Abstract
Spermatogenesis involves extremely marked cellular, genetic and chromatin changes resulting in the generation of the highly specialized sperm cell. Proteomics allows the identification of the proteins that compose the spermatogenic cells and the study of their function. The recent developments in mass spectrometry (MS) have markedly increased the throughput to identify and to study the sperm proteins. Catalogs of thousands of testis and spermatozoan proteins in human and different model species are becoming available, setting up the basis for subsequent research, diagnostic applications and possibly the future development of specific treatments. The present review intends to summarize the key genetic and chromatin changes at the different stages of spermatogenesis and in the mature sperm cell and to comment on the presently available proteomic studies.
Keywords: epigenetic, imprinting, protamine, proteome, spermatozoa
Introduction
Spermatogenesis involves extremely marked cellular, genetic and chromatin changes resulting in the generation of the highly specialized sperm cell (Figure 1). Spermatogonial stem cells replicate and differentiate into primary spermatocytes that undergo genetic recombination to give rise to haploid round spermatids.1, 2, 3, 4 Round spermatids then undergo a differentiation process called spermiogenesis where marked cellular, epigenetic and chromatin remodeling takes place.2, 5, 6, 7, 8, 9, 10, 11, 12 The nucleosomes are disassembled and the histones are removed and replaced by the high positively charged protamines forming tight toroidal complexes, organizing 85–95% of the human sperm DNA (Figure 1). At the cellular level, most of the cytoplasm is removed, and a large flagella and the acrosomal vesicle are assembled (Figure 1). Finally, the spermatozoon undergoes a maturation process through its transit in the epididymis where the chromatin is further compacted through the formation of disulfide bonds and zinc bridges among protamines, and the acquirement of different membrane and cellular functionalities.13, 14, 15 Once in the female tract, the spermatozoon must be capacitated, a process involving many signaling changes and the attainment of hyperactivated motility.16, 17, 18, 19 Before the sperm cell penetrates the oocyte, the sperm–oocyte recognition and the acrosomal reaction must take place.20 Finally, once in the oocyte, the male pronucleus must undergo another extremely marked chromatin remodeling process where the nucleoprotamine structure is disassembled and a new nucleosomal and chromatin structure is assembled (Figure 1). The accessibility of the spermatozoon has facilitated the study of its composition and mechanisms involved in its function and makes this cell particularly well suited for proteomic analysis.21 In addition, dissecting the differentiation process of spermatogenesis through proteomic analysis provides important potential biomedical applications in regenerative medicine,22, 23 in the identification of the genetic basis of male infertility,24, 25, 26, 27, 28 in understanding the origin of genetic and epigenetic mutation,5, 9, 10, 26, 29, 30, 31, 32 in reproductive toxicology33 and in the development of potential contraceptive strategies.34, 35 Different studies have investigated the genetic and protein changes and the mechanisms involved in the different stages of spermatogenesis and function of spermatozoa. The present review intends to complement different recent reviews focusing on the proteomics of the mature sperm cell,21, 36, 37, 38, 39, 40, 41, 42, 43 on testicular proteomics44, 45 or on the proteomic changes upon epididymal maturation and capacitation.46 To reach this goal, the structure followed will be to describe the key genetic and chromatin changes at the different stages of spermatogenesis (Figure 1), with indication of the related proteomic studies being performed based on large-scale mass spectrometry (MS) identification of proteins.
Figure 1.
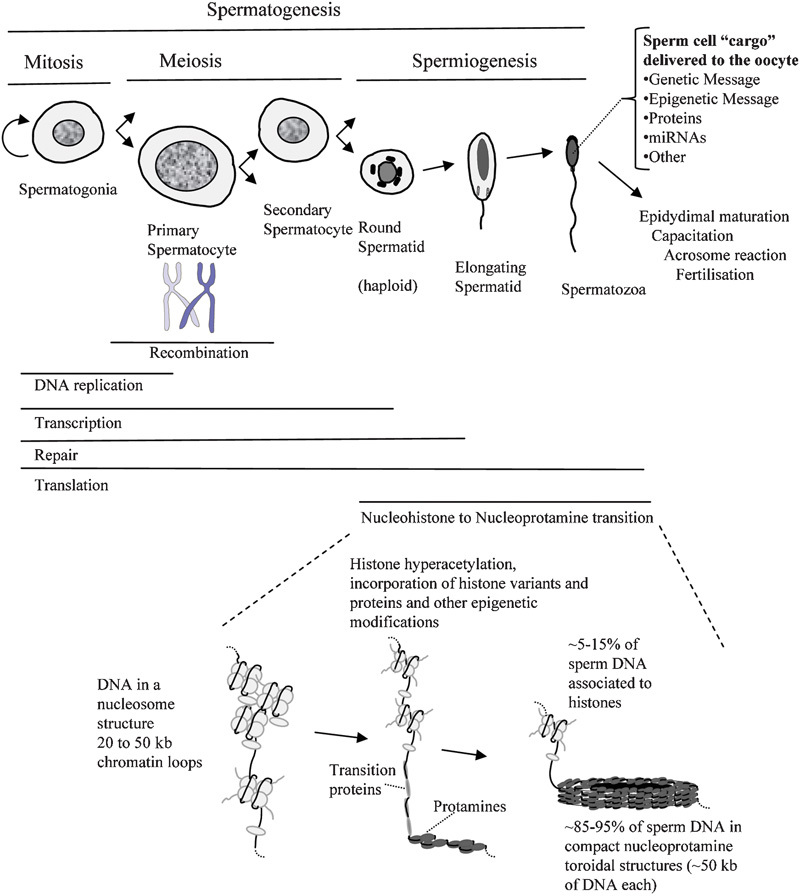
Cellular, genetic and chromatin changes at the different stages of spermatogenesis and sperm cell maturation. At the top of the figure, the different cellular stages of spermatogenesis are represented along with the genetic activities.2, 60 At the bottom of the image, a chromatin model of the mammalian nucleohistone to nucleoprotamine transition is shown.2, 5, 11, 21
Testicular proteomics: spermatogonial stem cells, spermatocytes and spermatids
One of the initial approaches applied to identify proteins present in the different stages of spermatogenesis exploited the changes in cellular abundance during testis development. Using this approach, the two-dimensional (2D) proteome profile changes during mouse testis development led to the identification using matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) of 44 proteins with substantial changes in protein abundance during development.47 Subsequent application of MALDI-TOF/TOF using a similar approach allowed identification of 257 different proteins that clustered into six different expression patterns.48 A limitation of the analysis of the entire testis is the existence of mixed cellular population including the presence of somatic cells. Therefore, other approaches have been used such as the isolation of the different cellular components of the testis or the study of cultured germ cells.
