Abstract
Mutations that truncate the C-terminal non-catalytic moiety of TTBK2 (tau tubulin kinase 2) cause the inherited, autosomal dominant, SCA11 (spinocerebellar ataxia type 11) movement disorder. In the present study we first assess the substrate specificity of TTBK2 and demonstrate that it has an unusual preference for a phosphotyrosine residue at the +2 position relative to the phosphorylation site. We elaborate a peptide substrate (TTBKtide, RRKDLHDDEEDEAMSIYpA) that can be employed to quantify TTBK2 kinase activity. Through modelling and mutagenesis we identify a putative phosphate-priming groove within the TTBK2 kinase domain. We demonstrate that SCA11 truncating mutations promote TTBK2 protein expression, suppress kinase activity and lead to enhanced nuclear localization. We generate an SCA11-mutation-carrying knockin mouse and show that this leads to inhibition of endogenous TTBK2 protein kinase activity. Finally, we find that, in homozygosity, the SCA11 mutation causes embryonic lethality at embryonic day 10. These findings provide the first insights into some of the intrinsic properties of TTBK2 and reveal how SCA11-causing mutations affect protein expression, catalytic activity, localization and development. We hope that these findings will be helpful for future investigation of the regulation and function of TTBK2 and its role in SCA11.
Keywords: movement disorder, neurodegeneration, protein kinase, signal transduction, tau tubulin kinase peptide substrate (TTBKtide)
INTRODUCTION
Previous studies have revealed that mutations within TTBK2 (tau tubulin kinase 2) cause a serious autosomal dominant inherited movement disorder termed SCA11 (spinocerebellar ataxia type 11) [1,2]. SCA11 is characterized by progressive cerebellar ataxia, pyramidal features, peripheral neuropathy and, on occasion, dystonia, with age of onset from the early teens to the mid-20s [3]. TTBK2 is closely related to a neuronal specific protein kinase termed TTBK1, which was discovered in 1995 as a protein kinase in bovine brain extract that phosphorylated tau and tubulin [4]. TTBK2 consists of 1244 residues and apart from an N-terminal serine/threonine protein kinase domain (residues 20–280) possesses no distinctive functional domains or motifs.
Thus far four distinct SCA11 families have been identified, each possessing a mutation leading to premature termination of the TTBK2 protein at around residue 450. This leaves the kinase catalytic domain intact, but eliminates most of the non-catalytic portion of the enzyme (Figure 1A). One of the affected families from Devon, U.K., stretches over eight generations (family 1 in Figure 1A) [1]. All affected individuals of this family displayed progressive cerebellar ataxia, abnormal eye signs and pyramidal features, as well as cerebellar atrophy visible upon magnetic resonance imaging [1]. Neuropathological examination of the brain blocks of one affected family-1 individual revealed gross atrophy of the cerebellum accompanied by severe and near-complete loss of Purkinje cells, with a marked preservation of the basket cells, and a significant loss of cerebellar granule cells [1]. There were also changes consistent with pathological aging, with some tangles and β-amyloid-positive plaques observed in the neocortex, hippocampus, transentorhinal, entorhinal and insular cortex [1].
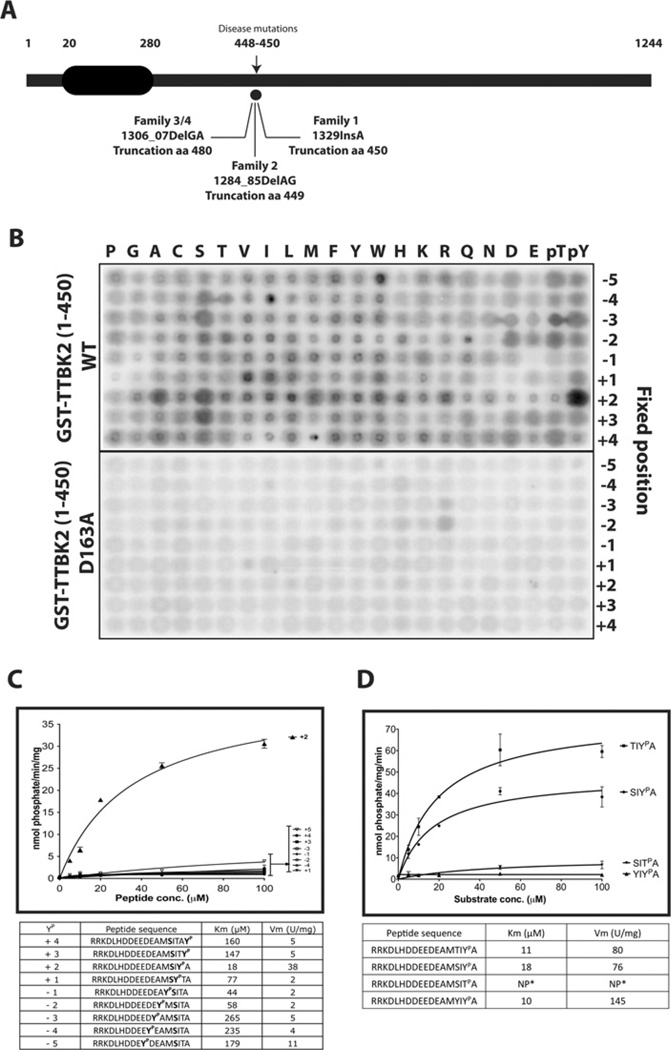
Figure 1. Domain structure and substrate specificity of TTBK2.
(A) Schematic representation of the domain structure of TTBK2 showing the location of the three reported SCA11-causing mutations. Numbering of residues corresponds to human TTBK2. (B) Recombinant HEK-293 purified GST–TTBK2-(1–450) wild-type (WT) and catalytically inactive GST–TTBK2[D163A]-(1–450) were used to screen a positional scanning peptide library consisting of 189 biotinylated peptide libraries in individual kinase assays. Reaction products were bound to streptavidin-coated membrane and, after washing, phosphorylation was visualized by phospho-imaging. (C) Peptides with various positions of the phosphotyrosine residue from +4 to −5 relative to the phosphorylated serine residue were synthesized and analysed for their ability to phosphorylate GST–TTBK2-(1–450) WT purified from HEK-293 cells. Km and Vmax values were derived by non-linear regression analysis as described in the Materials and methods section. (D) Three TTBKtide variants with serine, threonine and tyrosine residues at the phospho-acceptor position and a TTBKtide variant with a phosphothreonine at the +2 position were synthesized, and the kinetics of their phosphorylation by GST–TTBK2-(1–450) was analysed. Km and Vmax values were derived by non-linear regression analysis. NP* denotes that the peptide was phosphorylated too poorly to undertake accurate kinetic analysis. Results in (C) and (D) are means±S.D. for three independent experiments.
TTBK2 belongs to the CK1 (casein kinase 1) group of eukaryotic protein kinases composed of CK1 isoforms (CK1α, CK1α2, CK1δ, CK1ε, CK1γ1, CK1γ2 and CK1γ3), VRK (vaccinia-related kinase) isoforms (VRK1, VRK2 and VRK3) and TTBK isoforms (TTBK1 and TTBK2) [5]. CK1 differs from most other protein kinases by the presence of the sequence S-I-N motif instead of A-P-E in kinase domain region VIII [6]. TTBK1 and TTBK2 possess a P-P-E motif at this site. TTBK2 possesses 38% identity with CK1δ within the kinase domain, but there is no obvious homology beyond the catalytic moiety of TTBK to CK1 or VRK isoforms. TTBK1 and TTBK2 share 84% identity within the catalytic domain and also display significant homology in the non-catalytic region.
TTBK isoforms have been found in many species including, mouse, zebrafish, Caenorhabditis elegans and Drosophila [7]. TTBK1 is mainly expressed in neuronal tissues [8], whereas TTBK2 is more widely expressed and has been observed in tissues including heart, muscle, liver, thymus, spleen, lung, kidney, testis and ovary [9,10]. In situ hybridization showed that TTBK2 mRNA was present in all brain regions in human, rat and mouse [1]. The highest expression of TTBK2 mRNA was observed in the cerebellum Purkinje cells, granular cell layer, hippocampus, midbrain and substantia nigra [1]. Lower expression was seen in the cortex of human, rat and mouse brains [1].
