of target cells captured when compared to the number of target cells passing through the separation medium. In the case of negative selection, the separation efficiency relates to negative cell retention, not total cell retention. The separation efficiency is expressed as
| (2) |
where ncaptured is the number of cells retained in the separation medium and nintroduced is the total cell number introduced into the system. In systems where a set cell sample volume is introduced, the efficiency measurement is straightforward. However, in flow-through systems, where the sample is continually introduced, the separation efficiency can decrease over time if the medium becomes saturated with captured cells. In the same case, the capture purity could also decrease when saturation occurred.
High separation efficiency is less important in most applications than a high separation purity. A system that was only 50% efficient in positive selection, but resulted in 100% purity, would be of more use than a system with 80% efficiency and 80% purity. However, it is best to increase both whenever possible, especially for rare cell separations. While different research groups have emphasized varying levels of importance on the separation purity, efficiency, and enrichment ratio, the separation purity is the best figure of merit to use. The separation purity dictates if additional purification or sample handling steps are needed, and also sets limits on the accuracy of any subsequent analyses.
3. Separations Based on Physical Properties
Cell separations using physical attributes of cells vary widely. The earliest physical approaches used differences in cell size or density to achieve separation. Blood cell analysis is an example of these types of separations. Many leukocyte analyses require removal of the more abundant erythrocyte population prior to testing. Erythrocytes can be removed via selective lysis, or via centrifugation in a Ficoll-Hypaque gradient. The latter approach can also be used to remove granulocytes, producing a population of mononuclear cells. Leukocytes and erythrocytes have relatively large differences in size and morphology; separation of cell types that share similar physical attributes becomes difficult. Numerous external forces can be used to separate cells based on their physical properties, including acoustic, hydrodynamic, electric, and magnetic fields.7 Since these techniques often do not require labels, they are attractive methods when affinity ligands are not available. They are also desirable approaches because they can be performed with high throughput under continuous flow conditions, with minimal sample preparation before or after separation.
3.1 Acoustophoretic Sorting
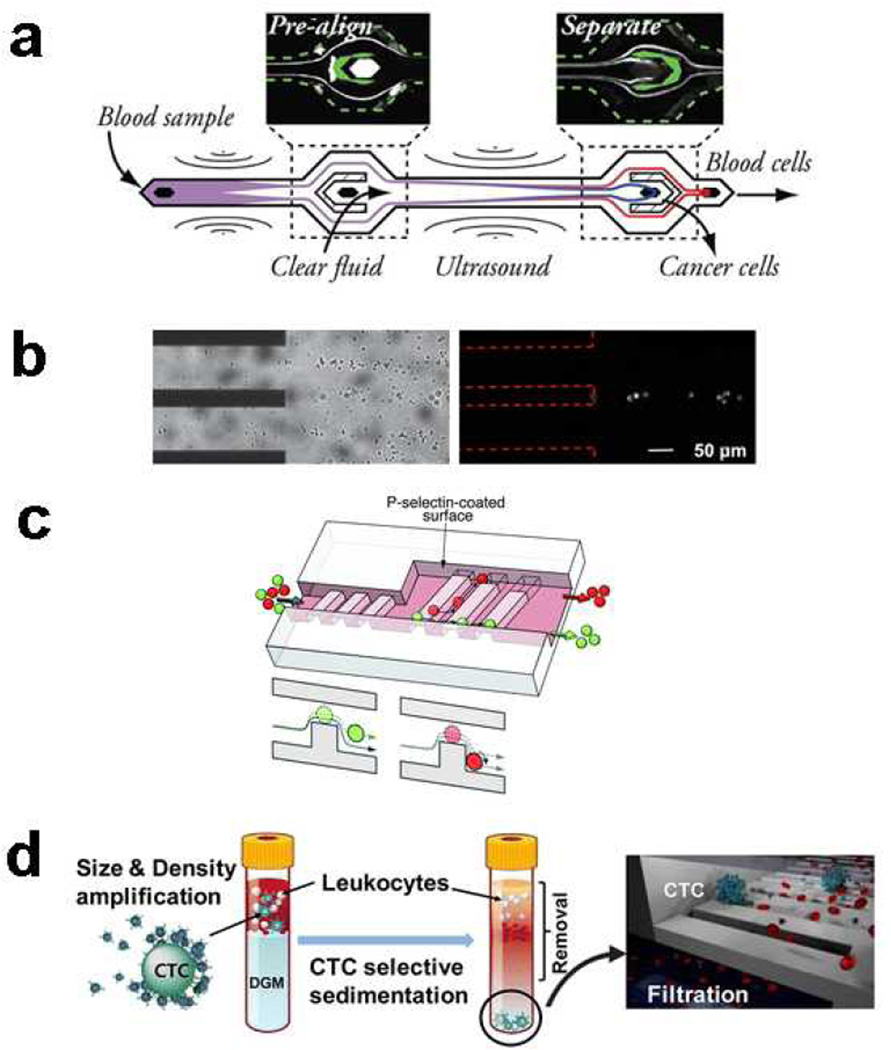
Acoustic fields have been used for cell focusing in flow cytometry, finding use in the Attune flow cytometer. Ultrasonic waves can also be used to generate an external field to separate cells in a microfluidic device. The approach is gentle on cells and is label-free. A recent example used acoustophoresis to sort live and dead MCF-7 breast tumor cells. This separation was primarily size-based, as apoptotic cells were found to be smaller than normal cells.16 The separation purity reached 90% after optimization of piezoelectric voltage amplitude and sample flow rates. Efforts to improve the separation resolution during acoustophoresis have included pre-focusing approaches,17 where a prealignment stage focuses cells before a second acoustic stage separates cells (Figure 1a). This prealignment stage resulted in purities approaching 99.7% with cell recoveries approaching 97.9%. Acoustophoretic systems are easily tunable with respect to the acoustic field, making them more adaptable than some microfluidic approaches that require redesign of the chip when changes are needed.