One of the testicular cells studied using proteomics is the spermatogonial stem cell.49, 50, 51 The interest in the study of the spermatogonial stem cells is multiple. Pathological perturbation of the stem cell is suspected as the origin of certain types of testicular cancers and male infertility, so that identification of the mechanisms involved would facilitate the development of preventive or treatment options.52, 53 In addition, the discovery of pluripotent stem cells within the testis raises important biomedical applications in regenerative medicine.23 Important issues in stem cell research are the identification of key genes and proteins needed to maintain the pluripotent state or that can be used as markers for their identification.49 A very elegant application of a comparative proteomics approach has been applied to demonstrate that after comparison of the proteomic profiles of cultured mouse multipotent adult germ line stem cells with embryonic stem cells, only 18 proteins were detected as differentially expressed out of a total of 409 proteins identified using 2D separation of the proteins followed by MS.22 The interpretation of this result was that the proteomes of multipotent adult germ line stem cells were highly similar to those of embryonic stem cells.22
A different approach to characterize the proteome of the spermatogonial stem cell exploited the peculiarities of the testis developmental biology in the dogfish Scyliorhinus canicula.54 These authors isolated, under stereomicroscope and dissection, the testicular germinative zone, highly enriched in spermatogonial stem cells, and used 2D and MALDI-TOF/TOF to identify 16 proteins and to also demonstrate the feasibility of this model to study the stem cell niche.54 Still, a different set of studies has used testicular cell sorting to obtain enriched cellular fractions with which proteomic analysis is performed. This approach was applied to separate spermatogonia from 9-day-old rats followed by protein 2D analysis and identification of the proteins using MALDI-TOF.55 More recently, the same group applied a similar procedure on immature and mature rat testis combined with sedimentation at unit gravity or elutriation to obtain highly enriched fractions of spermatogonia, spermatocytes and early spermatids.56 Subsequently, 2D difference in gel electrophoresis allowed identification of the relative abundance of 1274 proteins of which 265 differed significantly in the three groups of cell types. MALDI-TOF/TOF was then used to identify 123 non-redundant proteins clustering into the clades of mitotic, meiotic and post-meiotic cell types.56 It is also important to consider the close relationship between the Sertoli cell and the spermatogenic cells. Recently, the effect of the loss of Dicer in the Sertoli cell, required for microRNA biogenesis, on the testicular proteome, has been studied.57
Once the diploid spermatogonium is committed, it divides mitotically to produce two diploid intermediate cells called primary spermatocytes. Each primary spermatocyte then duplicates its DNA and subsequently undergoes meiosis I to produce two haploid secondary spermatocytes (Figure 1). Very importantly, this stage involves genetic recombination of homologous chromosomes to increase the genetic variability of the gamete. Many of the genetic causes of male infertility stem from meiotic anomalies. For instance, an important proportion of cases of male infertility are due to meiotic arrest.25, 26, 28 Also, many chromosomal structural anomalies result in incorrect pairing at meiosis and in the generation of chromosomally unbalanced gametes responsible for embryo lethality or severe anomalies in the offspring.58 Well-known causes and risk factors of male infertility such as the presence of Y-chromosome microdeletions also result in a variety of phenotypes which may include Sertoli cell-only syndrome, spermatogenic arrest and hypospermatogenesis resulting in azoospermia or oligospermia.24, 27, 59 Thus, proteomics, through the indicated above strategies, allows the identification of the proteins involved in the meiotic stage of spermatogenesis with the potential to contribute to the identification of the involved pathogenic mechanisms associated to male infertility in these cases.47, 48, 56
After the completion of the meiosis, the haploid round spermatids are generated (Figure 1). Haploid round spermatids are still transcriptionally active.60 However, another aspect of the biology of the spermatogenesis that deserves consideration is that each cell division from a spermatogonium to a spermatid is incomplete. The cells remain connected to one another by bridges of cytoplasm to allow synchronous development. It has been proposed that these cellular bridges allow the exchange of proteins and gene products so that, even though the round spermatids are genetically haploid, they may express proteins as if they were diploid.60 The haploid spermatids have been the focus of different proteomic studies. Fluorescence-activated cell sorting sorting has been applied to isolate haploid mouse spermatids followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) to identify 2116 proteins, 299 of which were testis specific and 155 were novel.61 Interestingly, the analysis of the chromosomal distribution of the haploid identified genes showed an underrepresentation of the X chromosome, interpreted as owing to meiotic X-chromosome inactivation, and overrepresentation of chromosome 11 upon expansion of the gene families.61
In a different approach, the proteomic analysis in testis biopsies of testosterone-treated men allowed the identification of proteins potentially related to the induced testis regression.62 As an experimental model, the effect of hyperthermia on mouse spermatogenesis has also been studied.63 The same group also investigated the whole human testis, identifying 462 unique proteins.64, 65 Evidence for protein heterogeneity was concluded upon the identification of 180 different proteins in more than one protein spot. Also, phosphoprotein staining allowed identification of 52 phosphorylated proteins.65 Proteins related to altered fertility in fish have also been identified using a whole-testis proteomic approach.66
The proteomic studies during spermiogenesis are highly relevant in the identification of all the proteins involved and the mechanisms of the nucleohistone-to-nucleoprotamine transition, which probably represents the most marked chromatin change that cells may undergo (Figure 1). One of the initial chromatin changes in the nucleohistone-to-nucleoprotamine transition is the incorporation of histone variants.67, 68, 69, 70, 71, 72 Another important early event is histone hyperacetylation that occurs during spermiogenesis just before the nucleosome disassembly.73, 74, 75, 76, 77, 78 It was postulated that histone hyperacetylation and rapid turnover of acetyl groups could rapidly and reversibly expose binding sites in chromatin for subsequent binding of chromosomal proteins.74 More recently, it was also shown in vitro that histone hyperacetylation facilitated nucleosome disassembly and histone displacement by protamines.79, 80 Also, hyperacetylated nucleosomes were shown to appear in a more relaxed structure upon binding to electron microscopy grids.80, 81 It has been shown that the testis-specific bromodomain-containing protein binds to hyperacetylated histone-4, triggering a reorganization of the chromatin.3, 10, 82, 83 Impaired histone-4 hyperacetylation has been detected in infertile patients.84, 85
Once the nucleosomes are disassembled, transition proteins are incorporated.2, 86 Transition proteins are then finally replaced by protamines to form a highly compact nucleoprotamine complex (Figure 1).2, 3, 6, 7, 10, 86, 87, 88, 89, 90 It is known that protamines are phosphorylated before binding to DNA and that a substantial dephosphorylation takes place concomitant to nucleoprotamine maturation.2, 91, 92, 93 The dynamics of binding of the protamines to DNA has also been studied.94, 95, 96 After binding to DNA, the formation of interdisulfide bonds between protamines further stabilizes the nucleoprotamine complex.15, 97 Different models for the structure of nucleoprotamine have been proposed.98, 99, 100, 101, 102, 103, 104 These chromatin changes during spermiogenesis take place in the context of a marked metamorphosis of the sperm cell and shaping of the head and associated structures such as the perinuclear theca and manchette.8, 105 However, despite substantial amount of information available, the identification of the molecular mechanisms governing the nucleohistone-to-nucleoprotamine transition and all the sperm cell changes still requires substantial effort. The available catalogs that are becoming available as derived from the proteomic projects represent a very important first step in the complete identification of the proteins and mechanisms involved.