Biochemical investigations initially identified a proteolytic fragment of the catalytic domain of TTKB2, encompassing residues 1–316, in various brain extracts that was capable of phosphorylating tau at two sites (Ser208 and Ser210) [9,10]. Phosphorylation of these residues primes tau for phosphorylation by isoforms of glycogen synthase kinase-3, an enzyme that has also been suggested to modulate tau pathology [11]. TTBK1 was reported to phosphorylate tau at four residues (Ser198, Ser199, Ser202 and Ser422) [8]. More recently, overexpression of TTBK1 in mouse was reported to enhance phosphorylation and oligomerization of tau in the brain [12]. Whether endogenous TTBK1 and/or TTBK2 regulate phosphorylation of endogenous tau has not been established. Recent genetic studies have also implicated TTBK1 in Alzheimer’s disease and in tangle formation [13,14]. Thus far only one brain from a SCA11 family-1 patient has been analysed and it displayed some tau deposition [1]. This may suggest that TTBK2 could be involved in regulating tau, but much further investigation is required to substantiate this conclusion.
Despite this previous work, little is understood about TTBK2 substrate specificity and how SCA11 truncating mutations affect protein expression, kinase activity, stability and localization. We have therefore undertaken an initial analysis of TTBK2 substrate specificity and shown that it has a conspicuous preference for a phosphotyrosine residue at the +2 position relative to the phosphorylation site. We exploit this information to develop an optimized peptide substrate to assess TTBK2 catalytic activity. Furthermore, we demonstrate that SCA11 truncating mutations markedly enhance TTBK2 protein expression, lead to inhibition of TTBK2 kinase activity and promote nuclear localization. We generate TTBK2-knockin mice expressing an SCA11 disease causing mutation and confirm that this results in the inhibition of endogenous protein kinase activity. We find that, in homozygosity, the SCA11 mutation causes embryonic lethality. These findings will help with further investigations into the function and regulation of TTBK2, as well as its role in SCA11.
MATERIALS AND METHODS
Reagents and general methods
Lysis buffer contained 50 mM Tris/HCl, pH 7.5, 1 mM EGTA, 1 mM EDTA, 1 mM sodium orthovanadate, 10 mM sodium-2-glycerophosphate, 50 mM sodium fluoride, 5 mM sodium pyrophosphate, 0.27 M sucrose, 1 mM benzamidine and 2 mM PMSF, supplemented with either 1%(v/v) Triton X-100 or 0.5% NP-40 (Nonidet P40) and 150 mM NaCl as indicated. Buffer A contained 50 mM Tris/HCl, pH 7.5, 50 mM NaCl, 0.1 mM EGTA and 0.27 M sucrose. Tissue-culture reagents were from Life Technologies and [γ-32P]ATP was from PerkinElmer. P81 phosphocellulose paper was from Whatman. The Flp-in T-REx system was purchased from Invitrogen and stable cell lines were generated following the manufacturer instructions by selection with hygromycin.
The coding region of full-length human TTBK2 was amplified by PCR from IMAGE clone 4829013 (NCBI accession number BC071556) and subcloned as a NotI-NotI fragment into several expression vectors. Site-directed mutagenesis was carried out using the QuikChange® method (Stratagene) with KOD polymerase (Novagen) in the presence of DMSO. All DNA constructs were verified by DNA sequencing, which was performed by the DNA Sequencing Service, School of Life Sciences, University of Dundee, Dundee, Scotland, U.K.
Antibodies
A fragment of the mouse TTBK2 fusion protein encompassing residues 314–385 with an N-terminal GST (glutathione transferase) tag was expressed in bacteria and purified by glutathione–Sepharose chromatography. After cleavage of the GST tag, TTBK2-(314–385) was used as an immunogen to raise a sheep polyclonal antibody (S572C). Antibodies were affinity purified from antisera using the TTBK2-(314–385) protein immunogen. An anti-GST antibody was raised in sheep against the GST protein (S902A, first bleed). An anti-GFP (green fluorescent protein) antibody (S268B) was raised in sheep against recombinant GFP protein and affinity purified against the antigen. Secondary antibodies coupled to horseradish peroxidase were obtained from Thermo Scientific and fluorescent secondary antibodies used for the Odyssey Infrared System were from Rockland.
Immunological procedures
Cell lysates (20–30 µg) and immunoprecipitates were subjected to SDS/PAGE electrophoresis (10% gels) and transferred on to nitrocellulose membranes. Membranes were blocked for 20 min at room temperature (20 °C) in 10%(w/v) non-fat dried skimmed milk powder in TBS-T (Tris-buffered saline with 1%Tween 20). All primary antibodies used for immunoblotting were used at 1 µg/ml in 5% (w/v) non-fat dried skimmed milk in TBS-T. Secondary antibodies used were either horseradish peroxidase-conjugated or fluorescent antibodies. For immunoprecipitations, the anti-TTBK2 antibody was covalently coupled to Protein G–Sepharose beads at a ratio of 1 µg of antibody/µl of beads using a DMP (dimethyl pimelimidate) procedure, or GFP-antibody-coupled beads were used. The indicated amount of cell lysates were immunoprecipitated using a 5 µl bead volume of coupled antibody for 1 h, washed twice with lysis buffer containing 0.5 M NaCl and twice with Buffer A. Immunoprecipitates were either subjected to protein kinase activity as described below or to immunoblot analysis.
TTBK2 immunoprecipitation kinase assays
Peptide kinase assays were set up in a total volume of 50 µl in 50 mM Tris/HCl, pH 7.5, 0.1 mM EGTA, 10 mM MgCl2 and 0.1 mM [γ-32P] ATP (~500–1000 c.p.m./pmol) in the presence of the indicated concentration of peptide substrate. The assays were performed for 20 min at 30 °C. When purified proteins were used, reactions were terminated by applying 40 µl of the reaction mixture on to P81 phosphocellulose paper. In the case of immunoprecipitated TTBK2, reactions were terminated by adding 10 µl of 0.2 MEDTA and gentle mixing. After a short spin, 50 µl of the reaction was spotted on to P81 phosphocellulose paper. After extensive washing in 50 mM phosphoric acid, reaction products were quantified by Cerenkov counting. Km and Vmax parameters were calculated using the Michaelis–Menten model of non-linear regression with the GraphPad Prism V5 program.
Fluorescence microscopy
HEK (human embryonic kidney)-293 Flp-in T-REx stable cell lines harbouring GFP-tagged wild-type and mutant forms of TTBK2 were generated using standard protocols. Cells were grown on coverslips and induced with 0.1 µg/ml doxycycline for 16 h. Cells were fixed for 10 min with 4% (v/v) PFA (paraformaldehyde) at room temperature and washed twice for 5 min. After permeabilization in PBS containing 1%(v/v) Triton X-100 for 10 min at room temperature, cells were incubated for 10 min with donkey serum (diluted 1:10; Sigma) and then for 1 h with 0.5 µg/ml anti-TTBK2 antibody. Cells were washed three times with PBS and incubated for 30 min with the secondary antibody (Alexa Fluor® 594) diluted 1:500 in PBS. After three further washes with PBS, cells were incubated with DAPI (4′,6-diamidino-2-phenylindole) for 10 min and mounted on to slides using Hydromount (National Diagnostics). Images were collected on a Zeiss LSM 700 confocal microscope using an α Plan-Apochromat ×100 objective.
Quantification of GFP–TTBK2 localization
GFP-expressing cell lines were fixed and stained with DAPI to identify their nuclei. Fields of cells selected on the basis of the DAPI staining were chosen at random, and single optical sections through the nucleus and cytoplasm in the DAPI and GFP channels were collected sequentially on the Zeiss LSM 700 confocal microscope using the α Plan-Apochromat ×100 objective. These images were quantified using the image-processing software Volocity (PerkinElmer). The nuclei were identified using the DAPI channel and the average GFP nuclear intensity was measured for each cell. Whole cells including the nuclei were identified from the GFP channel and the area of the previously identified nuclei was removed from these areas. The average intensity in the remaining cytoplasm was then measured for each cell. The nuclear intensity was then expressed as a ratio of the cytoplasmic intensity in order to avoid variability in protein expression by the cells. For each cell line, between 260 and 460 cells were counted over ten fields of cells.