Fig. 1.
Examples of the many approaches to separating cells based on physical or chemical differences. (a) Acoustic fields can be generated in microfluidic chips to sort cells. In this example (ref 17, published with permission from the American Chemical Society), a two-stage acoustophoresis chip separated prostate cancer cells from blood. (b) Electric and hydrodynamic forces can be combined to isolate cancer cells (ref 20, published with permission from the Royal Society of Chemistry). (c)Affinity and hydrodynamic forces can be combined to generate deterministic cell rolling (ref 25, published with permission from the Royal Society of Chemistry). (d) Sedimentation can be used to isolate cell types, and in some cases labels can be used to enhance the sedimentation process (ref 27, published with permission from the American Chemical Society)
3.2 Dielectrophoresis
Dielectrophoretic cell separations take advantage of differences in electrical properties of cells, as well as their size. Cell membrane capacitance and cell diameter affect the crossover frequency and result in separation. Thus cells of similar size can be separated based on differences in electrical properties. Dielectrophoresis can be operated with discrete sample introduction or continuous flow approaches. A recent advance in dielectrophoresis incorporates a metal oxide semiconductor field-effect transistor (MOSFET) as a Coulter-type counter in a dielectrophoresis system.18 The microfluidic device achieved sized-based separation and performed cell sizing at the same time, demonstrating that cell separations can be integrated into chips with additional cell handling or analysis functionalities.
A recent advance in continuous-flow dielectrophoresis used a periodic array of microelectrodes for cell separation.19 The resulting electric fields were three-dimensional and nonuniform, and eliminated the need to remove cells adsorbed to the electrodes. Separation of live cells from a live/dead mixture of NIH-3T3 cells reached a purity greater than 87%. An electrokinetic approach was developed to isolate MDA-MB-231 breast adenocarcinoma cells from leukocyte background cells.20 Target cells (breast adenocarcinoma) and background cells (leukocytes) were focused to different positions in the channel using both dielectrophoretic and hydrodynamic drag forces (Figure 1b).
3.3 Magnetophoresis
Unlike MACS, magnetophoretic cell separations employ differential migration of cells in a magnetic field to isolate cells. While the cells are unlabeled, paramagnetic salts are added to the medium to enhance migration. In the separation of U-937 cells from red blood cells, the concentration of the paramagnetic salt gadolinium diethylenetriamine pentaacetic acid was found to be a critical parameter for optimization of the separation. Separations with greater than 90% purity were possible with a sample throughput of 105 cells/hour.21
3.4 Hydrodynamic Separations
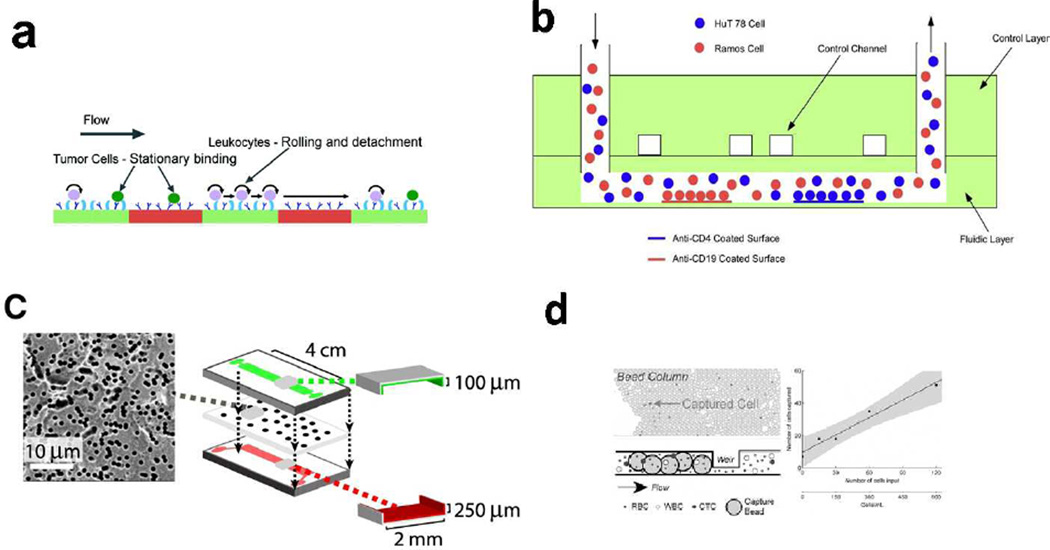
Hydrodynamic forces play a major role in many types of cell separations, including affinity methods (Section 4). Hydrodynamic separations rely on cell-fluid interactions for sorting and isolation of cell types. Using an inertial focusing mechanism, adrenal cortical progenitor cells were separated from digestions of murine adrenal glands. Under the influence of a combination of wall-effects and shear-gradient lift forces, larger cells were directed to the channel center while smaller cells were directed to the walls. Cell throughputs were as high as 24,000 cells/minute.22 A hydrodynamic centrifuge-on-a-chip was developed for cell mixture separation and sample preparation. MCF-7 cells in diluted human blood were trapped in fluid vortices and blood cells were removed after a wash step. The resulting enrichment ratio was 3.4×106, with a purity of 40% and capture efficiency of 20%. Sequential labeling can be conducted by reagent delivery afterwards for automated sample