Epididymal maturation
Testicular spermatozoa are haploid, have completed the nucleohistone-to-nucleoprotamine transition and have acquired most phenotypic features but they are not yet functional (Figure 1). Motility and the ability to bind to the oocyte are lacking and the sperm chromatin still needs to further complete maturation through the formation of disulfide bonds and zinc bridges in the nucleoprotamine (Figure 1). All these functionalities are acquired during the transit of the sperm cell through the epididymis.13, 14, 15, 106, 107 Proteomics has also been applied to the study of the sperm cell during epididymal maturation. 2D difference in gel electrophoresis has been used to isolate and to identify 60 protein spots, significantly modified as sperm traverse the epididymis.46 In one of the proteins, the change was found to represent a serine phosphorylation.46 The proteomic changes in mammalian spermatozoa during epididymal maturation have also been the subject of review.13
Epididymal secretions may also determine many aspects of the physiology of the spermatozoa. There is evidence that some of the proteins present in the fully mature ejaculated sperm cell may have been acquired during the epididymal transit.108 Therefore, several proteomic initiatives have also focused their efforts on the identification of the proteins present in the fluid secretions, which surround the sperm cell during its passage through the male genital tract. The human and stallion epididymal secretome and the human seminal plasma proteome have been reported.109, 110, 111, 112, 113 Also, the proteomic component of epididymosomes and accessory gland fluid has been characterized in several species.108, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123
Proteomics of mature spermatozoa and the sperm cell genome, epigenome and proteome delivered to the oocyte
The most extensive report on the identification of mature human sperm proteins used LC-MS/MS to identify 1056 gene products.124 This catalog is complementary to additional catalogs obtained using the 2D approaches.21, 125, 126, 127 In other mammals, spermatogenic proteomic profiles are also available.21, 128, 129, 130, 131 In invertebrates, MS identification of sperm proteins has been applied to the fruit fly.36, 132, 133 and the worm Caenorhabditis elegans134 among several others.21 One of the aspects amenable to study by proteomic analysis is the identification of the evolution and intensified selection of some sperm proteins.131, 135, 136, 137
Knowledge of the sperm cell proteome is also relevant to understand the different functions of the sperm cell. It is well known that the most essential function of the sperm cell is to deliver an intact paternal genomic DNA sequence to the oocyte, and that alterations in DNA integrity are a cause of male infertility, failed assisted reproduction and pregnancy loss.127, 138, 139, 140 More recently, it has been recognized that, in addition to the DNA sequence, the existence of imprints determined by the DNA methylation status is also important for a proper embryonic development, and that infertile patients have an aberrant DNA methylation at specific loci.29, 30, 31, 32 The analysis of the proteins identified in the different mature sperm proteomic projects has more recently identified proteins that may have a role in fertilization. For example, transcription factors, DNA-binding proteins and proteins involved in chromatin metabolism have been identified in cells that are transcriptionally inactive and that have most of its DNA tightly packaged with protamine.5, 21, 124 It will be interesting to determine whether these nuclear proteins are marking some regions of the male genome and may have an epigenetic function.5 An alternative explanation for the presence of some of these proteins is that they could represent leftovers from spermiogenesis, although in this case, the identification of these proteins could be useful as they could represent a ‘window' to the later stages of spermatogenesis with potential clinical implications. Recent reports indicate the presence of a complex chromatin organization of the genes in sperm, with an appreciable fraction containing both nucleohistone and nucleoprotamine domains that is suggested to be of potential relevance for embryo development.141, 142, 143 In addition to the sperm chromatin proteins, it will also be interesting to consider the possibility of additional sperm proteins having a potential role in fertilization.21, 37, 144
Proteomic alterations of ejaculated sperm cells in infertile patients
The presence of proteomic anomalies in infertile patients has been assessed by comparing the proteome of abnormal sperm samples from infertile patients with the proteome of control normozoospermic samples from fertile donors. One of the initial reports that demonstrated the potential of 2D proteomics in the study of sperm defects reported the proteomic mapping of a patient who experienced a failure in in vitro fertilization, where 20 different proteins were identified as compared with controls.145 Subsequently, different protein differences associated with asthenozoospermia have also been identified.146, 147, 148 Following a similar approach, the abundance of the proteins present in the sperm cells from 47 sperm samples from infertile patients and from 10 semen donors were analyzed in our laboratory by 2D polyacrylamide gel electrophoresis.127 In each of the 2D maps, the intensity of 101 spots previously identified by MALDI-MS analysis was measured. In addition, other parameters related to male infertility such as the protamine content and DNA integrity were also determined in each independent sample. Several interesting proteins such as transcription factors, prohibitin, heat shock and proteasome proteins were identified and linked to altered DNA integrity and abnormal protamination.127 Proteomics has also been applied to the analysis of round-headed spermatozoa by 2D fluorescence difference in gel electrophoresis, resulting in the identification of 35 protein spots (out of 61 identified) exhibiting significant changes in expression (9 proteins upregulated and 26 proteins downregulated) between normal and round-headed spermatozoa.149 It will be interesting to extend the analysis of patients through differential proteomics to incorporate the use of more robust methods based on non-isotopic labeling of the proteins and LC-MS/MS identification of the proteins.19
Capacitation
Capacitation is the activation process that leads to hyperactivated motility facilitating the sperm–oocyte interaction, binding and preparation for the acrosome reaction to penetrate the zona pellucida.16, 17, 18, 20, 150 The fact that most of the male genome is heavily condensed by protamines and transcription is completely blocked in the nucleoprotamine domains, together with the loss of most of the sperm cytoplasm, has classically favored the hypothesis that any protein changes concomitant to sperm capacitation had to be because of post-translational modification (Figure 1). However, some evidence suggests that the mature sperm cell is capable of translation of nuclear mRNA by mitochondrial-type ribosomes.151 Further support for this hypothesis of translation by mitochondrial-type ribosomes during sperm capacitation has been provided using a proteomic approach.17 In this study, differential proteomics was applied to identify 44 proteins with lower expression in D-chloramphenicol (a specific inhibitor of mitochondrial translation)-treated sperm cells in comparison with capacitated sperm. In addition, evidence was provided that 26 of 44 of these proteins were involved in critical processes for the sperm–egg interaction.17
However, in addition to the potential of de novo protein syntheses, the most widely accepted hypothesis is that most of the protein changes concomitant to capacitation are likely to be because of the post-translational modification of existing proteins. One of the most common post-translational modifications is phosphorylation. This modification was studied in capacitated human sperm resulting in the mapping of 60 sites of phosphorylation.152 The protein profiles of capacitated versus ejaculated human sperm have also been studied after 2D separation of the proteins.18 This study resulted in the identification of the 25 most abundant spots in ejaculated sperm, the 23 most abundant spots in capacitated sperm and the identification of proteins with substantial variation between uncapacitated and capacitated sperm.18 The role of the nitric oxide as an inducer of capacitation has also been studied to identify 240 S-nitrosylated human sperm proteins.153 A focused approach applied to the detergent-resistant membranes in capacitated sperm allowed the identification of 100 proteins, many of which were implicated in sperm–oocyte interaction.154
Isotopic labeling has also been applied to analyze the capacitation-associated changes in 42 different phosphopeptides.155 A radically different and robust approach has been recently applied to identify the proteomic changes associated with sperm capacitation through the combined use of immobilized pH gradient-strip prefractionation followed by LC-MS/MS analysis.19 Using this approach, label-free quantitative analysis of proteomic changes associated with capacitation identified 71 peptides corresponding to 52 proteins changing during capacitation many of which had not been previously implicated in this process.150
Conclusion
Proteomics applied to sperm cell research has so far led to the generation of catalogs of thousands of proteins present in the testis and in the mature sperm cell. This information is already being applied to the identification of the molecular mechanisms involved in spermatogonial stem cell physiology, meiotic recombination and in chromatin condensation, function and evolution of the sperm cell. Proteomics is also applied to identify the post-translational protein modifications occurring during epididymal maturation and capacitation, and to identify the complete proteomic complement of the sperm cell chromatin delivered to the oocyte. Furthermore, it is also being applied to the identification of the proteins involved in male fertility and infertility leading to the identification potential infertility markers and additional biomedical applications. However, there are still different issues and challenges that must be considered. Methodologically, it will be necessary for laboratories to keep up with the constant improvement in throughput of the MS equipment. The potential development of efficient spermatogenic in vitro culture systems, allowing synchronous differentiation of relatively pure cellular stages, would be a major accomplishment to potentiate the proteomic study of the sperm cell differentiation mechanisms. The alternative of separating and sorting testicular cells for proteomic analysis must take into account the cellular purity of the resulting cellular fractions. Even considering only the mature sperm cell, apparently a single cell type, it turns out to be a complex heterogeneous mixture of different quality sperm cells with substantial biochemical, functional and morphological differences. This fact together with the variation present in independent individuals and the physiological changes that the spermatozoa undergo upon ejaculation generates an enormous potential for variation, which must be taken into account in proteomic studies. Ultimately, the proteomic information will be very valuable in the context of the genetic, genomic, transcriptomic and metabolomic information. Altogether, we are now at an exciting and challenging momentum in proteomic sperm cell research with lots of work still to be done.
Acknowledgments
This work was supported by grants from the Ministerio de Educación y Ciencia (BFU2009-07118), fondos FEDER, Spain.
The authors declare no competing financial interests.