Animals and the generation of the TTBK2-knockin mouse
Animal studies were approved by the University of Dundee Ethics Committee and performed under a U.K. Home Office project license. Animals were obtained from Harlan and Taconic Artemis and maintained under specific pathogen-free conditions. The knockin mice were generated by Taconic Artemis on an inbred C57BL/6J background as described in Figure 5. Genotyping was performed by PCR on DNA extracted from ears or embryonic membranes. Primers 1 (5′-CATTTGTTGGCATTATTTCAAAGG-3′) and 2 (5′-AGTAGTAGTAGTAGTAGTAACATGG-3′) were used to detect the wild-type and knockin alleles. The PCR programme consisted of 2 min at 95 °C, 15 s at 95 °C, 30 s at 60 °C and 10 s at 72 °C for 35 cycles.
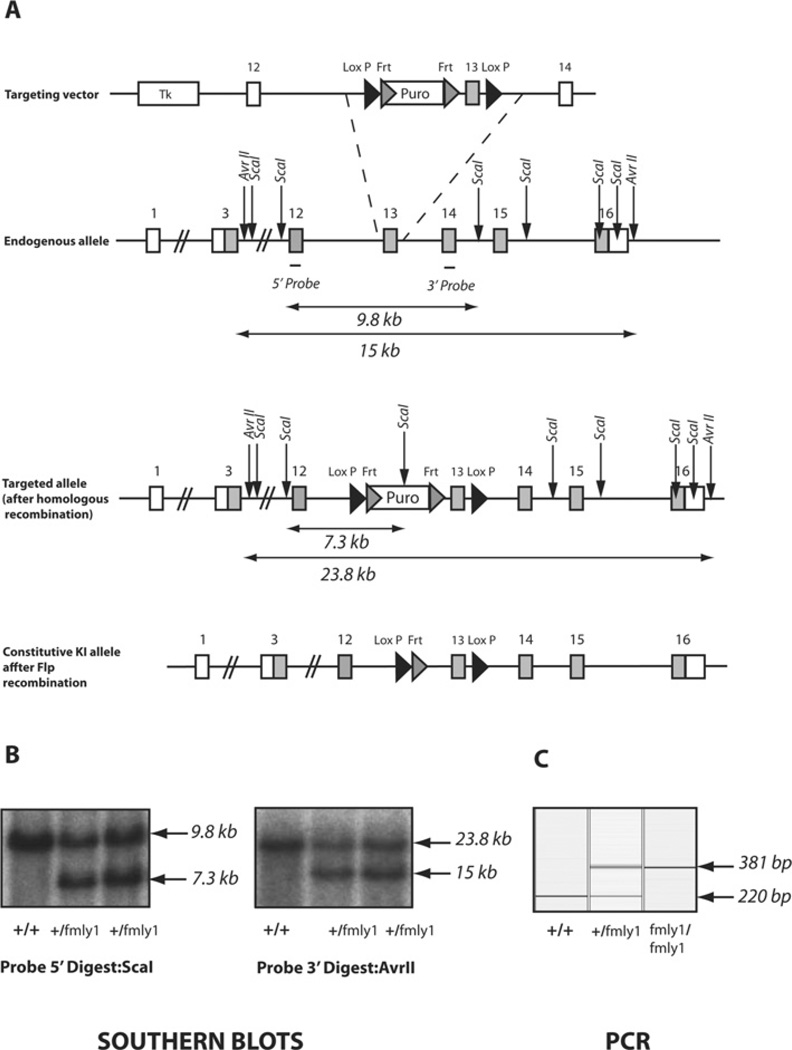
Figure 5. Targeting strategy used to generate TTBK2-knockin mutations.
(A) Diagram describing the knockin construct, the endogenous allele containing exon 13 and the targeted allele with the puromycin (Puro) cassette removed by Flp recombinase. The grey rectangles represent TTBK2 exons. The grey and black triangles represent Frt (Flp recognition target) and LoxP sites respectively. The positions of the TTBK2 primers (P1 and P2) used for genotyping are represented as short black lines with arrowheads. KI, kinase inactive; Tk, thymidine kinase. (B) Genomic DNA purified from the targeted embryonic stem cells of the indicated genotypes was digested with either ScaI or AvrI and subjected to Southern blot analysis with the corresponding DNA probes (positions shown). In the case of the 5′ probe, the wild-type allele produces a 9.8 kb fragment and the conditional knockin allele generates a 7.3 kb fragment. Similarly, the 3′ probe detects a fragment of 23.8 kb from the wild-type allele and a 15 kb fragment from the conditional knockin allele. (C) Genomic DNA was PCR-amplified with TTBK2 primers P1 and P2. The wild-type allele (detected using P1 and P2) generates a 220 bp product and the knockin allele generates a 381 bp product.
Embryos and MEF (mouse embryonic fibroblast) culture
To generate embryos, TTBK2 heterozygous mice were crossed and embryonic development was estimated considering the day of vaginal plug observation as 0.5 d.p.c. (days post coitum). Pregnant females were killed and uterine horns were collected in PBS. The embryos were dissected from the decidua in M2 medium (Sigma) at room temperature and fixed in 4%(v/v) PFA for 2 h and washed and stored in PBS for phenotypic analysis. MEFs were generated from embryos at 9.5–10.5 d.p.c. Embryos were removed from the M2 medium and placed in trypsin for 10 min at 37 °C. From this stage, embryos were cultured in advanced DMEM (Dulbecco’s modified Eagle’s medium)/F12 (Invitrogen) supplemented with 10%(v/v) FBS (fetal bovine serum), 1×MEM(minimal essential medium) non-essential amino acids, 2 mM l-glutamine and 0.1 mM 2-mercaptoethanol.
RESULTS
Analysis of substrate specificity of TTBK2 by a positional scanning peptide library approach
We initially used the positional scanning peptide library approach [15,16] to probe the substrate specificity preferences of TTBK2. This assay employs 198 biotinylated peptide libraries that each contain a 1:1mixture of serine and threonine at the central position and one additional position fixed to one of the 20 amino acids, as well as phosphothreonine or phosphotyrosine. Phosphothreonine and phosphotyrosine were included to allow substrate specificity analysis of kinases such as CK1, to which TTBK2 is related, that have preferences for priming phosphorylation sites. All other positions contained an equimolar degenerate mixture of natural amino acids (except serine, threonine and cysteine). We screened the 198 libraries using [γ-32P]ATP and recombinant TTBK2-(1–450) or kinase-inactive TTBK2[D163A]-(1–450) expressed in HEK-293 cells. Biotinylated peptides were then captured on a streptavidin-coated membrane. The relative amino acid preference at each position was determined by quantifying 32P-radioactivity incorporation following phospho-imaging. The key striking preference observed from this analysis was for a phosphotyrosine residue at the +2 position downstream from the site of phosphorylation (Figure 1B). For experiments undertaken with the kinase-inactive TTBK2[D163A], vastly lower overall levels of phosphorylation were observed with no preference for a +2 phosphotyrosine.
Elaboration of TTBKtide as an optimal peptide substrate
As TTBK2 is a member of the CK1 family, we decided to verify whether TTBK2 would phosphorylate a model peptide substrate termed CK1tide that is frequently employed to assess CK1 isoform activity [17]. CK1δ phosphorylates CK1tide (RRKDLHDDEEDEAMS*ITA) at the serine residue marked with an asterisk [17], and we confirmed that TTBK2-(1–450) also phosphorylates CK1tide specifically at this serine residue (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/437/bj4370157add.htm). Next, we investigated the effects of introducing a phosphotyrosine residue at every position from the −5 to the +4 residue. This confirmed the findings of the positional scanning peptide library approach, revealing that introducing a phosphotyrosine residue at the +2 position, but not at any other position, strongly promoted peptide phosphorylation, principally by decreasing the Km value approximately 8-fold (Figures 1C and 1D). The CK1tide peptide with a phosphotyrosine residue at the +2 position was named TTBKtide (RRKDLHDDEEDEAMSIYpA). Introducing a phosphothreonine rather than a phosphotyrosine residue at the +2 position failed to enhance peptide phosphorylation (Figure 1D). Replacing the phosphorylated serine residue in TTBKtide with a threonine residue did not effect phosphorylation by TTBK2, but replacement with a tyrosine residue abolished phosphorylation (Figure 1D).