preparation.23 A recent development from Lieu et al. used fluid traps as hydrodynamic tweezers for particle trapping.24 This approach is passive, in that the geometry affects fluid flow and results in particle trapping. In the future, it would be possible to trap and possibly separate cells based on size or other parameters. Recent work with microfluidic systems for deterministic cell rolling were able to separate HL60 cells from K562 cells, each with ~95% cell purity.25 Deterministic cell rolling requires a surface ligand compatible with a cell surface antigen; however, traditional affinity capture and immobilization does not take place. The presence of a matching affinity ligand alters the rolling behavior of the target cell, resulting in separation (Figure 1c). Cell elution is straightforward, as cells are not immobilized, and are separated in a flow-through manner. In other hydrodynamic separation work, tumor cells were isolated from blood using a double-spiral channel. The separation purity was greater than 88% with collection efficiencies greater than 90%.26
3.5 Sedimentation, Microfiltration, and Other Physical Methods
Sedimentation methods were some of the first cell separation approaches and continue to be used in microfluidic cell separations. However, sedimentation methods can suffer from poor separation purity in some cases. One approach to increase the separation abilities of sedimentation is to tag cells with labels that affect size and density.27 In this work, size-density amplification microbeads were used to label MCF-7 cells. Centrifugation resulted in isolated CTCs, followed by microfiltration to collect the cells (Figure 1d).
Sedimentation in flowing fields has been used for some time, particularly in field-flow fractionation of cells. Sedimentation field-flow fractionation has been used to sort cancer stem cells from colorectal cells,28 and deterministic lateral displacement has been used to sort cells based on several physical parameters.29 Based on differences in particle sizes, a constriction channel design with three differing heights was successful in separating microbes30 and mouse pancreatic islets.31
4. Affinity Labeling and Affinity Separations
Affinity ligands for cell surface markers can be used either to provide a force for separation, such as in cell affinity chromatography, or a label in FACS or MACS. Affinity ligands often offer more selectivity for a given cell type when compared with physical separations. However, ligand availability and performance continues to be a limitation for cell affinity separations. Since most affinity-based methods require binding of cells to antibodies or other ligands, nonspecific binding is also an issue that must be minimized for successful separations. Affinity separations are particularly well suited to cell types that are physically similar to the background cells in the sample. In addition, a growing number of capture molecule types drives innovations in cell separations and sorting.
4.1 Fluorescence Activated Cell Sorting
FACS instruments have been in use for decades, and have been miniaturized into microfluidic systems in recent years. Antibodies conjugated to fluorescent dyes typically are used to the sorting event; the affinity label provides the selectivity for the separation. The final purity is affected not only by the ligand-cell labeling, but also by erroneous sorting. Like conventional FACS, microfluidic FACS can achieve high separation purity and efficiency. The sorting process typically deflects cells from an analysis stream to a collection stream. In conventional, large-scale instruments, this sorting can be droplets in air or fluidic stream manipulation. In microfluidic systems, deflection of cells into different streams or channels is the most common approach. Both positive and negative cell selection can be employed, and in some cases multiple cell types can be diverted to different collection streams.