References
- Baccetti B, Afzelius BA. The biology of the sperm cell. Monogr Dev Biol. 1976;10:1–254. [PubMed] [Google Scholar]
- Oliva R, Dixon GH. Vertebrate protamine genes and the histone-to-protamine replacement reaction. Prog Nucleic Acid Res Mol Biol. 1991;40:25–94. doi: 10.1016/s0079-6603(08)60839-9. [DOI] [PubMed] [Google Scholar]
- Gaucher J, Reynoird N, Montellier E, Boussouar F, Rousseaux S, et al. From meiosis to postmeiotic events: the secrets of histone disappearance. FEBS J. 2010;277:599–604. doi: 10.1111/j.1742-4658.2009.07504.x. [DOI] [PubMed] [Google Scholar]
- Ward WS. Function of sperm chromatin structural elements in fertilization and development. Mol Hum Reprod. 2010;16:30–6. doi: 10.1093/molehr/gap080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva R. Protamines and male infertility. Hum Reprod Update. 2006;12:417–35. doi: 10.1093/humupd/dml009. [DOI] [PubMed] [Google Scholar]
- Mezquita C. Chromatin composition, structure and function in spermatogenesis. Revis Biol Celular. 1985;5:1–124. [PubMed] [Google Scholar]
- Poccia D. Remodeling of nucleoproteins during gametogenesis, fertilization, and early development. Int Rev Cytol. 1986;105:1–65. doi: 10.1016/s0074-7696(08)61061-x. [DOI] [PubMed] [Google Scholar]
- Kierszenbaum AL, Tres LL. The acrosome–acroplaxome–manchette complex and the shaping of the spermatid head. Arch Histol Cytol. 2004;67:271–84. doi: 10.1679/aohc.67.271. [DOI] [PubMed] [Google Scholar]
- Kimmins S, Sassone-Corsi P. Chromatin remodelling and epigenetic features of germ cells. Nature. 2005;434:583–9. doi: 10.1038/nature03368. [DOI] [PubMed] [Google Scholar]
- Rousseaux S, Caron C, Govin J, Lestrat C, Faure AK, et al. Establishment of male-specific epigenetic information. Gene. 2005;345:139–53. doi: 10.1016/j.gene.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Balhorn R. The protamine family of sperm nuclear proteins. Genome Biol. 2007;8:227. doi: 10.1186/gb-2007-8-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaman JA, Yamauchi Y, Ward WS. Function of the sperm nuclear matrix. Arch Androl. 2007;53:135–40. doi: 10.1080/01485010701329378. [DOI] [PubMed] [Google Scholar]
- Aitken RJ, Nixon B, Lin M, Koppers AJ, Lee YH, et al. Proteomic changes in mammalian spermatozoa during epididymal maturation. Asian J Androl. 2007;9:554–64. doi: 10.1111/j.1745-7262.2007.00280.x. [DOI] [PubMed] [Google Scholar]
- Bjorndahl L, Kvist U. Human sperm chromatin stabilization: a proposed model including zinc bridges. Mol Hum Reprod. 2010;16:23–9. doi: 10.1093/molehr/gap099. [DOI] [PubMed] [Google Scholar]
- de Mateo S, Ramos L, van der Vlag J, de Boer P, Oliva R.Improvement in chromatin maturity of human spermatozoa selected through density gradient centrifugation Int J Androle-pub ahead of print 20 June 2010; doi: 10.1111/j.1365-2605.2010.01080.x. [DOI] [PubMed]
- Austin CR. The capacitation of the mammalian sperm. Nature. 1952;170:326. doi: 10.1038/170326a0. [DOI] [PubMed] [Google Scholar]
- Zhao C, Guo XJ, Shi ZH, Wang FQ, Huang XY, et al. Role of translation by mitochondrial-type ribosomes during sperm capacitation: an analysis based on a proteomic approach. Proteomics. 2009;9:1385–99. doi: 10.1002/pmic.200800353. [DOI] [PubMed] [Google Scholar]
- Secciani F, Bianchi L, Ermini L, Cianti R, Armini A, et al. Protein profile of capacitated versus ejaculated human sperm. J Proteome Res. 2009;8:3377–89. doi: 10.1021/pr900031r. [DOI] [PubMed] [Google Scholar]
- Baker MA, Reeves G, Hetherington L, Aitken RJ. Analysis of proteomic changes associated with sperm capacitation through the combined use of IPG-strip pre-fractionation followed by RP chromatography LC-MS/MS analysis. Proteomics. 2010;10:482–95. doi: 10.1002/pmic.200900574. [DOI] [PubMed] [Google Scholar]
- Dan JC. Morphogenetic aspects of acrosome formation and reaction. Adv Morphog. 1970;8:1–39. doi: 10.1016/b978-0-12-028608-9.50005-3. [DOI] [PubMed] [Google Scholar]
- Oliva R, de Mateo S, Estanyol JM. Sperm cell proteomics. Proteomics. 2009;9:1004–17. doi: 10.1002/pmic.200800588. [DOI] [PubMed] [Google Scholar]
- Dihazi H, Dihazi GH, Nolte J, Meyer S, Jahn O, et al. Multipotent adult germline stem cells and embryonic stem cells: comparative proteomic approach. J Proteome Res. 2009;8:5497–510. doi: 10.1021/pr900565b. [DOI] [PubMed] [Google Scholar]
- Simon L, Hess RA, Cooke PS. Spermatogonial stem cells, in vivo transdifferentiation and human regenerative medicine. Expert Opin Biol Ther. 2010;10:519–30. doi: 10.1517/14712591003614731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva R, Margarit E, Ballesca JL, Carrio A, Sanchez A, et al. Prevalence of Y chromosome microdeletions in oligospermic and azoospermic candidates for intracytoplasmic sperm injection. Fertil Steril. 1998;70:506–10. doi: 10.1016/s0015-0282(98)00195-2. [DOI] [PubMed] [Google Scholar]
- Walsh TJ, Pera RR, Turek PJ. The genetics of male infertility. Semin Reprod Med. 2009;27:124–36. doi: 10.1055/s-0029-1202301. [DOI] [PubMed] [Google Scholar]
- Carrell DT. Contributions of spermatozoa to embryogenesis: assays to evaluate their genetic and epigenetic fitness. Reprod Biomed Online. 2008;16:474–84. doi: 10.1016/s1472-6483(10)60454-3. [DOI] [PubMed] [Google Scholar]
- Krausz C, Giachini C, Xue Y, O'Bryan MK, Gromoll J, et al. Phenotypic variation within European carriers of the Y-chromosomal gr/gr deletion is independent of Y-chromosomal background. J Med Genet. 2009;46:21–31. doi: 10.1136/jmg.2008.059915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan RI, O'Bryan MK. Clinical Review#: state of the art for genetic testing of infertile men. J Clin Endocrinol Metab. 2010;95:1013–24. doi: 10.1210/jc.2009-1925. [DOI] [PubMed] [Google Scholar]
- Marques CJ, Costa P, Vaz B, Carvalho F, Fernandes S, et al. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol Hum Reprod. 2008;14:67–74. doi: 10.1093/molehr/gam093. [DOI] [PubMed] [Google Scholar]
- Butler MG. Genomic imprinting disorders in humans: a mini-review. J Assist Reprod Genet. 2009;26:477–86. doi: 10.1007/s10815-009-9353-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poplinski A, Tuttelmann F, Kanber D, Horsthemke B, Gromoll J. Idiopathic male infertility is strongly associated with aberrant methylation of MEST and IGF2/H19 ICR1. Int J Androl. 2010;33:642–9. doi: 10.1111/j.1365-2605.2009.01000.x. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Purwar J, Pflueger C, Cairns BR, Carrell DT. Alterations in sperm DNA methylation patterns at imprinted loci in two classes of infertility. Fertil Steril. 2010;94:1728–33. doi: 10.1016/j.fertnstert.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Codrington AM, Hales BF, Robaire B. Exposure of male rats to cyclophosphamide alters the chromatin structure and basic proteome in spermatozoa. Hum Reprod. 2007;22:1431–42. doi: 10.1093/humrep/dem002. [DOI] [PubMed] [Google Scholar]