Molecular basis for phosphate priming
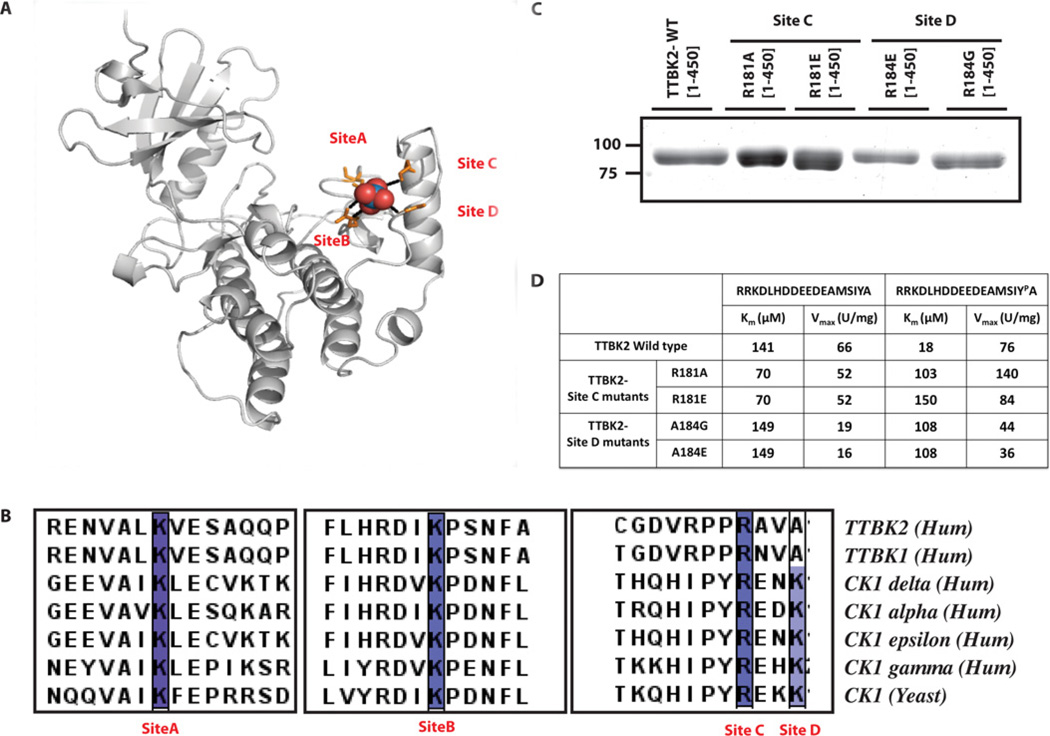
CK1 isoforms possess a well-known requirement for priming phosphorylation [18–20]. The crystal structures of yeast CK1 [21] (PDB code 1EH4) and human CK1δ [22] (PDB code 1CKI) reveal the presence of a sulfate-binding groove on the small lobe of the kinase domain, predicted to function as the phosphate-priming site (Figure 2A). Four highly conserved basic lysine and arginine residues on CK1 isoforms form ionic interactions with the sulfate residue (Figure 2A). Three of these residues are conserved in TTBK1 and TTBK2 (Lys50, Lys143 and Arg181). The fourth residue is an alanine in TTBK2 (Ala184) as well as in TTBK1 (Figure 2B). To verify whether these residues on TTBK2 could be involved in enhancing phosphorylation of +2-phosphotyrosinecontaining substrates, we studied the effect that mutation of Lys50, Lys143, Arg181 and Ala184 had on the phosphorylation of TTBKtide. Mutation of Lys50 or Lys143 to either glutamic acid or alanine inactivated/destabilized TTBK2 (see Supplementary Figure S2A at http://www.BiochemJ.org/bj/437/bj4370157add.htm) and equivalent mutations in CK1δ had similar effects (Supplementary Figure S2B). In contrast, mutation of Arg181 or Ala184 did not affect stability (Figure 2C) or the intrinsic TTBK2 kinase activity as judged by the ability of these mutants to phosphorylate a peptide lacking the +2 phosphotyrosine residue (Figure 2D). However, the ability of TTBK2[R181A or R181E] or TTBK2[A184G or A184E] mutants to phosphorylate tyrosine-phosphorylated TTBKtide was markedly impaired (Km values of 100–150 µM compared with 18 µM for wild-type TTBK2) (Figure 2D). These results suggest that TTBK2 possesses a phosphate priming groove similar to CK1, except that it is involved in recognizing +2 phosphotyrosine residues rather than N-terminal phosphoserine/phosphothreonine.
Figure 2. Molecular basis for phosphate priming.
(A) High-resolution structure of CK1δ protein showing the sulfate-binding site on the C-lobe of the kinase domain predicted to function as the phosphate-priming region [22]. Lys38 (site A), Lys130 (site B), Arg168 (site C) and Lys171 (site D) form ionic interactions with the sulfate molecule and are shown in orange. (B) Sequence alignment of the indicated species of TTBK1, TTBK2 and CK1 family enzymes showing the sequence conservation of the sulfate-binding residues. (C) Coomassie-Blue-stained gel of the GST–TTBK2-(1–450) truncated form of the indicated wild-type (WT) and mutant proteins expressed and purified from HEK-293 cells. The molecular mass in kDa is indicated on the left-hand side. (D) The indicated GST–TTBK2-(1–450) truncated forms expressed in (C) were tested for their ability to phosphorylate TTBKtide (RRKDLHDDEEDEAMSIYPA) and a variant of TTBKtide (RRKDLHDDEEDEAMSIYA) where the tyrosine at the +2 position was not phosphorylated. Km and Vmax values were derived by non-linear regression analysis.
Disease-causing TTBK2 mutations markedly increase protein expression and inhibit kinase activity
Next, we investigated how disease-causing truncation mutations affect TTBK2 protein expression as well as kinase activity. We generated HEK-293 cells that stably express GFP-tagged versions of full-length TTBK2, family-1 and family-2 disease variants of TTBK2 (see Figure 1) as well as a TTBK2-(1–450) truncation mutant. We observed that both family-1 and family-2 mutations and residue-450-truncation mutants were expressed at higher levels than full-length wild-type or kinase-inactive GFP–TTBK2[D163A] (Figure 3A). Similarly, we also observed increased expression of the truncation mutants of TTBK2 possessing FLAG (see Supplementary Figure S3A at http://www.BiochemJ.org/bj/437/bj4370157add.htm) or GST (Supplementary Figure 3B) tags.
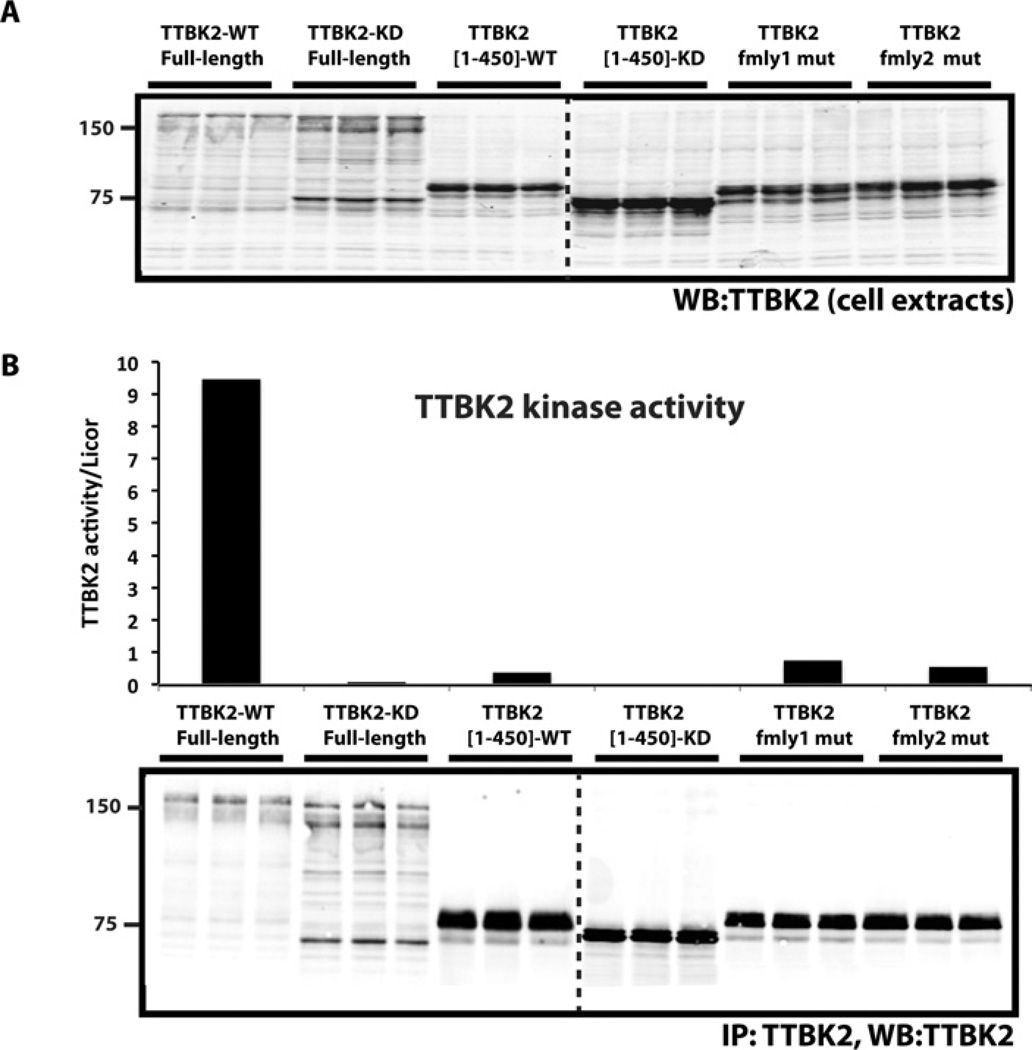
Figure 3. Truncated forms of TTBK2 are less active, but are expressed at higher levels than the full-length kinase.