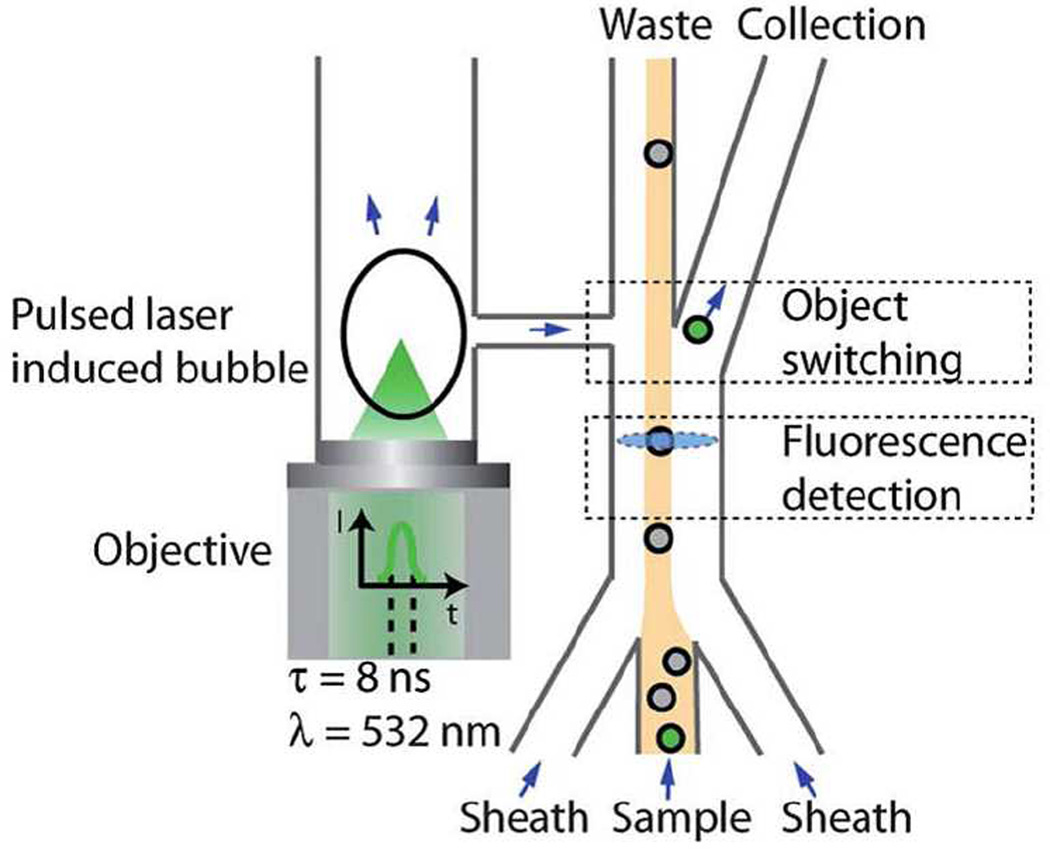
The throughput of microfluidic FACS systems is lower than commercial, large-scale instruments, however additional functionalities can be incorporated into microfluidic systems. In an effort to improve the switching time—a bottleneck to cell throughput—a pulsed laser triggered FACS system was developed to reduce the switching time to 30 µs. This pulse laser triggered fluorescence activated cell sorter (PLACS) was able to increase cell throughput using faster switching.32 Upon detecting a target cell, the laser is fired, creating a cavitation in the channel and deflecting the cell (Figure 2). The cavitation is generated in a neighboring channel joined by a bridge, thus minimizing pressure effects on the cells. The separation purity of the PLACS system was greater than 90%. The throughput of any microfluidic FACS system will also be affected by the flow rates needed to pass cells through the measurement volume, and currently represents a challenge to obtaining higher throughput.
Fig. 2.
A pulsed laser triggered fluorescence activated cell sorter. The laser generates a cavitation event in a neighboring channel, deflecting the target cell into a side channel for collection. The switching speed is an improvement in cell throughput for microfluidic cell sorting (ref 32, published with permission from the Royal Society of Chemistry).
4.2 Magnetic Activated Cell Sorting
MACS analysis requires affinity ligands attached to ferromagnetic beads of varying sizes. Many MACS labels used in macro-scale separations can be adapted for microfluidic applications, making adoption of these flow-through approaches more appealing. MACS, by nature of the external magnetic field applied for separation, is a high-efficiency process.33 The purity in MACS is limited largely by the affinity ligands and nonspecific binding to the particles. To date, the only FDA approved rare cell separation device is based on MACS technology. However, like any labeling technique, the label may interfere with later analysis steps.