- Shetty J, Diekman AB, Jayes FC, Sherman NE, Naaby-Hansen S, et al. Differential extraction and enrichment of human sperm surface proteins in a proteome: identification of immunocontraceptive candidates. Electrophoresis. 2001;22:3053–66. doi: 10.1002/1522-2683(200108)22:14<3053::AID-ELPS3053>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Sipila P, Jalkanen J, Huhtaniemi IT, Poutanen M. Novel epididymal proteins as targets for the development of post-testicular male contraception. Reproduction. 2009;137:379–89. doi: 10.1530/REP-08-0132. [DOI] [PubMed] [Google Scholar]
- Karr TL. Fruit flies and the sperm proteome. Hum Mol Genet. 2007;16:R124–33. doi: 10.1093/hmg/ddm252. [DOI] [PubMed] [Google Scholar]
- Oliva R, Martinez-Heredia J, Estanyol JM. Proteomics in the study of the sperm cell composition, differentiation and function. Syst Biol Reprod Med. 2008;54:23–36. doi: 10.1080/19396360701879595. [DOI] [PubMed] [Google Scholar]
- Barratt CL. The human sperm proteome: the potential for new biomarkers of male fertility and a transformation in our understanding of the spermatozoon as a machine: commentary on the article ‘Identification of proteomic differences in asthenozoospermic sperm samples' by Martinez et al. Hum Reprod. 2008;23:1240–1. doi: 10.1093/humrep/den019. [DOI] [PubMed] [Google Scholar]
- Aitken RJ, Baker MA. The role of proteomics in understanding sperm cell biology. Int J Androl. 2008;31:295–302. doi: 10.1111/j.1365-2605.2007.00851.x. [DOI] [PubMed] [Google Scholar]
- Sutovsky P. Proteomic analysis of mammalian gametes and sperm–oocyte interactions. Soc Reprod Fertil Suppl. 2009;66:103–16. [PubMed] [Google Scholar]
- Baker MA, Aitken RJ. Proteomic insights into spermatozoa: critiques, comments and concerns. Expert Rev Proteomics. 2009;6:691–705. doi: 10.1586/epr.09.76. [DOI] [PubMed] [Google Scholar]
- Oxenham SK.Sperm proteomics: thinking outside the collision cell J Androle-pub ahead of print 10 June 2010; doi: 10.2164/jandrol.110.010231. [DOI] [PubMed]
- Brewis IA, Gadella BM. Sperm surface proteomics: from protein lists to biological function. Mol Hum Reprod. 2010;16:68–79. doi: 10.1093/molehr/gap077. [DOI] [PubMed] [Google Scholar]
- Wu TF, Chu DS. Sperm chromatin: fertile grounds for proteomic discovery of clinical tools. Mol Cell Proteomics. 2008;7:1876–86. doi: 10.1074/mcp.R800005-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvel P, Rolland AD, Jegou B, Pineau C. Testicular postgenomics: targeting the regulation of spermatogenesis. Philos Trans R Soc Lond B Biol Sci. 2010;365:1481–500. doi: 10.1098/rstb.2009.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MA, Witherdin R, Hetherington L, Cunningham-Smith K, Aitken RJ. Identification of post-translational modifications that occur during sperm maturation using difference in two-dimensional gel electrophoresis. Proteomics. 2005;5:1003–12. doi: 10.1002/pmic.200401100. [DOI] [PubMed] [Google Scholar]
- Paz M, Morin M, del Mazo J. Proteome profile changes during mouse testis development. Comp Biochem Physiol Part D Genomics Proteomics. 2006;1:404–15. doi: 10.1016/j.cbd.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Huang XY, Guo XJ, Shen J, Wang YF, Chen L, et al. Construction of a proteome profile and functional analysis of the proteins involved in the initiation of mouse spermatogenesis. J Proteome Res. 2008;7:3435–46. doi: 10.1021/pr800179h. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Griswold MD.Regulation of spermatogoniaIn: Melton D, Girard L, editors. StemBook [Internet] Cambridge, MA: Harvard Stem Cell Institute; 2008 [PubMed] [Google Scholar]
- Oatley JM, Brinster RL. Regulation of spermatogonial stem cell self-renewal in mammals. Annu Rev Cell Dev Biol. 2008;24:263–86. doi: 10.1146/annurev.cellbio.24.110707.175355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips BT, Gassei K, Orwig KE. Spermatogonial stem cell regulation and spermatogenesis. Philos Trans R Soc Lond B Biol Sci. 2010;365:1663–78. doi: 10.1098/rstb.2010.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen N, Meyts ER, Main KM, Skakkebaek NE. Testicular dysgenesis syndrome comprises some but not all cases of hypospadias and impaired spermatogenesis. Int J Androl. 2010;33:298–303. doi: 10.1111/j.1365-2605.2009.01050.x. [DOI] [PubMed] [Google Scholar]
- Wyns C, Curaba M, Vanabelle B, van Langendonckt A, Donnez J. Options for fertility preservation in prepubertal boys. Hum Reprod Update. 2010;16:312–28. doi: 10.1093/humupd/dmp054. [DOI] [PubMed] [Google Scholar]
- Loppion G, Lavigne R, Pineau C, Auvray P, Sourdaine P. Proteomic analysis of the spermatogonial stem cell compartment in dogfish Scyliorhinus canicula L. Comp Biochem Physiol Part D Genomics Proteomics. 2010;5:157–64. doi: 10.1016/j.cbd.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Guillaume E, Evrard B, Com E, Moertz E, Jegou B, et al. Proteome analysis of rat spermatogonia: reinvestigation of stathmin spatio-temporal expression within the testis. Mol Reprod Dev. 2001;60:439–45. doi: 10.1002/mrd.1108. [DOI] [PubMed] [Google Scholar]
- Rolland AD, Evrard B, Guitton N, Lavigne R, Calvel P, et al. Two-dimensional fluorescence difference gel electrophoresis analysis of spermatogenesis in the rat. J Proteome Res. 2007;6:683–97. doi: 10.1021/pr060436z. [DOI] [PubMed] [Google Scholar]
- Papaioannou MD, Lagarrigue M, Vejnar CE, Rolland AD, Kuhne F, et al. Loss of Dicer in Sertoli cells has a major impact on the testicular proteome of mice Mol Cell Proteomics 2010. Epub ahead of print [DOI] [PMC free article] [PubMed]
- Anton E, Blanco J, Egozcue J, Vidal F. Sperm studies in heterozygote inversion carriers: a review. Cytogenet Genome Res. 2005;111:297–304. doi: 10.1159/000086903. [DOI] [PubMed] [Google Scholar]
- de Llanos M, Ballesca JL, Gazquez C, Margarit E, Oliva R. High frequency of gr/gr chromosome Y deletions in consecutive oligospermic ICSI candidates. Hum Reprod. 2005;20:216–20. doi: 10.1093/humrep/deh582. [DOI] [PubMed] [Google Scholar]
- Oliva R, Mezquita J, Mezquita C, Dixon GH. Haploid expression of the rooster protamine mRNA in the postmeiotic stages of spermatogenesis. Dev Biol. 1988;125:332–40. doi: 10.1016/0012-1606(88)90216-3. [DOI] [PubMed] [Google Scholar]
- Guo X, Shen J, Xia Z, Zhang R, Zhang P, et al. Proteomic analysis of proteins involved in spermiogenesis in mouse. J Proteome Res. 2010;9:1246–56. doi: 10.1021/pr900735k. [DOI] [PubMed] [Google Scholar]
- Cui Y, Zhu H, Zhu Y, Guo X, Huo R, et al. Proteomic analysis of testis biopsies in men treated with injectable testosterone undecanoate alone or in combination with oral levonorgestrel as potential male contraceptive. J Proteome Res. 2008;7:3984–93. doi: 10.1021/pr800259t. [DOI] [PubMed] [Google Scholar]
- Zhu YF, Cui YG, Guo XJ, Wang L, Bi Y, et al. Proteomic analysis of effect of hyperthermia on spermatogenesis in adult male mice. J Proteome Res. 2006;5:2217–25. doi: 10.1021/pr0600733. [DOI] [PubMed] [Google Scholar]
- Huo R, He Y, Zhao C, Guo XJ, Lin M, et al. Identification of human spermatogenesis-related proteins by comparative proteomic analysis: a preliminary study. Fertil Steril. 2008;90:1109–18. doi: 10.1016/j.fertnstert.2007.07.1342. [DOI] [PubMed] [Google Scholar]