HEK-293 cells were transiently transfected with the wild-type (WT) and indicated mutants of FLAG-tagged (A and B). (A) Cells were lysed and subjected to immunoblotting (WB) with the anti-TTBK2 antibody and the other indicated antibodies. (B) TTBK2 was immunoprecipitated (IP) from 30 µg of cell extract and subjected to a kinase-activity assay (upper panel) followed by immunoblot analysis (lower panel). TTBK2 kinase activity was quantified using 30 µM TTBKtide, and specific activity was calculated by correcting the amount of phosphate incorporation for protein levels in the immunoprecipitate using quantitative immunoblotting with the Odyssey system and is presented as c.p.m./absorbance units (c.p.m./LICOR arbitrary units). Results are means of duplicate experiments that were repeated four separate times with similar results. Dotted lines indicates that blots were on separate gels.
Monitoring the rate of degradation following pulse-labelling of cells with [35S]methionine revealed that both wild-type and disease-mutant family-1 TTBK2 displayed a similar ~24 h half-life in HEK-293 cells, indicating that there was not a significant difference in the rate at which these proteins were degraded in cells (see Supplementary Figure S4 at http://www.BiochemJ.org/bj/437/bj4370157add.htm).
Next, we immunoprecipitated GFP–TTBK2 variants and quantified their intrinsic activity employing the TTBKtide peptide substrate. We calculated specific activity of wild-type and mutant forms of TTBK2 by normalizing for differences in protein expression using quantitative LICOR immunoblot analysis. This revealed that full-length wild-type TTBK2 possessed approximately 10-fold higher specific activity than the family-1 and family-2 disease mutants or the TTBK2-(1–450) mutant (Figure 3A). The kinase-inactive full-length TTBK2[D163A] or truncated TTBK2[D163A]-(1–450) displayed no detectable activity, establishing that activity of TTBK2, rather than that of a contaminating kinase, was being measured (Figure 3A). Similar analysis of the specific activity of wild-type and mutants of TTBK2 variants with FLAG (Supplementary Figure S3A) or GST (Supplementary Figure S3B) epitope tags confirmed that SCA11 truncating mutations markedly suppressed kinase activity.
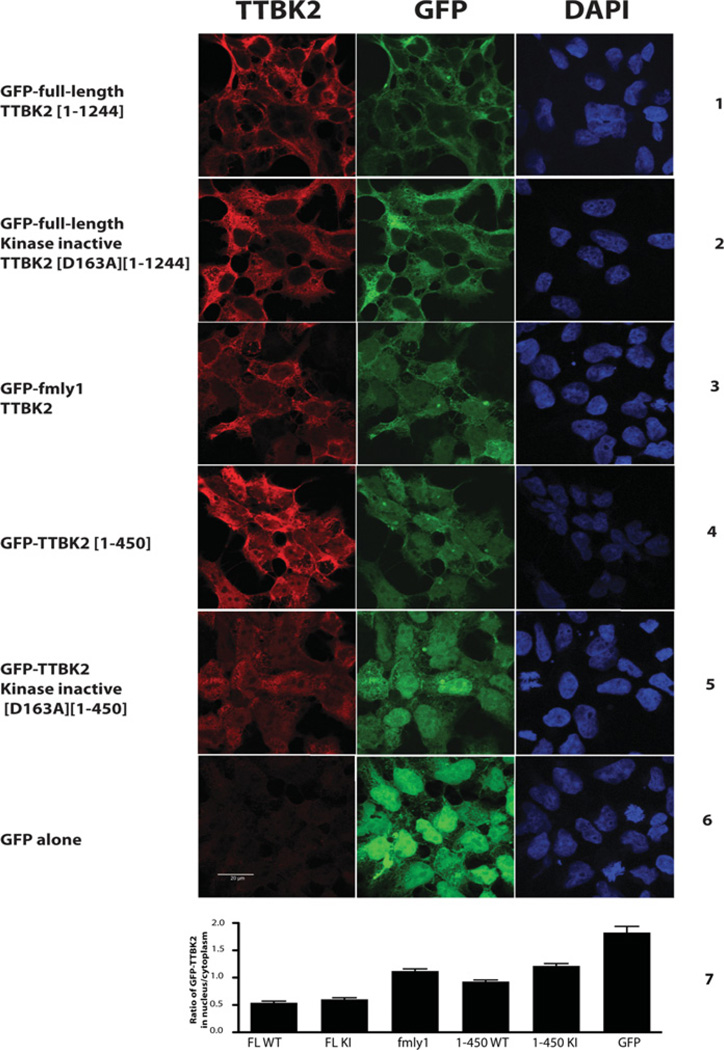
Disease-causing TTBK2 mutations promote nuclear localization
We analysed the cellular localization of stably expressed variants of GFP–TTBK2 in HEK-293 cells. This revealed that full-length wild-type TTBK2 (Figure 4, panel 1) or full-length kinase inactive TTBK2[D163A] (Figure 4, panel 2) was expressed at higher levels in the cytosol than the nucleus. However, TTBK2-(family-1 mutation) (Figure 4, panel 3) or truncated TTBK2-(1–450) (Figure 4, panels 4 and 5) were expressed at similar levels in both the cytosol and the nucleus. Quantification of the ratios of nuclear compared with cytosolic TTBK2 established that an approximately 2-fold higher proportion of TTBK2-(family-1 mutation) and truncated TTBK2-(1–450) is localized in the nucleus compared with full-length TTBK2 (Figure 4, panel 7).
Figure 4. SCA11 disease mutations promote TTBK2 nuclear localization.
HEK-293 cells stably expressing the indicated forms of GFP–TTBK2 were treated with 1 µg/ml tetracycline for 16 h to induce the expression of TTBK2 (panels 1–6). Cells were fixed with 4%PFA and stained with anti-TTBK2 antibody and with DAPI for nuclear staining. Fluorescent imaging was performed on a confocal microscope. Similar results were obtained in four independent experiments. Quantification of nuclear compared with cytoplasmic levels of TTBK2 (panel 7) was undertaken as described in the Materials and methods section. For each cell line, between 260 and 460 cells were counted over ten fields of cells; results are means±S.E.M. Scale bar, 20 µm. FL, full-length; KI, kinase inactive; WT, wild-type.