Cells can either be retained by a magnet or deflected into different collection streams in microfluidic systems. An approach using superparamagnetic beads in a deflection-based separation resulted in 90% purity of sorted MCF-7 cells isolated from whole blood. The separation efficiency was 85%.34 The same study reported a 96% efficiency for separating endothelial progenitor cells and hematopoietic stem cells from blood using anti-CD133-conjugated magnetic beads. Computational methods to optimize channel geometry have been performed for MACS separations, allowing 3D fluid modeling to direct future device design.35
4.3 Cell Affinity Separations
Cell Affinity Separations or Chromatography involves selective retention of one cell type while another is passed through the device. Alternately, spotted arrays can be used to isolate cells, although subsequent collection of the separated cells is difficult. Affinity separations can operate under positive or negative selection, and are directly amenable to most microfluidic systems. While affinity ligands are used on the chip surface for cell capture, the majority of the cell surface is unlabeled. It is possible to label cells after capture without steric hindrance since capture molecules do not completely cover the cell surface. Like other affinity techniques, ligand selection is critical. Affinity molecules for cell affinity separations include proteins,36 antibodies,37 and aptamers.38 If affinity bonds are formed between the cell and the capture surface, then the cell is held in place by the sum of all the bonds formed. The shear stress in the channel acts to remove cells from the surface, and only cells with binding strengths greater than the shear force are retained. While cells are retained in the chip, they can be eluted by a variety of methods for collection and additional analysis.38, 39
4.3.1 Geometry Effects
The geometry of the chip affects the cell flow and cell capture. Many microfluidic approaches use a vertical inlet, where a tubing or capillary connection is made orthogonal to the channel. Vertical inlets are convenient chip interfaces, and are easier to fabricate than parallel inlets, where the outer tubing connection is parallel to the channel.40 The choice of inlet affects the cell capture as well as the overall chip assembly. Vertical inlets were found to produce more binding at the inlet relative to downstream in the same chip, as well as relative to parallel inlets. Increased dead volume resulted in lower linear flow rates, as determined by tracking single molecules in flow using Fluorescence Correlation Spectroscopy.40 While the enhanced capture near vertical inlets can be problematic, especially when cell capture is supposed to take place in another portion of the chip, it is possible to take advantage of this inlet effect for cell capture. A chip containing a three-dimensional channel with multiple inlets was developed for enhanced capture efficiency in negative selection.41
Adding structures in the microchannel can enhance separation performance. In work by Nagrath et al., micropillars coated with anti-EpCAM antibodies yielded CTC separations with 47% purity from clinical samples.14 In subsequent work, a herringbone chaotic mixer was used for CTC separations, identifying CTCs in 93% of patients.42
4.3.2 Surface Coating Strategies
The chip surface coating forms the affinity surface and is an important aspect of experiment design. Wang et al. reported that using Protein G as the first layer of coating oriented capture antibodies in the optimal position for capture of HIV subtype virus.43 Their approach resulted in 70% capture efficiency of HIV subtypes from whole blood. The same approach can be adapted for other cell isolation methods. Tumor cells were isolated by patterning an affinity surface containing both anti-EpCAM and E-selectin.44 Tumor cells were isolated from leukocytes using this approach (Figure 3a).
Fig. 3.
Surface effects in cell separations. (a) Localized patterning of anti-EpCAM and E-selectin improved tumor cell isolation by reducing leukocyte capture (ref 44, published with permission from the American Chemical Society). (b) Multi parameter cell affinity chromatography was developed to capture multiple cell types in the same channel, or for evaluating different capture molecules against a single cell type (ref 45, published with permission from the American Chemical Society). (c) A porous polycarbonate membrane built into an affinity channel directs cell flow for improved cell capture (ref 48, published with permission from Biophysical Society). (d) While most microfluidic cell separations are open-walled, packed beds can also be used to isolate cells (ref 49, published with permission from the Royal Society of Chemistry).
In addition to ligand orientation and patterning, the type of ligand is also important for separations. A multi-region chip was developed with different separation zones in a single separation channel.45 Pneumatic control channels were used to define multiple antibody zones that could be used to compare several ligands against one cell type. The chip could also be used for separating multiple cell types. Two different capture antibodies were tested against a cell line to determine the best capture molecule. In the same study, CD19- and CD4-positive leukocytes were isolated from blood into two separate zones of the channel with greater than 97% purity (Figure 3b). Other surface strategies include altering the surface density of affinity ligands for selective capture of two phenotypically similar cell types.46 In this manner, cells with strong affinity can be separated in one affinity region, while cells with the same antigen, but expressed at lower levels can be captured in a second region of the chip. Sheng et al. used an aptamer to capture rare cells from whole blood using a series of micropillars.47 The increase in surface area and the flow profile generated by the micropillars resulted in increased capture efficiency of colorectal carcinoma cells.
4.3.3 Shear Effects
Shear effects, which are necessary for differential capture of cells in affinity separations, can also be detrimental to cell survival in some cases. An in-channel membrane approach was developed to reduce shear stress to cells and increase cell interaction with the capture surface.48 The membrane was oriented so that cells rolled along the affinity surface (Figure 3c). Other approaches for increasing surface area, such as the use of affinity beads in a packed channel, can improve cell capture as well (Figure 3d).49
4.3.4 Surface Chemistry for Efficient Cell Recovery
Surface chemistry continues to play a role in the innovation of cell separations. Selective cell capture using anti-EpCAM antibodies attached to an alginate hydrogel resulted in high release efficiency (99% cell release). The process employed selective release of the captured cells using alginate lyase.50 Other approaches using hydrogels have also achieved facile elution with high separation purity.51 Photocleavable ligands have also been developed to facilitate cell elution.52
4.3.5 Alternative Chip Materials
Surface treatments for non-glass affinity surfaces have also been developed for cell affinity separations. Si nanopillars have been employed to achieve 40–70% capture efficiency of CTCs using anti-EpCAM.53 Poly (methyl methacrylate) (PMMA) chips have also been developed for separation and enumeration of CTCs. In this work, colorectal cancer cells spiked into blood were isolated, enriched, and enumerated prior to DNA profiling.54
Conclusion
With applications ranging from preparatory to clinical, there is a need to develop systems with high separation purity and efficiency. Microfluidic systems continue to serve as a platform for many cell separation approaches, and often drive new innovations due to the advantages of integration, ease of fabrication, and flow control. No single cell separation approach will meet all current and future needs, so continued development across multiple fronts is required. Indeed, innovations in one area may lead to major advances in another, as flow geometries, surface chemistries, and separation strategies are continuously improved.