- Guo X, Zhao C, Wang F, Zhu Y, Cui Y, et al. Investigation of human testis protein heterogeneity using two-dimensional electrophoresis. J Androl. 2010;31:419–429. doi: 10.2164/jandrol.109.007534. [DOI] [PubMed] [Google Scholar]
- Forne I, Agulleiro MJ, Asensio E, Abian J, Cerda J. 2-D DIGE analysis of Senegalese sole (Solea senegalensis) testis proteome in wild-caught and hormone-treated F1 fish. Proteomics. 2009;9:2171–81. doi: 10.1002/pmic.200800696. [DOI] [PubMed] [Google Scholar]
- Prigent Y, Muller S, Dadoune JP. Immunoelectron microscopical distribution of histones H2B and H3 and protamines during human spermiogenesis. Mol Hum Reprod. 1996;2:929–35. doi: 10.1093/molehr/2.12.929. [DOI] [PubMed] [Google Scholar]
- Prigent Y, Troalen F, Dadoune JP. Immunoelectron microscopic visualization of intermediate basic proteins HPI1 and HPI2 in human spermatids and spermatozoa. Reprod Nutr Dev. 1998;38:417–27. doi: 10.1051/rnd:19980406. [DOI] [PubMed] [Google Scholar]
- Govin J, Caron C, Lestrat C, Rousseaux S, Khochbin S. The role of histones in chromatin remodelling during mammalian spermiogenesis. Eur J Biochem. 2004;271:3459–69. doi: 10.1111/j.1432-1033.2004.04266.x. [DOI] [PubMed] [Google Scholar]
- Churikov D, Zalenskaya IA, Zalensky AO. Male germline-specific histones in mouse and man. Cytogenet Genome Res. 2004;105:203–14. doi: 10.1159/000078190. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Iguchi N, Isotani A, Kitamura K, Toyama Y, et al. HANP1/H1T2, a novel histone H1-like protein involved in nuclear formation and sperm fertility. Mol Cell Biol. 2005;25:7107–19. doi: 10.1128/MCB.25.16.7107-7119.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loppin B, Bonnefoy E, Anselme C, Laurencon A, Karr TL, et al. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature. 2005;437:1386–90. doi: 10.1038/nature04059. [DOI] [PubMed] [Google Scholar]
- Candido EP, Dixon GH. Trout testis cells. 3. Acetylation of histones in different cell types from developing trout testis. J Biol Chem. 1972;247:5506–10. [PubMed] [Google Scholar]
- Oliva R, Mezquita C. Histone H4 hyperacetylation and rapid turnover of its acetyl groups in transcriptionally inactive rooster testis spermatids. Nucleic Acids Res. 1982;10:8049–59. doi: 10.1093/nar/10.24.8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes SR, Jr, Henderson N. Hyperacetylation of histone H4 in rat testis spermatids. Exp Cell Res. 1984;152:91–7. doi: 10.1016/0014-4827(84)90232-5. [DOI] [PubMed] [Google Scholar]
- Meistrich ML, Trostle-Weige PK, Lin R, Bhatnagar YM, Allis CD. Highly acetylated H4 is associated with histone displacement in rat spermatids. Mol Reprod Dev. 1992;31:170–81. doi: 10.1002/mrd.1080310303. [DOI] [PubMed] [Google Scholar]
- Hazzouri M, Rousseaux S, Mongelard F, Usson Y, Pelletier R, et al. Genome organization in the human sperm nucleus studied by FISH and confocal microscopy. Mol Reprod Dev. 2000;55:307–15. doi: 10.1002/(SICI)1098-2795(200003)55:3<307::AID-MRD9>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Marcon L, Boissonneault G. Transient DNA strand breaks during mouse and human spermiogenesis new insights in stage specificity and link to chromatin remodeling. Biol Reprod. 2004;70:910–8. doi: 10.1095/biolreprod.103.022541. [DOI] [PubMed] [Google Scholar]
- Oliva R, Mezquita C. Marked differences in the ability of distinct protamines to disassemble nucleosomal core particles in vitro. Biochemistry. 1986;25:6508–11. doi: 10.1021/bi00369a025. [DOI] [PubMed] [Google Scholar]
- Oliva R, Bazett-Jones D, Mezquita C, Dixon GH. Factors affecting nucleosome disassembly by protamines in vitro. Histone hyperacetylation and chromatin structure, time dependence, and the size of the sperm nuclear proteins. J Biol Chem. 1987;262:17016–25. [PubMed] [Google Scholar]
- Oliva R, Dixon GH. Vertebrate protamine gene evolution I. Sequence alignments and gene structure. J Mol Evol. 1990;30:333–46. doi: 10.1007/BF02101888. [DOI] [PubMed] [Google Scholar]
- Pivot-Pajot C, Caron C, Govin J, Vion A, Rousseaux S, et al. Acetylation-dependent chromatin reorganization by BRDT, a testis-specific bromodomain-containing protein. Mol Cell Biol. 2003;23:5354–65. doi: 10.1128/MCB.23.15.5354-5365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriniere J, Rousseaux S, Steuerwald U, Soler-Lopez M, Curtet S, et al. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461:664–8. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- Sonnack V, Failing K, Bergmann M, Steger K. Expression of hyperacetylated histone H4 during normal and impaired human spermatogenesis. Andrologia. 2002;34:384–90. doi: 10.1046/j.1439-0272.2002.00524.x. [DOI] [PubMed] [Google Scholar]
- Faure AK, Pivot-Pajot C, Kerjean A, Hazzouri M, Pelletier R, et al. Misregulation of histone acetylation in Sertoli cell-only syndrome and testicular cancer. Mol Hum Reprod. 2003;9:757–63. doi: 10.1093/molehr/gag101. [DOI] [PubMed] [Google Scholar]
- Meistrich ML, Mohapatra B, Shirley CR, Zhao M. Roles of transition nuclear proteins in spermiogenesis. Chromosoma. 2003;111:483–8. doi: 10.1007/s00412-002-0227-z. [DOI] [PubMed] [Google Scholar]
- Green GR, Balhorn R, Poccia DL, Hecht NB. Synthesis and processing of mammalian protamines and transition proteins. Mol Reprod Dev. 1994;37:255–63. doi: 10.1002/mrd.1080370303. [DOI] [PubMed] [Google Scholar]
- Dadoune JP. The nuclear status of human sperm cells. Micron. 1995;26:323–45. doi: 10.1016/0968-4328(95)00007-0. [DOI] [PubMed] [Google Scholar]
- Hecht NB. Molecular mechanisms of male germ cell differentiation. Bioessays. 1998;20:555–61. doi: 10.1002/(SICI)1521-1878(199807)20:7<555::AID-BIES6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Grootegoed JA, Siep M, Baarends WM. Molecular and cellular mechanisms in spermatogenesis. Baillieres Best Pract Res Clin Endocrinol Metab. 2000;14:331–43. doi: 10.1053/beem.2000.0083. [DOI] [PubMed] [Google Scholar]
- Ingles CJ, Dixon GH. Phosphorylation of protamine during spermatogenesis in trout testis. Proc Natl Acad Sci USA. 1967;58:1011–8. doi: 10.1073/pnas.58.3.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marushige Y, Marushige K. Phosphorylation of sperm histone during spermiogenesis in mammals. Biochim Biophys Acta. 1978;518:440–9. doi: 10.1016/0005-2787(78)90162-4. [DOI] [PubMed] [Google Scholar]
- Papoutsopoulou S, Nikolakaki E, Chalepakis G, Kruft V, Chevaillier P, et al. SR protein-specific kinase 1 is highly expressed in testis and phosphorylates protamine 1. Nucleic Acids Res. 1999;27:2972–80. doi: 10.1093/nar/27.14.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto MC, Maki AH, Balhorn R. Analysis of DNA-protamine interactions by optical detection of magnetic resonance. Biochemistry. 1997;36:11944–51. doi: 10.1021/bi971061l. [DOI] [PubMed] [Google Scholar]