Generation of TTBK2-(family-1 mutation)-knockin mice
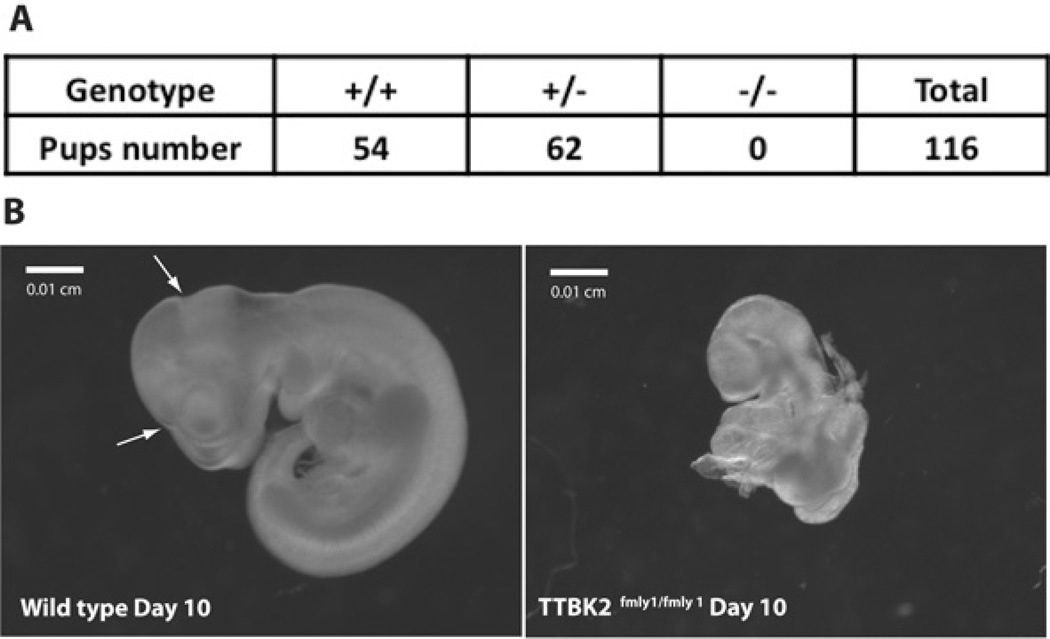
Knockin mice that precisely mimic the one-base insertion of adenosine in exon 13 at nucleotide 1329, observed in SCA11 family 1 [1], were generated as shown in Figure 5. We observed that heterozygous TTBK2fmly1/+ (where fmly1 is family 1) mice were viable and fertile. The oldest TTBK2fmly1/+ mice we have thus far analysed are ~1 year of age and display no overt phenotype. Anatomical dissection of 1-year-old mouse brains also revealed no discernable abnormality. Breeding of heterozygous TTBK2fmly1/+ mice resulted in no homozygous TTBK2fmly1/fmly1 mice being born (Figure 6A), indicating that the homozygous mutation resulted in embryonic lethality. We therefore dissected embryos at different stages of development and found that at E10 (embryonic day 10) TTBK2fmly1/fmly1 embryos were detected at the expected Mendelian frequency, but displayed major abnormalities compared with littermate TTBK2fmly1/+ or TTBK2+/+ embryos. By stage E11, no homozygous TTBK2fmly1/fmly1 embryos were observed, suggesting that these embryos perished before this stage. Homozygous E10-TTBK2fmly1/fmly1 embryos were developmentally delayed, lacking prominent subdivisions of the developing brain, and were smaller than wild-type littermates (Figure 6B). These embryos failed to complete embryonic turning movements or undergo normal caudal extension of the body; in several cases the rudimentary caudal body, which contained some somites (future vertebrae, muscle and dermis) and spinal cord, remained folded back on more rostral tissues (Figure 6B). This distortion and underdevelopment of the main body axis (Figure 6B) are the probable causes of embryonic lethality.
Figure 6. Embryonic lethality and knockin embryo description.
(A) TTBK2fmly1/+ mice were mated and the progeny genotyped as described in the Materials and methods section. The number of mice obtained is indicated for each genotype. (B) Wild-type and homozygous TTBK2fmly1/fmly1 embryos at E10 were detected at the expected Mendelian frequency. Mutant embryos are smaller and developmentally delayed, lacking prominent subdivisions of the brain (arrows on the wild-type embryo). Incomplete embryonic turning movements may result in failure to extend the body axis. A total of 27 separate E10 TTBK2fmly1/fmly1 embryos were analysed and similar phenotypes were observed.
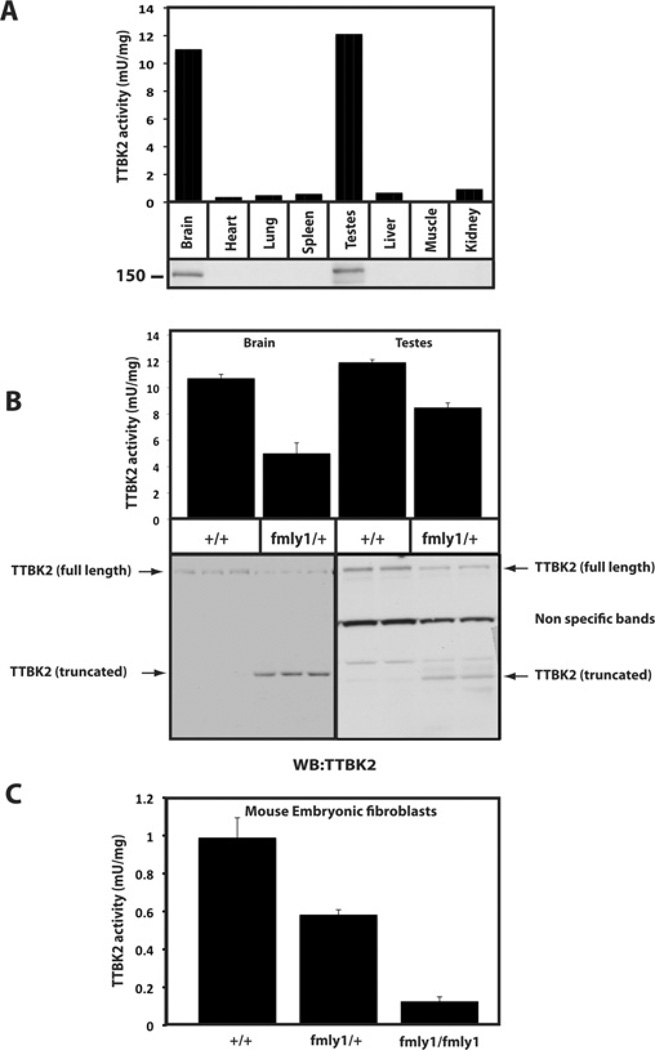
Expression and activity of mutant TTBK2 in tissues and fibroblasts derived from knockin mice
Next, we analysed endogenous TTBK2 expression and activity in various mouse tissues. This revealed significantly higher levels of TTBK2 protein and activity in brain and testes than other tissues investigated (Figure 7A). In wild-type TTBK2+/+ brain and testes, TTBK2 migrated on an SDS/PAGE gel at the expected ~150 kDa size. As predicted, in tissues of the heterozygous TTBK2fmly1/+ mice the level of full-length TTBK2 was reduced by ~50%and a truncated form of TTBK2 migrating at ~50 kDa that was not seen in the wild-type mice was observed (Figure 7B). We also analysed endogenous TTBK2 catalytic activity after immunoprecipitation from brain and testes extracts and found that TTBK2 activity was reduced ~40–50% in tissues derived from heterozygous TTBK2fmly1/+ compared with wild-type littermate animals, consistent with the truncating mutation suppressing TTBK2 catalytic activity (Figure 7B).
Figure 7. Study of TTBK2 in wild-type and TTBK2fmly1/+ knockin mice.
(A) The indicated tissue extracts were generated from wild-type mice. Extracts were immunoblotted for TTBK2 (lower panel) or TTBK2 was immunoprecipitated and subjected to a TTBK2 kinase assay employing the TTBKtide peptide substrate (upper panel). Results are means of duplicate experiments that were repeated four separate times with similar results. (B) Brain and testes lysates were generated from TTBK2+/+ and TTBK2fmly1/+ mice and subjected to immunoblot or TTBK2 kinase assay analysis, as in (A). (C) MEFs were generated from TTBK2+/+, TTBK2fmly1/+ and TTBK2fmly1/fmly1 E10 embryos as described in the Materials and methods section. TTBK2 activity was assessed following immunoprecipitation as in (A). Owing to the low levels of TTBK2 protein expressed in MEFs and high antibody background in immunoprecipitates, we were unable to detect expression of TTBK2 by immunoblot analysis. Results in (B) and (C) are means±S.D.