Table 1.
Comparison and Summary of Cell Separation Methods
| Method | Figure(s) of Merit | Cell Types Isolated | Label | Ref |
|---|---|---|---|---|
| Acoustophoresis | 90% purity | Viable/nonviable MCF-breast cancer cells | No labeling | 16 |
| Acoustophoresis | 99.7% purity | Prostate cancer cells | No labeling | 17 |
| Dielectrophoresis | 87% specificity | 4T1 tumor cells/murine bone marrow cells | No labeling | 18 |
| Dielectrophoresis | 87% purity | Live/dead NIH-3T3 cells | No labeling | 19 |
| Electrokinetic | 98% efficiency | MDA-MB-231 breast adenocarcinoma cells/ leukocytes | No labeling | 20 |
| Magnetophoresis | >90% purity | U-937 cells/red blood cells | No labeling | 21 |
| Hydrodynamic Separations | N/A | Adrenal cortical progenitor cells/digestions of murine adrenal glands | No labeling | 22 |
| Hydrodynamic Separations | 40% | MCF-7 cells in diluted human blood | No labeling | 23 |
| Hydrodynamic Separations | 95% purity | HL60 cells/K562 cells | No labeling | 25 |
| Hydrodynamic Separations | >88% purity and >90%efficiency | MCF-7 and Hela | No labeling | 26 |
| Sedimentation | N/A | MCF-7 cells and DMS-79 small cell lung cancer cells/blood | No labeling | 27 |
| Microfiltration | N/A | Cancer stem cells | Labeled | 28 |
| Size | 97% efficiency | Microbes | No labeling | |
| Size | N/A | Mouse pancreatic islets | No labeling | 31 |
| FACS | >90% purity | Nam-6, Ramos | Labeled | 32 |
| MACS | 90% purity | MCF-7 cells/ whole blood | Labeled | 34 |
| MACS | 96% efficiency | Colo205 cells/red blood cells | Labeled | 35 |
| Affinity Separations | 96% purity | Ramos/Hut 78 | Labeled | 41 |
| Affinity Separations | 47% purity | CTCs | Labeled | 14 |
| Affinity Separations | 91% efficiency | PC3, CTCs | Labeled | 42 |
| Affinity Separations | 70% efficiency | HIV subtype | Labeled | 43 |
| Affinity Separations | N/A | MCF-7 cells, HL60 | Labeled | 44 |
| Affinity Separations | >97% purity | CD19 and CD4 positive leukocytes | Labeled | 45 |
| Affinity Separations | 97% efficiency | HUVECs/HMVECs/ | Labeled | 46 |
| Affinity Separations | 95% efficiency and 81% purity | CCRF-CEM cells/Ramos | Labeled | 47 |
| Affinity Separations | N/A | PC3 | Labeled | 48 |
| Affinity Separations | 30–90% efficiency | MCF-7 cells/ whole blood | Labeled | 49 |
| Affinity Separations | >99% release efficiency | PC3/ whole blood | Labeled | 50 |
| Affinity Separations | 92+/−11% | EPCs/ HSCs | Labeled | 51 |
| Affinity Separations | 40–70% efficiency | MCF7, PC3, T24/blood | Labeled | 53 |
| Affinity Separations | N/A | SP2/O | Labeled | 52 |
| Affinity Separations | 96%+/− 4% efficiency | SW620 and HT29/whole blood | Labeled | 54 |
Acknowledgments
The authors acknowledge grants from the National Institutes of Health (Grants RR025782 and GM103550) and the Robert A. Welch Foundation (Grant D-1667) for support.