- Brewer LR, Corzett M, Balhorn R. Protamine-induced condensation and decondensation of the same DNA molecule. Science. 1999;286:120–3. doi: 10.1126/science.286.5437.120. [DOI] [PubMed] [Google Scholar]
- Brewer L, Corzett M, Lau EY, Balhorn R. Dynamics of protamine 1 binding to single DNA molecules. J Biol Chem. 2003;278:42403–8. doi: 10.1074/jbc.M303610200. [DOI] [PubMed] [Google Scholar]
- Balhorn R, Corzett M, Mazrimas JA. Formation of intraprotamine disulfides in vitro. Arch Biochem Biophys. 1992;296:384–93. doi: 10.1016/0003-9861(92)90588-n. [DOI] [PubMed] [Google Scholar]
- Balhorn R. A model for the structure of chromatin in mammalian sperm. J Cell Biol. 1982;93:298–305. doi: 10.1083/jcb.93.2.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MJ, Lee C, Lee JD IV, Pogany GC, Balooch M, et al. Atomic force microscopy of mammalian sperm chromatin. Chromosoma. 1993;102:623–30. doi: 10.1007/BF00352310. [DOI] [PubMed] [Google Scholar]
- Allen MJ, Bradbury EM, Balhorn R. AFM analysis of DNA–protamine complexes bound to mica. Nucleic Acids Res. 1997;25:2221–6. doi: 10.1093/nar/25.11.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raukas E, Mikelsaar RH. Are there molecules of nucleoprotamine. Bioessays. 1999;21:440–8. doi: 10.1002/(SICI)1521-1878(199905)21:5<440::AID-BIES11>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Hud NV, Allen MJ, Downing KH, Lee J, Balhorn R. Identification of the elemental packing unit of DNA in mammalian sperm cells by atomic force microscopy. Biochem Biophys Res Commun. 1993;193:1347–54. doi: 10.1006/bbrc.1993.1773. [DOI] [PubMed] [Google Scholar]
- Vilfan ID, Conwell CC, Hud NV. Formation of native-like mammalian sperm cell chromatin with folded bull protamine. J Biol Chem. 2004;279:20088–95. doi: 10.1074/jbc.M312777200. [DOI] [PubMed] [Google Scholar]
- Biegeleisen K. The probable structure of the protamine–DNA complex. J Theor Biol. 2006;241:533–40. doi: 10.1016/j.jtbi.2005.12.015. [DOI] [PubMed] [Google Scholar]
- Oko R, Sutovsky P. Biogenesis of sperm perinuclear theca and its role in sperm functional competence and fertilization. J Reprod Immunol. 2009;83:2–7. doi: 10.1016/j.jri.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Cooper TG. Sperm maturation in the epididymis: a new look at an old problem. Asian J Androl. 2007;9:533–9. doi: 10.1111/j.1745-7262.2007.00285.x. [DOI] [PubMed] [Google Scholar]
- Jones RC, Dacheux JL, Nixon B, Ecroyd HW. Role of the epididymis in sperm competition. Asian J Androl. 2007;9:493–9. doi: 10.1111/j.1745-7262.2007.00284.x. [DOI] [PubMed] [Google Scholar]
- Frenette G, Girouard J, Sullivan R. Comparison between epididymosomes collected in the intraluminal compartment of the bovine caput and cauda epididymidis. Biol Reprod. 2006;75:885–90. doi: 10.1095/biolreprod.106.054692. [DOI] [PubMed] [Google Scholar]
- Fouchecourt S, Metayer S, Locatelli A, Dacheux F, Dacheux JL. Stallion epididymal fluid proteome: qualitative and quantitative characterization; secretion and dynamic changes of major proteins. Biol Reprod. 2000;62:1790–803. doi: 10.1095/biolreprod62.6.1790. [DOI] [PubMed] [Google Scholar]
- Bourgeon F, Evrard B, Brillard-Bourdet M, Colleu D, Jegou B, et al. Involvement of semenogelin-derived peptides in the antibacterial activity of human seminal plasma. Biol Reprod. 2004;70:768–74. doi: 10.1095/biolreprod.103.022533. [DOI] [PubMed] [Google Scholar]
- Dacheux JL, Belghazi M, Lanson Y, Dacheux F. Human epididymal secretome and proteome. Mol Cell Endocrinol. 2006;250:36–42. doi: 10.1016/j.mce.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Dacheux JL, Belleannee C, Jones R, Labas V, Belghazi M, et al. Mammalian epididymal proteome. Mol Cell Endocrinol. 2009;306:45–50. doi: 10.1016/j.mce.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Pilch B, Mann M. Large-scale and high-confidence proteomic analysis of human seminal plasma. Genome Biol. 2006;7:R40. doi: 10.1186/gb-2006-7-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utleg AG, Yi EC, Xie T, Shannon P, White JT, et al. Proteomic analysis of human prostasomes. Prostate. 2003;56:150–61. doi: 10.1002/pros.10255. [DOI] [PubMed] [Google Scholar]
- Chaurand P, Fouchecourt S, DaGue BB, Xu BJ, Reyzer ML, et al. Profiling and imaging proteins in the mouse epididymis by imaging mass spectrometry. Proteomics. 2003;3:2221–39. doi: 10.1002/pmic.200300474. [DOI] [PubMed] [Google Scholar]
- Gatti JL, Metayer S, Belghazi M, Dacheux F, Dacheux JL. Identification, proteomic profiling, and origin of ram epididymal fluid exosome-like vesicles. Biol Reprod. 2005;72:1452–65. doi: 10.1095/biolreprod.104.036426. [DOI] [PubMed] [Google Scholar]
- Strzezek J, Wysocki P, Kordan W, Kuklinska M. Proteomics of boar seminal plasma—current studies and possibility of their application in biotechnology of animal reproduction. Reprod Biol. 2005;5:279–90. [PubMed] [Google Scholar]
- Burden HP, Holmes CH, Persad R, Whittington K. Prostasomes—their effects on human male reproduction and fertility. Hum Reprod Update. 2006;12:283–92. doi: 10.1093/humupd/dmi052. [DOI] [PubMed] [Google Scholar]
- Walker MJ, Rylett CM, Keen JN, Audsley N, Sajid M, et al. Proteomic identification of Drosophila melanogaster male accessory gland proteins, including a pro-cathepsin and a soluble gamma-glutamyl transpeptidase. Proteome Sci. 2006;4:9. doi: 10.1186/1477-5956-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veveris-Lowe TL, Kruger SJ, Walsh T, Gardiner RA, Clements JA. Seminal fluid characterization for male fertility and prostate cancer: kallikrein-related serine proteases and whole proteome approaches. Semin Thromb Hemost. 2007;33:87–99. doi: 10.1055/s-2006-958467. [DOI] [PubMed] [Google Scholar]
- Moura AA, Chapman DA, Koc H, Killian GJ. A comprehensive proteomic analysis of the accessory sex gland fluid from mature Holstein bulls. Anim Reprod Sci. 2007;98:169–88. doi: 10.1016/j.anireprosci.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Cornett DS, Reyzer ML, Chaurand P, Caprioli RM. MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nat Methods. 2007;4:828–33. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- Thimon V, Frenette G, Saez F, Thabet M, Sullivan R. Protein composition of human epididymosomes collected during surgical vasectomy reversal: a proteomic and genomic approach. Hum Reprod. 2008;23:1698–707. doi: 10.1093/humrep/den181. [DOI] [PubMed] [Google Scholar]
- Baker MA, Reeves G, Hetherington L, Muller J, Baur I, et al. Identification of gene products present in Triton X-100 soluble and insoluble fractions of human spermatozoa lysates using LC-MS/MS analysis. Proteomics Clin Appl. 2007;1:524–32. doi: 10.1002/prca.200601013. [DOI] [PubMed] [Google Scholar]
- Martinez-Heredia J, Estanyol JM, Ballesca JL, Oliva R. Proteomic identification of human sperm proteins. Proteomics. 2006;6:4356–69. doi: 10.1002/pmic.200600094. [DOI] [PubMed] [Google Scholar]
- Li LW, Fan LQ, Zhu WB, Nien HC, Sun BL, et al. Establishment of a high-resolution 2-D reference map of human spermatozoal proteins from 12 fertile sperm-bank donors. Asian J Androl. 2007;9:321–9. doi: 10.1111/j.1745-7262.2007.00261.x. [DOI] [PubMed] [Google Scholar]