We also analysed TTBK2 activity in MEFs derived from wild-type TTBK2+/+, heterozygous TTBK2fmly1/+ and homozygous TTBK2fmly1/fmly1 littermate stage-E10 embryos. TTBK2 activity derived from homozygous TTBK2fmly1/fmly1 cells was ~90%lower than activity observed in wild-type TTBK2+/+ fibroblasts (Figure 7C). The heterozygous TTBK2fmly1/+ displayed intermediate TTBK2 activity.
DISCUSSION
We have undertaken the first characterization of TTBK2 catalytic activity and substrate specificity, and we find that it possesses a marked preference for a phosphotyrosine residue at the +2 position from the site of phosphorylation (Figure 1). Sequence comparisons indicate that TTBK2 possesses a phosphate-priming binding site in its catalytic domain equivalent to that in related CK1 isoforms. We find that mutating two key conserved residues within this putative phosphate-binding groove (Arg181 or Ala184) did not affect the ability of TTBK2 to phosphorylate a nontyrosine-phosphorylated peptide, but inhibited phosphorylation of a tyrosine-phosphorylated peptide (Figure 2). The residues making up this putative phosphate-binding pocket are conserved on TTBK1, suggesting that this isoform may also possess a preference for +2-residue phosphotyrosine priming. The key difference in the putative phosphate-binding groove between CK1 and TTBK1/TTBK2 is the presence of a non-basic alanine residue in TTBK1 and TTBK2 rather than a basic lysine residue in all CK1 isoforms. It would be interesting to explore whether this contributes towards the preference of TTBK2 for phosphotyrosine rather than phosphoserine/phosphothreonine priming in CK1 isoforms.
One of the most-studied consensus phosphorylation sites for CK1 isoforms is S/Tp-X-X-S/T, where S/Tp refers to a phosphoserine or phosphothreonine, X refers to any amino acid and the underlined residues refer to the target site [18–20]. In the case of CK1, the priming phosphorylation site is at the −3 position rather than at the +2 position for TTBK2. If both TTBK2 and CK1 use a similar phosphate-binding site within their catalytic domain, there would need to be marked differences in the mechanism and orientation at which primed phosphorylated substrates interact with CK1 and TTBK2. In future work, it would be interesting to co-crystallize the catalytic domain of TTBK2 with a +2-tyrosine-phosphorylated peptide to define the molecular mechanism by which TTBK2 interacts with such substrates and establish whether this putative phosphate-binding groove we have analysed is involved. To our knowledge, the structure of CK1 bound to an S/Tp-X-X-S/T-motif-containing peptide has not been reported, but would be of interest to compare with that of TTBK2. It would also be important to mine phosphorylation-site databases for proteins phosphorylated on Sp/Tp-X-Yp motifs and determine whether any of these proteins might comprise physiological substrates for TTBK2.
Our analysis has enabled us to elaborate a reasonable peptide substrate for assessing TTBK2 activity (TTBKtide-RRKDLHDDEEDEAMSIYpA) that is phosphorylated with a Km of 18 µM and a Vmax of 76 units/mg. We have used this assay to investigate how SCA11 truncating mutations affect TTBK2 activity and demonstrated that two familial SCA11 mutations analysed induce an ~10-fold reduction in TTBK2 protein kinase activity. Truncation of TTBK2 at residue 450, in close proximity to where the familialTTBK2truncatingmutations occur, similarly suppressed TTBK2 activity. The observations that TTBK2 immunoprecipitated from TTBK2fmly1/+ mice possessed ~40%less activity (Figure 7B) and TTBK2 immunoprecipitated from TTBK2fmly1/fmly1 MEFs (Figure 7C) had ~10-fold lower activity also support the notion that SCA11 mutations drastically reduce endogenous TTBK2 activity. Thus these findings indicate that loss of TTBK2 activity could underpin the development of SCA11 in patients.
It will be important to study in more detail the mechanism by which the SCA11 truncating mutations lead to suppression of TTBK2 protein kinase activity. The SCA11 truncating mutations leave the kinase catalytic domain intact, removing a C-terminal non-catalytic region possessing no obvious functional domains (Figure 1A). The removal of the C-terminal non-catalytic region of TTBK2 might delete a motif required for kinase activation, activity or proper folding of the catalytic domain. Depending upon the mechanism by which SCA11 mutations suppress TTBK2 catalytic activity, it may be worth undertaking a drug screen to determine whether it is possible to identify compounds that could lead to the re-activation of truncated TTBK2 mutants. Such compounds could have potential for treating patients with SCA11.
The truncated TTBK2 mutants were expressed in HEK-293 cells at significantly higher levels than the full-length wild-type enzyme (Figure 3 and Supplementary Figure S3). However, in the testes and brain of TTBK2fmly1/+ mice, more similar levels of expression of the endogenous truncated and full-length forms of TTBK2 were observed (Figure 6). Although the relative expression of full-length and truncated mutants of TTBK2 in patients with SCA11 mutations has not yet been investigated, TTBK2 mRNA levels from lymphoblasts of affected individuals were significantly lower compared with lymphoblast mRNA from unaffected individuals [1]. This suggests that the premature stop codon in the TTBK2 mRNA leads to nonsense-mediated decay of mutated mRNA. It is still likely that some of the truncated SCA11 protein is expressed in SCA11 patients, as the mutant transcript was still detected [1]. Treating lymphoblasts from SCA11-affected individuals with cycloheximide, an inhibitor of nonsense-mediated decay, increased TTBK2 mRNA to levels that are similar in the normal transcript [1]. Taking these data into consideration, it is therefore possible that the truncated TTBK2 protein is indeed expressed at much higher levels than the full-length protein, but due to nonsense-mediated decay, less of the truncated protein is produced. This could account for why similar levels of wild-type and truncated mutant protein are observed in the TTBK2fmly1/+ mice. In future work it will be important to analyse the relative levels of wild-type and mutant Ttbk2 mRNA in TTBK2fmly1/+ mice. We also detected enhanced nuclear localization of the truncated forms of TTBK2, indicating that inappropriate localization within the nucleus could potentially contribute to the mechanism by which mutations in TTBK2 lead to SCA11. Potentially, the truncated TTBK2 mutants result in the inappropriate phosphorylation of nuclear proteins.
The developmental delay and small size of homozygous TTBK2fmly1/fmly1 embryos are consistent with a role for TTBK2 activity in the regulation of fundamental cellular processes, such as cell proliferation. The failure of embryonic turning and underdevelopment of the body axis might also lead to a poor placental connection and hence embryonic lethality. TTKB1 expression is prominently localized in cortical and hippocampal neurons [8]. Further insight into the TTKB2 phenotype and its role in spinocerebellar ataxia would be gained by establishing its expression pattern, particularly with respect to its localization in the cerebellum. The TTBK2fmly1/+ mice as well as TTBK2fmly1/fmly1 MEFs will be useful in the validation of potential TTBK2 substrates that are identified.
In conclusion, our present study provides some initial insights into substrate specificity of TTBK2 and how SCA11-causing mutations affect kinase activity and localization. In future studies, it will be important to define the substrates that TTBK2 phosphorylates and investigate whether reduced phosphorylation of these targets contributes to the development of SCA11. In particular, it would be interesting to determine whether physiological TTBK2 substrates are primed with a +2 phosphotyrosine and how regulation of tyrosine phosphorylation is coupled to the control of these TTBK2 targets. Identifying the key targets of TTBK2 could provide vital new insights into the molecular mechanism underpinning the development of spinocerebellar ataxia and result in new ideas as to how this debilitating disease might be better treated in the future.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Gail Fraser for genotyping of the mice, the DNA Sequencing Service (School of Life Sciences, University of Dundee, Dundee, Scotland) and the protein production and antibody purification teams [Division of Signal Transduction Therapy (DSTT), University of Dundee] co-ordinated by Hilary McLauchlan and James Hastie.
FUNDING
This work was supported by the Medical Research Council and the European Regional Development Fund [Innovation Pipeline for Translational Science grant number LUPS/ERDF/2008/2/1/0429]. AstraZeneca, Boehringer-Ingelheim, GlaxoSmithKline, Merck KgaA and Pfizer support the DSST Unit. N.E. is funded by an MRC-UK Studentship. E.F. is a Ph.D. student supported by the Wellcome Trust, and K.G.S. is funded by an MRC programme grant [grant number G0600234]. M.J.B. and L.C.C. are supported by the National Institutes of Health [grant number GM56203].