References
- 1.Pappas D, Wang K. Analytica Chimica Acta. 2007;601:26–35. doi: 10.1016/j.aca.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 2.Jiao LR, Apostolopoulos C, Jacob J, Szydlo R, Johnson N, Tsim N, Habib NA, Coombes RC, Stebbing J. Journal of Clinical Oncology. 2009;27:6160–6165. doi: 10.1200/JCO.2009.24.5837. [DOI] [PubMed] [Google Scholar]
- 3.Li P, Stratton ZS, Dao M, Ritz J, Huang TJ. Lab on a Chip. 2013;13:602–609. doi: 10.1039/c2lc90148j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arora A, Simone G, Salieb-Beugelaar GB, Kim JT, Manz A. Analytical Chemistry. 2010;82:4830–4847. doi: 10.1021/ac100969k. [DOI] [PubMed] [Google Scholar]
- 5.Kovarik ML, Ornoff DM, Melvin AT, Dobes NC, Wang Y, Dickinson AJ, Gach PC, Shah PK, Allbritton NL. Analytical Chemistry. 2013;85:451–472. doi: 10.1021/ac3031543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Ali J, Sorger PK, Jensen KF. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 7.Kim SM, Lee SH, Suh KY. Lab on a Chip. 2008;8:1015–1023. doi: 10.1039/b800835c. [DOI] [PubMed] [Google Scholar]
- 8.Baret J-C, Miller OJ, Taly V, Ryckelynck M, El-Harrak A, Frenz L, Rick C, Samuels ML, Hutchison JB, Agresti JJ, Link DR, Weitz DA, Griffiths AD. Lab on a Chip. 2009;9:1850–1858. doi: 10.1039/b902504a. [DOI] [PubMed] [Google Scholar]
- 9.Saliba A-E, Saias L, Psychari E, Minc N, Simon D, Bidard F-C, Mathiot C, Pierga J-Y, Fraisier V, Salamero J, Saada V, Farace F, Vielh P, Malaquin L, Viovy J-L. Proceedings of the National Academy of Sciences. 2010;107:14524–14529. doi: 10.1073/pnas.1001515107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang K, Cometti B, Pappas D. Analytica Chimica Acta. 2007;601:1–9. doi: 10.1016/j.aca.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 11.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. The Journal of Cell Biology. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darzynkiewicz Z, Robinson JP, Crissman HA. Flow cytometry. Academic Press; 1994. [Google Scholar]
- 13.Wachtel SS, Shulman LP, Sammons D. Clinical Genetics. 2001;59:74–79. doi: 10.1034/j.1399-0004.2001.590202.x. [DOI] [PubMed] [Google Scholar]
- 14.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smirnov DA, Zweitzig DR, Foulk BW, Miller MC, Doyle GV, Pienta KJ, Meropol NJ, Weiner LM, Cohen SJ, Moreno JG, Connelly MC, Terstappen LWMM, O'Hara SM. Cancer Research. 2005;65:4993–4997. doi: 10.1158/0008-5472.CAN-04-4330. [DOI] [PubMed] [Google Scholar]
- 16.Yang AHJ, Soh HT. Analytical Chemistry. 2012;84:10756–10762. doi: 10.1021/ac3026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Augustsson P, Magnusson C, Nordin M, Lilja H, Laurell T. Analytical Chemistry. 2012;84:7954–7962. doi: 10.1021/ac301723s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun J, Gao Y, Isaacs RJ, Boelte KC, Lin PC, Boczko EM, Li D. Analytical Chemistry. 2012;84:2017–2024. doi: 10.1021/ac203212g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ling SH, Lam YC, Chian KS. Analytical Chemistry. 2012;84:6463–6470. doi: 10.1021/ac300079q. [DOI] [PubMed] [Google Scholar]
- 20.Gao J, Riahi R, Sin MLY, Zhang S, Wong PK. Analyst. 2012;137:5215–5221. doi: 10.1039/c2an35707k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen F, Hwang H, Hahn YK, Park J-K. Analytical Chemistry. 2012;84:3075–3081. doi: 10.1021/ac201505j. [DOI] [PubMed] [Google Scholar]
- 22.Hur SC, Brinckerhoff TZ, Walthers CM, Dunn JCY, Di Carlo D. PLoS ONE. 2012;7:e46550. doi: 10.1371/journal.pone.0046550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mach AJ, Kim JH, Arshi A, Hur SC, Di Carlo D. Lab on a Chip. 2011;11:2827–2834. doi: 10.1039/c1lc20330d. [DOI] [PubMed] [Google Scholar]
- 24.Lieu VH, House TA, Schwartz DT. Analytical Chemistry. 2012;84:1963–1968. doi: 10.1021/ac203002z. [DOI] [PubMed] [Google Scholar]
- 25.Choi S, Karp JM, Karnik R. Lab on a Chip. 2012;12:1427–1430. doi: 10.1039/c2lc21225k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun J, Li M, Liu C, Zhang Y, Liu D, Liu W, Hu G, Jiang X. Lab on a Chip. 2012;12:3952–3960. doi: 10.1039/c2lc40679a. [DOI] [PubMed] [Google Scholar]
- 27.Park J-M, Lee J-Y, Lee J-G, Jeong H, Oh J-M, Kim YJ, Park D, Kim MS, Lee HJ, Oh JH, Lee SS, Lee W-Y, Huh N. Analytical Chemistry. 2012;84:7400–7407. doi: 10.1021/ac3011704. [DOI] [PubMed] [Google Scholar]