- de Mateo S, Martinez-Heredia J, Estanyol JM, Domiguez-Fandos D, Vidal-Taboada JM, et al. Marked correlations in protein expression identified by proteomic analysis of human spermatozoa. Proteomics. 2007;7:4264–77. doi: 10.1002/pmic.200700521. [DOI] [PubMed] [Google Scholar]
- Baker MA, Hetherington L, Reeves G, Muller J, Aitken RJ. The rat sperm proteome characterized via IPG strip prefractionation and LC-MS/MS identification. Proteomics. 2008;8:2312–21. doi: 10.1002/pmic.200700876. [DOI] [PubMed] [Google Scholar]
- Baker MA, Hetherington L, Reeves GM, Aitken RJ. The mouse sperm proteome characterized via IPG strip prefractionation and LC-MS/MS identification. Proteomics. 2008;8:1720–30. doi: 10.1002/pmic.200701020. [DOI] [PubMed] [Google Scholar]
- Peddinti D, Nanduri B, Kaya A, Feugang JM, Burgess SC, et al. Comprehensive proteomic analysis of bovine spermatozoa of varying fertility rates and identification of biomarkers associated with fertility. BMC Syst Biol. 2008;2:19. doi: 10.1186/1752-0509-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorus S, Wasbrough ER, Busby J, Wilkin EC, Karr TL. Sperm proteomics reveals intensified selection on mouse sperm membrane and acrosome genes. Mol Biol Evol. 2010;27:1235–46. doi: 10.1093/molbev/msq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorus S, Busby SA, Gerike U, Shabanowitz J, Hunt DF, et al. Genomic and functional evolution of the Drosophila melanogaster sperm proteome. Nat Genet. 2006;38:1440–5. doi: 10.1038/ng1915. [DOI] [PubMed] [Google Scholar]
- Karr TL. Application of proteomics to ecology and population biology. Heredity. 2008;100:200–6. doi: 10.1038/sj.hdy.6801008. [DOI] [PubMed] [Google Scholar]
- Chu DS, Liu H, Nix P, Wu TF, Ralston EJ, et al. Sperm chromatin proteomics identifies evolutionarily conserved fertility factors. Nature. 2006;443:101–5. doi: 10.1038/nature05050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramm SA, McDonald L, Hurst JL, Beynon RJ, Stockley P. Comparative proteomics reveals evidence for evolutionary diversification of rodent seminal fluid and its functional significance in sperm competition. Mol Biol Evol. 2009;26:189–98. doi: 10.1093/molbev/msn237. [DOI] [PubMed] [Google Scholar]
- Eirin-Lopez JM, Ausio J. Origin and evolution of chromosomal sperm proteins. Bioessays. 2009;31:1062–70. doi: 10.1002/bies.200900050. [DOI] [PubMed] [Google Scholar]
- Findlay GD, Swanson WJ. Proteomics enhances evolutionary and functional analysis of reproductive proteins. Bioessays. 2010;32:26–36. doi: 10.1002/bies.200900127. [DOI] [PubMed] [Google Scholar]
- Dominguez-Fandos D, Camejo MI, Ballesca JL, Oliva R. Human sperm DNA fragmentation: correlation of TUNEL results as assessed by flow cytometry and optical microscopy. Cytometry A. 2007;71:1011–8. doi: 10.1002/cyto.a.20484. [DOI] [PubMed] [Google Scholar]
- Zini A, Boman JM, Belzile E, Ciampi A. Sperm DNA damage is associated with an increased risk of pregnancy loss after IVF and ICSI: systematic review and meta-analysis. Hum Reprod. 2008;23:2663–8. doi: 10.1093/humrep/den321. [DOI] [PubMed] [Google Scholar]
- Aitken RJ, de Iuliis GN, McLachlan RI. Biological and clinical significance of DNA damage in the male germ line. Int J Androl. 2009;32:46–56. doi: 10.1111/j.1365-2605.2008.00943.x. [DOI] [PubMed] [Google Scholar]
- Arpanahi A, Brinkworth M, Iles D, Krawetz SA, Paradowska A, et al. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res. 2009;19:1338–49. doi: 10.1101/gr.094953.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–8. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, et al. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 2010;17:679–87. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- Zimmerman S, Sutovsky P. The sperm proteasome during sperm capacitation and fertilization. J Reprod Immunol. 2009;83:19–25. doi: 10.1016/j.jri.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Pixton KL, Deeks ED, Flesch FM, Moseley FL, Bjorndahl L, et al. Sperm proteome mapping of a patient who experienced failed fertilization at IVF reveals altered expression of at least 20 proteins compared with fertile donors: case report. Hum Reprod. 2004;19:1438–47. doi: 10.1093/humrep/deh224. [DOI] [PubMed] [Google Scholar]
- Zhao C, Huo R, Wang FQ, Lin M, Zhou ZM, et al. Identification of several proteins involved in regulation of sperm motility by proteomic analysis. Fertil Steril. 2007;87:436–8. doi: 10.1016/j.fertnstert.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Martinez-Heredia J, de Mateo S, Vidal-Taboada JM, Ballesca JL, Oliva R. Identification of proteomic differences in asthenozoospermic sperm samples. Hum Reprod. 2008;23:783–91. doi: 10.1093/humrep/den024. [DOI] [PubMed] [Google Scholar]
- Siva AB, Kameshwari DB, Singh V, Pavani K, Sundaram CS, et al. Proteomics-based study on asthenozoospermia: differential expression of proteasome alpha complex. Mol Hum Reprod. 2010;16:452–62. doi: 10.1093/molehr/gaq009. [DOI] [PubMed] [Google Scholar]
- Liao TT, Xiang Z, Zhu WB, Fan LQ. Proteome analysis of round-headed and normal spermatozoa by 2-D fluorescence difference gel electrophoresis and mass spectrometry. Asian J Androl. 2009;11:683–93. doi: 10.1038/aja.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MA, Smith ND, Hetherington L, Taubman K, Graham ME, et al. Label-free quantitation of phosphopeptide changes during rat sperm capacitation. J Proteome Res. 2010;9:718–29. doi: 10.1021/pr900513d. [DOI] [PubMed] [Google Scholar]
- Gur Y, Breitbart H. Protein synthesis in sperm: dialog between mitochondria and cytoplasm. Mol Cell Endocrinol. 2008;282:45–55. doi: 10.1016/j.mce.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Ficarro S, Chertihin O, Westbrook VA, White F, Jayes F, et al. Phosphoproteome analysis of capacitated human sperm. Evidence of tyrosine phosphorylation of a kinase-anchoring protein 3 and valosin-containing protein/p97 during capacitation. J Biol Chem. 2003;278:11579–89. doi: 10.1074/jbc.M202325200. [DOI] [PubMed] [Google Scholar]
- Lefievre L, Chen Y, Conner SJ, Scott JL, Publicover SJ, et al. Human spermatozoa contain multiple targets for protein S-nitrosylation: an alternative mechanism of the modulation of sperm function by nitric oxide. Proteomics. 2007;7:3066–84. doi: 10.1002/pmic.200700254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon B, Bielanowicz A, McLaughlin EA, Tanphaichitr N, Ensslin MA, et al. Composition and significance of detergent resistant membranes in mouse spermatozoa. J Cell Physiol. 2009;218:122–34. doi: 10.1002/jcp.21575. [DOI] [PubMed] [Google Scholar]
- Platt MD, Salicioni AM, Hunt DF, Visconti PE. Use of differential isotopic labeling and mass spectrometry to analyze capacitation-associated changes in the phosphorylation status of mouse sperm proteins. J Proteome Res. 2009;8:1431–40. doi: 10.1021/pr800796j. [DOI] [PMC free article] [PubMed] [Google Scholar]