Abbreviations used
- CK1
casein kinase 1
- DAPI
4′,6-diamidino-2-phenylindole
- DMEM
Dulbecco’s modified Eagle’s medium
- d.p.c.
days post coitum
- E10
embryonic day 10
- GFP
green fluorescent protein
- GST
glutathione transferase
- HEK
human embryonic kidney
- MEF
mouse embryonic fibroblast
- PFA
paraformaldehyde
- SCA11
spinocerebellar ataxia type 11
- TBS-T
Tris-buffered saline with 1% Tween-20
- TTBK2
tau tubulin kinase 2
- VRK
vaccinia-related kinase
Footnotes
AUTHOR CONTRIBUTION
Michale Bouskila undertook most of the experiments shown in Figures 3, 4, 6 and 7. Noor Esoof performed analysis of substrate specificity of TTBK2 (Figures 1 and 2) and helped with the analysis of Figure 5. Mike Begley and Lewis Cantley performed and analysed positional scanning peptide library experiments (Figure 1B). Laurie Gay undertook initial studies on TTBK2.Maria Deak undertook cloning. Alan Prescott undertook cellular localization studies (Figure 4). Emily Fang and Kate Storey undertook embryo dissection and analysis of embryonic development (Figure 6). Mike Begley, Noor Esoof and Dario Alessi planned experiments and analysed the data. Michale Bouskila and Dario Alessi wrote the manuscript.
REFERENCES
- 1.Houlden H, Johnson J, Gardner-Thorpe C, Lashley T, Hernandez D, Worth P, Singleton AB, Hilton DA, Holton J, Revesz T, et al. Mutations in TTBK2, encoding a kinase implicated in tau phosphorylation, segregate with spinocerebellar ataxia type 11. Nat. Genet. 2007;39:1434–1436. doi: 10.1038/ng.2007.43. [DOI] [PubMed] [Google Scholar]
- 2.Bauer P, Stevanin G, Beetz C, Synofzik M, Schmitz-Hubsch T, Wullner U, Berthier E, Ollagnon-Roman E, Riess O, Forlani S, et al. Spinocerebellar ataxia type 11 (SCA11) is an uncommon cause of dominant ataxia among French and German kindreds. J. Neurol. Neurosurg. Psychiatry. 2010;81:1229–1232. doi: 10.1136/jnnp.2009.202150. [DOI] [PubMed] [Google Scholar]
- 3.Houlden H. Spinocerebellar ataxia type 11. In: Pagon RA, Bird TD, Dolan CR, Stephens K, editors. GeneReviews. Seattle: University of Washington; 2008. PMID 20301723. [Google Scholar]
- 4.Takahashi M, Tomizawa K, Sato K, Ohtake A, Omori A. A novel tau-tubulin kinase from bovine brain. FEBS Lett. 1995;372:59–64. doi: 10.1016/0014-5793(95)00955-9. [DOI] [PubMed] [Google Scholar]
- 5.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 6.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 7.Manning G, Plowman GD, Hunter T, Sudarsanam S. Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 2002;27:514–520. doi: 10.1016/s0968-0004(02)02179-5. [DOI] [PubMed] [Google Scholar]
- 8.Sato S, Cerny RL, Buescher JL, Ikezu T. Tau-tubulin kinase 1 (TTBK1), a neuron-specific tau kinase candidate, is involved in tau phosphorylation and aggregation. J. Neurochem. 2006;98:1573–1584. doi: 10.1111/j.1471-4159.2006.04059.x. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi M, Tomizawa K, Ishiguro K, Takamatsu M, Fujita SC, Imahori K. Involvement of tau protein kinase I in paired helical filament-like phosphorylation of the juvenile tau in rat brain. J. Neurochem. 1995;64:1759–1768. doi: 10.1046/j.1471-4159.1995.64041759.x. [DOI] [PubMed] [Google Scholar]
- 10.Tomizawa K, Omori A, Ohtake A, Sato K, Takahashi M. Tau-tubulin kinase phosphorylates tau at Ser-208 and Ser-210, sites found in paired helical filament-tau. FEBS Lett. 2001;492:221–227. doi: 10.1016/s0014-5793(01)02256-6. [DOI] [PubMed] [Google Scholar]
- 11.Noble W, Planel E, Zehr C, Olm V, Meyerson J, Suleman F, Gaynor K, Wang L, LaFrancois J, Feinstein B, et al. Inhibition of glycogen synthase kinase-3 by lithium correlates with reduced tauopathy and degeneration in vivo. Proc. Natl. Acad. Sci. U.S.A. 2005;102:6990–6995. doi: 10.1073/pnas.0500466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J, Sato S, Okuyama S, Swan RJ, Jacobsen MT, Strunk E, Ikezu T. Tau-tubulin kinase 1 enhances prefibrillar tau aggregation and motor neuron degeneration in P301L FTDP-17 tau-mutant mice. FASEB J. 2010;24:2904–2915. doi: 10.1096/fj.09-150144. [DOI] [PubMed] [Google Scholar]
- 13.Vazquez-Higuera JL, Martinez-Garcia A, Sanchez-Juan P, Rodriguez-Rodriguez E, Mateo I, Pozueta A, Frank A, Valdivieso F, Berciano J, Bullido MJ, Combarros O. Genetic variations in tau-tubulin kinase-1 are linked to Alzheimer’s disease in a Spanish case-control cohort. Neurobiol. Aging. 2011;32:e555–e559. doi: 10.1016/j.neurobiolaging.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 14.Yu NN, Yu JT, Xiao JT, Zhang HW, Lu RC, Jiang H, Xing ZH, Tan L. Tau-tubulin kinase-1 gene variants are associated with Alzheimer’s disease in Han Chinese. Neurosci. Lett. 2011;491:83–86. doi: 10.1016/j.neulet.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 15.Hutti JE, Jarrell ET, Chang JD, Abbott DW, Storz P, Toker A, Cantley LC, Turk BE. A rapid method for determining protein kinase phosphorylation specificity. Nat. Methods. 2004;1:27–29. doi: 10.1038/nmeth708. [DOI] [PubMed] [Google Scholar]
- 16.Turk BE, Hutti JE, Cantley LC. Determining protein kinase substrate specificity by parallel solution-phase assay of large numbers of peptide substrates. Nat. Protoc. 2006;1:375–379. doi: 10.1038/nprot.2006.57. [DOI] [PubMed] [Google Scholar]
- 17.Marin O, Meggio F, Pinna LA. Design and synthesis of two new peptide substrates for the specific and sensitive monitoring of casein kinases-1 and -2. Biochem. Biophys. Res. Commun. 1994;198:898–905. doi: 10.1006/bbrc.1994.1128. [DOI] [PubMed] [Google Scholar]
- 18.Flotow H, Graves PR, Wang AQ, Fiol CJ, Roeske RW, Roach PJ. Phosphate groups as substrate determinants for casein kinase I action. J. Biol. Chem. 1990;265:14264–14269. [PubMed] [Google Scholar]
- 19.Flotow H, Roach PJ. Synergistic phosphorylation of rabbit muscle glycogen synthase by cyclic AMP-dependent protein kinase and casein kinase I. Implications for hormonal regulation of glycogen synthase. J. Biol. Chem. 1989;264:9126–9128. [PubMed] [Google Scholar]
- 20.Nakielny S, Campbell DG, Cohen P. The molecular mechanism by which adrenalin inhibits glycogen synthesis. Eur. J. Biochem. 1991;199:713–722. doi: 10.1111/j.1432-1033.1991.tb16175.x. [DOI] [PubMed] [Google Scholar]
- 21.Xu RM, Carmel G, Sweet RM, Kuret J, Cheng X. Crystal structure of casein kinase-1, a phosphate-directed protein kinase. EMBO J. 1995;14:1015–1023. doi: 10.1002/j.1460-2075.1995.tb07082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longenecker KL, Roach PJ, Hurley TD. Three-dimensional structure of mammalian casein kinase I: molecular basis for phosphate recognition. J. Mol. Biol. 1996;257:618–631. doi: 10.1006/jmbi.1996.0189. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.