- 28.Melin C, Perraud A, Akil H, Jauberteau M-O, Cardot P, Mathonnet M, Battu S. Analytical Chemistry. 2012;84:1549–1556. doi: 10.1021/ac202797z. [DOI] [PubMed] [Google Scholar]
- 29.Beech JP, Holm SH, Adolfsson K, Tegenfeldt JO. Lab on a Chip. 2012;12:1048–1051. doi: 10.1039/c2lc21083e. [DOI] [PubMed] [Google Scholar]
- 30.Nam K-H, Eddington DT. Journal of Microelectromechanical Systems. 2010;19:375–383. [Google Scholar]
- 31.Nam K-H, Yong W, Harvat T, Adewola A, Wang S, Oberholzer J, Eddington DT. Biomedical Microdevices. 2010;12:865–874. doi: 10.1007/s10544-010-9441-2. [DOI] [PubMed] [Google Scholar]
- 32.Wu T-H, Chen Y, Park S-Y, Hong J, Teslaa T, Zhong JF, Di Carlo D, Teitell MA, Chiou P-Y. Lab on a Chip. 2012;12:1378–1383. doi: 10.1039/c2lc21084c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zborowski M, Chalmers JJ. Analytical Chemistry. 2011;83:8050–8056. doi: 10.1021/ac200550d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plouffe BD, Mahalanabis M, Lewis LH, Klapperich CM, Murthy SK. Analytical Chemistry. 2012;84:1336–1344. doi: 10.1021/ac2022844. [DOI] [PubMed] [Google Scholar]
- 35.Hoshino K, Chen P, Huang Y-Y, Zhang X. Analytical Chemistry. 2012;84:4292–4299. doi: 10.1021/ac2032386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green JV, Murthy SK. Lab on a Chip. 2009;9:2245–2248. doi: 10.1039/b906768j. [DOI] [PubMed] [Google Scholar]
- 37.Wang K, Solis-Wever X, Aguas C, Liu Y, Li P, Pappas D. Analytical Chemistry. 2009;81:3334–3343. doi: 10.1021/ac900277y. [DOI] [PubMed] [Google Scholar]
- 38.Phillips JA, Xu Y, Xia Z, Fan ZH, Tan W. Analytical Chemistry. 2009;81:1033–1039. doi: 10.1021/ac802092j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang K, Marshall MK, Garza G, Pappas D. Analytical Chemistry. 2008;80:2118–2124. doi: 10.1021/ac702553w. [DOI] [PubMed] [Google Scholar]
- 40.Li P, Tian Y, Pappas D. Analytical Chemistry. 2011;83:774–781. doi: 10.1021/ac102975g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li P, Gao Y, Pappas D. Analytical Chemistry. 2011;83:7863–7869. doi: 10.1021/ac201752s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stott SL, Hsu C-H, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP, Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. Proceedings of the National Academy of Sciences. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang S, Esfahani M, Gurkan UA, Inci F, Kuritzkes DR, Demirci U. Lab on a Chip. 2012;12:1508–1515. doi: 10.1039/c2lc20706k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Launiere C, Gaskill M, Czaplewski G, Myung JH, Hong S, Eddington DT. Analytical Chemistry. 2012;84:4022–4028. doi: 10.1021/ac2033408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li P, Gao Y, Pappas D. Analytical Chemistry. 2012;84:8140–8148. doi: 10.1021/ac302002a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vickers DAL, Chory EJ, Murthy SK. Lab on a Chip. 2012;12:3399–3407. doi: 10.1039/c2lc40290d. [DOI] [PubMed] [Google Scholar]
- 47.Sheng W, Chen T, Kamath R, Xiong X, Tan W, Fan ZH. Analytical Chemistry. 2012;84:4199–4206. doi: 10.1021/ac3005633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mittal S, Wong I, Deen W, Toner M. Biophysical journal. 2012;102:721–730. doi: 10.1016/j.bpj.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kralj JG, Arya C, Tona A, Forbes TP, Munson MS, Sorbara L, Srivastava S, Forry SP. Lab on a Chip. 2012;12:4972–4975. doi: 10.1039/c2lc41048f. [DOI] [PubMed] [Google Scholar]
- 50.Shah AM, Yu M, Nakamura Z, Ciciliano J, Ulman M, Kotz K, Stott SL, Maheswaran S, Haber DA, Toner M. Analytical Chemistry. 2012;84:3682–3688. doi: 10.1021/ac300190j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hatch A, Pesko DM, Murthy SK. Analytical Chemistry. 2012;84:4618–4621. doi: 10.1021/ac300496q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ariyasu S, Hanaya K, Watanabe E, Suzuki T, Horie K, Hayase M, Abe R, Aoki S. Langmuir. 2012;28:13118–13126. doi: 10.1021/la302393p. [DOI] [PubMed] [Google Scholar]
- 53.Wang S, Liu K, Liu J, Yu ZTF, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee Y-K, Chung LWK, Huang J, Rettig M, Seligson D, Duraiswamy KN, Shen CKF, Tseng H-R. Angewandte Chemie International Edition. 2011;50:3084–3088. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dharmasiri U, Njoroge SK, Witek MA, Adebiyi MG, Kamande JW, Hupert ML, Barany F, Soper SA. Analytical Chemistry. 2011;83:2301–2309. doi: 10.1021/ac103172y. [DOI] [PMC free article] [PubMed] [Google Scholar]